Abstract
Inositol polyphosphate binding specificity of the jasmonate receptor is largely determined by the F-box protein COI1.
Recent findings that receptor complexes for auxin and jasmonate bind inositol polyphosphates stimulated the idea that plant hormone perception is regulated by inositol-derived molecules (Tan et al., 2007; Sheard et al., 2010). Inositol polyphosphates regulate critical cellular functions in eukaryotic cells (Munnik and Nielsen, 2011; Munnik and Vermeer, 2010; Gillaspy, 2013; Tsui and York, 2010; Kuo et al., 2014; Lee et al., 2015), and the discovery that these molecules bind to plant hormone receptors provides an interesting case model to study plant hormone perception. For instance, the ASK1-TIR1 component of the auxin receptor complex was copurified and cocrystallized with insect cell-derived inositol hexakisphosphate (InsP6; Tan et al., 2007). TIR1 mutants defective in InsP6 binding failed to interact with the IAA7 transcriptional repressor in the presence of auxin in yeast two-hybrid assays and in pull-down experiments using tagged-recombinant Aux/IAA protein (Calderón Villalobos et al., 2012), suggesting that InsP6 binding might be important for auxin receptor function. Interestingly, the ASK1-COI1 component of the jasmonate receptor complex also copurified with inositol polyphosphate (Sheard et al., 2010). Here, NMR analyses revealed that insect cell-purified, nondialyzed protein contained either d- and/or l-myo-inositol-1,2,4,5,6-pentakisphosphate (Sheard et al., 2010), also referred to as Ins(1,2,4,5,6)P5 or short InsP5 [3-OH] and Ins(2,3,4,5,6)P5 or short InsP5 [1-OH], respectively. Unfortunately, NMR cannot discriminate between enantiomers; therefore, the structure of the insect-purified InsP5 isomer remains unresolved. Dialyzed ASK1-COI1 protein depleted of inositol polyphosphate failed to reconstitute the jasmonate receptor complex in vitro, while addition of InsP5 [3-OH] robustly stimulated complex formation (Sheard et al., 2010). Interestingly, Ins(1,4,5,6)P4 and InsP6 also stimulated complex formation, although InsP6 stimulated with lower efficiency (Sheard et al., 2010). Other InsP5 isomers (including the possible alternative InsP5 [1-OH] enantiomer) were not tested in this study.
In plants, three InsP5 species with distinct chromatographic mobilities have been identified (Stevenson-Paulik et al., 2005; Hanke et al., 2012; Laha et al., 2015; Brearley and Hanke, 1996). Among them, only the isomeric nature of the symmetrical molecule InsP5 [2-OH] was determined, while the identity of the other two InsP5 isomers remains unknown (Stevenson-Paulik et al., 2005; Brearley and Hanke, 1996). Independent work in amoeba and in a pancreatoma cell line showed that inositol polyphosphates can be further phosphorylated at an existing phosphate position to give rise to inositol pyrophosphates, molecules such as InsP7 and InsP8 that contain energy-rich diphosphate bonds and have important cellular functions in amoeba, animal, and yeast cells (Menniti et al., 1993; Stephens et al., 1993; Shears et al., 2012; Mulugu et al., 2007; Wilson et al., 2013; Thota and Bhandari, 2015). Inositol pyrophosphates have also been detected in different plant species (Desai et al., 2014; Lemtiri-Chlieh et al., 2000; Brearley and Hanke, 1996; Laha et al., 2015), and recent work suggests an important function of these molecules in regulating jasmonate-dependent responses (Laha et al., 2015).
Jasmonate perception is regulated by COI1, the F-box component of an SCF ubiquitin E3 ligase complex. COI1 recruits Jasmonate ZIM-domain (JAZ) transcriptional repressors upon binding to the bioactive jasmonic acid (JA) conjugate JA-Ile, resulting in polyubiquitylation and proteasomal degradation of the JAZ repressors and subsequent activation of jasmonate-dependent gene expression (Chini et al., 2007; Thines et al., 2007; Katsir et al., 2008; Pauwels and Goossens, 2011). A combinatorial approach analyzing InsP8-deficient vih2 mutant plants and using in vitro reconstitution and in silico molecular docking experiments suggested that coincidence detection (i.e. simultaneous detection) of active jasmonate and the inositol pyrophosphate InsP8 by the ASK1-COI1-JAZ receptor complex is critical for the activation of defense gene expression and for defenses against insect herbivores and necrotrophic fungi (Laha et al., 2015). Another study proposed InsP5 [2-OH] to be involved in jasmonate perception (Mosblech et al., 2011). Collectively, these reports raise the question whether the jasmonate receptor shows selectivity for distinct inositol polyphosphates.
COMPETITIVE BINDING ASSAYS REVEAL LARGE DIFFERENCES IN RELATIVE BINDING AFFINITIES OF DISTINCT INSP5 ISOMERS TO THE JASMONATE RECEPTOR COMPLEX
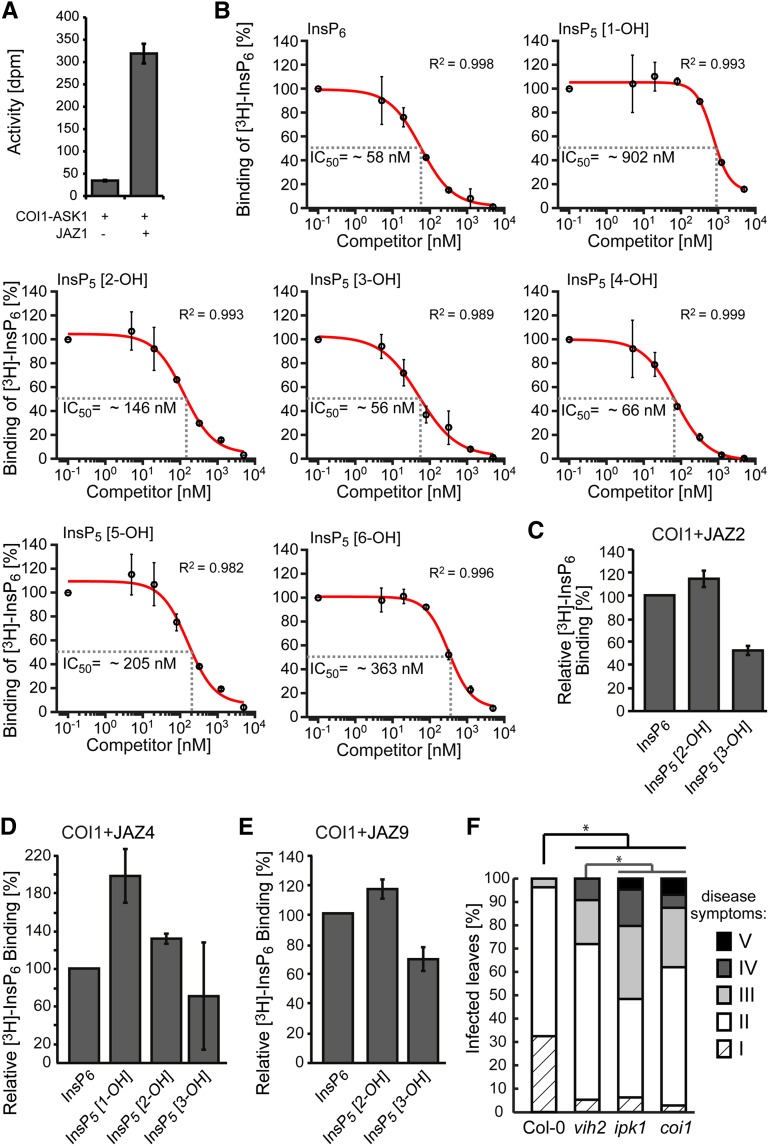
To investigate inositol polyphosphate binding specificity, we performed in vitro reconstitution experiments with insect cell-purified ASK1-COI1, recombinant JAZ proteins, the JA-Ile mimic coronatine, and [3H]InsP6 to determine IC50 values (50% inhibition of radioligand binding) for different InsP5 isomers. This approach was chosen because radiolabeled InsP5 isomers are not commercially available. A similar strategy was recently employed to investigate relative binding affinities of mammalian casein kinase-2 to InsP6, 5-InsP7, and a nonhydrolyzable InsP7 derivative (Rao et al., 2014). We used His8-tagged recombinant JAZ protein to pull down ASK1-COI1 in the presence of coronatine via Ni-NTA affinity chromatography and then determined [3H]InsP6-derived activity (see “Supplemental Data”). For JAZ1, the following relative order of effectiveness of InsP6 and the various InsP5 isomers in competing with [3H-InsP6] binding was observed (Fig. 1, A and B): InsP5 [3-OH] (IC50: 56 nm) ≥ InsP6 (IC50: 58 nm) > InsP5 [4-OH] (IC50: 66 nm) > InsP5 [2-OH] (IC50: 146 nm) > InsP5 [5-OH] (IC50: 205 nm) > InsP5 [6-OH] (IC50: 363 nm) > InsP5 [1-OH] (IC50: 902 nm). The data suggest strong differences in the relative binding affinity of different InsP5 isomers (including enantiomers) to the jasmonate receptor complex. For instance, the IC50 value of InsP5 [1-OH] is 16-fold higher than that of InsP5 [3-OH], suggesting a much higher affinity of the jasmonate receptor to InsP5 [3-OH]. This is remarkable as both isomers are enantiomers that are chemically indistinguishable and for which a method to determine enantiomer identity has not yet been developed. Furthermore, the IC50 value for InsP5 [2-OH], an isomer previously suggested to play a role in the activation of the jasmonate receptor (Mosblech et al., 2011), is 2.5-fold higher than that of InsP5 [3-OH] and InsP6, suggesting it is less effective in potentiating jasmonate receptor assembly (Fig. 1B).
Figure 1.
COI1 determines the inositol polyphosphate binding specificity of the jasmonate receptor complex. A, JAZ-dependent binding of [3H]InsP6 to ASK1-COI. Insect cell-purified ASK1-COI1 was incubated with recombinant His8-MBP-JAZ1 and [3H]InsP6 in the presence of 1 µm coronatine. The complex was then purified by immobilized Ni2+ affinity chromatography (taking advantage of JAZ1’s N-terminal His8 tag), and the immobilized activity was determined by scintillation counting. A reaction in the absence of JAZ protein served as a negative control. Values show background-subtracted means ± se. B, Competitive binding assays with [3H]InsP6 and unlabeled inositol polyphosphates as indicated. Results are presented as percentage of total binding. Nonlinear regression analysis was employed to fit data to a sigmoidal model, which allowed the determination of IC50 values. R2 values given in the plots provide estimations for goodness of fit. Error bars represent ±se. C to E, Relative [3H]InsP6 binding to the ASK1-COI1 complex in the presence of 1 µm coronatine and different InsP5 isomers and JAZ proteins as depicted. For the JAZ2 experiment, all competing InsP species were at 150 nm; for JAZ4, we used 80 nm of all InsP species; and for JAZ9, all competing InsP were at 50 nm. The average of [3H]InsP6 binding to the jasmonate receptor complex in the absence of unlabeled inositol polyphosphate was set to 100%. The experiment was repeated with similar results. Error bars denote ±se. F, Compromised defenses of vih2-4, ipk1-1, and coi1-t against a necrotrophic fungus corroborates a role of higher inositol polyphosphates (≥InsP6) in COI1-dependent responses. All genotypes were treated with 5 µL of an A. brassicicola spore suspension (1 × 106 spores/mL). Disease symptoms were scored in a double-blinded manner after 10 d of spore inoculation and categorized as different classes. Classes are defined as follows: Class I, light brown spots at the site of infection; Class II, dark brown spots on the site of infection; Class III, spreading necrosis; Class IV, leaf maceration; Class V, sporulation. The distribution of data were analyzed with a χ2 test (no. of leaves, n ≥ 29 classes contained at least 2.5% of total scorings per genotype), * P < 0.05. The experiments were repeated independently with similar results.
COI1 LARGELY DETERMINES THE INOSITOL POLYPHOSPHATE BINDING SPECIFICITY
To investigate the contribution of the JAZ protein to the inositol polyphosphate binding specificity, we performed similar experiments as described above with JAZ2, JAZ4, and JAZ9 (Fig. 1, C–E) with InsP6 and selected InsP5 isomers at a fixed concentration of “cold” inositol polyphosphate. The effectiveness of InsP5 isomers to compete with [3H]InsP6 binding largely recapitulated the observations from the experiment using the JAZ1 protein, showing binding affinities in the following order: InsP5 [3-OH] ≥ InsP6 > InsP5 [2-OH]. Binding experiments with JAZ4 further indicate that, as in the case of JAZ1, InsP5 [1-OH] has the weakest affinity, suggesting that the jasmonate receptor complex retains its ability to discriminate between the two enantiomers (1/3-OH) when using another JAZ protein. Altogether these data corroborate the idea that COI1, not the JAZ protein, determines inositol polyphosphate binding specificity.
The observation that InsP5 [2-OH] has a weaker relative affinity than InsP6 was surprising, as an increase of InsP5 [2-OH] at the cost of InsP6 in the Arabidopsis (Arabidopsis thaliana) ipk1-1 mutant was previously proposed to activate COI1 function and to cause increased resistance to Plutella xylostella caterpillars (Mosblech et al., 2011).
INCREASE OF INSP5 [2-OH] BY DEACTIVATION OF THE INOSITOL 1,3,4,5,6-PENTAKISPHOSPHATE 2-KINASE (IPK1) DOES NOT GLOBALLY ACTIVATE COI1 FUNCTIONS IN ARABIDOPSIS
To investigate whether findings by Mosblech et al. (2011) reflect a global role of InsP5 [2-OH] in increasing COI1 functions, we analyzed the resistance of ipk1-1 against Alternaria brassicicola, a fungal necrotroph that plants contain by COI1-dependent defenses (Leon-Reyes et al., 2010). In agreement with previous observations suggesting that InsP8 (which is strongly reduced in ipk1-1) is critical for COI1 activation (Laha et al., 2015), the ipk1-1 line showed a severe increase in susceptibility in this assay, similar to coi1 mutant plants (Fig. 1F; Supplemental Fig. S1A). This is also in agreement with a previous report showing increased susceptibility of the ipk1-1 line to Botrytis cinerea, another fungal necrotroph, in an assay where whole plants were sprayed with fungal spores and analyzed for plant survival (Murphy et al., 2008). We have repeated this assay with a complementary approach in which we spotted fungal spores onto the leaf surface and subsequently classified disease symptoms 72 h postinoculation. We again found increased susceptibility of the ipk1-1 line (Supplemental Fig. S1B) in complete agreement with Murphy et al. (2008). Collectively, these data question the idea that InsP5 [2-OH] globally activates COI1 functions in vivo.
ANISOTROPIC COORDINATION OF THE COI1 SOLENOID BY INOSITOL POLYPHOSPHATE SUGGESTS ACTIVATION OF THE JASMONATE RECEPTOR BY AN ALLOSTERIC SWITCH
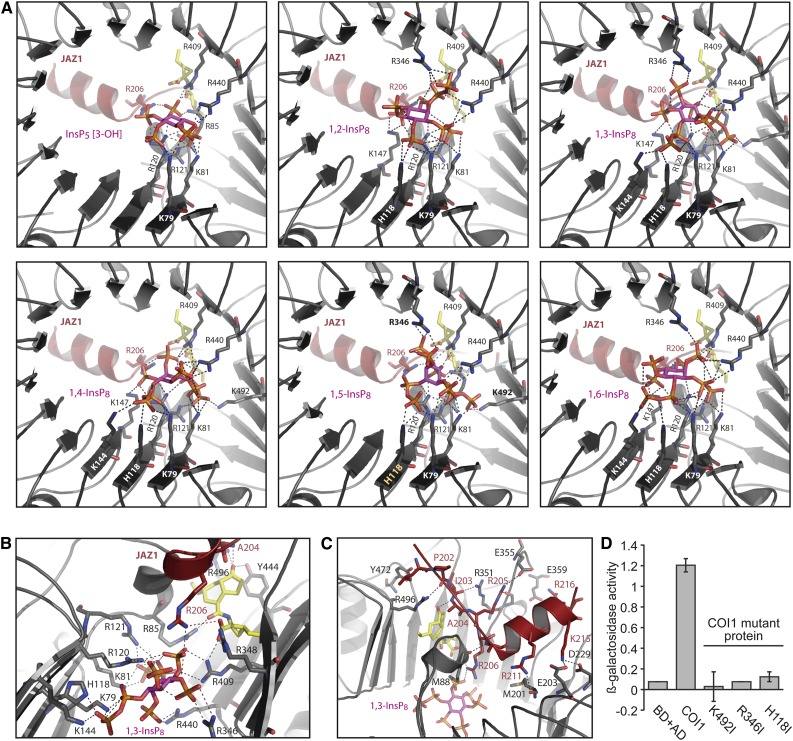
Our findings that the JAZ component of the jasmonate receptor has little, if any, effect on the relative inositol polyphosphate binding affinities (Fig. 1, B–E) suggest that COI1 largely determines the inositol polyphosphate binding specificity of the jasmonate receptor. This may be explained by in silico molecular docking experiments, which predicted the solenoid-fold of the F-box protein COI1 to provide an intricate network of electrostatic interactions engaging in inositol polyphosphate coordination (Laha et al., 2015). These docking experiments also predicted 1,5-InsP8 to be a better ligand of the jasmonate receptor complex as compared with InsP5 [3-OH], supporting a physiological role of InsP8 jasmonate perception in agreement with defective defense gene expression and defective defenses against insect herbivores and necrotrophic fungi in plants compromised in InsP8 synthesis (Laha et al., 2015). Unfortunately, the isomer identity of plant InsP8 remains unknown. While our previous work indicates that VIH proteins convert 5-InsP7 to 1,5-InsP8 in yeast and thus have the ability to catalyze 1-PP bond formation (Laha et al., 2015), the isomer identity of VIH-dependent InsP8 in plants remains elusive. This is mainly because the isomeric nature of plant InsP7 is unknown. Since plant genomes do not encode Kcs1/IP6K enzymes (which are responsible for 5-InsP7 production in nonplant eukaryotes) and since plant InsP7 synthetases have not yet been identified, the structure identification of plant InsP7 and InsP8 remains a challenging task for future research. In addition, low amounts of these molecules in plant extracts complicate a thorough analysis. Assuming that VIH proteins retain their 1-PP synthetase activity independent of the InsP7 substrate, we also performed in silico docking experiments with the remaining 1,X-InsP8 isomers. All 1,X-InsP8 isomers occupy largely overlapping sites of the presumptive inositol polyphosphate binding pocket (Fig. 2A; Supplemental Table S1). As we have previously seen for 1,5-InsP8 and InsP5 [3-OH], all inositol polyphosphates are coordinated by a single electrostatic interaction with the JAZ1 degron residue Arg-206 (Fig. 2, A and B; Supplemental Table S1). All 1,X-InsP8 isomers are furthermore predicted to form extensive interactions with the highly basic concave surface of the COI1 solenoid. Interestingly, these interactions stabilize and hold together the two faces of the inner wall of the Leu-rich repeat (LRR) solenoid that are distal and proximal to the hormone binding pocket (Fig. 2, A and B). At the distal face, the following COI1 residues are predicted to coordinate 1,X InsP8: Lys-79, Lys-81, His-118, Arg-120, and Arg-121. Additional residues at the distal face are Arg-85 (for 1,3-InsP8; 1,4-InsP8; 1,5-InsP8), Lys-144 (for 1,3-InsP8; 1,4-InsP8; 1,6-InsP8), and Lys-147 (for 1,2-InsP8; 1,3-InsP8; 1,4-InsP8; 1,6-InsP8). COI1 residues at the proximal face near the hormone binding site that are predicted to coordinate inositol polyphosphate are Arg-409, Arg-440, and additionally Arg-346 (for 1,2-InsP8; 1,3-InsP8; 1,5-InsP8; 1,2-InsP8) and Arg-492 (for 1,3-InsP8; 1,4-InsP8; 1,5-InsP8; Fig. 2, A and B; Supplemental Table S1). The anisotropic nature of these interactions (which are partially compensated for by four single phosphate ions in the inositol polyphosphate-free crystal structure; PDB ID: 3OGM) is likely to have a strong effect on the elliptical shape of the LRR solenoid. Coronatine forms a salt bridge and hydrogen bond network with COI1 residues Arg-85, Arg-348, Arg-409, Tyr-444 and Arg-496. Two of these residues coordinate all (Arg-409) or most (Arg-85) inositol polyphosphate isomers and further stabilize the shape of the solenoid (Fig. 2, A and B). The elliptical shape in turn is likely critical for efficient recruitment of the JAZ1 degron to the top surface of the carboxy-terminal LRR domain: besides hydrophobic packing, a number of polar interactions stabilize the COI1-JAZ1 interface. For instance, strong interactions are mediated by a hydrogen bond formed between the backbone carbonyl of Ala-207 in JAZ1 and the backbone amide of COI1 residue Met-88, by the hydrogen bond interaction of Tyr-472 (COI1) with the backbone carbonyl of Leu-201 (JAZ1), a hydrogen bond donated by COI1 residue Arg-351 to the JAZ1 backbone carbonyl of Ile-203, a salt bridge formed between the side chain of JAZ1 residue Arg-205 and the carboxyl group of Glu-355 (COI1), a hydrogen bond donated by the same JAZ1 residue to the backbone carbonyl of Gly-352, a salt bridge formed between side chains of COI1 residue Glu-359 and Arg-216 in JAZ1, salt bridges formed between COI1 residues Glu-203/Asp-229 and JAZ1 residue Lys-215, hydrogen bonds between the COI1 backbone carbonyl of Met-203 and the side chain of JAZ residue Arg-211, as well as hydrogen bonds between the backbone carbonyl of Pro-202 in JAZ1 and the coronatine-interacting COI1 residue Arg-496 (Fig. 2C; Supplemental Fig. S2). The interaction is further stabilized by a hydrogen bond between the backbone amide group of JAZ1 residue Ala-204 and the keto moiety of the hormone mimic coronatine, as well as by the interaction between JAZ1 residue Arg-206 and a phosphate group of the inositol polyphosphate ligand as mentioned above (Fig. 2C; Supplemental Fig. S2). Because of the involvement of several strong polar backbone interactions (eight in total), it seems likely that small changes in the elliptical shape of the COI1 solenoid will have a strong effect on JAZ recruitment because backbone interactions cannot adjust easily compared with interactions mediated by amino acid side chains. We therefore propose that inositol polyphosphate might induce a conformational change or allosteric switch of the COI1 carboxy-terminal LRR solenoid that, with the help of coronatine/JA-Ile, allows docking of the JAZ degron.
Figure 2.
Anisotropic coordination of the COI1 solenoid by inositol polyphosphate suggests activation of the jasmonate receptor by an allosteric switch. A to C, Structural snapshots of the COI1-ASK1-JAZ1 degron complex bound to coronatine and different inositol polyphosphates (as indicated) generated from in silico docking experiments. COI1 (gray), JAZ1 degron (dark red), inositol polyphosphates (magenta stick), and coronatine (yellow stick) are presented. Residues employed for mutagenesis and yeast two-hybrid assays are depicted in bold (Arg-346, Lys-492) or orange (His-118) in the 1,5-InsP8 structure. Bottom views (A) of InsP5 [3-OH] and 1,X InsP8 structures, as well as side view (B) and top view (C) of the presumed 1,3-InsP8 and coronatine binding pockets, are shown. Dashed gray lines represent strong polar contacts; dashed firebrick lines mark a weak polar contact between the side chain of JAZ1 residue Arg-206 and the carboxy group of coronatine. D, JAZ12 interaction with wild-type or mutant COI1 in yeast was evaluated in the presence of 50 μm coronatine by coexpression of pGBKT7-COI1 (and mutated versions as indicated) with pGADT7-JAZ12 in yeast strain Y187 (Clontech) and subsequent quantification of β-galactosidase-mediated hydrolysis of ortho-nitrophenyl-β-d-galactopyranoside. Values represent means of two independent biological replicas ±se. BD+AD, yeast strain harboring the empty vector controls pGBKT7 (expressing the Gal4 DNA-binding domain) and pGADT7 (expressing the Gal4 activation domain).
To distinguish InsP5- and InsP8-dependent interaction of COI1-JAZ in the yeast system (in which, based on the catalytic activities of Kcs1/IP6K and Vip1/PPIP5K enzymes, the identity of InsP8 is likely to represent 1,5-InsP8; Wang et al., 2011; Draskovic et al., 2008), we have previously engineered single COI1 mutant proteins affected in residues His-118, Lys-492, and Arg-346. These residues were chosen since all three are predicted to interact with 1,5-InsP8, but for geometrical reasons, not all three residues can interact simultaneously with an InsP5 molecule (irrespective of InsP5 isomer identity). The observation that all single mutant COI1 proteins failed to interact with JAZ1 in a yeast two-hybrid assay suggested that, at least in yeast, InsP5 isomers are not critically involved in COI1-JAZ1 interaction (Laha et al., 2015). We have now extended these analyses to the interaction between COI1 and JAZ12. We have chosen JAZ12 because it is, together with JAZ11, most distantly related to JAZ1 (Cuéllar Pérez et al., 2014). While wild-type COI1 interacted robustly with JAZ12, all three single Ile substitutions of COI1 residues His-118, Lys-492, and Arg-346 strongly compromised COI1-JAZ interaction despite stable protein being made in all cases (Fig. 2D; Supplemental Fig. S3). These results suggest that for the interaction of COI1 with JAZ12, like JAZ1, in yeast, InsP5 isomers are unlikely to play a major role, providing further evidence that COI1, not the JAZ partner, determines inositol polyphosphate binding specificity.
FUTURE TASKS
We envisage that the ability of the jasmonate receptor to discriminate between inositol polyphosphate enantiomers might be employed as a tool to reveal isomer identity of these molecules in biological samples, independent of their precise role in activating jasmonate perception. To address more directly the role of inositol pyrophosphates in triggering an allosteric switch of the COI1 carboxy-terminal LRR solenoid, we propose molecular dynamics simulations and/or crystallization of various ASK1-COI1 complexes in the presence and absence of ligands, as well as traditional biochemical measurements of affinity. The latter two approaches are currently complicated by the lack of commercially available InsP8 isomers and, more importantly, because the isomer identity of plant InsP8 remains unknown. It will be a major task for future research to develop technologies to determine the structure of inositol polyphosphates when present in only small amounts in biological extracts. Additionally, it will be important to identify the proteins that generate InsP7 in plants so as to allow in vitro reactions to produce sufficient amounts of InsP7 and InsP8 for proper structure determination.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: VIH2 (At3g01310), JAZ1 (At1g19180), JAZ2 (At1g74950), JAZ4 (At1g48500), JAZ9 (At1g70700), JAZ12 (At5g20900), ASK1 (At1g75950), COI1 (At2g39940), and IPK1 (At5g42810). Accession numbers for T-DNA insertion lines are as follows: vih2-4 (GK-080A07), ipk1-1 (SALK_065337C), and coi1-t (SALK_035548).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Increased susceptibility of the Arabidopsis ipk1-1 mutant to fungal necrotrophs.
Supplemental Figure S2. Polar backbone interactions between JAZ1 and COI1 suggest strong influence of COI1 carboxy-terminal LRR solenoid shape on JAZ recruitment.
Supplemental Figure S3. Immunoblots of soluble lysates prepared from yeast transformants.
Supplemental Table S1. List of presumptive electrostatic interactions between inositol polyphosphate and the jasmonate receptor complex; script for plotting sigmoidal curves.
Acknowledgments
We thank Birgit Kemmerling for providing spores of A. brassicicola, Elke Sauberzweig and Louis-Philippe Maier for their excellent help with plant work, Charles Brearley and Marília K.F. de Campos for helpful discussions, and Kristina E. Ile for critically reading the manuscript. No conflict of interest declared.
Glossary
- JA
jasmonic acid
- LRR
Leu-rich repeat
References
- Brearley CA, Hanke DE (1996) Inositol phosphates in barley (Hordeum vulgare L.) aleurone tissue are stereochemically similar to the products of breakdown of InsP6 in vitro by wheat-bran phytase. Biochem J 318: 279–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Cuéllar Pérez A, Nagels Durand A, Vanden Bossche R, De Clercq R, Persiau G, Van Wees SC, Pieterse CM, Gevaert K, De Jaeger G, Goossens A, et al. (2014) The non-JAZ TIFY protein TIFY8 from Arabidopsis thaliana is a transcriptional repressor. PLoS One 9: e84891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M, Rangarajan P, Donahue JL, Williams SP, Land ES, Mandal MK, Phillippy BQ, Perera IY, Raboy V, Gillaspy GE (2014) Two inositol hexakisphosphate kinases drive inositol pyrophosphate synthesis in plants. Plant J 80: 642–653 [DOI] [PubMed] [Google Scholar]
- Draskovic P, Saiardi A, Bhandari R, Burton A, Ilc G, Kovacevic M, Snyder SH, Podobnik M (2008) Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem Biol 15: 274–286 [DOI] [PubMed] [Google Scholar]
- Gillaspy GE. (2013) The role of phosphoinositides and inositol phosphates in plant cell signaling. Adv Exp Med Biol 991: 141–157 [DOI] [PubMed] [Google Scholar]
- Hanke DE, Parmar PN, Caddick SEK, Green P, Brearley CA (2012) Synthesis of inositol phosphate ligands of plant hormone-receptor complexes: pathways of inositol hexakisphosphate turnover. Biochem J 444: 601–609 [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HF, Chang TY, Chiang SF, Wang WD, Charng YY, Chiou TJ (2014) Arabidopsis inositol pentakisphosphate 2-kinase, AtIPK1, is required for growth and modulates phosphate homeostasis at the transcriptional level. Plant J 80: 503–515 [DOI] [PubMed] [Google Scholar]
- Laha D, Johnen P, Azevedo C, Dynowski M, Weiß M, Capolicchio S, Mao H, Iven T, Steenbergen M, Freyer M, et al. (2015) VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in Arabidopsis. Plant Cell 27: 1082–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Lee DH, Cho HK, Kim SH, Auh JH, Pai HS (2015) InsP6-sensitive variants of the Gle1 mRNA export factor rescue growth and fertility defects of the ipk1 low-phytic-acid mutation in Arabidopsis. Plant Cell 27: 417–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F, MacRobbie EA, Brearley CA (2000) Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc Natl Acad Sci USA 97: 8687–8692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Reyes A, Van der Does D, De Lange ES, Delker C, Wasternack C, Van Wees SCM, Ritsema T, Pieterse CMJ (2010) Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 232: 1423–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menniti FS, Miller RN, Putney JW Jr, Shears SB (1993) Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J Biol Chem 268: 3850–3856 [PubMed] [Google Scholar]
- Mosblech A, Thurow C, Gatz C, Feussner I, Heilmann I (2011) Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J 65: 949–957 [DOI] [PubMed] [Google Scholar]
- Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, Dollins DE, Haystead TA, Ribeiro AA, York JD (2007) A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316: 106–109 [DOI] [PubMed] [Google Scholar]
- Munnik T, Nielsen E (2011) Green light for polyphosphoinositide signals in plants. Curr Opin Plant Biol 14: 489–497 [DOI] [PubMed] [Google Scholar]
- Munnik T, Vermeer JE (2010) Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant Cell Environ 33: 655–669 [DOI] [PubMed] [Google Scholar]
- Murphy AM, Otto B, Brearley CA, Carr JP, Hanke DE (2008) A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J 56: 638–652 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Goossens A (2011) The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, Cha J, Xu J, Xu R, Vandiver MS, Tyagi R, Tokhunts R, Koldobskiy MA, Fu C, Barrow R, et al. (2014) Inositol pyrophosphates mediate the DNA-PK/ATM-p53 cell death pathway by regulating CK2 phosphorylation of Tti1/Tel2. Mol Cell 54: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB, Ganapathi SB, Gokhale NA, Schenk TM, Wang H, Weaver JD, Zaremba A, Zhou Y (2012) Defining signal transduction by inositol phosphates. Subcell Biochem 59: 389–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens L, Radenberg T, Thiel U, Vogel G, Khoo KH, Dell A, Jackson TR, Hawkins PT, Mayr GW (1993) The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s). J Biol Chem 268: 4009–4015 [PubMed] [Google Scholar]
- Stevenson-Paulik J, Bastidas RJ, Chiou ST, Frye RA, York JD (2005) Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc Natl Acad Sci USA 102: 12612–12617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Thota SG, Bhandari R (2015) The emerging roles of inositol pyrophosphates in eukaryotic cell physiology. J Biosci 40: 593–605 [DOI] [PubMed] [Google Scholar]
- Tsui MM, York JD (2010) Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv Enzyme Regul 50: 324–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Falck JR, Hall TM, Shears SB (2011) Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat Chem Biol 8: 111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MSC, Livermore TM, Saiardi A (2013) Inositol pyrophosphates: between signalling and metabolism. Biochem J 452: 369–379 [DOI] [PubMed] [Google Scholar]