Rice root genetic regions determining root architectural plasticity can be used for selection of improved adaptability to variable conditions.
Abstract
Future rice (Oryza sativa) crops will likely experience a range of growth conditions, and root architectural plasticity will be an important characteristic to confer adaptability across variable environments. In this study, the relationship between root architectural plasticity and adaptability (i.e. yield stability) was evaluated in two traditional × improved rice populations (Aus 276 × MTU1010 and Kali Aus × MTU1010). Forty contrasting genotypes were grown in direct-seeded upland and transplanted lowland conditions with drought and drought + rewatered stress treatments in lysimeter and field studies and a low-phosphorus stress treatment in a Rhizoscope study. Relationships among root architectural plasticity for root dry weight, root length density, and percentage lateral roots with yield stability were identified. Selected genotypes that showed high yield stability also showed a high degree of root plasticity in response to both drought and low phosphorus. The two populations varied in the soil depth effect on root architectural plasticity traits, none of which resulted in reduced grain yield. Root architectural plasticity traits were related to 13 (Aus 276 population) and 21 (Kali Aus population) genetic loci, which were contributed by both the traditional donor parents and MTU1010. Three genomic loci were identified as hot spots with multiple root architectural plasticity traits in both populations, and one locus for both root architectural plasticity and grain yield was detected. These results suggest an important role of root architectural plasticity across future rice crop conditions and provide a starting point for marker-assisted selection for plasticity.
The emerging problems of increased food demand, declining water tables, and increasingly unpredictable growing environments due to climate change require increasingly adaptable varieties in order to maintain high rice (Oryza sativa) yields under variable conditions. Although genotype × environment variation has typically been viewed as a challenge to plant breeding efforts (Basford and Cooper, 1998; Cooper et al., 1999), the variation across environments known as adaptive phenotypic plasticity is likely to be an important trait for future crop plants, as it increases plant fitness and survival (Nicotra and Davidson, 2010). In some future growing seasons, rice may face edaphic stresses such as drought stress (due to low rainfall or reduced availability of irrigation) and lower nutrient availability (due to decreased fertilizer or water availability), whereas in other seasons, the growing conditions may remain optimal. Specialized root architectures, although effective for a specific stress-prone environment, can be functionally maladaptive in different conditions (Ho et al., 2005; Poot and Lambers, 2008). Therefore, increased plasticity in root traits in terms of allocational, morphological, anatomical, or developmental plasticity (Sultan, 2000) could improve crop performance across future growing seasons (Aspinwall et al., 2015).
A number of previous studies have reported that plasticity in certain root traits conferred improved plant performance under stress or variable growth conditions to which the crop may be exposed. Under different types of drought stress, plasticity in root length density or total root length (Kano-Nakata et al., 2011; Tran et al., 2015) and lateral root length and/or branching (Suralta et al., 2010; Kano et al., 2011; Kano-Nakata et al., 2013) has been observed to improve shoot biomass, water uptake, and photosynthesis under drought in rice. Plasticity in the level of root aerenchyma development (measured as root porosity) was reported to result in higher shoot dry matter (Niones et al., 2013) and grain yield (Niones et al., 2012) under transient drought stress in rice, and plasticity in other anatomical traits has been hypothesized as a major reason for wheat (Triticum aestivum) being more drought tolerant than rice (Kadam et al., 2015). In a set of 42 native and crop species, plasticity in root depth was a better predictor of shoot response to drought than absolute root depth (Reader et al., 1993). Under low nitrogen, plasticity in specific root area, specific root length, and root tissue density conferred the least reduction in relative growth rate in 10 perennial herbaceous species (Useche and Shipley, 2010), and plasticity in maize (Zea mays) root growth angle improved yield (Trachsel et al., 2013). These examples provide strong evidence that root phenotypic plasticity can result in improved plant performance across variable conditions that include edaphic stress and would be an effective target for crop improvement efforts.
Deciphering the genetic and molecular mechanisms controlling root phenotypic plasticity will be necessary for effective selection and crop breeding efforts. Despite the likely genetic complexity behind the regulation of trait expression according to environmental conditions, phenotypic plasticity is heritable and selectable (for review, see Nicotra and Davidson, 2010). Genetic regions identified to be related to root phenotypic plasticity traits in crops include quantitative trait loci (QTLs) for root hair length plasticity in maize under low phosphorus (Zhu et al., 2005a), lateral root number plasticity in maize under low phosphorus (Zhu et al., 2005b), plasticity in aerenchyma development in response to drought stress in rice (Niones et al., 2013), and plasticity in lateral root growth in response to drought stress in rice (Niones et al., 2015). In wheat translocation lines, a plastic response of increased root biomass to drought was located to chromosome 1BS (Ehdaie et al., 2011). These identified genetic regions can be used in selection for the development of stress-tolerant crops.
Future rice crops will likely experience a range of soil conditions including prolonged aerobic periods, drought stress (progressive or intermittent), low soil fertility, and flooding. Rice may be established by either transplanting or direct seeding depending upon the amount and duration of initial rainfall. Therefore, the identification of root phenotypic plasticity traits suitable for adaptability to the particular range of conditions faced by rice crops, as well as the genetic regions responsible for those plasticity traits, may facilitate selection for wide adaptation of rice genotypes to variable conditions to confer stable yield. To address these needs, this study was conducted to identify the rice root phenotypic plasticity traits conferring adaptability across variable growth conditions by comparing contrasting genotypes from crosses between traditional and modern varieties. Our aim was to effectively quantify root architectural plasticity in order to identify which root traits may play the most important roles in rice adaptability. We hypothesized that the most plastic genotypes may show the most stable yields across environments.
RESULTS
Water, Seedling Establishment, and Phosphorus Treatments Imposed in Lysimeter, Field, and Rhizoscope Studies
A series of experiments was conducted in order to identify and select appropriate genotypes with plasticity traits, to investigate the role of root phenotypic plasticity in adaptation to a range of seedling establishment, nutrient, and drought stress conditions, and to identify the genetic regions related to the observed responses. The experiments were conducted (1) in a greenhouse lysimeter facility, in which plants were grown in approximately 1-m-tall cylinders and weighed and imaged weekly to monitor growth and water uptake; (2) in the field under well-watered and drought stress treatments in upland and lowland conditions to evaluate agronomic or physiological parameters; and (3) in controlled-environment Rhizoscope studies, in which root growth was imaged through transparent boxes filled with glass beads to measure the response to a low-phosphorus treatment (Table I; refer to Table I legend for abbreviations not defined in the text or figure legends). Subsets of genotypes from two populations (the Aus 276 population and the Kali Aus population) that contrasted for yield in an initial field evaluation were used for root sampling (16 genotypes in the field physiology experiment and 40 genotypes in the greenhouse lysimeter and Rhizoscope experiments) to evaluate root architectural plasticity.
Table I. Experiments conducted and traits considered in this study.
Ave root diam, Average root diameter; CT, canopy temperature; DTF, days to 50% flowering; gS, stomatal conductance; GY, grain yield; L, lowland; LDS, lowland drought stress; %LR, percentage lateral root (>60 cm); LRW, lowland rewatered; NS, nonstress (control); PHT, plant height; Plas, plasticity; RDW, root dry weight; RGR, relative growth rate from 24 to 31 d after sowing (DAS); RL, root length; RLD, root length density; R/S, root-shoot ratio; S, stress; SDW, shoot dry weight; SL, shoot length; TWU, total water uptake; U, upland; UDS, upland drought stress; URW, upland rewatered.
| Study | Treatment | Env. Code | Date of Planting/Transplanting | Observed Traits |
|---|---|---|---|---|
| Lysimeter | Lowland drought stress | LDS stress | August 17, 2012/August 29, 2012 | TWU, maximum root depth, SDW, RDW (0–20, 20–40, 40–60, and >60 cm), R/S, Ave root diam (>60 cm), RLD (>60 cm), %LR (>60 cm), forks (>60 cm), Plas RDW (0–20, 20–40, 40–60, and >60 cm), Plas %LR (0–20, 20–40, 40–60, and >60 cm) |
| Lowland rewatered | LRW stress | August 17, 2012/August 29, 2012 | ||
| Lowland control (for LDS) | LDS control | August 17, 2012/August 29, 2012 | TWU, maximum root depth, SDW, RDW (0–20, 20–40, 40–60, and >60 cm), R/S, Ave root diam (>60 cm), RLD (>60 cm), %LR (>60 cm), forks (>60 cm) | |
| Lowland control (for LRW) | LRW control | August 17, 2012/August 29, 2012 | ||
| Direct-seeded upland drought stress | UDS stress | August 17, 2012 | TWU, maximum root depth, SDW, RDW (0–20, 20–40, 40–60, and >60 cm), R/S, Ave root diam (>60 cm), RLD (>60 cm), %LR (>60 cm), forks (>60 cm), Plas RDW (0–20, 20–40, 40–60, and >60 cm), Plas %LR (0–20, 20–40, 40–60, and >60 cm) | |
| Direct-seeded upland rewatered | URW stress | August 17, 2012 | ||
| Direct-seeded upland control (for UDS) | UDS control | August 17, 2012 | TWU, maximum root depth, SDW, RGR, RDW (0–20, 20–40, 40–60, and >60 cm), R/S, Ave root diam (>60 cm), RLD (>60 cm), %LR (>60 cm), forks (>60 cm) | |
| Direct-seeded upland control (for URW) | URW control | August 17, 2012 | ||
| Field agronomic | Direct-seeded upland nonstress | 2012UNS | January 3, 2012 | GY, DTF, PHT |
| Direct-seeded upland stress | 2012US | January 6, 2012 | ||
| Direct-seeded upland nonstress | 2013UNS | December 20, 2012 | ||
| Direct-seeded upland stress | 2013US | January 22, 2013 | ||
| Lowland control | 2012LNS | December 10, 2011/January 5, 2012 | ||
| Lowland stress | 2012LS | December 20, 2011/January 14, 2012 | ||
| Lowland stress (Kali Aus population only) | 2013LS | December 19, 2012/January 7, 2013 | ||
| Field physiology | Direct-seeded upland nonstress | 2013UNS phys | December 17, 2013 | RLD, %LR, forks, Ave root diam, RDW (0–15, 15–30, 30–45, and 45–60 cm), GY |
| Direct-seeded upland stress | 2013US phys | December 17, 2013 | CT, gS, soil water potential, RLD, %LR, forks, Ave root diam, RDW (0–15, 15–30, 30–45, and 45–60 cm), Plas RLD (0–15, 15–30, 30–45, and 45–60 cm), Plas %LR (0–15, 15–30, 30–45, and 45–60 cm), Plas RDW (0–15, 15–30, 30–45, and 45–60 cm), GY | |
| Rhizoscope | Control phosphorus | P | July 8, 2013 | RL in rhizobox, root number >30 cm, root number >20 cm, shoot length, tiller number, SDW, RDW (0–15, 15–30, and >30 cm), R/S, Ave root diam, root angle |
| Low phosphorus | P/8 | July 8, 2013 | RL in rhizobox, RL, root number >30 cm, root number >20 cm, tiller number, SL, SDW, RDW (0–15, 15–30, and >30 cm), R/S, Ave root diam, root angle, Plas RL in rhizobox, Plas RL, Plas root number >30 cm, Plas root number >20 cm, Plas tiller number, Plas SL, Plas SDW, Plas RDW (0–15, 15–30, and >30 cm), Plas total root dry weight, Plas R/S, Plas Ave root diam, Plas root angle |
The stress treatments applied in all experiments were severe enough to elicit a plastic response in multiple root architectural parameters. The treatments applied in the lysimeter experiment had significant effects on all traits measured, and significant genetic variability was observed in TWU, RGR, RDW at the depth of 0 to 20 cm, SDW, and water use efficiency (WUE; Supplemental Table S1). In the Aus 276 population, SDW was reduced by 24.7% and 35.3% and WUE was increased by 11.7% and 14.9% by drought stress in the lowland and upland treatments, respectively. Similarly, SDW was reduced by 27.8% and 17.3% and WUE was increased by 10.5% and 20.6% by drought stress in the lowland and upland treatments, respectively, in the Kali Aus population (Supplemental Table S1). The RDW below 60 cm was increased by 21.8% and 48.3% in the Aus 276 populations and by 12.5% and 43.1% in the Kali Aus population under upland drought and rewatered conditions, respectively (Supplemental Table S1).
The effects of stress in the field agronomic trials varied by season, with the soil in the upland trials typically drying more rapidly than in the lowland trials based on visual observation. Likewise, the effects of stress on grain yield were most severe in the upland stress trials (Supplemental Table S2). The drought treatment in the field physiology trial showed a progressive dry down throughout the experiment (Supplemental Fig. S1) that was only interrupted by rainfall or rewatering at 65 and 81 d after sowing. Significant genotypic variation was observed for percentage lateral roots at depths of 0 to 15 cm and 15 to 30 cm in the stress treatment in the Aus 276 population and at depths of 15 to 30 cm and 30 to 45 cm in the Kali Aus population (Supplemental Table S3), as well as for soil moisture, canopy temperature, and stomatal conductance (Supplemental Fig. S2). Drought stress reduced the root diameter by 28.8% and 21.9% in the Aus 276 population and by 31.4% and 27.5% in the Kali Aus population at depths of 30 to 45 cm and 45 to 60 cm, respectively (Supplemental Table S3).
In the Rhizoscope study, the low-phosphorus treatment led to 18% and 26% increases in RDW at 0 to 15 cm and 15 to 30 cm, respectively (Supplemental Table S4). Although the RDW below 30 cm was not significantly affected by the low-phosphorus treatment, the number of roots below 30 cm was increased 47% by the low-phosphorus treatment (Supplemental Table S4).
Root Phenotypic Plasticity
Root phenotypic plasticity was calculated as the relative change in each root trait under stress as compared with the control treatment. In general, most root architectural traits at depth showed positive plasticity values in the field and lysimeter studies (i.e. root growth at depth increased under drought; Table II), although significant genotypic variation ranging from positive to negative plasticity values were observed (Tables II and III). Root growth at shallow depths in the Rhizoscope study showed positive plasticity values in response to low phosphorus (Table IV). Some root phenotypic plasticity traits were correlated with each other in the field and lysimeter experiments, and a high degree of colinearity was observed among root phenotypic plasticity traits in the Rhizoscope experiment (Supplemental Tables S5–S10). In the Aus 276 population, root architectural plasticity values at depth in the field studies were correlated with root architectural plasticity values at certain depths in the lysimeter and Rhizoscope studies (Supplemental Table S11). In both populations, negative correlations were observed between plasticity values in the field and lysimeter drought studies (mid to deeper depths) and plasticity values at deeper depths in the Rhizoscope low-phosphorus study. That is, genotypes that showed increased growth at depth under drought showed decreased root growth at depth under low phosphorus.
Table II. Genotypic differences in root architectural plasticity at different soil depths under different water and seedling establishment treatments in the greenhouse lysimeter experiment in the Aus276 and Kali Aus populations.
Values shown are mean plasticity values for all genotypes calculated from the listed drought stress treatments and each respective well-watered treatment. Significant differences among genotypes are indicated by asterisks: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. See Table I for abbreviation definitions.
| Experiment | Treatment | Trait | Aus 276 | Kali Aus |
|---|---|---|---|---|
| Lysimeter | LDS | Plas RDW 0–20 cm | 0.0536 | 0.063 |
| Plas RDW 20–40 cm | 0.207* | 0.151 | ||
| Plas RDW 40–60 cm | 0.545 | 0.216 | ||
| Plas RDW >60 cm | 2.429 | 2.072 | ||
| LRW | Plas RDW 0–20 cm | −0.153 | −0.165 | |
| Plas RDW 20–40 cm | 0.310 | −0.044 | ||
| Plas RDW 40–60 cm | 1.050 | 0.603 | ||
| Plas RDW >60 cm | 1.518 | 1.413 | ||
| UDS | Plas RDW 0–20 cm | −0.124 | −0.209 | |
| Plas RDW 20–40 cm | 0.241 | 0.139 | ||
| Plas RDW 40–60 cm | 0.975 | 0.403 | ||
| Plas RDW >60 cm | 2.070 | 1.949 | ||
| URW | Plas RDW 0–20 cm | −0.117 | −0.029 | |
| Plas RDW 20–40 cm | 0.358 | 0.680 | ||
| Plas RDW 40–60 cm | 0.886** | 0.889** | ||
| Plas RDW >60 cm | 1.986 | 1.893 | ||
| Field physiology | 2013U phys | Plas RLD 0–15 cm | 0.457 | 0.468* |
| Plas RLD 15–30 cm | 0.0095** | 0.219 | ||
| Plas RLD 30–45 cm | −0.759** | −0.418*** | ||
| Plas RLD 45–60 cm | 0.0029* | 0.278* | ||
| Plas %LR 0–15 cm | 0.0117** | −0.0068 | ||
| Plas %LR 15–30 cm | −0.052* | −0.036 | ||
| Plas %LR 30–45 cm | −0.154* | −0.178 | ||
| Plas %LR 45–60 cm | −0.072 | −0.057 |
Table III. Genotypic differences in root, shoot, and root architectural plasticity traits in the Aus 276 and Kali Aus populations under control and low-phosphorus stress treatments in the Rhizoscope study.
Values shown are mean trait or plasticity values for each treatment. Significant differences among genotypes are indicated by asterisks: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
| Trait | Aus 276 |
Kali Aus |
Plasticity |
|||
|---|---|---|---|---|---|---|
| Control | Stress | Control | Stress | Aus 276 | Kali Aus | |
| Tiller number | 5 | 5 | 5 | 6 | ||
| Shoot length (cm) | 64.82 | 62.50 | 64.06 | 64.06* | ||
| SDW (g) | 0.706* | 0.752 | 0.731 | 0.846* | ||
| Root length in rhizobox (cm) | 29.288 | 30.73 | 29.92 | 31.95 | 0.0482 | 0.087** |
| Root length without rhizobox beads (cm) | 30.603 | 31.57 | 31.33 | 32.96 | 0.0260 | 0.070*** |
| RDW 0–15 cm (g) | 0.121 | 0.144 | 0.131 | 0.161* | 0.186 | 0.331*** |
| RDW 15–30 cm (g) | 0.017 | 0.019 | 0.020 | 0.161 | 0.187 | 0.626* |
| RDW below 30 cm (g) | 0.00011 | 0.00043 | 0.00057 | 0.00082* | 2.522* | 5.512*** |
| Total root dry weight (g) | 0.139 | 0.163 | 0.151 | 0.191** | 0.181 | 0.382*** |
| Root-shoot ratio | 0.204* | 0.225** | 0.209*** | 0.232 | 0.106 | 0.086 |
| Root number >20 cm | 10* | 11 | 11* | 12* | 0.035 | 0.332*** |
| Root number >30 cm | 2 | 3 | 3 | 4* | 0.469 | 0.767*** |
| Root diameter (cm) | 0.056 | 0.062*** | 0.060 | 0.068 | 0.084* | 0.180*** |
| Root cone angle (°) | 71.94 | 74.45 | 80.56 | 74.44 | 0.053 | −0.012 |
Table IV. Multivariate linear regression equations of root architectural plasticity traits in each experiment related to the grain yield stability coefficient of each genotype.
TRL, Total root length; refer to Table I legend for other abbreviation definitions.
| Population/Experiment | P | r2 | Model (Yield Stability Coefficient) |
|---|---|---|---|
| Aus 276 | |||
| Field physiology | 0.0324 | 0.7456 | 1.15 − (1.23 × Plas RLD 0–15 cm) + (0.22 × Plas RLD 30–45 cm) + (0.35 × Plas RLD 45–60 cm) |
| Lowland lysimeter | 0.0125 | 0.4629 | 0.6434 − (0.0169 × Plas TRL >60 cm LDS) − (0.0380 × Plas TRL >60 cm LRW) − (0.0513 × Plas RDW >60 cm LDS) |
| Upland lysimeter | 0.0384 | 0.2904 | 0.3778 + (0.0597 × Plas TRL >60 cm UDS) − (0.5055 × Plas RDW 20–40 cm UDS) |
| Rhizoscope | 0.1485 | 0.1015 | 0.2543 + (0.6108 × Plas diameter) |
| Kali Aus | |||
| Field physiologya | 0.0864 | 0.9703 | 3.45 + (1.33 × Plas RLD 0–15 cm) − (4.82 × Plas RLD 15–30 cm) − (1.89 × Plas RLD 45–60 cm) + (6.23 × Plas %LR 15–30 cm) + (5.74 × Plas %LR 30–45 cm) + (4.10 × Plas %LR 45–60 cm) |
| Lowland lysimeterb | 0.055 | 0.1896 | 1.29 − (0.34 × Plas RDW 20–40 cm LRW) |
| Upland lysimetera | 0.0067 | 0.5907 | 1.50 − (0.16 Plas TRL >60 cm URW) + (0.53 × Plas %LR >60 cm URW) + (0.5 × Plas RDW 0–20 cm UDS) − (0.15 × Plas RDW 20–40 cm URW) |
| Rhizoscope | 0.1764 | 0.0894 | 1.25 + (0.82 × Plas angle) |
Excluding genotypes IR 92801-371-B and IR 92801-434-B with yield stability coefficients > 2.
Excluding parents Kali Aus and MTU1010.
Yield Stability across Field Experiments with Different Establishment Methods and Drought Stress Treatments
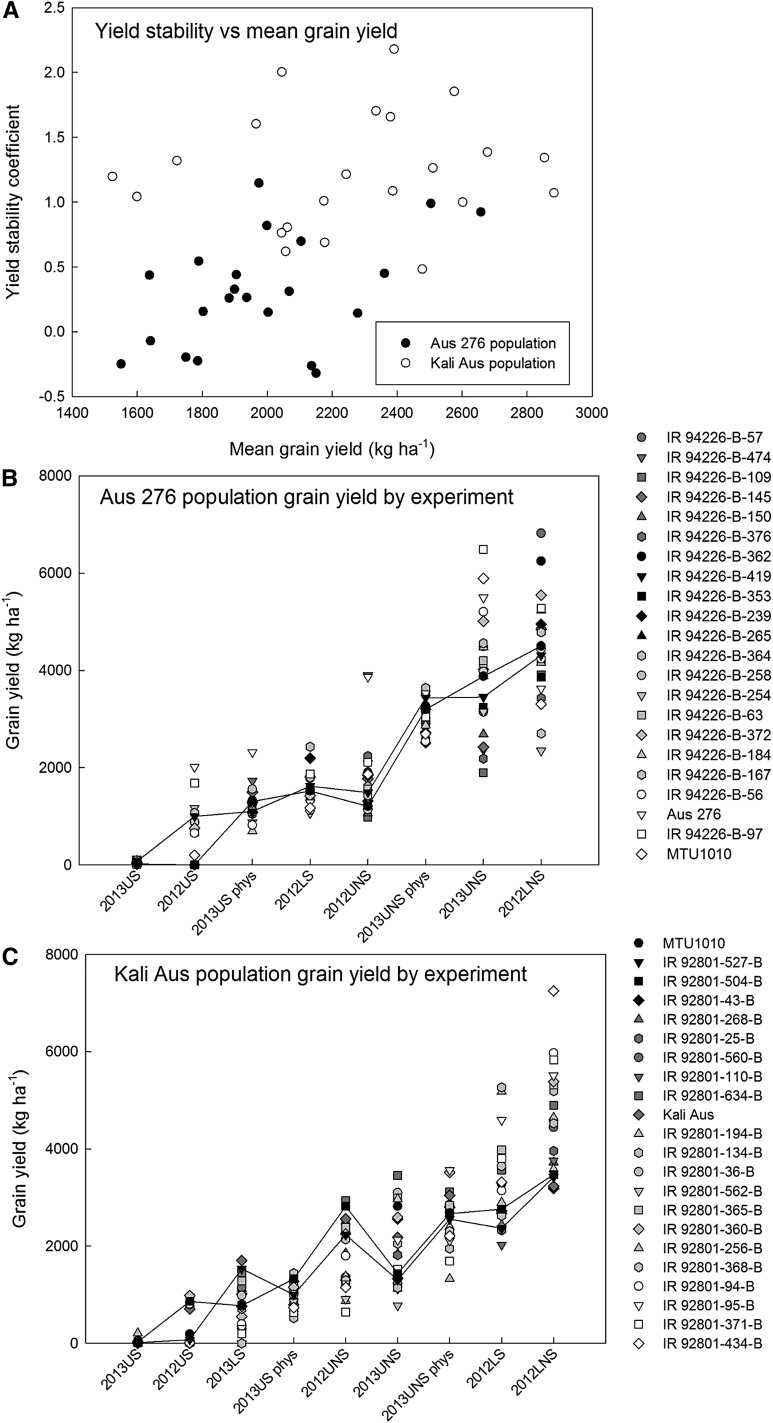
The goal of this study was to identify the root phenotypic plasticity traits that contributed to yield stability by conferring adaptability across different cultivation systems. An additive main effects and multiplicative interaction (AMMI-1) analysis showed large genotypic variation in yield stability across the multiple field environments evaluated (Fig. 1; Supplemental Table S12). In the Aus 276 population, IR 94226-B-362, IR 94226-B-419, IR 94226-B-239, IR 94226-B-353, and IR 94226-B-265 showed relatively small coefficients, indicating lower sensitivity to environmental changes or above-average stability. The negative AMMI-1 yield stability coefficients observed in the Aus 276 population implies a yield depression for those genotypes under favorable environmental conditions. Of all genotypes in the Aus 276 population that showed small yield stability coefficients, IR 94226-B-419 and IR 94226-B-265 also were identified as stable yielding by the Eberhart-Russell model (Supplemental Table S12). In the Kali Aus population, MTU1010, IR 92801-527-B, IR 92801-504-B, and IR 92801-43-B had relatively low coefficients, indicating lower sensitivity to environmental changes or above-average stability. Yield stability was generally independent of mean yield across field experiments (Fig. 1A). The stable-yielding genotypes listed above showed moderate yield across experiments compared with the other genotypes (Fig. 1, B and C) and were selected for further analysis.
Figure 1.
Yield stability and grain yield across field trials. A, Plots of yield stability coefficients from the AMMI-1 analysis in relation to the mean yield across all field trials. B, Grain yield in the Aus 276 population. C, Grain yield in the Kali Aus population. Each point represents one genotype. Yield stability coefficients close to zero represent genotypes with the best yield stability, and negative yield stability coefficients indicate a decrease in yield in the high-yielding environments. The x axes of B and C indicate field trials in order of mean trial yield. In B and C, genotypes are listed in order of yield stability coefficient, and black symbols indicate lines with the most stable grain yield. Selected genotypes IR 94226-B-265 and IR 94226-B-419 in the Aus 276 population and IR 92801-504-B and IR 92801-527-B in the Kali Aus population are indicated by the lines shown.
Relationships among Yield Stability, Root Architectural Plasticity, and Drought Response
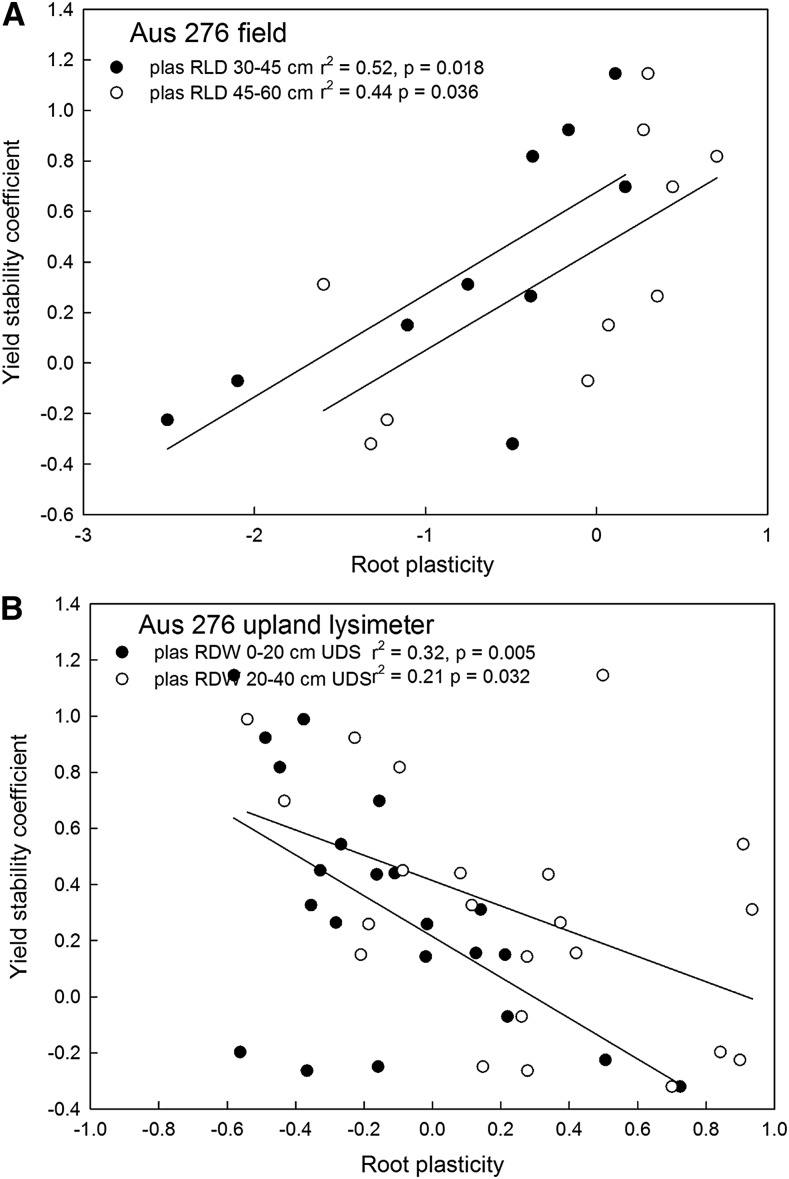
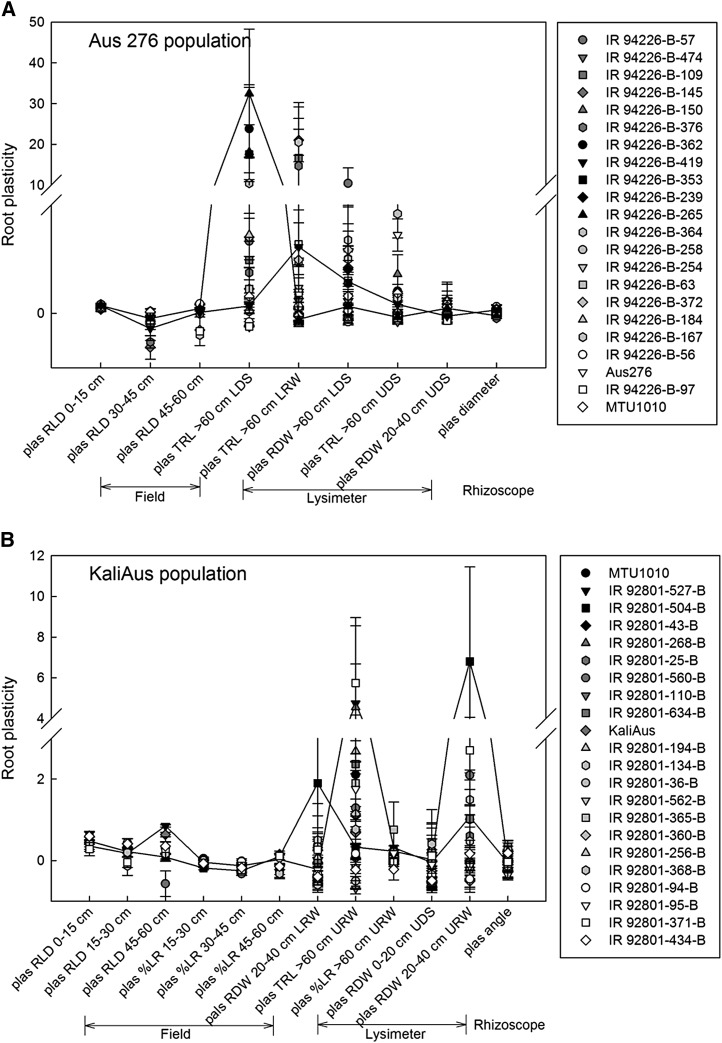
Individual plasticity traits related to yield stability were deep root length density in the field (a positive relationship) and shallow root dry weight in the upland lysimeter study (a negative relationship) in the Aus 276 population (Fig. 2; Supplemental Tables S5 and S6). In both populations, significant relationships between combinations of root plasticity traits and yield stability were indicated by multiple regression (Table IV). Among the root plasticity traits related to yield stability, the selected stable-yielding genotypes generally showed a higher degree of root architectural plasticity in some traits compared with the medium and unstable-yielding genotypes (Fig. 3). Genotypes IR 94226-B-265 and IR 94226-B-419 in the Aus 276 population and IR 92801-504-B and IR 92801-527-B in the Kali Aus population showed a greater degree of architectural plasticity than other genotypes in terms of root length density and total root length, especially at the deepest depths measured. Equations relating yield stability to plasticity in the root diameter and angle in the Rhizoscope study, although not significant (P = 0.1 and P = 0.09), also were determined by step-wise multiple regression (Table IV; Fig. 2). Furthermore, the stable-yielding genotypes showed more dispersed root growth (e.g. a larger root cone angle) in the low-phosphorus treatment compared with the control treatment in the Rhizoscope study (Fig. 4).
Figure 2.
Correlations of yield stability coefficients for each genotype in the Aus populations with plasticity in deep RLD in the field physiology trial (A) and plasticity in shallow RDW in the upland lysimeter experiment (B).
Figure 3.
Genotypic variation in root architectural plasticity traits in the Aus 276 population (A) and the Kali Aus population (B). The plasticity traits shown are those related to grain yield stability in multiple linear regressions for each root experiment. The grain yield stability coefficient was determined by an AMMI-1 analysis, where values close to zero indicate more stable grain yield across field experiments. Genotypes are listed in order of yield stability coefficient, and black symbols indicate lines with the most stable grain yield. Selected genotypes IR 94226-B-265 and IR 94226-B-419 in the Aus 276 population and IR 92801-504-B and IR 92801-527-B in the Kali Aus population are indicated by the lines shown.
Figure 4.
Root architecture images of the most yield-stable genotypes in the Aus 276 and Kali Aus populations and the parents under control (P; A–G) and low-phosphorus (P/8; H–N) stress conditions in the Rhizoscope study. Genotypes shown are the selected stable-yielding genotypes from each population (A–D and H–K) and the parents (E–G and L–N).
A functional role of root architectural plasticity related to yield stability, however, was less straightforward. In the Aus 276 population, yield-stable genotypes IR 94226-B-265 and IR 94226-B-419 showed generally lower canopy temperature in the field (Supplemental Fig. S2) and higher water uptake in the lysimeters compared with MTU1010 (Supplemental Fig. S3), but these genotypic differences were not observed in the stomatal conductance measurement (Supplemental Fig. S2). Likewise, stable-yielding genotype IR 92801-504-B in the Kali Aus population showed slightly lower soil moisture levels in the field (Supplemental Fig. S2F), indicating greater water uptake, but these observations were not confirmed by the other water-uptake parameters measured. Nevertheless, these functional responses were independent of the number of days to 50% flowering, which was similar between the yield-stable genotypes and the parents (Supplemental Table S13).
Genomic Regions Related to Root Architectural Plasticity and Grain Yield
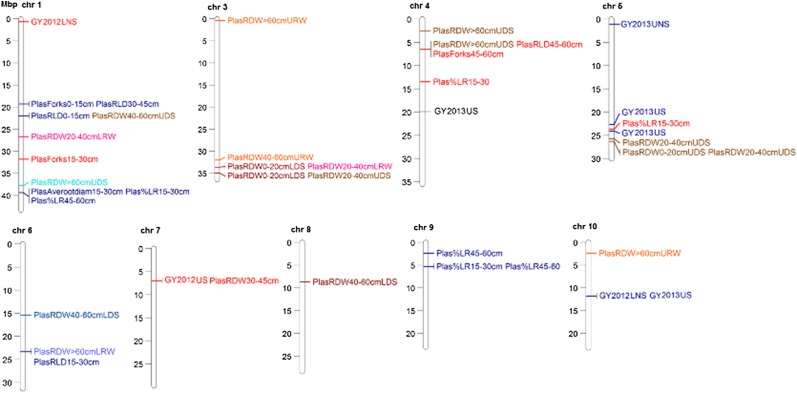
To facilitate the implementation of an efficient selection strategy for the observed root architectural plasticity traits, we aimed to identify the genomic regions related to those traits. Since a relatively small set of 20 genotypes was used for the root phenotypic plasticity analysis due to the labor required for those measurements, we used a two-step approach to the identification of genomic regions in order to increase our confidence in the loci identified. First, a single-nucleotide polymorphism (SNP) marker analysis was conducted in which a total of 235 and 219 SNP markers distributed on all 12 chromosomes showed polymorphisms in the Aus 276 and Kali Aus populations, respectively.
Out of these SNPs, 113 (lysimeter) and 68 (field) loci were found to be significantly related to traits measured in the Aus 276 population, and 88 (lysimeter) and 101 (field) markers were significantly related to traits measured in the Kali Aus population. Then, a marker class analysis was conducted to test if the trait values differed significantly at each marker locus. The marker class analysis for the respective traits was significantly different at 37 (lysimeter) and 23 (field) loci out of 113 (lysimeter) and 68 (field) loci in the Aus 276 population and for 50 (lysimeter) and 11 (field) loci out of 88 (lysimeter) and 101 (field) loci in the Kali Aus population (Fig. 5; Supplemental Tables S14–S17). No significant loci were identified among the traits measured in the seedling-stage Rhizoscope study, perhaps due to the age of the plants and the limited number of replicates.
Figure 5.
Chromosome map of significant loci for grain yield and root architectural plasticity detected under lysimeter and field conditions in the Aus 276 (blue shades) and Kali Aus (red shades) populations. LNS, Lowland nonstress; UNS, upland nonstress; US, upland stress; refer to Table I legend for other abbreviation definitions.
Two genomic loci (id1024972 for the Aus 276 population and id4002562 for the Kali Aus population) stood out as hot spots where three root architectural plasticity traits were correlated with the same SNP marker (Fig. 5; Supplemental Tables S15–S17). At locus id7001156, grain yield and root architectural plasticity traits were correlated with the same SNP marker. Some of the alleles for root architectural plasticity and grain yield were contributed by the traditional donor parents (Aus 276 or Kali Aus) and some were contributed by the recipient parent MTU1010.
DISCUSSION
In selected progeny from crosses with traditional rice varieties and a prominent variety cultivated over a large area in south Asia (MTU1010), we observed that the most yield-stable genotypes were generally those that showed the greatest degree of root architectural plasticity in the field or lysimeters across drought-stressed and well-watered experiments under both transplanted and direct-seeded conditions. The yield stability conferred by the root architectural plasticity traits explored in this study would be desirable from a rice farmer’s perspective, since they would result in more consistent performance across seasons with unpredictable environmental conditions.
Yield and yield stability showed different relationships with root phenotypic plasticity. We observed direct relationships between individual root architectural plasticity and yield stability as well as significant relationships between combinations of root architectural plasticity traits and yield stability (Figs. 2 and 3). Although the stable lines with plasticity traits were not the lowest yielding across experiments, the most root-plastic/yield-stable genotypes also were not the highest yielding, indicating a tradeoff or undesirable linkages between root architectural plasticity and the highest yield in particular environments. Agronomic tradeoffs to root architectural plasticity may occur when multiple resources are limited (Ho et al., 2004); this may be the case in this study, in which multiple rice establishment methods were evaluated, including dry direct seeding (which is typically prone to more edaphic stresses than flooded transplanted conditions; Kumar and Ladha, 2011). Furthermore, it is possible that the tradeoff in high yield potential among genotypes showing the greatest degree of root architectural plasticity may be due to genetic linkage rather than functional tradeoffs. Unfavorable linkages between high yield under drought and undesirable traits such as tall plant height and very early flowering have been reported previously, and the linkages were successfully broken through breeding to develop high-yielding, medium-duration drought-tolerant rice varieties (Swamy et al., 2013; Vikram et al., 2015). If this is the case in our study, such undesirable linkages could be broken through precise identification and fine-mapping of the genomic regions governing the root plasticity traits identified here.
The positive plasticity values observed in response to stress indicate that the growth of that particular root trait was increased by the stress applied. This response is distinct from an allometric response, in which larger root mass is related to larger shoot size because, although root growth increased under stress, shoot growth decreased under stress (Supplemental Tables S1, S2, and S4). One limitation to the approach of analyzing plasticity in this study was that no plasticity value could be calculated at soil depths at which roots were present in the stress treatment but not in the control treatment. Another common approach is to calculate a plasticity coefficient based on the slope of the genotypic value versus the environment mean (as described by Sadras et al., 2009), but this approach would require root architectural plasticity to be evaluated in a greater number of experiments than in our study.
The positive correlations in plasticity values for root architectural traits between the two drought experiment systems (field and lysimeter) suggest that the most root-plastic genotypes would consistently show a plastic response in different drought environments (i.e. after transplanting or direct seeding or in different soil types). The negative correlations in plasticity values under drought (field and lysimeter experiments) with plasticity values under low phosphorus (Rhizoscope study) reflect the contrasting root growth response to these two stresses based on the availability of the limiting resource (at deeper soil depths in the case of drought and at shallow soil depths in the case of low phosphorus). Furthermore, these negative relationships with the low-phosphorus Rhizoscope study indicate that the most root-plastic genotypes under drought also would show a relatively greater degree of plasticity under low phosphorus, although at different soil depths.
The combination of plasticity in RDW, RLD, and percentage lateral roots as related to yield stability may indicate different and complementary functional roles of those traits for the acquisition of different soil resources at different locations in the soil profile. The correlations of yield stability coefficients and root plasticity traits (Fig. 2) and genomic colocation of grain yield with root plasticity traits (Fig. 5) further support the role of plasticity in improving rice yield stability. Combinations of multiple root plasticity traits in response to drought and/or low-phosphorus stress (rather than the absolute values of those traits in a single condition) have been related to genotypic variation for adaptation to contrasting environments (Fort et al., 2015). Such trait complementarities may result in improved performance beyond the sum of that conferred by the individual traits, resulting in trait synergism (York et al., 2013). In this study, no single functional parameter was strongly related to trends in yield or root architectural plasticity (Supplemental Figs. S2 and S3). The functional complexity of root phenotypic plasticity is reflected by the different trends in water uptake patterns indicated by canopy temperature, soil moisture, and lysimetric measurements, which also are likely influenced by shoot characteristics.
In addition to root architectural plasticity, plasticity in traits such as root anatomy, WUE, and phenology has been reported to be related to more stable plant performance across varying environments in various species (Sadras et al., 2009; Niones et al., 2012, 2013; Kenney et al., 2014). In the case of rice, phenological plasticity in response to drought may be difficult to assess because rice shows delayed flowering under drought, and this delay can be reduced by plasticity in root architectural traits that improve water uptake (i.e. an interaction between different plasticity traits can affect the overall response to drought). Nevertheless, further research on plasticity in other drought-response traits could complement the adaptability conferred by root architectural plasticity.
Although hundreds of rice root/drought QTLs have been reported (for review, see Kamoshita et al., 2008; Courtois et al., 2009; Gowda et al., 2011), few genomic regions for root architectural plasticity have been identified. Although the set of genotypes used in this study was small, we were able to identify genomic regions related to root architectural plasticity traits using a marker class analysis approach, some of which colocate with previously reported QTLs related to phenotypic plasticity in rice. The effectiveness of this approach using just 20 genotypes per population in the field studies is evidenced by the identification of a locus for grain yield at the same locus on chromosome 10 as a QTL for grain yield reported by Sandhu et al. (2015) from a population of 300 genotypes. In terms of plasticity, the region on chromosome 1 (loci id1023892 and id1024972) is located near qDTY1.1, a major-effect drought-yield QTL that has been observed to confer plastic responses to drought, including increased root growth at depth and regulation of shoot growth (Vikram et al., 2015), as well as a hot spot for root traits including an increased proportion of deep roots by drought in the OryzaSNP panel (Wade et al., 2015). No plasticity traits from this study were colocated with the recently identified QTL qLLRN-12 for plasticity in lateral root growth under soil moisture fluctuation stress (Niones et al., 2015), although a locus at which root traits were detected in the Aus 276 population (id12001321 on chromosome 12) colocates with qLLRN-12.
The genotypic differences in root architectural plasticity and related genomic regions reported here support the idea that phenotypic plasticity is under genetic control and is selectable. The contribution of most root phenotypic plasticity loci by the traditional donor parents (Aus 276 and Kali Aus) is likely due to the variable stress-prone environments under which these genotypes were developed compared with the more optimal environments under which MTU1010 was bred. We used MTU1010 as the recipient parent in our study because it is one of the few newly developed rice varieties that have shown consistent performance across locations and variable conditions in India; this may explain why MTU1010 also contributed some of the alleles governing plasticity traits in this study. Although some genetic regions for root phenotypic plasticity traits have now been identified, the mode of genetic control of phenotypic plasticity remains largely unknown (although it has been hypothesized to be regulated by transcription factors, stress-inducible promoters, or epigenetics; Juenger, 2013; Zhang et al., 2013). Much more work is necessary in terms of fine-mapping of identified genomic regions and molecular characterization to understand how phenotypic plasticity is regulated in response to stress. Meanwhile, the SNPs identified to be related to root architectural plasticity traits in this study can be used in marker-assisted breeding to improve the yield stability of rice for future variable growing environments.
CONCLUSION
Traditional rice varieties Aus 276 and Kali Aus contributed genetic loci related to root architectural plasticity when crossed with improved variety MTU1010. Root architectural plasticity was related to yield stability across different water and crop establishment treatments. Such stable yield across environments will be increasingly desirable for rice farmers in the face of climate change-related variability. The SNPs related to root architectural plasticity traits can be used in marker-assisted breeding to improve the yield stability of rice for future variable growing environments.
MATERIALS AND METHODS
Plant Material
Two rice (Oryza sativa) breeding populations from the crosses Kali Aus/2 × MTU1010 (BC1F4) and Aus 276/3 × MTU1010 (BC2F4) that were developed through selfing and bulking of BC1F1 and BC2F1 populations, respectively, were used in this study. The recipient parent MTU1010 is a moderately drought-tolerant rice variety that is widely grown in India based on its high-yielding ability, adaptability to transplanted as well as direct-seeded conditions in both wet seasons and dry seasons, desirable quality traits, and acceptable tolerance to major biotic stresses. This variety was chosen as the recipient parent to investigate the potential for real improvement of one of the best available varieties that can be achieved through the incorporation of root plasticity traits. Of the donor parents, Kali Aus is a medium-duration drought-tolerant aus line from India and drought tolerance donor (Sandhu et al., 2014), and Aus 276 is an early-maturing drought-tolerant aus pure line from Bangladesh that has been characterized to contribute traits beneficial for direct-seeded conditions (Sandhu et al., 2015). From a set of 294 genotypes from the Kali Aus/2 × MTU1010 (BC1F4; Kali Aus population) and 300 genotypes from the Aus 276/3 × MTU1010 (BC2F4; Aus 276 population) mapping populations grown in the 2012 dry season under reproductive stage stress conditions, the 10 highest and 10 lowest yielding genotypes from each population were selected for the lysimeter, field agronomic, and Rhizoscope studies. For the field physiological studies, a set of eight genotypes from each population was selected to include a range of genotypes on the basis of water uptake and dry root biomass data from the lysimeter experiment.
Greenhouse Lysimeter Experiment
A lysimeter experiment was conducted during the 2012 wet season at the International Rice Research Institute (IRRI) in Los Banos, Laguna, Philippines (14°10′11.81′′N, 121°15′39.22′′E). Forty genotypes were planted (20 from each population; 10 previously classified as high yielding and 10 previously classified as low yielding) along with the three parents (Kali Aus, Aus 276, and MTU1010) in three water treatments (well-watered control, drought stress, and rewatered following drought stress) with two establishment treatments (direct-seeded upland and transplanted lowland conditions). A separate well-watered control treatment was included for each stress treatment for comparison and calculation of plasticity. The stress treatments were as follows: LDS, UDS, LRW, and URW. The lysimeters were arranged in a split-plot design, with water treatments as the main plots and genotypes as subplots. Six concrete tanks (1.35-m depth, 3.5-m width, and 6.8-m length) within the greenhouse were used, with one replicate of four to five treatments in each tank and a total of 1,204 lysimeters in the experiment.
The plants were grown in lysimeters constructed from 0.95-m-long × 0.18-m-diameter PVC cylinders with plastic liners and filled with 23 and 27 kg of dry, sieved soil (bulk density, 1.1 g cm–3) from the IRRI upland farm in the lowland and upland treatments, respectively. The soil was manually compressed with a metal plate after each 5 kg of soil was added, to a height of 70 cm for the lowland treatments and to 90 cm for the upland treatments. Wet lowland paddy soil that had been cleaned of debris was then added on top of the upland soil for the lowland treatment, leaving a clearance of 5 cm at the top of the cylinder. Three holes were drilled at the bottom of the cylinders and plastic liners allowed drainage, but these were sealed with a rubber plug to maintain flooded conditions in the lowland well-watered treatment, in the lowland stress treatments before the initiation of stress, and in the rewatered treatment after rewatering.
Seeds of each genotype were germinated in petri dishes, and three seeds per genotype were transferred to the lysimeters at 4 DAS for upland treatments. For the lowland treatments, germinated seeds were first transferred to seedling trays at 4 DAS, and then three seedlings were transplanted in each lysimeter at 16 DAS. The three plants per lysimeter were thinned to two seedlings at 24 DAS and then to one seedling at 31 DAS. Shoots of the thinned seedlings were dried in an oven at 60°C for 3 d, and early vegetative vigor was calculated in terms of RGR: [ln (dry shoot weight at sampling 2) – ln (dry shoot weight at sampling 1)]/(date of sampling 2 − date of sampling 1).
In the upland and lowland drought and rewatered treatments, the stress was initiated at 33 DAS by draining water from the lysimeters and by withholding the addition of water to those lysimeters. All lysimeters were then covered with polythene sheets sealed around the base of each plant to minimize direct evaporation, in order to ensure that only water loss by transpiration was measured. In the well-watered control treatment, the exact amount of water transpired was replaced on each weighing date to maintain the soil near field capacity (upland) or flooded (lowland) for the duration of the experiment. Following the initial drought stress period, the rewatered treatment was maintained flooded after rewatering at 72 DAS. Ambient conditions outside the greenhouse during the study period averaged 27.6°C, 85.9% relative humidity, and 14.2 MJ d−1 solar radiation.
Plant water uptake was monitored by weighing the lysimeters at 36, 43, 50, 57, 64, and 71 DAS. Water use was calculated as the difference between the initial lysimeter weight and the weight at each respective weighing date; these values were summed to calculate cumulative water use and total water uptake. Weighing was conducted according to the procedure described by Kijoji et al. (2013) using a cylinder-lifting system that consisted of an electric hoist (Shopstar Electric Chain Hoist; Columbus McKinnon) attached to a custom-built gantry crane that rolls along the top of the cement tank walls. Cylinders were lifted one at a time and placed on a weighing balance (KERN SCE-3.0; Kern and Sohn) that was connected to a laptop computer to record the weights.
Harvest of the shoots was carried out at 74 DAS for the drought treatment (LDS and UDS) and respective well-watered controls and at 89 DAS for the rewatered treatment (LRW and URW) and respective well-watered controls. Plants were cut at the stem base and oven dried to measure the shoot dry weight. WUE was calculated as the ratio of the total shoot biomass at harvest to the amount of water transpired by the plant over the entire drought stress period. After the shoot harvest, the plastic liners were pulled out of the lysimeters and the soil columns were cut into four depths (0–20 cm, 20–40 cm, 40-60 cm, and below 60 cm). The soil/root samples were stored at 4°C until washing within 2 to 3 weeks. Roots from each soil depth were washed carefully by repeatedly mixing the soil with water in a container and pouring the root water suspension over a 1-mm-diameter plastic sieve to separate living roots from soil and debris. The cleaned roots below 60 cm were then stored in 70% ethanol until they were scanned (Epson V700), and the scanned images were analyzed for architectural attributes using WinRhizo version 2007 d (Régent Instruments). Diameter classes of less than 0.05 mm, 0.05 to 0.1 mm, 0.1 to 0.2 mm, 0.2 to 0.5 mm, 0.5 to 1 mm, and greater than 1 mm were used, of which those less than 0.2 mm were considered lateral roots and used to calculate the percentage of total root length as lateral roots (percentage lateral roots). Root branching was determined by the forks parameter in WinRhizo. All root samples were then oven dried at 75°C for 3 d and weighed to determine the root dry weight at each soil depth sampled. The root-shoot ratio was determined by dividing total root weight by the shoot dry weight per plant.
Field Studies
Agronomic Trials
Upland and lowland field trials for agronomic traits (2012DUN, 2012DUS, 2013DUN, 2013DUS, 2012DLN, 2012DLS, 2013DLN, and 2013DLS) were conducted in 2012 and 2013 at the IRRI (Table I) on a set of approximately 300 genotypes each from the Kali Aus (BC1F4:F5) and Aus 276 (BC2F4:5) populations. For these field trials, the term lowland refers to flooded, puddled, transplanted, and anaerobic conditions, and direct seeded upland refers to directly sown, nonpuddled, nonflooded, aerobic conditions in leveled fields.
Each agronomic field trial was planted in an α-lattice design with two replications and 30 blocks in each replication. Each block included 10 randomized entries with single-row plots of 2 m and with border rows planted on the edges of each block. The three parents (Aus 276, Kali Aus, and MTU1010) were included as checks. For all lowland trials, seeds were sown in a raised-bed nursery, and 26-d-old (2012) and 20-d-old (2013) seedlings were transplanted to the main field with row spacing of 0.2 m and hill spacing of 0.2 m. In all upland trials, seeds were sown directly into the soil with a row spacing of 0.2 m. The well-watered control treatments were maintained flooded in the lowland trials and were sprinkler irrigated twice weekly in the upland trials. The stress trials were irrigated during establishment and early vegetative growth, but irrigation was stopped at 30 d after transplanting/51 DAS in the case of lowland trials, at 56 and 40 DAS in the 2012 and 2013 upland trials, respectively. Plots were reirrigated periodically when most genotypes were wilted and exhibited leaf rolling and drying in reproductive stage treatments. This type of cyclical stress is considered to be efficient for screening of populations consisting of genotypes with a broad range of growth durations to ensure that genotypes of all durations are stressed during reproductive development. Soil water potential was measured in the upland trials with tensiometers at a soil depth of 30 cm until crop maturity.
Plant height was recorded as the mean height of three random plants in each plot, measured from the base of the plant to the tip of the panicle during maturity stage. The number of days to flowering was recorded when 50% of the plants in the plot exerted their panicles. The plants were harvested at physiological maturity or when 80% to 85% of the panicles turned to golden yellow and the panicles at the base were already at the hard dough stage; harvested grains (from an area of 0.4 m2 per plot) were threshed and oven dried for 3 d at 50°C, moisture content was measured using a grain moisture meter (Riceter; Kett Electric Laboratory), and grain weight data were normalized to a moisture content of 14% to determine grain yield (kg ha−1).
Physiology Trials
The same 40 genotypes and checks were planted in the field trials for physiological traits at the IRRI during the 2013 dry season (Table I) in an α-lattice design in four replicates. Each block within a replicate consisted of 11 plots, with four rows per plot of 3 m length and an interrow spacing of 0.25 m under direct-seeded upland flooded and direct-seeded upland drought stress conditions. Irrigation was supplied using an overhead sprinkler during the first week after sowing and then by surface flooding every 3 d. The direct-seeded upland stress trial was drained at 46 DAS. Based on the 2012 greenhouse lysimeter experiment data, a total of eight genotypes from each population were selected with different combinations of high and low water uptake and root dry weight for root measurement in the field as well as characterization of aboveground response to drought and agronomic traits.
To monitor soil moisture levels, four tensiometers (at a 30-cm soil depth) and 57 PVC tubes (4 cm diameter and 1 m long) were installed as soon as the soil was near field capacity after draining the stress treatment. Volumetric soil moisture content was measured through the PVC tubes at 10-cm increments to a depth of 70 cm using a Diviner 2000 (Sentek Sensor Technologies), and water table depth from a 1-m-deep perforated tube was monitored up to three times per week. The PVC tubes for soil moisture readings were installed within the plots of 16 selected genotypes and the three parental genotypes and were located at the mid point between rows and hills, approximately 30 cm from the edge of the plot. Stomatal conductance was measured three times weekly using a Delta T AP4 porometer (Delta-T Devices). Measurements were taken on the abaxial side of the last fully elongated leaf.
Grain yield in the physiology trial was recorded as in the agronomic trials from a harvested area of 1.5 m2. Soil samples for root measurements were acquired with a 4-cm-diameter core in both the drought and well-watered treatments to a depth of 60 cm at 82 DAS. Cores were sampled at the mid point between rows and hills. The samples were separated into depth increments of 15 cm, and three subreplicate samples were collected per plot. The root samples from each depth were washed, scanned, and analyzed for root length density, percentage lateral root length, average root diameter, branching, and root dry weight as described for the lysimeter study.
Rhizoscope Study
A Rhizoscope study was conducted at CIRAD, Montpellier, France (43°37 N, 03°52 E), in July 2013. The Rhizoscope is a high-throughput screening system for phenotyping rice root traits (Courtois et al., 2013). Two-dimensional nail board rhizoboxes made of Plexiglas (50 cm × 20 cm × 2 cm) were filled with transparent soda-glass beads to simulate soil conditions and bathed with a recirculating nutrient solution in four large tanks with 48 rhizoboxes each, for a total of 192 rhizoboxes in the experiment. The beads were homogenously arranged in the rhizoboxes without applying external pressure. After pregerminating seeds of all 40 genotypes (20 from each population) and four checks (Kali Aus, Aus 276, MTU1010, and IR64) at 28°C for 3 d, one well-developed seedling per rhizobox was planted on the top of the beads, with two replications of each genotype in two treatments (control and low phosphorus). To apply the two treatments, separate modified Yoshida nutrient solutions for the control and stress (low-phosphorus) conditions were used (Supplemental Table S18) that were maintained at pH 5.5. The photoperiod was 12 h, and air temperature inside the growth room averaged 23°C/28°C (night/day), humidity averaged 60%, photosynthetically active radiation averaged 350 mol m−2 s−1 at the plant level, and the solution temperature averaged 25°C. Plants were grown for 21 d, at which time the root systems were imaged. Tiller numbers were counted manually, and shoot and root length in the rhizobox before the removal of beads was measured using a centimeter scale. The deepest point reached by the roots also was measured after the plants were removed from the rhizobox. The numbers of crown roots reaching below depths of 20 and 30 cm were counted. Then, the root system was carefully washed to remove the remaining beads and cut into three segments (0–15 cm, 15–30 cm, and below 30 cm). Shoots and roots were dried in an oven at 72°C for 3 d. The total root dry weight was determined as the sum of the dry root weight in all three segments. Shoot dry weight, total root dry weight, root-shoot ratio, and root dry biomass in all three segments were measured. The angles of the most external crown roots to the left and right of the root-shoot junction were measured with ImageJ (Abràmoff, 2004). The sum of these two angles was used as the angle of the root cone in the analyses. Average root diameter was measured for the nodal roots reaching below 20 cm using ImageJ.
Root Phenotypic Plasticity Calculations and Statistical Analysis
Calculation of Root Phenotypic Plasticity
In this study, root phenotypic plasticity was defined as the response of root growth to a stress treatment compared with the control conditions. The method for calculating and analyzing root phenotypic plasticity focused on architectural traits that were measured in both stress and control conditions. From the lysimeter study, root architectural plasticity was calculated using root dry weight (0–20, 20–40, 40–60, and below 60 cm), proportion of lateral roots (below 60 cm), number of root branches/forks (below 60 cm), and average root diameter (below 60 cm). From the field physiology trial, root architectural plasticity was calculated using the data for root length density, proportion of lateral roots, branching/forks, average root diameter, and root dry weight at depths of 0 to 15 cm, 15 to 30 cm, 30 to 45 cm, and 45 to 60 cm from the soil core. For the Rhizoscope study, root architectural plasticity was calculated for root length in the rhizobox (before separating from the glass beads), number of roots below 20 and 30 cm, root length, number of tillers, shoot length, root dry weight 0 to 15 cm, 15 to 30 cm, and below 30 cm, total root dry weight, root diameter, root angle, and root-shoot ratio.
For each root architectural trait, plasticity was calculated using single replicates from the stress treatment and mean values from the control treatment:
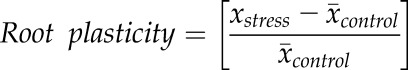 |
This approach to quantifying root plasticity allowed for standardization of the data and the use of all single replicates for statistical analysis. ANOVA was performed to compare genotypes for each plasticity calculation. This analysis was conducted in R version 3.1.2 (R Core Team, 2012).
Yield Stability Analysis
Grain yield data across all agronomic trials were compiled and analyzed to determine the most stable and high-yielding genotypes across trials. After harvest, each trial was classified for observed drought stress intensity based on yield reduction compared with the well-watered control according to Kumar et al. (2009): mild stress, less than 30%; moderate stress, 30% to 65%; severe stress, 65% to 85%; and overstressed, greater than 85% (Supplemental Table S2). Trials classified as having mild stress or being overstressed were excluded from the yield stability analysis due to poor expression of genetic variability. To conduct the yield stability analysis, the grain yield results were embedded in a mixed-model framework where environments were random factors and treatments were fixed factors. The variance components from these models were interpreted as measures of stability according to Piepho (1999). The choice of the appropriate covariance structure was based on the model with the lowest Akaike information criterion value. The best-fitting model for both populations was AMMI-1, in which a relatively small coefficient indicates lower sensitivity (above-average stability) to changing environmental conditions. The mean grain yield for the two populations was then plotted against the stability coefficients from the AMMI-1 model.
Correlating Root Architectural Plasticity Traits with Yield Stability
After calculating the plasticity for each root architectural trait, correlations among plasticity traits within each experiment were examined in a correlation matrix for each population by Pearson two-sided correlation analysis in R (Supplemental Tables S5–S10). Since the genetic variation for root architectural plasticity was colinear among some traits within an experiment (especially in the Rhizoscope study), the data sets were parsed to include only plasticity traits that were not correlated with each other in order to conduct a multiple linear regression analysis. Step-wise linear regression was performed using STAR version 2.0.1 to obtain an equation relating combinations of root architectural plasticity traits with the yield stability coefficient for each genotype.
Genotyping
Fresh leaf samples were collected from each genotype of both mapping populations grown in the greenhouse in the 2013 wet season at 21 DAS, and leaves were lyophilized. DNA was extracted using the modified CTAB protocol (Murray and Thompson, 1980). The agarose gel electrophoresis method was used to check the quality and quantity of DNA with a reference λDNA. The DNA samples were diluted with 1× TE into an equal concentration of 50 ng µL−1 and were submitted to the Genotyping Service Laboratory at the IRRI for genotyping using the GoldenGate 384-plex SNP Genotyping Assay on the BeadXpress platform (Illumina).
Single-Marker and Class Analysis
Given the limitations of quantifying root phenotypic plasticity in large numbers of mature plants, we conducted a genotypic analysis that was adapted to a small population (i.e. 20 genotypes per population), in which an SNP marker analysis was followed by a marker class analysis for confirmation of the most significant loci related to each trait. A total of 219 and 235 polymorphic SNP markers distributed on all 12 chromosomes were identified for Kali Aus and Aus 276, respectively. Single-marker regression analysis was carried out to identify significant markers associated with traits using Windows QTL Cartographer version 2.5 (Wang et al., 2011). The single-marker analysis can be described by the model
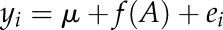 |
where yi is the trait value for the ith individual in the population, μ is the population mean, f(A) is a function of marker genotype A, and ei is the residual associated with the ith individual. When a gene Q is located near marker A, the trait controlled by Q can be modeled by the marker A.
Then, to confirm that the significant marker was really linked to that particular trait, class analysis for each significant marker corresponding to the trait was carried out using the asreml script in R version 3.0.1 using the model
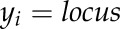 |
where yi is the trait value for the ith individual in the population. The class analysis provided the means of the marker classes (lines having the Aus 276 parent allele, lines having the MTU1010 allele, lines having the Kali Aus allele, and lines with a heterozgote allele) as well as the F test for comparing the marker classes to know if the marker classes differed significantly for each particular trait. For each marker, each allele was considered a class, and all of the members of the population with that genotype were considered an observation for that class. Data were typically pooled over replications to obtain a single quantitative trait value for each genotype. If the variance among marker classes was significant, only then was the marker reported to be associated with the respective trait. The markers significantly associated with each trait were plotted on the rice chromosome cv Nipponbare genome map using MapChart version 2.2 (Voorrips, 2002).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Soil water potential measured by tensiometers in field studies at the soil depth of 30 cm.
Supplemental Figure S2. Functional comparison of the parents with the root-plastic, yield-stable genotypes in the field physiology study.
Supplemental Figure S3. Cumulative water uptake in the lysimeter experiment for the genotypes showing highest yield stability across field studies.
Supplemental Table S1. Mean values of all genotypes and significance levels of genotypes for each treatment for root, shoot, and water-uptake traits in the greenhouse lysimeter study.
Supplemental Table S2. Field trial means for grain yield, days to flowering, and plant height as well as the stress classification based on trial grain yield means.
Supplemental Table S3. Root, shoot, and water-uptake traits measured in the field physiology experiment.
Supplemental Table S4. Mean values of all genotypes and significance levels of the treatment effect on different root and shoot traits in the Rhizoscope experiment.
Supplemental Table S5. Correlation matrix of root architectural plasticity traits measured in the Aus 276 population in the field physiology study.
Supplemental Table S6. Correlation matrix of root architectural plasticity traits measured in the Aus 276 population in the lysimeter study.
Supplemental Table S7. Correlation matrix of root architectural plasticity traits measured in the Aus 276 population in the Rhizoscope study.
Supplemental Table S8. Correlation matrix of root architectural plasticity traits measured in the Kali Aus population in the field physiology study.
Supplemental Table S9. Correlation matrix of root architectural plasticity traits measured in the Kali Aus population in the lysimeter study.
Supplemental Table S10. Correlation matrix of root architectural plasticity traits measured in the Kali Aus population in the Rhizoscope study.
Supplemental Table S11. Correlations of plasticity in RDW among the field, lysimeter, and Rhizoscope studies.
Supplemental Table S12. Grain yield stability in the Aus 276 and Kali Aus populations across experiments, according to the analyses showing the best-fitting variance-covariance structure according to the Akaike information criterion.
Supplemental Table S13. Days to 50% flowering in the yield-stable, root-plastic genotypes and the parents in field agronomic trials.
Supplemental Table S14. Significant loci identified for different traits under different lysimeter treatments in the Aus 276 population.
Supplemental Table S15. Significant loci identified for different traits under field conditions in the Aus 276 population.
Supplemental Table S16. Significant loci identified for different traits under different lysimeter treatments in the Kali Aus population.
Supplemental Table S17. Significant loci identified for different traits under field conditions in the Kali Aus population.
Supplemental Table S18. Composition of the modified Yoshida nutrient solutions used in the Rhizoscope study.
Supplementary Material
Acknowledgments
We thank M. Teresa Sta Cruz, Paul Cornelio Maturan, Leonardo Holongbayan, Eleanor Mico, Lesly Satioquia, Allan Los Añes, and Nicanor Turingan for assistance with experimental measurements; Jocelyn Guevarra and RuthErica Carpio for assistance with seed preparation; Violeta Bartolome, Avigail Cosico, and Emilie Thomas for assistance in data analysis; Alexandre Grondin for suggestions on the plasticity calculations; and the IRRI Genotyping Services Laboratory for SNP genotyping.
Glossary
- QTL
quantitative trait locus
- TWU
total water uptake
- RGR
relative growth rate
- RDW
root dry weight
- SDW
shoot dry weight
- WUE
water use efficiency
- AMMI-1
additive main effects and multiplicative interaction
- SNP
single-nucleotide polymorphism
- IRRI
International Rice Research Institute
- DAS
days after sowing
- UDS
upland drought stress
- URW
upland rewatered
- LRW
lowland rewatered
- LDS
lowland drought stress
Footnotes
This work was supported by the Monsanto Beachell Borlaug International Scholarship Programme.
Articles can be viewed without a subscription.
References
- Abràmoff M. (2004) Image processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Aspinwall MJ, Loik ME, Resco de Dios V, Tjoelker MG, Payton PR, Tissue DT (2015) Utilizing intraspecific variation in phenotypic plasticity to bolster agricultural and forest productivity under climate change. Plant Cell Environ 38: 1752–1764 [DOI] [PubMed] [Google Scholar]
- Basford KE, Cooper M (1998) Genotype × environment interactions and some considerations of their implications for wheat breeding in Australia. Aust J Agric Res 49: 153–174 [Google Scholar]
- Cooper M, Fukai S, Wade L (1999) How can breeding contribute to more productive and sustainable rainfed lowland rice systems? Field Crops Res 64: 199–209 [Google Scholar]
- Courtois B, Ahmadi N, Khowaja F, Price AH, Rami JF, Frouin J, Hamelin C, Ruiz M (2009) Rice root genetic architecture: meta-analysis from a drought QTL database. Rice (N Y) 2: 115–128 [Google Scholar]
- Courtois B, Audebert A, Dardou A, Roques S, Ghneim-Herrera T, Droc G, Frouin J, Rouan L, Gozé E, Kilian A, et al. (2013) Genome-wide association mapping of root traits in a japonica rice panel. PLoS ONE 8: e78037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehdaie B, Layne AP, Waines JG (2011) Root system plasticity to drought influences grain yield in bread wheat. Euphytica 186: 219–232 [Google Scholar]
- Fort F, Cruz P, Catrice O, Delbrut A, Luzarreta M, Stroia C, Jouany C (2015) Root functional trait syndromes and plasticity drive the ability of grassland Fabaceae to tolerate water and phosphorus shortage. Environ Exp Bot 110: 62–72 [Google Scholar]
- Gowda VRP, Henry A, Yamauchi A, Shashidhar HE, Serraj R (2011) Root biology and genetic improvement for drought avoidance in rice. Field Crops Res 122: 1–13 [Google Scholar]
- Ho M, Rosas J, Brown K, Lynch J (2005) Root architectural tradeoffs for water and phosphorus acquisition. Funct Plant Biol 32: 737–748 [DOI] [PubMed] [Google Scholar]
- Ho MD, McCannon BC, Lynch JP (2004) Optimization modeling of plant root architecture for water and phosphorus acquisition. J Theor Biol 226: 331–340 [DOI] [PubMed] [Google Scholar]
- Juenger TE. (2013) Natural variation and genetic constraints on drought tolerance. Curr Opin Plant Biol 16: 274–281 [DOI] [PubMed] [Google Scholar]
- Kadam NN, Yin X, Bindraban PS, Struik PC, Jagadish KSV (2015) Does morphological and anatomical plasticity during the vegetative stage make wheat more tolerant of water deficit stress than rice? Plant Physiol 167: 1389–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoshita A, Babu RC, Boopathi NM, Fukai S (2008) Phenotypic and genotypic analysis of drought-resistance traits for development of rice cultivars adapted to rainfed environments. Field Crops Res 109: 1–23 [Google Scholar]
- Kano M, Inukai Y, Kitano H, Yamauchi A (2011) Root plasticity as the key root trait for adaptation to various intensities of drought stress in rice. Plant Soil 342: 117–128 [Google Scholar]
- Kano-Nakata M, Gowda VRP, Henry A, Serraj R, Inukai Y, Fujita D, Kobayashi N, Suralta RR, Yamauchi A (2013) Functional roles of the plasticity of root system development in biomass production and water uptake under rainfed lowland conditions. Field Crops Res 144: 288–296 [Google Scholar]
- Kano-Nakata M, Inukai Y, Wade L, Siopongco JD, Yamauchi A (2011) Root development, water uptake, and shoot dry matter production under water deficit conditions in two CSSLs of rice: functional roles of root plasticity. Plant Prod Sci 14: 307–317 [Google Scholar]
- Kenney AM, McKay JK, Richards JH, Juenger TE (2014) Direct and indirect selection on flowering time, water-use efficiency (WUE, δ (13)C), and WUE plasticity to drought in Arabidopsis thaliana. Ecol Evol 4: 4505–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijoji A, Nchimbi-Msolla S, Kanyeka Z, Klassen S, Serraj R, Henry A (2013) Water extraction and root traits in Oryza sativa × Oryza glaberrima introgression lines under different soil moisture regimes. Funct Plant Biol 40: 54–66 [DOI] [PubMed] [Google Scholar]
- Kumar A, Verulkar S, Dixit S, Chauhan B, Bernier J, Venuprasad R, Zhao D, Shrivastava MN (2009) Yield and yield-attributing traits of rice (Oryza sativa L.) under lowland drought and suitability of early vigor as a selection criterion. Field Crops Res 114: 99–107 [Google Scholar]
- Kumar V, Ladha J (2011) Direct seeding of rice: recent developments and future research needs. Adv Agron 111: 297–413 [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra A, Davidson A (2010) Adaptive phenotypic plasticity and plant water use. Funct Plant Biol 37: 117–127 [Google Scholar]
- Niones JM, Inukai Y, Suralta RR, Yamauchi A (2015) QTL associated with lateral root plasticity in response to soil moisture fluctuation stress in rice. Plant Soil 391: 63–75 [Google Scholar]
- Niones JM, Suralta RR, Inukai Y, Yamauchi A (2012) Field evaluation on functional roles of root plastic responses on dry matter production and grain yield of rice under cycles of transient soil moisture stresses using chromosome segment substitution lines. Plant Soil 359: 107–120 [Google Scholar]
- Niones JM, Suralta RR, Inukai Y, Yamauchi A (2013) Roles of root aerenchyma development and its associated QTL in dry matter production under transient moisture stress in rice. Plant Prod Sci 16: 205–216 [Google Scholar]
- Piepho HP. (1999) Stability analysis using the SAS system. Agron J 9: 154–160 [Google Scholar]
- Poot P, Lambers H (2008) Shallow-soil endemics: adaptive advantages and constraints of a specialized root-system morphology. New Phytol 178: 371–381 [DOI] [PubMed] [Google Scholar]
- R Core Team (2012) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Reader R, Jalili A, Grime J, Spencer R, Matthews N (1993) A comparative study of plasticity in seedling rooting depth in drying soil. J Ecol 81: 543–550 [Google Scholar]
- Sadras VO, Reynolds MP, de la Vega J, Petrie PR, Robinson R (2009) Phenotypic plasticity of yield and phenology in wheat, sunflower and grapevine. Field Crops Res 110: 242–250 [Google Scholar]
- Sandhu N, Singh A, Dixit S, Sta Cruz MT, Maturan PC, Jain RK, Kumar A (2014) Identification and mapping of stable QTL with main and epistasis effect on rice grain yield under upland drought stress. BMC Genet 15: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu N, Torres RO, Sta Cruz MT, Maturan PC, Jain R, Kumar A, Henry A (2015) Traits and QTLs for development of dry direct-seeded rainfed rice varieties. J Exp Bot 66: 225–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SE. (2000) Phenotypic plasticity for plant development, function and life history. Trends Plant Sci 5: 537–542 [DOI] [PubMed] [Google Scholar]
- Suralta RR, Inukai Y, Yamauchi A (2010) Dry matter production in relation to root plastic development, oxygen transport, and water uptake of rice under transient soil moisture stresses. Plant Soil 332: 87–104 [Google Scholar]
- Swamy BPM, Ahmed HU, Henry A, Mauleon R, Dixit S, Vikram P, Tilatto R, Verulkar SB, Perraju P, Mandal NP, et al. (2013) Genetic, physiological, and gene expression analyses reveal that multiple QTL enhance yield of rice mega-variety IR64 under drought. PLoS ONE 8: e62795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP (2013) Maize root growth angles become steeper under low N conditions. Field Crops Res 140: 18–31 [Google Scholar]
- Tran TT, Kano-Nakata M, Suralta RR, Menge D, Mitsuya S, Inukai Y, Yamauchi A (2015) Root plasticity and its functional roles were triggered by water deficit but not by the resulting changes in the forms of soil N in rice. Plant Soil 386: 65–76 [Google Scholar]
- Useche A, Shipley B (2010) Plasticity in relative growth rate after a reduction in nitrogen availability is related to root morphological and physiological responses. Ann Bot (Lond) 106: 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikram P, Swamy BP, Dixit S, Singh R, Singh BP, Miro B, Kohli A, Henry A, Singh NK, Kumar A (2015) Drought susceptibility of modern rice varieties: an effect of linkage of drought tolerance with undesirable traits. Sci Rep 5: 14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE. (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93: 77–78 [DOI] [PubMed] [Google Scholar]
- Wade L, Bartolome V, Mauleon R, Vasant VD, Prabakar SM, Chelliah M, Kameoka E, Nagendra K, Reddy KR, Varma CM, et al. (2015) Environmental response and genomic regions correlated with rice root growth and yield under drought in the OryzaSNP panel across multiple study systems. PloS ONE 10: e0124127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng ZB (2011) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh [Google Scholar]
- York LM, Nord EA, Lynch JP (2013) Integration of root phenes for soil resource acquisition. Front Plant Sci 4: 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Fischer M, Colot V, Bossdorf O (2013) Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol 197: 314–322 [DOI] [PubMed] [Google Scholar]
- Zhu J, Kaeppler SM, Lynch JP (2005a) Mapping of QTL controlling root hair length in maize (Zea mays L.) under phosphorus deficiency. Plant Soil 270: 299–310 [Google Scholar]
- Zhu J, Kaeppler SM, Lynch JP (2005b) Mapping of QTLs for lateral root branching and length in maize (Zea mays L.) under differential phosphorus supply. Theor Appl Genet 111: 688–695 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.