Comparative sequence analysis delineates conserved noncoding sequences that are functionally relevant, a subset of which are conserved throughout green plants.
Abstract
Transcription factors (TFs) regulate gene expression by binding cis-regulatory elements, of which the identification remains an ongoing challenge owing to the prevalence of large numbers of nonfunctional TF binding sites. Powerful comparative genomics methods, such as phylogenetic footprinting, can be used for the detection of conserved noncoding sequences (CNSs), which are functionally constrained and can greatly help in reducing the number of false-positive elements. In this study, we applied a phylogenetic footprinting approach for the identification of CNSs in 10 dicot plants, yielding 1,032,291 CNSs associated with 243,187 genes. To annotate CNSs with TF binding sites, we made use of binding site information for 642 TFs originating from 35 TF families in Arabidopsis (Arabidopsis thaliana). In three species, the identified CNSs were evaluated using TF chromatin immunoprecipitation sequencing data, resulting in significant overlap for the majority of data sets. To identify ultraconserved CNSs, we included genomes of additional plant families and identified 715 binding sites for 501 genes conserved in dicots, monocots, mosses, and green algae. Additionally, we found that genes that are part of conserved mini-regulons have a higher coherence in their expression profile than other divergent gene pairs. All identified CNSs were integrated in the PLAZA 3.0 Dicots comparative genomics platform (http://bioinformatics.psb.ugent.be/plaza/versions/plaza_v3_dicots/) together with new functionalities facilitating the exploration of conserved cis-regulatory elements and their associated genes. The availability of this data set in a user-friendly platform enables the exploration of functional noncoding DNA to study gene regulation in a variety of plant species, including crops.
DNA sequences that are not actively transcribed and that are conserved across a large number of related species are called conserved noncoding sequences (CNSs). These regions are assumed to have biological relevance because nonfunctional sequences change at a higher rate during evolution compared with functional sequences (Tagle et al., 1988). The detection of CNSs in plants remains an ongoing challenge, because established methods applied in animals or fungi are not always compatible with the properties of plant genomes. The large phylogenetic distance between the currently sequenced dicot plant species hampers the use of lift overs, in which detected transcription factor binding sites (TFBSs) are transferred from one species to another through whole-genome alignments. The potential for this transfer is further decreased by the frequent occurrence of whole-genome duplications and genomic rearrangements in the genomes of flowering plants. Despite these challenges, within the Brassicaceae clade, a set of CNSs was successfully identified between closely related species (Haudry et al., 2013). Avoiding the step of whole-genome alignments and replacing it with a multiple pairwise alignment approach has proven to be a useful alternative method to detect CNSs in distantly related plants (Van de Velde et al., 2014). Various software tools also have been developed to identify regulatory regions without using sequence alignments but based on experimental features, such as coregulation, or using advanced computational methods (MacIsaac and Fraenkel, 2006). Although the naïve mapping of known or de novo-found binding sites to promoter regions is frequently used to explore cis-regulatory elements, this approach yields many false positives, because TFBSs often are short and typically contain some level of degeneracy in the binding motif (Tompa et al., 2005). The combination of alignment-free binding site detection combined with phylogenetic conservation of these regions has shown great promise, because the application of these methods shows significant overlap with experimental TFBSs (Van de Velde et al., 2014).
In the Brassicaceae, CNSs have been shown to be under a selective pressure that is comparable to that of protein-coding sequences (Haudry et al., 2013). CNSs also are enriched for regions of open chromatin in Arabidopsis (Arabidopsis thaliana) and, as such, provide a global perspective on possible protein binding to the genome (Van de Velde et al., 2014). In both of the above-mentioned studies, it was shown that CNSs greatly overlap with transcription factor (TF) chromatin immunoprecipitation sequencing (ChIP-Seq) bound regions. This is an important observation, because TFs play an important role in translating the genotypes of plants into their respective phenotypes by controlling the spatiotemporal expression of target genes through (combinatorial) binding on TFBSs. A direct application of this feature is the mapping of gene regulatory networks (GRNs) starting from CNSs (Kheradpour et al., 2007; Van de Velde et al., 2014). A GRN is a set of interactions between a TF and target genes and gives a global overview of how transcriptional control is steered in the cell through the activity of TFs.
In the past, much of the CNS research has been focused on Arabidopsis (Kaplinsky et al., 2002; Vandepoele et al., 2006, 2009; Freeling et al., 2007; Baxter et al., 2012; Haudry et al., 2013; Van de Velde et al., 2014) and grasses (Guo and Moose, 2003; Inada et al., 2003; Turco et al., 2013; De Witte et al., 2015), with the exception of the analysis of Baxter et al. (2012), where also footprints were obtained for grape (Vitis vinifera) and poplar (Populus trichocarpa). Given the limited and biased set of species with available CNSs, there is a great need for CNS detection in other plant species, because these CNSs offer a practical means to enhance the construction of GRNs in crops starting from well-studied model species. An exponent of these CNSs is called ultraconserved sequences, which are typically long stretches of sequences that are conserved across very large phylogenetic distances. In vertebrates, they are defined as regions that are at least 100 bp long and share 100% sequence identity (Stephen et al., 2008). A pioneering study in plant CNS research suggests that CNSs in grasses (plants) are smaller and far less frequent than those identified in mammalian genes (Kaplinsky et al., 2002). A recent attempt to identify very deeply conserved CNSs reported that sequences conserved throughout the Eudicot clade of flowering plants could be detected (Burgess and Freeling, 2014). The authors discovered that, based on 10 species, a subset of 37 CNSs could be found in all flowering plants. The detected CNSs were functionally similar to vertebrate CNSs, being highly associated with TF-encoding and developmental genes and also enriched in TFBSs (Burgess and Freeling, 2014).
We recently developed a phylogenetic footprinting approach to identify CNSs in Arabidopsis through the comparison with multiple dicot genomes. Comparator species were selected based on the presence of saturated substitution patterns, which means that noncoding regions that are not under functional constraint will have undergone, on average, one or more mutations. A combination of an alignment-based approach and a non-alignment-based approach was used to delineate CNSs. The alignment-based approach is best summarized as a multiple local alignment strategy, because local pairwise alignments are first identified and subsequently aggregated on the Arabidopsis reference genome in order to obtain multispecies footprints. The non-alignment-based approach, called comparative motif mapping (CMM), requires a candidate motif (e.g. a TFBS represented as a consensus sequence or position count matrix) as input and assesses the motif conservation in the promoter of an Arabidopsis gene. Conservation is scored based on the occurrence of the motif in the promoter regions of the orthologs from the query gene in other species, allowing for incomplete motif conservation (Van de Velde et al., 2014). Here, we applied this methodology to 10 dicot genomes and validated the functional importance of these regions by comparing them with experimentally determined TFBSs. We also show that a subset of these CNSs is very deeply conserved in the green plant lineage and can be applied to gain information about the function of TFs through functional enrichment of their predicted target genes.
RESULTS
Identification of CNSs in 10 Dicot Plant Genomes
A phylogenetic footprinting method that uses an alignment- and non-alignment-based approach was used to detect CNSs in 10 dicot species representative of eight plant families (Table I). For each query species, a set of comparator species was selected based on saturated substitution patterns in orthologous gene pairs (Supplemental Table S1). Comparator species also were selected with regard to the genome assembly and annotation quality (see “Materials and Methods”). Each query species was compared with a set of 13 comparator species: Arabidopsis, papaya (Carica papaya), cocoa tree (Theobroma cacao), rose gum (Eucalyptus grandis), peach (Prunus persica), melon (Cucumis melo), soybean (Glycine max), poplar, grape, tomato (Solanum lycopersicum), beet (Beta vulgaris), rice (Oryza sativa), and Amborella (Amborella trichopoda). Three different genomic sequence types were defined to identify CNSs (2 kb upstream, 1 kb downstream, and intron). In this analysis, upstream and downstream are used relative to the translation start site and translation stop site, respectively. This is done because it was shown previously that regulatory elements can be found in the 5′ and 3′ untranslated region (UTR; Chabouté et al., 2002; Liu et al., 2010; Wang and Xu, 2010). The second reason to include UTRs is that not all genes have information about their UTR available. Gene orthology information was retrieved with the PLAZA 3.0 integrative orthology method (Van Bel et al., 2012; Proost et al., 2015), which uses a combination of different detection methods to infer consensus orthology predictions, both for simple one-to-one gene relationships and for more complex many-to-many gene relationships (see “Materials and Methods”).
Table I. Overview of footprinting results for all species.
| Species | Plant Family | No. of Genes | Genome Size | No. of CNSs | Coverage | No. of Genes with CNSs | No. of CNSs per Gene | Percentage of Coding CNS | Percentage of CNSs within 500 bp | Median Length of CNS |
|---|---|---|---|---|---|---|---|---|---|---|
| Mb | Mb | bp | ||||||||
| Arabidopsis thaliana (Arabidopsis) | Brassicaceae | 33,602 | 120 | 74,381 | 1.0 | 19,474 | 3.82 | 0.09 | 62.00 | 11 |
| Brassica rapa (field mustard) | Brassicaceae | 40,998 | 284 | 92,578 | 1.3 | 29,277 | 3.16 | 0.34 | 60.17 | 11 |
| Eucalyptus grandis (rose gum) | Myrtaceae | 36,493 | 691 | 86,434 | 1.6 | 23,350 | 3.70 | 0.62 | 50.58 | 13 |
| Prunus persica (peach) | Rosaceae | 27,864 | 227 | 109,381 | 2.3 | 21,020 | 5.20 | 0.66 | 55.80 | 15 |
| Cucumis melo (melon) | Cucurbitaceae | 28,812 | 375 | 63,803 | 1.2 | 16,144 | 3.95 | 0.45 | 55.90 | 14 |
| Glycine max (soybean) | Fabaceae | 54,302 | 974 | 213,799 | 3.8 | 43,198 | 4.95 | 0.40 | 53.25 | 12 |
| Populus trichocarpa (poplar) | Salicaceae | 41,479 | 417 | 157,567 | 3.5 | 30,662 | 5.14 | 0.53 | 56.68 | 15 |
| Vitis vinifera (grape) | Vitaceae | 26,644 | 486 | 105,137 | 2.1 | 18,916 | 5.56 | 0.56 | 55.86 | 13 |
| Solanum lycopersicum (tomato) | Solanaceae | 34,859 | 824 | 63,428 | 1.2 | 19,721 | 3.22 | 0.88 | 53.57 | 13 |
| Solanum tuberosum (potato) | Solanaceae | 35,130 | 706 | 65,783 | 1.2 | 21,425 | 3.07 | 0.71 | 53.41 | 13 |
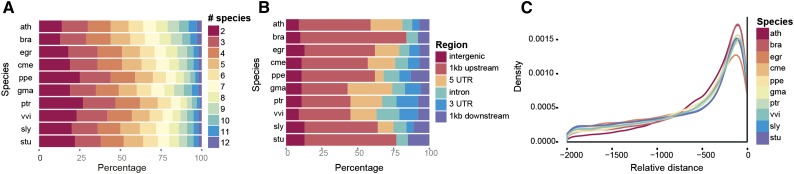
For the detection of CNSs, a multispecies alignment-based approach was applied using the Sigma aligner (Siddharthan, 2006). The CMM approach was used with an enlarged set of 1,211 input sequence motifs and positional weight matrices for 35 TF families (Supplemental Table S2; see “Materials and Methods”). The results of all footprinting analyses are reported in Table I. In total, 1,032,291 CNSs were detected for 243,187 genes (Supplemental Data Set S1). To determine whether any of the identified CNSs represent unannotated coding features, we performed a sequence similarity search of all CNSs against a large set of known plant proteins (see “Materials and Methods”). Across all species, only 5,223 CNSs, which corresponds to less than 1% of the total discovered set of CNSs, showed a significant BLASTX hit. These false-positive CNSs were discarded for downstream analysis. The largest number of CNSs was found in soybean (213,799), which also had the largest number of genes with a CNS. The smallest numbers of CNSs were found for tomato (63,428), potato (Solanum tuberosum; 65,783), and melon (63,803). Both soybean and poplar cover over 3 Mb in CNSs, which is 3.5 to 4 times as much as Arabidopsis, which has the smallest CNS sequence space (1 Mb). The mean number of CNSs per gene varies between 3.07 for potato and 5.56 for grape. The number of CNSs shows a strong correlation with the number of genes in the genome (r2 = 0.74), which is higher than the correlation with the genome size (r2 = 0.33). Because CNSs are detected per gene, this correlation is to be expected. The median length of CNSs per species varies between 11 and 15 bp (Table I). Whereas Arabidopsis and beet have the smallest median CNS length, peach and poplar have the largest. The median number of conserved orthologous species for each CNS is found between four and five comparator species (Fig. 1A), which shows that many CNSs are conserved in more than one comparator species and illustrates the multispecies nature of this approach. An evaluation of the location of the CNS relative to the query gene also was performed, revealing that the majority of CNSs are found on the 5′ side of the gene (Fig. 1B). Some species, such as grape, poplar, and peach, have a high fraction of CNSs that are found on the 3′ side compared with the other species analyzed. For poplar, this finding is supported by recently performed ChIP-Seq analysis for four TFs, in which three TFs were found to have 19% to 25% of the binding events occurring downstream of a gene (Liu et al., 2015a). In order to further investigate the positional differences of CNSs between species, a density distribution was made for all CNSs up to 2,000 bp upstream of a gene, showing three groups of CNS densities in the first 500 bp upstream (Fig. 1C). Arabidopsis and field mustard (Brassica rapa) show a high fraction (60% or greater), rose gum shows only 50%, and all other species are found to have a fraction of CNSs in the first 500 bp between 50% and 60%. There is a strong negative correlation (−0.8) between genome size and the percentage of CNSs found in the first 500 bp, which suggests that promoters of species with larger genomes tend to be more stretched out. This result is in agreement with a comparative analysis performed in grasses, where intergenic region expansions from the small Oropetium thomaeum to the larger Sorghum bicolor were observed (VanBuren et al., 2015).
Figure 1.
Overview of CNS properties for all query species. A, Overview of significantly conserved footprints in relation to the number of comparator species in which the footprint was conserved. B, Breakdown of CNSs over different structurally annotated genomic regions. C, Density of CNSs across the first 2 kb upstream of the translation start site.
Overlap with TF ChIP-Seq Data
To evaluate the functionality of the identified CNSs and to verify whether these conserved footprints can provide a template to computationally map TF target gene interactions, detailed comparisons of the CNSs were made with publicly available TF ChIP-Seq experiments from tomato, poplar, and soybean. The ASR1 TF ChIP-Seq data set from Ricardi et al. (2014) was used for tomato, a TF ChIP-Seq data set comprising two TFs (NAC and YABBY) was used from Shamimuzzaman and Vodkin (2013) for soybean, and two data sets from poplar were used, one containing the ARK1 TF (Liu et al., 2015b) and one containing four TFs (ARK2, PRE, PCN, and BLR; Liu et al., 2015a). The number of overlapping TF ChIP-Seq peaks for each set of CNSs of the corresponding species was determined with the requirement that a CNS had to completely overlap with a TF ChIP-Seq bound region. The overlap of CNSs with the respective TF ChIP-Seq bound regions is shown in Figure 2. In poplar, both ARK data sets show a high recovery (62%–64%) of chromatin immunoprecipitation peaks, as opposed to the recovery of PRE, which is rather low (11%). The recovery of the ASR1 data set is also very low (4%). Certain data sets have a very low (ASR1 and PRE) or high (ARK1 and YABBY) number of bound regions, compared with results from a recent overview study of TF ChIP-Seq analyses in Arabidopsis (Heyndrickx et al., 2014), which might have an influence on the results of the overlap analysis. Additionally, instead of determining the overlapping true-positive instances, we also estimated false positives by reshuffling the TF ChIP-Seq genomic locations 1,000 times across the genome and determining the overlap with CNSs detected for each species. The estimated number of false positives was used to determine the enrichment for known TF ChIP-Seq bound regions (observed number of elements over expected number of elements; see “Materials and Methods”). This approach does not guarantee that the reshuffled data set, which covers in essence randomly selected noncoding genomic regions that have no overlap with real bound regions, contains only true negatives. However, the shuffled data set can be used as a proxy to estimate the specificity. Although the recovery rate for individual TFs varied greatly, the enrichment analysis showed that for six out of eight TFs, the number of overlapping peaks was significantly higher compared with those expected by chance (P < 0.001; Table II).
Figure 2.
Recovery of TF ChIP-Seq bound regions using CNSs. The percentage of TF ChIP-Seq bound regions overlapping with a CNS for each individual TF is shown. Asterisks indicates that the P value of the enrichment was less than 0.001.
Table II. TF ChIP-Seq overlap and enrichment results.
| Species | ChIP-Seq Data | No. of Peaks | Observed Overlap | Expected Overlap | Enrichment Fold | P |
|---|---|---|---|---|---|---|
| Poplar | ARK1 | 14,463 | 8,833 | 2,286 | 3.742 | 0.001 |
| Poplar | ARK2 | 2,287 | 1,448 | 364 | 3.864 | 0.001 |
| Poplar | BLR | 5,674 | 1,564 | 593 | 2.564 | 0.001 |
| Poplar | PCN | 3,148 | 705 | 290 | 2.362 | 0.001 |
| Poplar | PRE | 658 | 67 | 61 | 1.063 | 0.243 |
| Soybean | NAC | 8,246 | 1,970 | 950 | 2.012 | 0.001 |
| Soybean | YABBY | 18,064 | 2,913 | 1,607 | 1.752 | 0.001 |
| Tomato | ARS1 | 225 | 8 | 5 | 1.600 | 0.114 |
Quantifying the Evolutionary Conservation of TF Target Gene Interactions
In order to obtain an overview of the evolutionary conservation of TF target gene interactions in the green plant lineage (Viridiplantae), the deep conservation of TFBSs was evaluated. Therefore, the CMM approach was repeated for Arabidopsis, but with a larger number of comparator species (Physcomitrella patens, Chlamydomonas reinhardtii, and Ostreococcus lucimarinus) sampling a larger number of plant families (Funariaceae, Chlamydomonadaceae, and Bathycoccaceae; Supplemental Data Set S2). This allowed the predicted target genes for each TFBS to be stratified into five phylogenetic clades. The analysis was performed using dicot species as a reference, so the first level of conservation was only within the dicots. If a TFBS also was conserved in rice, it was labeled angiosperms, indicating that the binding site was conserved in dicots and monocots. The label Magnoliophyta was given to interactions that were conserved in the flowering plants comprising dicots, monocots, and Amborella. The last two clades were Embryophyta and Viridiplantae, if the interactions were conserved in P. patens and C. reinhardtii or O. lucimarinus, respectively. There are 10,976 genes with at least one conserved element in dicots (44% of genes with conserved orthologs in dicots), 5,788 genes for the angiosperm clade (26%), 2,917 genes for the Magnoliophyta clade (13%), 1,568 genes for the Embryophyta clade (8%), and 501 genes for the Viridiplantae clade (4%). As expected, these 501 genes cover Gene Ontology (GO) terms related to basal functions such as transport, carbohydrate metabolism, and cell cycle. For all the above counts, full conservation in all clades was required, but not in all species of that clade. The median number of species in which a binding site was conserved ranged from 87% for Viridiplantae to 40% for dicots (Supplemental Table S3). These numbers illustrate that the early-diverging clades have a higher level of species conservation across all species than the younger clades. This could indicate that these evolutionarily deeply conserved binding sites play regulatory roles in essential biological processes, whereas less deeply conserved binding sites are more involved in clade-specific developmental or responsive processes.
In order to further study the evolutionary conservation of regulatory interactions, the evolutionary depth at which an ortholog of the TF that is linked to the conserved binding site could reliably be detected was taken into account. Adding this additional criterion greatly reduced the number of genes for which a conserved interaction could be detected in the distant clades (Supplemental Table S3). The Embryophyta clade contains 334 genes after filtering (365 interactions), and in the Viridiplantae clade, only eight genes remain (10 interactions). These results illustrate that reliably detecting orthologs over very large evolutionary distances is inherently difficult. An example of a regulatory interaction that is conserved in Viridiplantae is the interaction between the E2Fa TF and POL2A, a DNA polymerase ε catalytic subunit. This interaction is shown in Figure 3, together with other regulatory interactions for E2F TFs that are conserved in angiosperms, Embryophyta, or Viridiplantae. To illustrate the validity of these predictions, we compared the conserved TF target genes with tandem chromatin affinity purification bound target genes of E2Fa (Verkest et al., 2014) and differentially expressed genes upon overexpression of E2Fa (Naouar et al., 2009). This comparison revealed that, in total, 108 out of 119 predicted target genes for E2Fa are supported by experimental evidence (82 are bound and regulated, 100 are bound, and 90 are regulated). We also integrated the predicted target genes with a set of genes that were deemed to be involved in the cell cycle because they display peak expression during specific stages of the cell cycle (Menges et al., 2003). Six of the predicted target genes display this cell cycle-dependent expression pattern. Although the majority of these predicted deeply conserved target genes are known to be involved in cell cycle-related processes, several genes lack detailed functional annotation. AT4G33870, AT4G23860, AT1G77620, AT3G48540, AT1G61000, and AT3G27640 all are predicted deeply conserved target genes that also are supported by experimental evidence. These genes, however, lack information about the specific biological processes they are involved in, except for AT4G23860 and AT3G27640, which have been assigned to a functional module involved in DNA-dependent DNA replication (Heyndrickx and Vandepoele, 2012). Both the conserved E2F-binding sites and the integrated experimental data sets strongly suggest that these genes play an important role in cell cycle-related processes.
Figure 3.
A gene regulatory network of predicted conserved target genes for E2Fa, E2Fc, and E2Fe. All interactions that are conserved in angiosperms, Embryophyta, or Viridiplantae are shown. Interactions conserved up to angiosperms are shown in yellow, up to Magnoliophyta in red, up to Embryophyta in green, and up to Viridiplantae in black. Experimental evidence is indicated by the edge type: a solid line indicates that an interaction is supported by both tandem chromatin affinity purification sequence binding and differential expression upon TF perturbation; a dashed and dotted line indicates that the target gene was only differentially expressed; and a dotted line indicates that a prediction was not supported by experimental evidence or that no experimental evidence was available.
Obtaining Functional Annotation through GO Enrichment of Conserved Target Genes
Apart from focusing on deeply conserved CNSs, the large number of binding sites conserved in dicots also can be used to functionally characterize individual TFs, through GO enrichment of the associated conserved target genes (see “Materials and Methods”). Known functions from the literature were used to evaluate if the enriched GO terms were correct. MYB58 and MYB63 activate lignin biosynthesis in fibers and vessels (Zhou et al., 2009), and for both genes, we found the GO term lignin biosynthetic process to be enriched in target gene sets conserved in the dicot clade. Also, both MYB46 and MYB83 are known to be involved in secondary cell wall processes (Zhong and Ye, 2012; Kim et al., 2013). Many enriched GO terms of the target genes of these TFs were related to the regulation of lignin biosynthesis and to cellulose and xylan biosynthetic processes (Supplemental Table S4). MYB84 is part of the set of three regulators of axillary meristem genes that are partially redundant regulators of axillary meristem formation (Müller et al., 2006). In the set of target genes, we observed the axillary shoot meristem initiation and meristem maintenance GO terms, confirming this function (Supplemental Table S4). The TF MYB3 represses phenylpropanoid biosynthetic gene expression (Dubos et al., 2008), and indeed, we recovered the enriched GO term regulation of phenylpropanoid metabolic process for this gene set. A direct predicted target of MYB3 is MYB4, which, together with MYB32, can influence pollen development by changing the flux along the phenylpropanoid biosynthetic pathways, affecting the composition of the pollen wall (Preston et al., 2004). Both TFs showed enrichment toward the GO term regulation of phenylpropanoid metabolic process. MYB4 also has been shown to be involved in the production of UV light-protecting sunscreens in Arabidopsis in response to light stress (Jin et al., 2000). The GO term anthocyanin accumulation in tissues in response to UV light was representative for this proposed function. (Supplemental Table S4). The enriched GO terms positive regulation of flavonoid biosynthetic process and flavonol biosynthetic process for MYB111 were a validation of its role in the biosynthesis of flavonol (Stracke et al., 2007). Prevalent throughout the whole GO enrichment table for these MYB TFs were GO terms related to flavonoid biosynthetic processes or related to precursors of flavonoids. This finding suggests a link between MYB TFs and their role in stress response, which is supported by previous research showing that flavonoid biosynthesis is up-regulated in response to a wide range of abiotic stresses, such as cold, salinity, and drought (Supplemental Table S4; Ma et al., 2014).
Discovery and Exploration of Conserved Mini-Regulons
All previous analyses have focused on linking a conserved binding site of a TF to a target gene. In this section, we explore whether we can detect more complex transcriptional units, focusing on divergent gene pairs that have been shown to have a higher correlation in expression than random gene pairs (Krom and Ramakrishna, 2008). First, 6,501 divergent gene pairs were identified in the genome of Arabidopsis. Out of this total set of divergent gene pairs, 576 also had a shared conserved cis-regulatory element that was identified for each gene independently. We also checked whether the divergent orientation of these gene pairs was conserved in orthologous gene pairs across other genomes. There were 2,238 gene pairs that had their orientation conserved in orthologous gene pairs in one or more other genomes, and 174 of 2,238 gene pairs also had a shared conserved binding site conserved across these orthologous gene pairs. An example of a deep conserved gene pair with a shared conserved binding site is TOM5 (AT5G08040) and DUF1118 (AT5G08050), which have a conserved PIF1-binding site and conserved orientation in orthologous gene pairs of six other genomes, including rice. In a next step, the correlation in expression profile of gene pairs, part of these different categories of divergent gene pairs was evaluated using Pearson correlation coefficients (PCCs) based on an RNA sequencing (RNA-Seq) expression compendium (Supplemental Table S5; see “Materials and Methods”). A comparison of absolute PCCs for each of the four categories is shown in Figure 4A, which shows an increase in correlation between gene pairs when a conserved binding site is present. The difference is maximal when both binding site and divergent orientation are conserved across multiple genomes. A significant difference was observed between gene pairs with a conserved binding site and conserved orientation compared with basic divergent gene pairs and divergent gene pairs with conserved orientation. This finding hints at the existence of conserved mini-regulons, where the presence of a conserved regulatory element results in increased coexpression of flanking genes, suggesting tight coregulation. In order to analyze these 174 mini-regulons in more detail, PCCs also were calculated for the TF that is linked to the conserved binding site (Supplemental Table S6). In 12 out of these 174 cases, there also were striking similarities in the gene expression profiles of the TF and both gene pairs (see “Materials and Methods”). One example is shown for a conserved BES1-binding site between YLMG2 (AT5G21920) and PAA2 (AT5G21930) conserved in three genomes (cocoa tree, rose gum, and grape) and with strong positive PCCs between the genes and TF itself (PCC > 0.70; Fig. 4B). A second example is shown for a conserved PIF5-binding site between ANS (AT4G22880) and PGR5-LIKE A (AT4G22890) conserved in four other genomes (papaya, rose gum, poplar, and grape), also with a strong positive PCC between the flanking genes and the TF.
Figure 4.
Transcriptional coherence of divergent gene pairs with and without conserved binding sites. A, The distribution of absolute PCCs for all divergent gene pairs is split into four categories. These categories are divergent gene pair, divergent gene pair with orientation conservation, divergent gene pair with conserved binding site, and divergent gene pair with conserved binding site and orientation conservation. Asterisks indicate that the P value of the Wilcoxon rank-sum test was less than 0.05 (*) and 0.01 (**), respectively. B, Two examples of conserved mini-regulons. The conserved binding site is indicated in the center, together with the PCCs between the TF and the divergent genes. Below the divergent genes, the orthologous genes with conserved divergent configurations and the presence of a conserved binding site in other species are shown.
Exploration and Visualization of Plant CNSs through the PLAZA 3.0 Dicots Platform
The CNSs detected for all 10 dicot query species were uploaded to the PLAZA 3.0 Dicots database, and a number of new features were added to facilitate their exploration. On each gene page, a link was added to the toolbox to explore the conserved binding sites (CNSs overlaid with all TFBSs used in this study) for that gene. On this page, a complete overview per investigated region, upstream, downstream, or intron, is given for all retrieved binding sites per gene. Complementary, conserved binding sites also are visualized using the GenomeView genome browser for all 10 species (Abeel et al., 2012). On all Arabidopsis TF-encoding gene pages that have TFBS information, a tab was added containing the associated binding sites for that TF. Besides additions to the gene pages, a binding site page also was created for all motifs and position weight matrices used in this study. On these pages, a common name, description, and sequence logo are provided for each binding site, together with the total number of genes associated with this binding (Supplemental Fig. S1A). Breakdowns of the number of target genes per species and per investigated region are depicted as pie charts (Supplemental Fig. S1B). Different functionalities are provided in the toolbox section on the binding site page: there is the possibility to explore the associated gene families, as well as GO, MapMan, and InterPro functional annotations, based on the conserved target genes. The toolbox also contains the opportunity to look for binding sites with a similar binding profile (Supplemental Fig. S1C).
DISCUSSION
In this study, we applied a phylogenetic footprinting approach to identify CNSs in 10 dicot species. This approach uses both alignment-based and alignment-free techniques and combines different gene orthology prediction methods that do not rely on synteny information. In this manner, it circumvents the step of whole-genome alignment, which is difficult owing to the frequent nature of polyploidy and genome rearrangements in plant genomes. As such, our approach is well suited to incorporate more distantly related species, including many-to-many gene orthology relationships. A set of high-quality comparator species was selected for each query species, ensuring that a saturated substitution rate in the absence of selection was present. Across all experiments, 1,032,291 CNSs were detected for 243,187 genes. A strong correlation was detected between the number of CNSs and the total number of genes present in a genome, whereas no strong correlation could be detected between the total number of CNSs and the genome size. However, there is another manner in which genome size could be correlated with promoter architecture. The fraction of CNSs in the first 500 bp upstream of the translation start site compared with the total number of CNSs was inversely correlated with genome size. This indicates that smaller genomes have their functional elements packed more closely toward the translation start site of the gene compared with larger genomes and, as such, have smaller promoters. The detected CNSs were compared with TF ChIP-Seq data from poplar, soybean, and tomato. We found that CNSs were enriched for TF bound regions compared with randomly selected regions for six out of eight data sets, illustrating the functional regulatory character of these sequences. Furthermore, using a more extended phylogenetic sampling than used in previous research (Burgess and Freeling, 2014), we were able to discover 715 TFBSs for 501 genes that were conserved from dicots to Viridiplantae. Functions associated with this gene set comprise basal biological processes such as transport, carbohydrate metabolism, and cell cycle. The fact that these functions are not highly specialized for flowering plants is in concordance with the predicted age of these interactions. When the presence of the orthologous TF in the comparator species also was taken into account, we were able to discover deeply conserved interactions that showed strong experimental support for the E2Fa TF. Through GO enrichment of the target genes of TFs, we were able to predict putative functional annotations and confirm known functions for different sets of TFs. This process of assigning functions to the predicted target genes proves useful for genes for which little functional information is available. Assessing the functional coherence of target genes is an alternative manner to validate regulatory interactions and is based on the idea that genes that are part of the same biological pathway are regulated by similar sets of TFs (Marbach et al., 2012; Lindemose et al., 2014).
The idea that conserved binding sites exert a regulatory role on a bigger scale than only on the closest gene is largely unexplored in plants. To obtain possible mechanistic insights from the presence of conserved binding sites on gene regulation, we investigated the effect on the coexpression of a conserved binding site located between divergent gene pairs. Previously, Krom and Ramakrishna (2008) reported that specific regulatory elements were overrepresented in divergent or convergent gene pairs with a strong correlation in gene expression. In this analysis, we were able to show that the presence of a conserved binding site leads to a significant increase in transcriptional coherence compared with divergent gene pairs that did not share a conserved binding site. This effect became stronger when the divergent gene pair also was conserved in the corresponding genomes where the binding site was conserved. This cooccurrence of binding site conservation and divergent orientation conservation was called a mini-regulon. Finally, through a background model of randomly generated mini-regulons, several cases were discovered where the gene expression profile of the TF was strongly correlated with the divergent gene pair linked to the conserved binding site. These conserved mini-regulons represent examples of spatially conserved transcriptional units encompassing multiple target genes conserved in multiple plant genomes.
In order to get a better understanding of the organization as well as the function of TFs, it is crucial to study GRNs. CNSs have been shown to be important stepping stones for generating functionally relevant GRNs based on TFBSs (Kheradpour et al., 2007; Van de Velde et al., 2014). In the past, much of CNS research has focused on Arabidopsis and grasses. With the availability of CNSs for an increasing number of dicot species, it now becomes possible to leverage existing regulatory annotation approaches in nonmodel species. A widely used approach to elucidate the function of a TF is to perturb the given TF and compare expression profiles of the wild-type and perturbed states, leading to lists of differentially expressed genes on which de novo motif finding often is performed to obtain new insights on the regulation of these genes. The combination of these motifs with conservation analysis is a powerful approach to identify genome-wide bona fide target genes with these motifs and can help to unravel the underlying regulatory cascade, as was shown recently for leaf development in maize (Zea mays; Yu et al., 2015). Another approach in which CNSs can play a key role is the translation of existing knowledge of GRNs in model species into economically more interesting species, which is not trivial, owing to the occurrence of evolutionary changes. On the gene level, duplication and loss events play an important role. On the binding site level, the movement as well as the gain and loss of TFBSs can occur. Both of these types of events can lead to the disappearance or the creation of regulatory interactions (Dermitzakis and Clark, 2002). Given these obstacles, CNSs can provide a useful tool for guiding the delineation of GRNs.
The integration of this large data set in the PLAZA 3.0 Dicots platform opens up opportunities for plant scientists to quickly gain information about putative regulators of a gene of interest. It also allows for downstream analysis, such as the functional enrichment of target genes of a TF or the investigation of the associated gene families. The presentation of this CNS data set in an easy accessible form offers advantages for noncomputational scientists to access these data and generate new regulatory hypotheses in a diverse set of plant species.
MATERIALS AND METHODS
Sequence and Orthology Information
The 18 species used in this study were Arabidopsis (Arabidopsis thaliana [The Arabidopsis Information Resource 10]; Arabidopsis Genome Initiative, 2000), field mustard (Brassica rapa [FPsc version 1.3; DOE-JGI]; Wang et al., 2011), papaya (Carica papaya [Hawaii Agriculture Research Center]; Ming et al., 2008), soybean (Glycine max [JGI 1.0]; Schmutz et al., 2010), poplar (Populus trichocarpa [JGI 2.0]; Tuskan et al., 2006), cocoa tree (Theobroma cacao [CocoaGen version 1.0]; Argout et al., 2011), grape (Vitis vinifera [Genoscope version 1]; Jaillon et al., 2007), rose gum (Eucalyptus grandis [JGI 1.1]; Myburg et al., 2014), melon (Cucumis melo [Melonomics version 3.5]; Garcia-Mas et al., 2012), peach (Prunus persica [JGI 1.0]; International Peach Genome Initiative, 2013), tomato (Solanum lycopersicum [ITAG 2.3]; Tomato Genome Consortium, 2012), potato (Solanum tuberosum [ITAG 001]; Potato Genome Sequencing Consortium, 2011), beet (Beta vulgaris [RefBeet 1.1]; Dohm et al., 2014), rice (Oryza sativa [MSU RGAP 7]; International Rice Genome Sequencing Project, 2005), Amborella (Amborella trichopoda [Amborella version 1.0]; Amborella Genome Project, 2013), Physcomitrella patens (JGI 1.6; Rensing et al., 2008), Ostreococcus lucimarinus (JGI 2.0; Palenik et al., 2007), and Chlamydomonas reinhardtii (JGI 5.5; Merchant et al., 2007), and sequences were obtained from the PLAZA 3.0 database (Proost et al., 2015).
Three sequence types (i.e. upstream, downstream, and intronic) were used to identify CNSs. Upstream sequences were restricted to the first 1,000 or 2,000 bp upstream of the translation start site or to a shorter region if the adjacent upstream gene was located within a distance smaller than 1,000 or 2,000 bp. The 1,000- and 2,000-bp upstream sequences were processed as two independent runs. Downstream sequences were restricted to the first 1,000 bp downstream of the stop codon or to a shorter region if the adjacent downstream gene was within 1,000 bp. The intronic sequence type is defined as the complete gene locus starting from the translation start site with exons masked.
Orthologs for each gene were determined in 17 species using the PLAZA 3.0 integrative orthology method (Proost et al., 2015). The included orthology detection methods are OrthoMCL (Li et al., 2003), phylogenetic tree-based orthologs, and best-hit and in-paralogous families (Van Bel et al., 2012; Proost et al., 2015). Two orthology definitions were used. The first definition uses a simple best BLAST hit-derived method that includes in-paralogs, called best-hit and in-paralogous families, whereas the second definition, called integrative orthology, requires that at least two PLAZA detection methods confirm an orthologous gene relationship.
Species Selection
An average pairwise Ks matrix was created with the PLAZA 3.0 platform by calculating the Ks between all one-to-one collinear homologs of each species combination. Ks is defined as the number of synonymous substitutions per synonymous site. This was done to confirm that all included species have saturated substitution patterns (mean Ks > 1) when comparing orthologous gene pairs with one another (Proost et al., 2015). Saturated substitution patterns indicate that, in the absence of selection, the average position in a DNA sequence stretch has undergone at least one substitution. To detect CNSs in potato, tomato was removed as a comparator species, and Arabidopsis was removed when field mustard was analyzed. This was done because substitution rates are not saturated between the genomes of these two combinations of species. To make a more informed decision of which comparator species to include, two other metrics were calculated via the PLAZA 3.0 platform. The first metric was the percentage of protein-coding genes that were not complete (truncated) in the genome assembly. This percentage was assessed by counting for all gene families which genes were removed from the multiple sequence alignment used to generate the phylogenetic tree for each gene family (Proost et al., 2009). The second metric was the percentage of gene families for which a given species did not have a representative gene.
Detection of CNSs Using Comparative Motif Mapping and Alignment-Based Phylogenetic Footprinting
The comparative motif mapping algorithm was used as described by Van de Velde et al. (2014). Known binding sites were mapped on the regions covered by the three sequence types for all included species using DNA pattern allowing no mismatches (Thomas-Chollier et al., 2008). A total of 690 cis-regulatory elements were obtained from AGRIS (Yilmaz et al., 2011), PLACE (Higo et al., 1999), and AthaMap (Steffens et al., 2004). In addition, 44 positional count matrices were obtained from AthaMap, and for 15 TFs, positional count matrices were obtained from ChIP-Seq data (Heyndrickx et al., 2014). Finally, 108 and 623 positional weight matrices were obtained from protein-binding microarray studies performed by Franco-Zorrilla et al. (2014) and Weirauch et al. (2014), respectively. Positional count matrices were mapped genome wide using MatrixScan with a P value cutoff of less than 1e-05 (Thomas-Chollier et al., 2008).
The alignment-based approach was performed as described by Van de Velde et al. (2014), except that only the Sigma alignment tool (Siddharthan, 2006) was run, with the –x parameter set to 0.5. Pairwise alignments were generated between all query genes and their orthologous genes for all three sequence types. All experiments performed were filtered to retain only regions with a P value that corresponds to a false discovery rate of 10% or less.
Overlap of CNSs with Benchmarks
TF ChIP-Seq binding location data sets were obtained from the supplementary tables of the respective papers (Shamimuzzaman and Vodkin, 2013; Ricardi et al., 2014; Liu et al., 2015a, 2015b) for all TFs. The benchmark data set was formatted as a BED file, and the overlap was determined using the BEDTools function intersectBed with the –u parameter and the –f parameter set to 1 (Quinlan and Hall, 2010). This means that a TF bound region was considered correctly identified if a CNS was completely overlapped with it. False positives were determined by shuffling the TF bound data set 1,000 times using shuffleBed. The overlap with CNSs was determined for each shuffled file, and the median number of recovered elements over 1,000 shuffled files was used as a measure for the expected number of overlapping regions. This estimation was used to calculate the fold enrichment, defined as the ratio between observed overlap and expected overlap by chance. RepeatMasker (Smit et al., 2013) was run with default parameters on all three genomes for which TF ChIP-Seq data were available, and all identified repeat regions were excluded from the sequence space to shuffle the TF bound regions.
Deep Conservation and GO Enrichment
All TFs were categorized according to the TF families described in PlantTFDB 3.0 (Jin et al., 2014). The phylogenetic quantification of TF target genes in their respective TF families was performed based on these TF family annotations. GO annotations for Arabidopsis were obtained from the PLAZA 3.0 database (Proost et al., 2015). Per TF and per phylogenetic group, the enrichment of conserved target genes toward GO annotations (hypergeometric distribution + Bonferroni correction) was determined. The enriched GO terms were made nonredundant by removing enriched parental GO terms, considering the structure of the GO graph. For the gene-GO network, enriched GO terms needed to be supported by at least five target genes. Network visualizations were generated using Cytoscape 3 (Shannon et al., 2003).
Protein-Coding Potential of CNSs
The coding potential of a CNS was determined using BLASTX (Altschul et al., 1990) against the PLAZA 3.0 protein database, and all significant hits were removed. To establish an appropriate E value cutoff for a significant hit, we randomly permuted each sequence in our CNS data set and performed the BLASTX search using this set of sequences to obtain the distribution of E values for random sequences with the same length distribution (Baxter et al., 2012). We then performed the same BLASTX search on the real sequences, using the minimum E value from the random set (E < 0.001) as the cutoff for a significant hit.
RNA-Seq Compendium
The RNA-Seq expression compendium was built with public data sets from the National Center for Biotechnology Information’s Sequence Read Archive (SRA; Kodama et al., 2012). The compendium contains gene-level expression values for 40 manually selected samples (Supplemental Table S5) of different treatment and tissue combinations. SRA files for each sequencing run were downloaded from the SRA and converted to the FASTQ format using fastq-dump (version 2.4.4) from the SRA toolkit. FASTQ files from runs of the same sample were concatenated. Paired-end reads were unpaired by randomly selecting either the forward or reverse read and processing it as a single end. FastQC (version 0.9.1) was used to detect overrepresented adapter sequences, which were subsequently clipped with fastx_clipper from the FASTX toolkit (version 0.0.13). Nucleotides with Phred quality scores lower than 20 were trimmed with fastq_quality_trimmer from the FASTX toolkit. Reads shorter than 20 nucleotides after quality trimming were discarded. To obtain raw read counts for each transcript in The Arabidopsis Information Resource 10 annotation (Lamesch et al., 2012), Sailfish (version 0.6.3; Patro et al., 2014) was run with a k-mer length of 20. For genes with multiple transcripts, the raw read counts of transcripts were summed to get a gene-level read count. Counts were then normalized for the entire compendium with the Variance Stabilizing Transformation from the DESeq R package (version 1.14.0; Anders and Huber, 2010). Variance Stabilizing Transformation was chosen because it results in correlation coefficients between genes that are most comparable to those obtained with microarray data (Giorgi et al., 2013).
Detecting Transcriptionally Coherent Mini-Regulons
A background model of random mini-regulons was created by first sampling a divergent gene pair from the set of 6,183 divergent gene pairs for which gene expression data were available in our RNA-Seq compendium and randomly assigning a TF to this gene pair (this procedure was repeated 10,000 times). PCCs were determined using the RNA-Seq expression compendium between the divergent gene pair and between the assigned TFs for each random mini-regulon. The harmonic mean was calculated for the three PCCs of each randomly generated mini-regulon. The top 5% highest scores from the resulting distribution were used as a cutoff value (0.47) to identify mini-regulons showing strong TF coexpression.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Overview of the new motif page in the PLAZA 3.0 Dicots platform.
Supplemental Table S1. Ks matrix for selecting comparator species.
Supplemental Table S2. Overview of TF families and their members that have TFBS information used in this study.
Supplemental Table S3. Overview of conservation statistics per phylogenetic clade.
Supplemental Table S4. GO enrichment for all target genes of each member of the MYB TF family.
Supplemental Table S5. Overview of the RNA-Seq experiments used in the gene expression compendium.
Supplemental Table S6. Overview of the divergent gene pairs with conserved binding sites and orientations in other genomes.
Supplemental Data Set S1. ZIP archive containing CNSs for all 10 dicot species formatted as BED and FASTA files.
Supplemental Data Set S2. Resulting conserved elements of the deep conservation analysis.
Glossary
- CNS
conserved noncoding sequence
- TFBS
transcription factor binding site
- TF
transcription factor
- ChIP-Seq
chromatin immunoprecipitation sequencing
- GRN
gene regulatory network
- CMM
comparative motif mapping
- UTR
untranslated region
- GO
Gene Ontology
- PCC
Pearson correlation coefficient
- RNA-Seq
RNA sequencing
- SRA
Sequence Read Archive
Footnotes
This work was supported by the Multidisciplinary Research Partnership Bioinformatics: From Nucleotides to Networks Project of Ghent University (grant no. 01MR0410W) and by the Agency for Innovation by Science and Technology in Flanders (predoctoral fellowships to J.V.d.V. and D.V.).
Articles can be viewed without a subscription.
References
- Abeel T, Van Parys T, Saeys Y, Galagan J, Van de Peer Y (2012) GenomeView: a next-generation genome browser. Nucleic Acids Res 40: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Amborella Genome Project (2013) The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Argout X, Salse J, Aury JM, Guiltinan MJ, Droc G, Gouzy J, Allegre M, Chaparro C, Legavre T, Maximova SN, et al. (2011) The genome of Theobroma cacao. Nat Genet 43: 101–108 [DOI] [PubMed] [Google Scholar]
- Baxter L, Jironkin A, Hickman R, Moore J, Barrington C, Krusche P, Dyer NP, Buchanan-Wollaston V, Tiskin A, Beynon J, et al. (2012) Conserved noncoding sequences highlight shared components of regulatory networks in dicotyledonous plants. Plant Cell 24: 3949–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess D, Freeling M (2014) The most deeply conserved noncoding sequences in plants serve similar functions to those in vertebrates despite large differences in evolutionary rates. Plant Cell 26: 946–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabouté ME, Clément B, Philipps G (2002) S phase and meristem-specific expression of the tobacco RNR1b gene is mediated by an E2F element located in the 5′ leader sequence. J Biol Chem 277: 17845–17851 [DOI] [PubMed] [Google Scholar]
- Dermitzakis ET, Clark AG (2002) Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol 19: 1114–1121 [DOI] [PubMed] [Google Scholar]
- De Witte D, Van de Velde J, Decap D, Van Bel M, Audenaert P, Demeester P, Dhoedt B, Vandepoele K, Fostier J (2015) BLSSpeller: exhaustive comparative discovery of conserved cis-regulatory elements. Bioinformatics 31: 3758–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Minoche AE, Holtgräwe D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, Rupp O, Sörensen TR, Stracke R, Reinhardt R, et al. (2014) The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505: 546–549 [DOI] [PubMed] [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA 111: 2367–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Rapaka L, Lyons E, Pedersen B, Thomas BC (2007) G-boxes, bigfoot genes, and environmental response: characterization of intragenomic conserved noncoding sequences in Arabidopsis. Plant Cell 19: 1441–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, González VM, Hénaff E, Câmara F, Cozzuto L, Lowy E, et al. (2012) The genome of melon (Cucumis melo L.). Proc Natl Acad Sci USA 109: 11872–11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi FM, Del Fabbro C, Licausi F (2013) Comparative study of RNA-seq- and microarray-derived coexpression networks in Arabidopsis thaliana. Bioinformatics 29: 717–724 [DOI] [PubMed] [Google Scholar]
- Guo H, Moose SP (2003) Conserved noncoding sequences among cultivated cereal genomes identify candidate regulatory sequence elements and patterns of promoter evolution. Plant Cell 15: 1143–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, Forczek E, Joly-Lopez Z, Steffen JG, Hazzouri KM, et al. (2013) An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet 45: 891–898 [DOI] [PubMed] [Google Scholar]
- Heyndrickx KS, Vandepoele K (2012) Systematic identification of functional plant modules through the integration of complementary data sources. Plant Physiol 159: 884–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyndrickx KS, Van de Velde J, Wang C, Weigel D, Vandepoele K (2014) A functional and evolutionary perspective on transcription factor binding in Arabidopsis thaliana. Plant Cell 26: 3894–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada DC, Bashir A, Lee C, Thomas BC, Ko C, Goff SA, Freeling M (2003) Conserved noncoding sequences in the grasses. Genome Res 13: 2030–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Peach Genome Initiative (2013) The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 45: 487–494 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42: D1182–D1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplinsky NJ, Braun DM, Penterman J, Goff SA, Freeling M (2002) Utility and distribution of conserved noncoding sequences in the grasses. Proc Natl Acad Sci USA 99: 6147–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P, Stark A, Roy S, Kellis M (2007) Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res 17: 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WC, Kim JY, Ko JH, Kim J, Han KH (2013) Transcription factor MYB46 is an obligate component of the transcriptional regulatory complex for functional expression of secondary wall-associated cellulose synthases in Arabidopsis thaliana. J Plant Physiol 170: 1374–1378 [DOI] [PubMed] [Google Scholar]
- Kodama Y, Shumway M, Leinonen R (2012) The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res 40: D54–D56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krom N, Ramakrishna W (2008) Comparative analysis of divergent and convergent gene pairs and their expression patterns in rice, Arabidopsis, and Populus. Plant Physiol 147: 1763–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemose S, Jensen MK, Van de Velde J, O’Shea C, Heyndrickx KS, Workman CT, Vandepoele K, Skriver K, De Masi F (2014) A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Res 42: 7681–7693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ramsay T, Zinkgraf M, Sundell D, Street NR, Filkov V, Groover A (2015a) A resource for characterizing genome-wide binding and putative target genes of transcription factors expressed during secondary growth and wood formation in Populus. Plant J 82: 887–898 [DOI] [PubMed] [Google Scholar]
- Liu L, Zinkgraf M, Petzold HE, Beers EP, Filkov V, Groover A (2015b) The Populus ARBORKNOX1 homeodomain transcription factor regulates woody growth through binding to evolutionarily conserved target genes of diverse function. New Phytol 205: 682–694 [DOI] [PubMed] [Google Scholar]
- Liu WX, Liu HL, Chai ZJ, Xu XP, Song YR, Qu Q (2010) Evaluation of seed storage-protein gene 5′ untranslated regions in enhancing gene expression in transgenic rice seed. Theor Appl Genet 121: 1267–1274 [DOI] [PubMed] [Google Scholar]
- Ma D, Sun D, Wang C, Li Y, Guo T (2014) Expression of flavonoid biosynthesis genes and accumulation of flavonoid in wheat leaves in response to drought stress. Plant Physiol Biochem 80: 60–66 [DOI] [PubMed] [Google Scholar]
- MacIsaac KD, Fraenkel E (2006) Practical strategies for discovering regulatory DNA sequence motifs. PLOS Comput Biol 2: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach D, Roy S, Ay F, Meyer PE, Candeias R, Kahveci T, Bristow CA, Kellis M (2012) Predictive regulatory models in Drosophila melanogaster by integrative inference of transcriptional networks. Genome Res 22: 1334–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA (2003) Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53: 423–442 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, et al. (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D, Schmitz G, Theres K (2006) Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myburg AA, Grattapaglia D, Tuskan GA, Hellsten U, Hayes RD, Grimwood J, Jenkins J, Lindquist E, Tice H, Bauer D, et al. (2014) The genome of Eucalyptus grandis. Nature 510: 356–362 [DOI] [PubMed] [Google Scholar]
- Naouar N, Vandepoele K, Lammens T, Casneuf T, Zeller G, van Hummelen P, Weigel D, Rätsch G, Inzé D, Kuiper M, et al. (2009) Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J 57: 184–194 [DOI] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, Rouzé P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S, et al. (2007) The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci USA 104: 7705–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Mount SM, Kingsford C (2014) Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol 32: 462–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195 [DOI] [PubMed] [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW (2004) AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 40: 979–995 [DOI] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K (2009) PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21: 3718–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K (2015) PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res 43: D974–D981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Ricardi MM, González RM, Zhong S, Domínguez PG, Duffy T, Turjanski PG, Salgado Salter JD, Alleva K, Carrari F, Giovannoni JJ, et al. (2014) Genome-wide data (ChIP-seq) enabled identification of cell wall-related and aquaporin genes as targets of tomato ASR1, a drought stress-responsive transcription factor. BMC Plant Biol 14: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Shamimuzzaman M, Vodkin L (2013) Genome-wide identification of binding sites for NAC and YABBY transcription factors and co-regulated genes during soybean seedling development by ChIP-Seq and RNA-Seq. BMC Genomics 14: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddharthan R. (2006) Sigma: multiple alignment of weakly-conserved non-coding DNA sequence. BMC Bioinformatics 7: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P (2013) RepeatMasker Current Version: Open-4.0.5 (RMLib: 20140131 and Dfam: 1.3). http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker
- Steffens NO, Galuschka C, Schindler M, Bülow L, Hehl R (2004) AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res 32: D368–D372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen S, Pheasant M, Makunin IV, Mattick JS (2008) Large-scale appearance of ultraconserved elements in tetrapod genomes and slowdown of the molecular clock. Mol Biol Evol 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagle DA, Koop BF, Goodman M, Slightom JL, Hess DL, Jones RT (1988) Embryonic ε and γ globin genes of a prosimian primate (Galago crassicaudatus): nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J Mol Biol 203: 439–455 [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, Brohée S, van Helden J (2008) RSAT: regulatory sequence analysis tools. Nucleic Acids Res 36: W119–W127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al. (2005) Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol 23: 137–144 [DOI] [PubMed] [Google Scholar]
- Turco G, Schnable JC, Pedersen B, Freeling M (2013) Automated conserved non-coding sequence (CNS) discovery reveals differences in gene content and promoter evolution among grasses. Front Plant Sci 4: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Van Bel M, Proost S, Wischnitzki E, Movahedi S, Scheerlinck C, Van de Peer Y, Vandepoele K (2012) Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol 158: 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R, Bryant D, Edger PP, Tang H, Burgess D, Challabathula D, Spittle K, Hall R, Gu J, Lyons E, et al. (2015) Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature 527: 508–511 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Casneuf T, Van de Peer Y (2006) Identification of novel regulatory modules in dicotyledonous plants using expression data and comparative genomics. Genome Biol 7: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y (2009) Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol 150: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde J, Heyndrickx KS, Vandepoele K (2014) Inference of transcriptional networks in Arabidopsis through conserved noncoding sequence analysis. Plant Cell 26: 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Abeel T, Heyndrickx KS, Van Leene J, Lanz C, Van De Slijke E, De Winne N, Eeckhout D, Persiau G, Van Breusegem F, et al. (2014) A generic tool for transcription factor target gene discovery in Arabidopsis cell suspension cultures based on tandem chromatin affinity purification. Plant Physiol 164: 1122–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CT, Xu YN (2010) The 5′ untranslated region of the FAD3 mRNA is required for its translational enhancement at low temperature in Arabidopsis roots. Plant Sci 179: 234–240 [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E (2011) AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res 39: D1118–D1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CP, Chen SCC, Chang YM, Liu WY, Lin HH, Lin JJ, Chen HJ, Lu YJ, Wu YH, Lu MYJ, et al. (2015) Transcriptome dynamics of developing maize leaves and genomewide prediction of cis elements and their cognate transcription factors. Proc Natl Acad Sci USA 112: E2477–E2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2012) MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol 53: 368–380 [DOI] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]