Plant growth and development are strongly affected by small differences in temperature1. Current climate change has already altered global plant phenology and distribution2, 3, and projected increases in temperature pose a significant challenge to agriculture4. Despite the important role of temperature on plant development, the underlying pathways are unknown. It has previously been shown that thermal acceleration of flowering is dependent on the florigen, FLOWERING LOCUS T (FT)5, 6. How this occurs is however not understood, since the major pathway known to upregulate FT, the photoperiod pathway, is not required for thermal acceleration of flowering6. Here we demonstrate a direct mechanism by which increasing temperature causes the bHLH transcription factor PHYTOCHROME INTERACTING FACTOR4 (PIF4) to activate FT. Our findings provide a new understanding of how plants control their timing of reproduction in response to temperature. Flowering time is an important trait in crops as well as affecting the lifecycles of pollinator species. A molecular understanding of how temperature affects flowering will be important for mitigating the effects of climate change.
Arabidopsis thaliana, like many higher plants, responds to warmer ambient temperatures by increasing its growth rate and accelerating the floral transition1, 5, 7. Arabidopsis is a facultative long day plant, and plants grown under short photoperiods are dramatically delayed in flowering. Interestingly, late flowering in short days can be overcome by growth at higher temperatures6. The underlying mechanism is however unknown. The flowering response to temperature is dependent on the floral pathway integrator gene FT6 indicative of a thermosensory pathway that upregulates FT expression independently of daylength. Since the bHLH transcription factor PHYTOCHROME INTERACTING FACTOR4 (PIF4) has been shown to regulate architectural responses to high temperature8, 9, we tested if PIF4 is required for the induction of flowering at high temperature in short photoperiods. While pif4-101 is slightly delayed in flowering at 22 °C, pif4-101 mutants show a striking loss of thermal induction of flowering at 27 °C (Fig. 1a and b). To test if pif4-101 perturbed floral induction by affecting FT expression, we examined the thermal induction of FT in Col-0 and pif4-101. While FT expression is strongly thermally inducible in Col-0, this response is largely abolished in pif4-101 at 27 °C (Fig. 1c), indicating that PIF4 is necessary for the thermal acceleration of flowering in short days. By contrast, PIF4 is not required for the thermosensory induction of flowering under continuous light8, suggesting that the photoperiod pathway also interacts with the ambient temperature sensing pathway. The reduced role of PIF4 under continuous light likely reflects the instability of PIF4 in light10 coupled with the fact that the output of the photoperiod pathway, CONSTANS (CO) protein, is stabilised by light11, shifting the balance of floral induction from PIF4 to the photoperiod pathway. Since PIF4 is necessary for the thermal induction of flowering in short days, we tested if it is sufficient to trigger flowering when overexpressed. 35S::PIF4 causes extremely early flowering (Fig. 1d and 1e), similarly to the effect of overexpressing a related gene, PHYTOCHROME INTERACTING FACTOR512, suggesting that PIF4 is limiting for the acceleration of flowering at lower temperature in short photoperiods. Consistently, 35S::PIF4 plants show elevated levels of FT (Fig. 1f). Furthermore, 35S::PIF4 ft-3 shows a complete suppression of the early flowering phenotype, showing that the induction of flowering by 35S::PIF4 is dependent on FT (Fig. 1g and h). This activation of FT appears to be independent of the established photoperiod pathway since CO does not change in response to 35S::PIF4 (Fig. 1f). Finally, while co-9 mutants are late flowering6, 13, we find 35S::PIF4 co-9 plants are early flowering, indicating that PIF4 acts largely independently of CO (supplementary information Fig. S1), consistent with the thermal induction of flowering being independent of the photoperiod pathway (Fig. 1f)6.
Fig. 1. PIF4 is necessary for the thermal induction of flowering in short photoperiods.
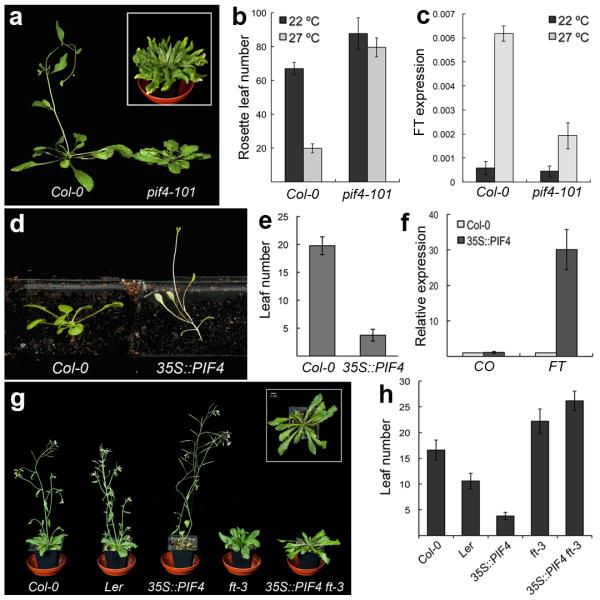
(a) pif4-101 plants do not show acceleration of flowering at 27 °C compared to Col-0. Inset shows a 16 week old pif4-101 plant grown at 27 °C. (b) rosette leaf numbers at flowering for Col-0 and pif4-101 grown at 22 °C and 27 °C in short photoperiod conditions (error bars are +/− SD, n=6). (c) FT expression as measured by Q-PCR in 4 week old plants at 22 °C and 27 °C under short photoperiods in a PIF4 dependent manner. (data from 3 biological replicates, Error bars are +/− SD) (d) 35S::PIF4 overexpression triggers very early flowering. (e) Leaf numbers at flowering for Col-0 and 35S::PIF4 in long photoperiods (error bars are +/− SD, n=5). (f) CO and FT gene expression data measured by Q-PCR in Col-0 and 35S::PIF4 at 21 °C in long photoperiods (Samples taken 2 weeks after sowing; data from 3 biological replicates. All error bars are +/− SD). (g) FT is required for the early flowering phenotype of 35S::PIF4 plants. When crossed into the ft-3 background, the early flowering of 35S::PIF4 is completely suppressed. Inset is a top view of the 35S::PIF4 ft-3 plant showing that petiole elongation growth is retained. (h) Flowering time data for Col-0, Ler, 35S::PIF4, ft-3, and 35S::PIF4 ft-3 plants (error bars are +/− SD, n=5).
Although PIF4 has been shown to be important for high temperature responses, long-term increases in either PIF4 transcript or PIF4 protein levels in response to higher ambient temperature that can account for the observed growth responses have not been detected8, 9. To examine if variation of PIF4 transcription under our experimental conditions might account for the increases in PIF4 activity with temperature, we measured PIF4 transcript levels at 12, 17, 22 and 27 °C in seedlings (Fig. 2a). PIF4 transcript levels increase from 12 °C to 22 °C, while the difference between 22 °C and 27 °C is not statistically significant. Plants at 27 °C, compared to 22 °C, show a very large PIF4-dependent response, suggesting that variation in the PIF4 transcript is not sufficient to account for the acceleration of flowering at 27 °C compared to 22 °C. To test whether temperature-mediated changes in PIF4 transcription are rate-limiting for the biological response, we analysed the behaviour of plants constitutively expressing PIF4. While 35S::PIF4 plants at 22 °C are extremely early flowering, this phenotype can be largely suppressed at 12 °C (Fig. 2b and Fig. S2), indicating that even when PIF4 transcript is abundant, lower temperatures are inhibitory for PIF4 activity. A possible explanation for this difference is that PIF4 protein is destabilised by low temperature. Indeed, PIF4 protein levels have already been shown to be strongly regulated by light10, and growth in red and blue photocycles destabilises PIF4 protein at low temperatures14. To test this, we examined the levels of PIF4:HA protein at 12 °C, 17 °C, 22 °C and 27 °C under the same light conditions used for our flowering time assays. Consistent with previous studies10 we see a strong accumulation of PIF4 at the end of the night period, which is subsequently degraded during the day. Despite the suppression of early flowering in 35S::PIF4 at 12 °C compared to 22 ° (Fig. 2b), we do not observe an appreciable difference in PIF4 protein levels at these two temperatures that is likely to account for these different phenotypes (Fig. 2c, Fig. S3). Slightly higher levels of PIF4:HA appear to be present at 27 °C (Fig. 2c), suggesting high temperature stabilisation of PIF4 may also contribute to higher PIF4 activity at 27 °C.
Fig. 2. Regulation of PIF4 by temperature.
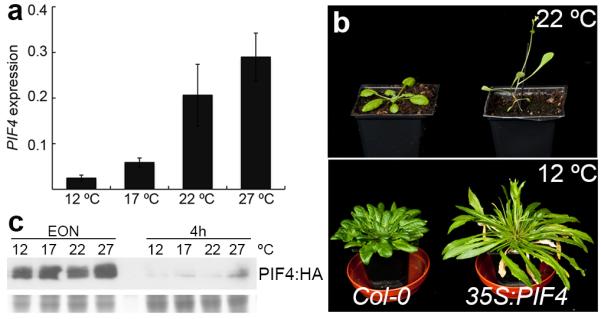
(a) Trancriptional regulation of PIF4 by temperature. 10 day-old Col-0 seedlings grown at 12, 17, 22 and 27 °C under short photoperiods were analyzed for PIF4 expression by Q-PCR. Data shown is from three biological replicates. Error bars are +/− SD. (b) Flowering phenotype of 35S::PIF4 is temperature dependent. While 35S::PIF4 plants (right) flower very early at 22 °C (upper panel) as compared to Col-0 (left), this phenotype is largely suppressed by growth at 12 °C (lower panel). (c) PIF4 protein levels in 35S::PIF4 plants are not affected by growth temperature. Seven day old 35S::PIF4:HA seedlings grown at 17 °C were transferred to 12, 17, 22 and 27 °C under short photoperiods for 2 days and samples were collected at the end of night (EON) before light and after 4 hours under illumination. While PIF4:HA protein levels are independent of growth temperatures, the protein is robustly degraded in presence of light. PIF4:HA protein was detected by HRP conjugated anti-HA antibody. Stained lower half of the gel used for immunoblot is shown as loading control.
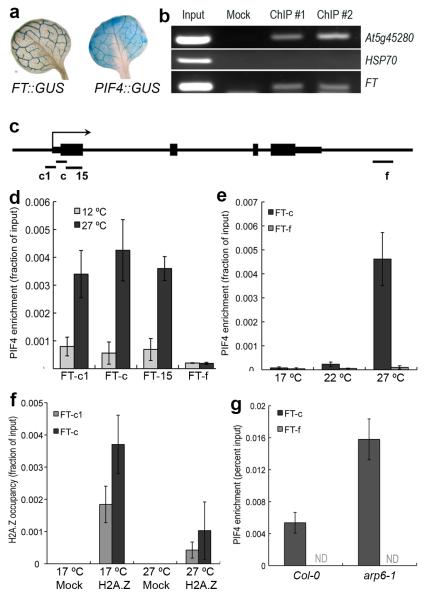
Taken together, these data indicate that PIF4 regulates FT in a temperature dependent manner. To determine if this is likely to be the case in planta, we analysed the spatial expression of FT and PIF4. FT has a distinctive pattern of expression in the vasculature of the leaf15, 16, and significantly PIF4 is expressed in the same domain (Fig. 3a). Since the regulation of FT by PIF4 could be either direct or indirect, we used chromatin immunopurification (ChIP) to analyse if PIF4 binds directly to the FT promoter proximal to the transcriptional start site (TSS). This region of the promoter was chosen since it has been shown to be both phylogenetically conserved and the site for light mediated regulation of FT expression16, 17. We observe robust enrichment of PIF4 near to the TSS (Fig. 3b), indicating that PIF4 binds this region in vivo to activate FT expression.
Fig. 3. PIF4 directly binds the FT promoter in a temperature dependent manner.
(a) GUS histochemical analysis of the expression domains of FT and PIF4 in rosette leaves. (b) ChIP analysis shows PIF4 binding to the FT locus in vivo in seedlings. The At5g45280 promoter is a positive control for PIF4 binding activity21, the HSP70 promoter has been used as a negative control. (c) Schematic summarising structure of the FT promoter and positioning of Q-PCR amplicons for ChIP analysis. (d) ChIP analysis of 35S::PIF4:HA at 12 °C and 27 °C (2 week old seedlings, short photoperiods). (e) ChIP analysis of PIF4::PIF4:ProA at 17, 22 and 27 °C (4 week old soil-grown plants, short photoperiods). (f) Analysis of H2A.Z occupancy at the FT locus at 17 °C and 27 °C (3 week old plate grown plants, short photoperiods). (g) ChIP analysis of PIF4 binding to FT in Col-0 and arp6-1 (3 week old soil-grown plants, 22 °C short photoperiod). (For all ChIP experiments, plant materials were collected at the end of dark period before lights come ON and were protected from light until frozen. All data presented are from two independent ChIP experiments; all error bars are +/− SD).
Given the striking effect of ambient temperature on PIF4 activity, which occurs even when PIF4 is constitutively expressed, we hypothesised that the ability of PIF4 to bind the FT promoter may be temperature dependent. To test this, we performed ChIP experiments using 35S::PIF4 plants grown at 12 °C and 27 °C with primers flanking an E-box in the FT promoter (Fig. 3c). Strikingly, we observe a very strong temperature dependence for this binding, with an approximately 5-fold increase in binding at 27 °C compared to 12 °C (Fig. 3d). This indicates that the later flowering of 35S::PIF4 at 12 °C is caused by a decrease in PIF4 binding to FT. Since the 35S promoter causes strong ectopic expression of PIF4, we sought to confirm that PIF4 protein expressed at endogenous levels displays similar temperature dependent binding to the FT promoter. We therefore performed ChIP experiments on a pif4-101 line complemented with PIF4pro::PIF4:ProteinA (Fig. S4). Consistent with the overexpression studies, we observe a strong increase in PIF4 binding to FT as a function of temperature. Reduced binding is observed at 17 °C, consistent with the very late flowering of plants under short days at low temperature, but this binding increases at 22 °C and is even higher at 27 °C (Fig. 3e). The temperature dependent binding of PIF4 to FT could be due to growth temperature influencing the affinity of the PIF4 transcription factor for its binding site or the efficiency of the ChIP could be affected by the temperature that tissues were grown at. To test these possibilities, we analysed another recently described PIF4 target locus18, CYP79B2 (At4g39950), which is up-regulated in 35S::PIF4 (Fig. S5a). We find PIF4 binding to occur constitutively at both 12 and 27 °C at a region in the first exon (Fig. S5b). Another region further upstream in the promoter shows a temperature dependent binding of PIF4, and in both cases, no enrichment is seen for a control locus (Fig. S5b). This indicates that the abundant PIF4 protein we observe at 12 °C is active and able to bind target sites and confirms that the ChIP method per se is not influenced by the temperature at which the sample is grown, consistent with other studies19. The ability of PIF4 to bind loci in a more temperature independent-manner might explain why 35S::PIF4 at 12 °C maintains hypocotyl and petiole elongation, while early flowering is strongly suppressed. We do not exclude that temperature may also influence PIF4 activity post-translationally.
Temperature signals are mediated through H2A.Z-nucleosomes in Arabidopsis20, suggesting that temperature may be increasing the accessibility of the PIF4 binding site at the FT promoter. Consistent with this hypothesis, we find that H2A.Z-nucleosomes are present at the PIF4 binding site in the FT promoter. Furthermore, we find that the levels of H2A.Z-nucleosomes at the FT promoter decrease with higher temperature (Fig. 3f). These results suggest that the presence of H2A.Z nucleosomes is limiting for PIF4 binding to FT, and that the PIF4 binding we observe at higher temperature is due to the greater accessibility of chromatin containing H2A.Z-nucleosomes at higher temperature. This suggests that in the absence of H2A.Z-nucleosomes, PIF4 should bind FT more strongly. We therefore compared the ability of PIF4 expressed under its own promoter to bind to the FT promoter in wild-type compared to arp6-1, a background lacking incorporation of H2A.Z-nucleosomes. Interestingly, we observe considerably greater binding of PIF4 in arp6-1 (Fig. 3g), indicating that H2A.Z-nucleosomes are rate-limiting for PIF4 to activate FT expression. The eviction of H2A.Z-nucleosomes by higher temperature therefore provides a direct mechanism for the temperature regulated expression of FT (Fig. 4c). Consistent with our previous results and the established role of H2A.Z in regulating temperature dependent gene expression, we find that there is increased PIF4 mRNA in arp6-1 background (Fig. S6). However, our results for 35S::PIF4 suppression by 12 °C indicate that transcriptional up-regulation of PIF4 is not the rate-limiting step in regulating PIF4 mediated flowering at higher temperatures.
Fig. 4. PIF4 integrates environmental signals.
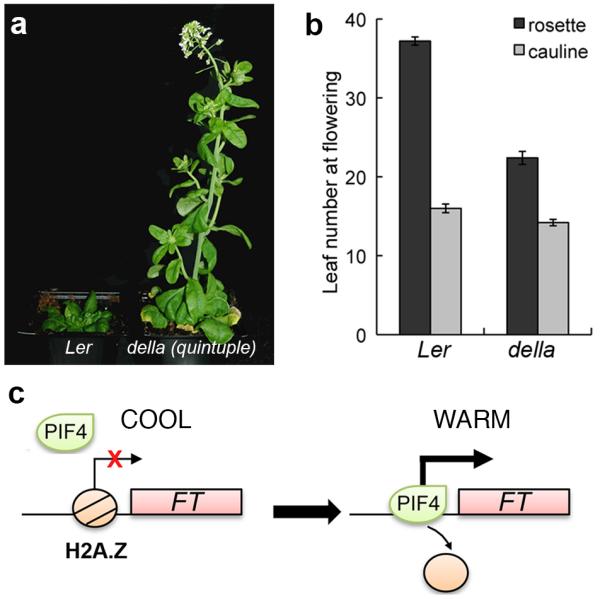
(a) Suppression of flowering at 12 °C is significantly repressed in the absence of DELLA mediated repression. (b) Leaf number at flowering for della global is reduced compared to Ler at 12 °C (error bars are +/− SD, n=6). (c) A schematic representation of temperature dependent FT regulation by PIF4. Temperature induced H2A.Z nucleosome dynamics can regulate PIF4 binding to target loci for transcriptional activation.
Our results indicate that the temperature dependent regulation of FT by PIF4 is controlled at the level of chromatin accessibility of the FT promoter and possibly at the level of PIF4 protein activity. PIF4 activity is controlled through the repressive activity of DELLA proteins that prevent PIF4 binding DNA21, 22. Consistently, plants having reduced or absent DELLA function are early flowering23. We hypothesised that delay in flowering at lower temperatures might at least in part be due to DELLA mediated repression of PIF4 activity. If so, it would be expected that absence of DELLAs should cause accelerated flowering at lower temperatures. In accord with this expectation, we found that a mutant lacking DELLAs flowers much earlier than wild-type when grown at 12 °C (Fig. 4a and 4b). The phytohormone gibberellin (GA) triggers DELLA protein degradation, and plays a key permissive role for FT induction, since in a GA deficient background, GA application increases FT expression 15-fold24. While it was proposed more than 50 years ago that gibberellins are upstream of florigen25, the mechanism has not been clear. As DELLA proteins have been shown to be key regulators by which GA influences PIF4, our finding that PIF4 is able to directly activate FT suggests a possible mechanism by which changes in GA levels may influence flowering.
Climate change has already caused measurable changes in plant phenology and behaviour2, and plants that incorporate temperature information into their lifecycles appear to be able to adapt to warmer conditions more effectively than those plants that primarily rely on photoperiod to synchronise their lifestyles3. The importance of the effects of climate change on yield are highlighted by the significant detrimental effects of increasing temperatures on yield4. PIF4 is a central integrator of environmental information in the plant and our finding that it activates FT at higher temperatures suggests it will be a key node for breeding crops resilient to climate change. This importance is suggested by the recent discovery that natural variation at PIF4 plays a major role in key ecological traits26.
ONLINE METHODS
1. Plant material and growth conditions
All plant lines used were in Col-0 background unless otherwise specified. pif4-101 mutant was provided by C. Fankhauser, HA tagged 35S::PIF4 by S. Prat12 All references to “35S::PIF” and “PIF:HA” refer to this line, i.e. 35S::PIF4:HA. FT::GUS was obtained from K.Goto15. phyb-9 ft-10 double mutants were generated by crossing respective single mutants. 35S::PIF4:HA co-9 was obtained by crossing. The crosses were genotyped for presence of the 35S::PIF4 construct by PCR on genomic DNA using primers 2362 and 2363, resulting in two products of different size representing the cDNA transgene and the genomic DNA fragment, respectively. ft-10 was genotyped using primers 1580 and 1582 for the insertion, or 1580 and 1581 to detect the wildtype fragment. co-9 was genotyped with primers 3650 and 3652 for insertion, and 3291 and 3292 for the wildtype fragment. For genotyping phyb-9, DNA was amplified using oligos 2137 and 2138 followed by Mnl I digestion to distinguish between wildtype and mutant alleles. The global della mutant is in the Ler background and was described previously8. PIF4::PIF4:PROTEINA and PIF4::PIF4:GUS were constructed by amplifying the genomic fragment of PIF4 including the promoter with oligos 1534 and 1535. The PCR product was cloned into pENTR/D-TOPO (Invitrogen) and inserted into the binary plasmids PW889 (C-terminal PROTEIN A) and PW395 (C-terminal GUS), respectively, using Gateway technology (Invitrogen). Transgenic plants were obtained by transforming pif4-101 by floral dip. For hypocotyl measurements seeds were surface sterilized, sown on ½ MS media, stratified for 2 days at 4°C in the dark and germinated for 24 h at 22°C. The plates were then transferred to short day conditions (8/16 h photoperiod) at 22°C and 27°C respectively and grown vertically for 10 days before being imaged and hypocotyl length measured using the ImageJ software (http://rsbweb.nih.gov/ij/). Oligonucleotide sequences are provided in Table S1.
2. Transcript analysis
Samples from plants grown in long days (16/8 h photoperiod) were harvested and total RNA was extracted using Trizol Reagent (Invitrogen). 2 μg of RNA were treated with DnaseI (Roche) and used for cDNA synthesis (First strand cDNA synthesis kit, Fermentas). cDNA was diluted 1:8 and used for quantitative PCR using a Roche Lightcycler 480 and the corresponding Sybr Green master mix. To detect FT transcript levels, oligos 3180 and 3181 were used, for CO oligos 2951 and 2952. PIF4 transcript levels were analyzed using oligos 3952 and 3953. Oligos 3247 and 3408 amplifying TUB6 (At5g12250) were used for normalization.
3. Immunoblot analysis
For analysing the possible effect of temperature on PIF4 protein stability, Plants overexpressing PIF4:HA (35S::PIF4:HA) were used. Seven day old 35S::PIF4:HA seedlings grown in short days at 17 °C were transferred to 12, 17, 22 and 27 °C in short days for 2 days. Samples were collected at end of night (EON) and thereafter 30 min, 1 hour and 4 hours under illumination. Protein samples were separated by SDS-PAGE and transferred on to nitrocellulose membrane. PIF4:HA was detected using HRP conjugated anti-HA antibody (Miltenyi Biotech) and visualised by chemiluminscent detection using Immobilon Chemiluminescent HRP substrate (Millipore).
4. GUS histochemical assay
For GUS-staining plants were grown on ½ MS plates in long days (16/8 h photoperiods) for 10 days and kept in the dark for 24 h before harvesting. Plants were stained in buffer containing 100 mM phosphate buffer, pH 7, 10 mM EDTA, 0.1% Triton-X100, 0.5 mM K-ferrocyanide and 1 mM X-gluc at 37°C for 24 h before destaining in ethanol.
5. Chromatin immunoprecipitation (ChIP)
ChIP was performed as described20 with minor modifications. 35S::PIF4:HA seedlings were grown on ½ MS plates for 10 days and kept in the dark for 24 h at respective temperatures before harvesting. 1.5 g of plant tissue and 4 μg of antibody (HA-tag antibody ab9110 from Abcam) were used for ChIP. To analyse the dynamics of PIF4::PIF4:PROTEINA, plants were grown in respective temperatures under short day conditions for four weeks. Aerial parts of the plants were collected and cross-linked before being used for chromatin preparations. ChIP was done using magnetic beads (Dynabeads M-270 Epoxy, Invitrogen) coated with rabbit IgG (Sigma, I5006) as described (http://www.ncdir.org/protocols/Rout/Conjugation%20of%20Dynabeads.pdf). To analyse H2A.Z dynamics at the FT locus in response to temperature, we used 3 week old seedlings of HTA11::HTA11:GFP grown at 17 °C and 27 °C. ChIP was done using anti-GFP antibody (Abcam, ab290). To analyze PIF4 binding in Col-0 and arp6-1 backgrounds, respective genotypes with PIF4::PIF4:3XFLAG were grown on soil at 22 °C under short photoperiods for three weeks before samples fixed by formaldehyde cross-linking. ChIP was performed using anti-FLAG M2 affinity gel (Sigma A2220). Immuno-complexes were eluted using 3X FLAG peptide (Sigma F4799) according to manufacturer’s instructions. Immunoprecipitated DNA was eluted after reverse cross-linking by boiling at 95 °C for 1 min in presence of 10 % Chelex (BioRad laboratories) followed by treatment with Proteinase K. Oligonucleotides 3255 and 3256 for FT-15 region, 3613 and 3614 for FT-c1, 3607 and 3608 for FT-c and 3261 and 3262 for FT-f were used for detecting PIF4 binding to the FT locus. As a positive control for PIF4 binding, At5g45280 was analysed using oligos 2857 and 2958. HSP70 was used as a negative control using oligos 1862 and 1865. For analysing PIF4 binding at At4g39950 oligonucleotides 4240 and 4241 was used for region 1; and oligonucleotides 4246 and 4247 were used for region 2. Oligos 1860 and 1861 were used for HSP70 as a negative control. Oligonucleotide sequences are provided in Table S1.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Salome Prat, Christian Fankhauser, Kerry Franklin, Koji Goto, George Coupland and Detlef Weigel for seeds. We are grateful to members of the Wigge lab for helpful discussions. This work was supported in part by Award No KUK-I1-002-03 (to NPH) made by King Abdullah University of Science and Technology (KAUST) and a BBSRC grant BB/I019022/1 (to SVK). DL was supported by an Erwin Schroedinger Fellowship from the Austrian Science Fund FWF. PAW was supported by start-up funds from the John Innes Centre and BBSRC, a BBSRC Grant (BB/D0100470/1) and a European Research Council Starting Grant (ERC 243140).
References
- 1.Samach A, Wigge PA. Ambient temperature perception in plants. Curr Opin Plant Biol. 2005;8:483–486. doi: 10.1016/j.pbi.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 2.Fitter AH, Fitter RS. Rapid changes in flowering time in British plants. Science. 2002;296:1689–1691. doi: 10.1126/science.1071617. [DOI] [PubMed] [Google Scholar]
- 3.Willis CG, Ruhfel B, Primack RB, Miller-Rushing AJ, Davis CC. Phylogenetic patterns of species loss in Thoreau’s woods are driven by climate change. Proc Natl Acad Sci U S A. 2008;105:17029–17033. doi: 10.1073/pnas.0806446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battisti DS, Naylor RL. Historical warnings of future food insecurity with unprecedented seasonal heat. Science. 2009;323:240–244. doi: 10.1126/science.1164363. [DOI] [PubMed] [Google Scholar]
- 5.Halliday KJ, Salter MG, Thingnaes E, Whitelam GC. Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 2003;33:875–885. doi: 10.1046/j.1365-313x.2003.01674.x. [DOI] [PubMed] [Google Scholar]
- 6.Balasubramanian S, Sureshkumar S, Lempe J, Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006;2:e106. doi: 10.1371/journal.pgen.0020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blazquez MA, Ahn JH, Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet. 2003;33:168–171. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- 8.Koini MA, et al. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 9.Stavang JA, et al. Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 2009;60:589–601. doi: 10.1111/j.1365-313X.2009.03983.x. [DOI] [PubMed] [Google Scholar]
- 10.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 11.Valverde F, et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 12.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 13.Wenkel S, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foreman J, et al. Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 65:441–452. doi: 10.1111/j.1365-313X.2010.04434.x. [DOI] [PubMed] [Google Scholar]
- 15.Takada S, Goto K. Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adrian J, et al. cis-Regulatory Elements and Chromatin State Coordinately Control Temporal and Spatial Expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell. 2010;22:1425. doi: 10.1105/tpc.110.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 18.Franklin KA, et al. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci U S A. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature. 476:105–108. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]
- 20.Kumar SV, Wigge PA. H2A.Z-Containing Nucleosomes Mediate the Thermosensory Response in Arabidopsis. Cell. 2010;140:136–140. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 21.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 22.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mutasa-Gottgens E, Hedden P. Gibberellin as a factor in floral regulatory networks. J Exp Bot. 2009;60:1979–1989. doi: 10.1093/jxb/erp040. [DOI] [PubMed] [Google Scholar]
- 24.Hisamatsu T, King RW. The nature of floral signals in Arabidopsis. II. Roles for FLOWERING LOCUS T (FT) and gibberellin. J Exp Bot. 2008;59:3821–3829. doi: 10.1093/jxb/ern232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brian PW. Role of gibberellin-like hormones in regulation of plant growth & flowering. Nature. 1958;181:1122–1123. doi: 10.1038/1811122a0. [DOI] [PubMed] [Google Scholar]
- 26.Brock MT, Maloof JN, Weinig C. Genes underlying quantitative variation in ecologically important traits: PIF4 (phytochrome interacting factor 4) is associated with variation in internode length, flowering time, and fruit set in Arabidopsis thaliana. Mol Ecol. 2010;19:1187–1199. doi: 10.1111/j.1365-294X.2010.04538.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.