Abstract
E-cadherin-mediated cell-cell adhesion and signaling plays an essential role in development and maintenance of healthy epithelial tissues. Adhesiveness is conferred by cadherin extracellular domains, and is regulated by an assembly of adaptors and enzymes associated with the cytoplasmic tail. Here, we employed proximity biotinylation and quantitative proteomics to isolate and identify 612 proteins in the vicinity of E-cadherin’s cytoplasmic tail. We used a structure-informed database of protein-protein interactions to construct the most comprehensive E-cadherin interactome to date, containing 89 known E-cadhesome components and 346 novel proteins. Moreover, through cloning and expression of GFP-tagged fusion proteins we localized 26 of the novel proteins to adherens junctions. Finally, employing calcium depletion and myosin inhibition we show the E-cadherin interactome to be remarkably robust to perturbation and essentially independent of cell-cell junctions or actomyosin contractility.
Introduction
The extracellular domains of “classical” cadherin adhesion receptors, such as epithelial (E)-cadherin, are sufficient to cluster and mediate cell-cell adhesion (1). However, connecting the cytoplasmic tail of cadherin with F-actin is crucial for cluster stability (2). Moreover, the coupling of cadherin with the force-generating actin cytoskeleton facilitates the integration of individual cell shape changes into tissue dynamics such as wound healing or morphogenesis (3). Linker proteins, most notably the catenins (4), mediate the physical linkage of cadherin with the actin cytoskeleton (5), and an array of signaling enzymes regulates their interactions through post-translational modifications (e.g.(6)). Additional regulatory proteins, localized at the cytoplasmic face of junctions, control actin dynamics and endocytosis (7, 8). Together, this ensemble of structural and regulatory proteins and their interactions has been coined the cadherin adhesome (“cadhesome”)(9). The cadhesome can be viewed as the “command and control” of cell-cell adhesion. Therefore, elucidating its components and their interactions is an essential step in understanding how cell adhesion is regulated in health and mis-regulated in disease.
Due to its intricate connections with the cytoskeleton the structural cadhesome is mostly insoluble, rendering it refractory to most biochemical assays, such as co-immunoprecipitation (10). Furthermore, many of the regulatory cadhesome components associate with it through transient and low affinity interactions. Nevertheless, work from many labs over the past 30 years has identified more than 170 proteins as cadhesome components (9). This number is comparable to the size of the literature-based integrin adhesome (11). In recent years, the integrin adhesome has been probed using quantitative mass spectrometry (MS)-based proteomics and the number of proteins identified by these approaches was significantly higher than what was previously described in the literature (12–14). As integrin- and cadherin-based adhesions have evolved in parallel and have similar functions, we anticipated the complexity of the cadhesome would be comparable to that of the integrin adhesome, and chose to probe this complexity using mass spectrometry.
Adhesive interactions between “classical” cadherin receptors in trans are calcium-dependent (15). Calcium ion binding rigidifies the crescent shape of the extracellular domain of cadherin (16, 17), stabilizes the X-dimer interface surface (18, 19) and energetically favors strand swapping (20). Calcium depletion was shown to dissociate trans-dimers and at the same time promote the formation of lateral dimers of cadherin (21). E-cadherin has been reported to undergo endocytosis following calcium depletion, into vesicles that remain in close proximity to the plasma membrane (22, 23). Several linker proteins, including p120-catenin, beta-catenin and vinculin, were shown to remain co-localized with endocytosed E-cadherin (22, 23). However, the fate of the entire cadhesome in response to calcium depletion and loss of cell-cell adhesion has not been addressed to date.
E-cadherin adhesions are intimately associated with actomyosin structures (24, 25) and a growing body of work shows that forces play an important role in regulating cadherin dynamics (26–28). While a number of proteins, most notably α-catenin, vinculin, and eplin, have been implicated in cadherin mechanotransduction (29–31), the global changes in the cadhesome elicited by inhibition of myosin-generated tension are still unknown.
Here, using E-cadherin proximity biotinylation proteomics followed by GFP-tagged protein localization and immunofluorescence microscopy we provide a comprehensive analysis of the molecular composition and localization of the E-cadherin interactome and its response to two perturbations: inhibition of myosin activity and calcium depletion. Our mapping of the E-cadherin vicinal proteins validates much of the literature-based cadhesome, identifies many novel components and makes numerous predictions regarding the structure and regulation of “classical” cadherin adhesions. Unexpectedly, we found the composition of the E-cadhesome to be mostly independent of trans-ligation of cadherin receptors (cell-cell adhesion) and of myosin-generated contractility.
Results
Isolation of E-cadherin proximal proteins with E-cad-BirA*
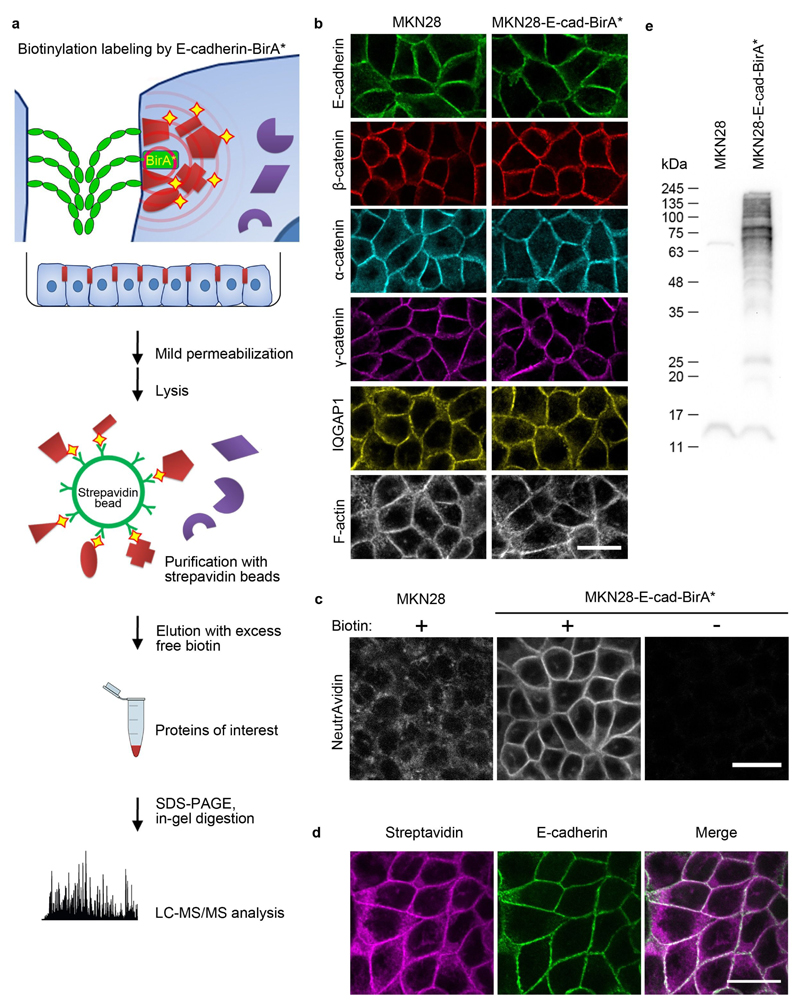
In order to specifically isolate and identify only proteins closely associated with E-cadherin we employed bioID, a recently developed technique for spatially restricted biotinylation based on a promiscuous mutant of the bacterial biotin ligase birA (32) (BirA*). Upon addition of biotin to the cell media, BirA* generates a small cloud of bioAMP, the active form of biotin that reacts with any primary amine it encounters. We fused BirA* to the C-terminus of E-cadherin (E-cad-BirA*) in order to biotinylate its neighboring proteins, whether they interact with it directly or indirectly. In order to enrich for proteins associated with E-cadherin specifically at cell-cell junctions we introduced a permeabilization and washing step before cells were lysed, and biotinylated proteins were purified using streptavidin and identified using quantitative MS (Fig 1a).
Figure 1. Isolation of E-cadherin proximal proteins enriched for cell-cell junctions with E-cad-BirA*.
(a) Schematic representation of the protocol for biotinylation, isolation and identification of proteins in close proximity to E-cadherin primarily within cell-cell junctions. Mass spectrometry was performed on four independent biological replicates. (b) MKN28 cells and MKN28 cells stably expressing the E-cadherin-BirA* fusion were grown to confluence and immunolabeled for E-cadherin, β-catenin, α-catenin, γ-catenin, IQGAP1, and phalloidin-stained for F-actin. Scale bar, 25 μm. (c) Native MKN28 cells and MKN28 cells stably expressing the E-cadherin-BirA* fusion were grown to confluence and labeled with streptavidin to show biotinylated proteins in the presence and absence of biotin in the media. Scale bar, 25 μm. (d) MKN28 cells expressing E-cad-BirA* co-stained for E-cadherin and biotinylated proteins (labeled with streptavidin). Scale bar, 25 μm. (e) Western blot probed with streptavidin-HRP of proteins eluted from streptavidin beads after incubation with lysate from native MKN28 cells or MKN28 cells stably expressing the E-cadherin-BirA* fusion.
We generated a stable line of MKN28, human gastric adenocarcinoma cells, expressing E-cad-BirA*. The fusion protein localized to cell-cell junctions along with endogenous E-cadherin and, as shown in figure 1b, these junctions were indistinguishable from control junctions with regard to the organization of E-cadherin, catenins, IQGAP and F-actin. Next, we ascertained where in the cell biotinylation occurs by staining cells with fluorescently labeled streptavidin (Fig. 1c). Incubation of parental MKN28 cells with biotin resulted in weak and diffuse streptavidin staining, whereas the E-cad-BirA* cells exposed to biotin showed robust streptavidin staining specifically at cell-cell junctions. Without the addition of biotin to the media nearly no streptavidin signal was detected, demonstrating the specificity of the labeling as well as the very low level of endogenous biotinylated proteins (Fig. 1c). Double staining with streptavidin and an E-cadherin antibody demonstrated nearly complete overlap between biotinylated proteins and E-cadherin at cell-cell junctions (Fig. 1d). Western blot analysis with streptavidin-HRP confirmed the existence of only a few biotinylated proteins in control cells and extensive biotinylation in E-cad-BirA* cells incubated with biotin (Fig. 1e).
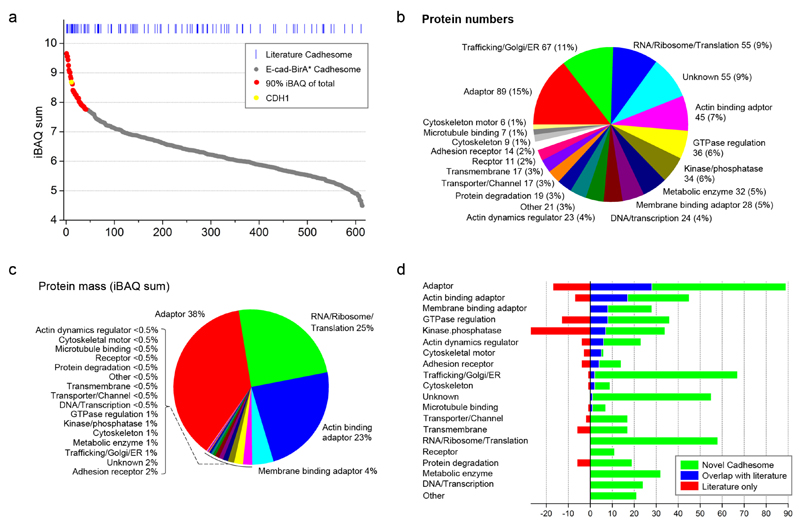
In light of these results we isolated the biotinylated proteins using streptavidin beads (following permeabilization and washing in order to remove cytosolic proteins). As will be described below, we repeated this procedure for cells treated with the calcium chelator EGTA or with the myosin inhibitor blebbistatin. As a negative control we used MKN28 cells that do not express E-cad-BirA*. Proteins enriched from quadruplicate experiments were analyzed by high resolution MS. After subtracting the proteins identified in negative control cells we reproducibly identified 612 proteins by using the label-free quantification algorithm available in MaxQuant (33) (Supplementary Table 1). As shown in figure 2a, the abundance of identified proteins, measured in terms of sum iBAQ intensity of 12 replicate experiments, spans five orders of magnitude. The 41 most abundant proteins alone account for more than 90% of the total mass of proteins quantified. Table 1 lists the 40 most abundant proteins (excluding ribosomal proteins) of which more than half are known components of the cadhesome or are paralogs of known components. However, literature-based cadhesome components are also readily found among the less abundant proteins identified (Fig. 2a), indicating that there is little correlation between the mass intensity of a protein and its likelihood of being a true cadhesome component. Altogether 89 of the 612 proteins were previously described as E-cadhesome components. An additional 37 proteins are paralogs or homologs of E-cadhesome components; 16 proteins were reported to interact with E-cadherin or catenins or to localize to cell-cell junctions; 17 proteins were reported as components of the integrin adhesome; and 12 proteins are known tight junction or desmosome components (Supplementary table 1). The remaining 442 are completely novel.
Figure 2. E-cadherin proximal proteins isolated from E-cadherin-birA* cells and identified by mass spectrometry.
(a) 612 proteins reproducibly identified and quantified in at least three replicate E-cad-BirA* experiments are plotted, ordered by their iBAQ intensity. The 41 most abundant proteins (red) account for 90% of the total mass of proteins quantified and are detailed in Table 1. Blue lines on top indicate the position of proteins that are found in the literature-based cadhesome. (b, c) The 612 proteins from (a) were manually annotated for molecular function based on information in Uniprot, Entrez Gene and the primary literature, and each protein was classified in one of 20 general functional categories. The percentage of proteins in each functional category is shown according to their number (b) or their mass (c). (d) The E-cad-BirA* proteins were classified in one of 20 functional categories and plotted together with literature-based cadhesome proteins from the same functional categories in a manner that highlights the overlap between the two sets of proteins (blue), novel proteins identified only in E-cad-BirA* (green), and literature-based cadhesome proteins that were not identified by E-cad-BirA* (red).
Table 1. Forty most abundant proteins in the E-cad-birA* MS-cadhesome (excluding ribosomal proteins).
| Mass relative to CDH1 | Gene names | Protein names | Functional category | Component of Literature-based cadhesome |
|---|---|---|---|---|
| 9.32 | CTNNA1 | α-E-catenin | Actin binding adaptor | Yes |
| 7.50 | JUP | Junction plakoglobin | Adaptor | Yes |
| 7.40 | AHNAK | Neuroblast differentiation-associated protein | Adaptor | Yes |
| 2.57 | CTNND1 | δ catenin; p120-catenin | Adaptor | Yes |
| 2.18 | FLNA | Filamin-A | Actin dynamics regulator | 1 |
| 1.54 | CTNNB1 | β-catenin | Adaptor | Yes |
| 1.00 | CDH1 | E-Cadherin; Cadherin-1 | Adhesion receptor | Yes |
| 0.94 | FAM129B | Niban-like protein 1 | Membrane binding adaptor | Yes |
| 0.86 | ANXA2 | Annexin A2 | Membrane binding adaptor | Yes |
| 0.53 | ERBB2IP | Erbb2 interacting protein | Adaptor | Yes |
| 0.51 | SKT | Sickle tail protein homolog | Unknown | No |
| 0.45 | CTNNA2 | α-N-catenin | Actin binding adaptor | Yes |
| 0.45 | CRKL | Crk-like protein | Adaptor | 2 |
| 0.42 | SCRIB | Protein scribble homolog | Adaptor | Yes |
| 0.38 | CTTN | Src substrate cortactin | Actin dynamics regulator | Yes |
| 0.31 | TJP1 | Tight junction protein ZO-1 | Actin binding adaptor | Yes |
| 0.30 | DLG1 | Disks large homolog 1 | Adaptor | Yes |
| 0.28 | KRT18 | Keratin, type I cytoskeletal 18 | Cytoskeleton | No |
| 0.28 | EGFR | Epidermal growth factor receptor | Receptor tyrosine kinase | Yes |
| 0.22 | TAGLN2 | Transgelin-2 | Actin binding adaptor | No |
| 0.21 | ANXA1 | Annexin A1 | Membrane binding adaptor | 1 |
| 0.20 | TNKS1BP1 | Tankyrase-1-binding protein | Adaptor | No |
| 0.19 | MKL2 | MKL/myocardin-like protein 2 | Transcription factor | No |
| 0.17 | NDRG1 | N-myc downstream regulated 1 | Adaptor | Yes |
| 0.17 | DSP | Desmoplakin | Adaptor | 3 |
| 0.16 | VAMP3 | Vesicle-associated membrane protein 3 | Vesicle docking/fusion | No |
| 0.15 | SLC3A2 | Solute carrier family 3, member 2 | Transmembrane transporter | 2 |
| 0.15 | SNAP29 | Synaptosomal-associated protein 29 | Vesicle docking/fusion | No |
| 0.15 | EPB41 | Protein 4.1 | Membrane binding adaptor | Yes |
| 0.14 | SHROOM3 | Protein Shroom3 | Actin binding adaptor | Yes |
| 0.12 | EFHD2 | EF-hand domain-containing protein D2 | Unknown | No |
| 0.12 | NUMB | Protein numb homolog | Adaptor | Yes |
| 0.12 | PDLIM1 | PDZ and LIM domain protein 1 | Actin binding adaptor | No |
| 0.12 | SNAP23 | Synaptosomal-associated protein 23 | Vesicle docking/fusion | No |
| 0.11 | ARHGAP32 | Rho GTPase-activating protein 32 | Rho GAP | Yes |
| 0.11 | MARK2 | microtubule affinity-regulating kinase 2 | Serine/Threonine Kinase | No |
| 0.11 | S100A11 | Protein S100-A11 | Adaptor | 1 |
| 0.11 | MLLT4 | Afadin | Actin binding adaptor | Yes |
| 0.10 | LYPLA2 | Acyl-protein thioesterase 2 | Metabolic enzyme | No |
| 0.10 | PPFIBP1 | Liprin-β-1 | Adaptor | No |
Notes: 1. A paralog exists in the literature cadhesome.
2. Component of the literature-based integrin adhesome.
3. Component of desmosomes.
We manually annotated the function of the 612 proteins based on UniProt (34), Entrez Gene (35) and the primary literature and classified each protein into one of 20 functional categories (Supplementary table 1). As shown in figure 2b, the largest group of proteins in the E-cad-BirA* cadhesome is adaptors (total of 170 proteins): 45 bind actin, 7 bind microtubules and 28 bind the plasma membrane. Other highly represented groups are transmembrane proteins (42, including 14 adhesion receptors), GTPase regulators (35), kinases and phosphatases (34), actin dynamics regulators (22), and cytoskeleton structural and motor proteins (15). Together, the aforementioned functional groups, which resemble the composition of the literature-based cadhesome, account for over 50% of the 612 proteins identified. The remaining proteins are involved in transcription (25), translation (58), trafficking (67), proteolysis (19) and metabolism (32), or they are of unknown function (55). When the estimated abundance of the identified proteins is taken into account it becomes evident that 65% of the mass of the identified E-cadhesome is made up of adaptors (including actin binding and membrane binding adaptors), 25% of translation-related proteins and all the other functional groups together make up the remaining 10% (Fig. 2c). Figure 2d delineates for each of the functional categories the number of novel proteins in the E-cad-BirA* MS (green), the degree of overlap with the literature-based cadhesome (blue) and the number of literature-based proteins absent from the E-cad-BirA* cadhesome (red). Evidently, a handful of literature-based cadhesome components, most notably adaptors, GTPase and phosphorylation regulators, were not detected by E-cadherin proximity biotinylation. Conversely, many novel components were identified across all categories, including completely new categories such as RNA/ribosome/translation and metabolic enzymes.
Identification of E-cadhesome components that localize at adherens junctions
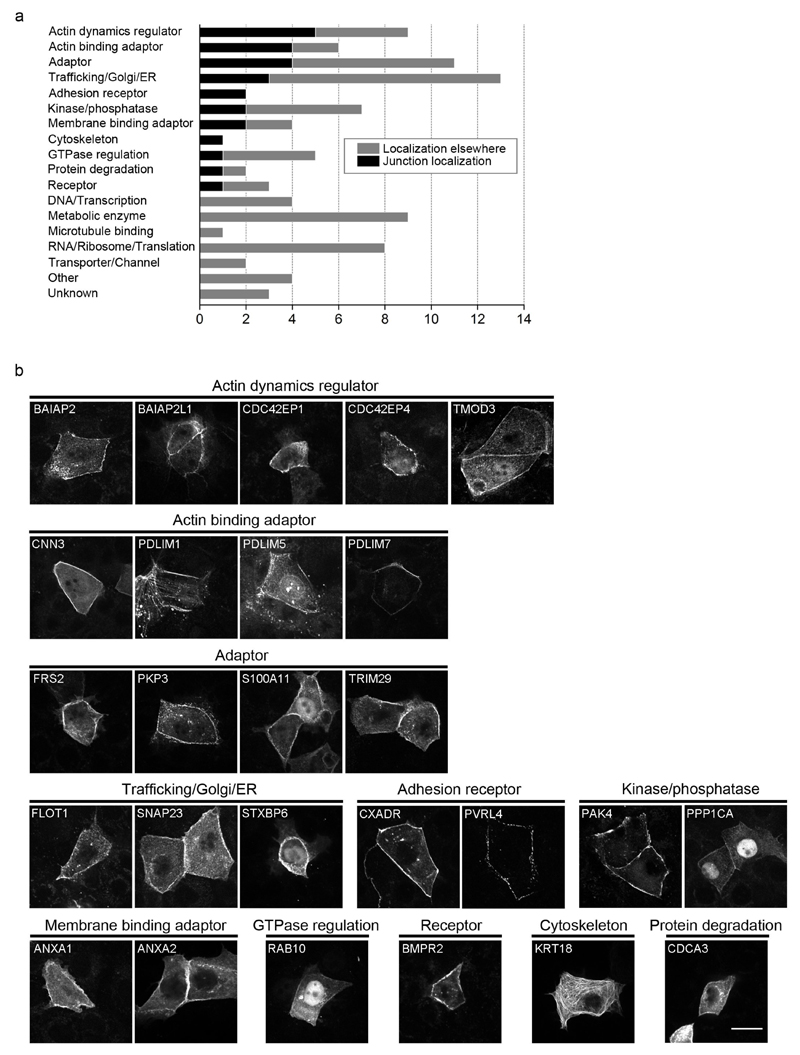
E-cad-BirA* is expected to actively biotinylate proteins in its vicinity regardless of whether it is localized at the junction or elsewhere in the cell (e.g. in a vesicle). While we made an effort to enrich for junctional proteins, the appearance of many transcription and translation proteins in the E-cad-BirA* results suggested that not all 612 proteins reside at adherens junctions. Therefore, we sought to examine the subcellular localization of some of the novel proteins by expressing C-terminal EGFP fusions of the proteins in MKN28 cells. As a test group we cloned 122 genes across all functional groups, for which we could obtain cDNA. Of the 122 engineered genes, 94 were expressed at detectable levels in MKN28 cells and were visually scored for their subcellular localization by confocal microscopy. As figure 3 shows, 26 of the proteins were found to localize to cell-cell junctions. Of the 68 non-junctional proteins 46 were cytoplasmic and 22 showed specific cellular localization, most often in or around the nucleus or in vesicles (Supplementary table 2). While the overall percentage of tested proteins exhibiting junctional localization was 28%, this proportion clearly varies between functional categories: none of the DNA/transcription, RNA/ribosome/translation, or metabolic enzymes tested showed junctional localization whereas among adaptors and actin dynamics regulators 50% of the tested proteins localized to cell-cell junctions. Junctional absence of a fusion protein does not rule out junctional localization of the endogenous protein, since the fusion of EGFP at the C-terminus might interfere with required interactions. However, it is more likely that many of the E-cadhesome proteins, especially from the transcription/translation groups, interact with E-cadherin in other locations in the cell.
Figure 3. Localization of novel E-cad-BirA* cadhesome proteins by GFP fusion.
(a) 94 candidates from the E-cad-BirA* MS, across various functional groups, were GFP-tagged and expressed in MKN28 cells, and the proteins were scored for junctional localization using confocal microscopy. (b) Confocal images of the 26 GFP-fusion proteins that exhibited localization at cell-cell junctions. Scale bar, 25 μm.
To confirm that proteins localizing to cell-cell junctions are specific to adherens junctions we fixed cells expressing the GFP-tagged proteins and immunolabled them for endogenous E-cadherin. Confocal microscopy imaging confirmed co-localization of the fusion protein with E-cadherin in a high proportion of cells examined (Supplementary Fig. 1).
Construction of an E-cadhesome network using known and predicted protein-protein interactions
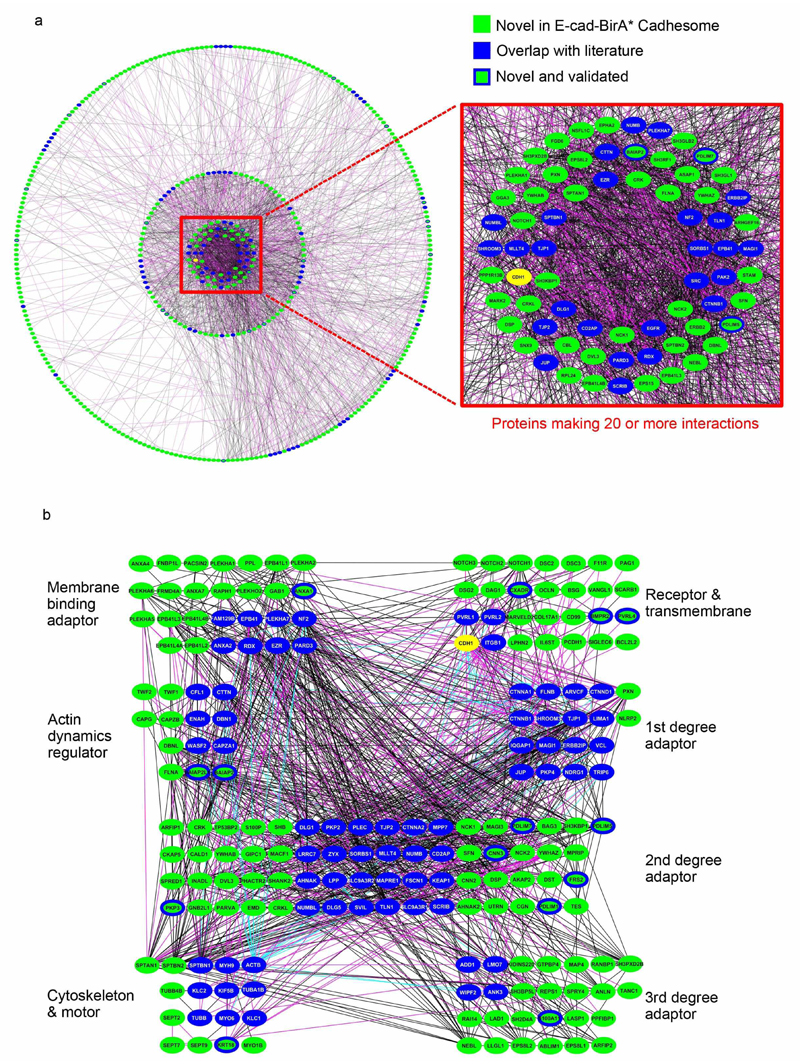
We employed PrePPI (36, 37), a recently developed structure-informed database of human protein-protein interactions, to map known and predicted interactions between the 612 proteins identified in the E-cad-BirA* cadhesome. The PrePPI database identified 2,288 highly likely interactions (probability ≥0.5) between 435 of the 612 proteins (Supplementary Table 3). The 177 proteins for which no interactions were identified within the protein set consisted primarily of metabolic enzymes, ribosomal and trafficking proteins, and proteins of unknown function. We used the network analyzer (38) in Cytoscape (39) to define the topology of the predicted cadhesome network and found it to follow the power law typical of biological networks (40) (Supplementary Fig. 2a). Moreover, based on its calculated clustering coefficient, average number of neighbors, and characteristic path length (0.313, 10.520, and 3.189, respectively), we found it to be denser than the signaling network of small GTPases (0.30, 4.9, and 4.04) (41) and similar to the structural/signaling network of the mitochondria (0.35, 15.1, and 3.22) (42). As a control, we also used PrePPI to predict the interactions between the 89 literature cadhesome proteins identified in the BirA* cadhesome and a set of 523 proteins randomly selected from the human proteome. PrePPI identified only 780 non-self interactions between 259 proteins in the control set, demonstrating the enrichment of predicted interactions in BirA* relative to control. Of the 780 interactions made by the control set, only 226 of these are between the literature cadhesome and the randomly selected proteins. This is in stark contrast to the BirA* cadhesome, in which 796 interactions are made between the literature cadhesome and the novel BirA* proteins. Furthermore, 1142 interactions are made between novel BirA* proteins, compared to only 204 interactions between the randomly selected proteins.
Figure 4a displays all 435 proteins as nodes, grouped in concentric circles according to how many interactions they make: from 40-80 interactions per protein in the inner most circle to 1-9 interactions per protein in the outer most circle. Protein nodes were colored green if they are novel, blue if they are found in the literature-based cadhesome (9), and green with a blue border if a GFP-tagged protein localized to cell-cell junctions in our experiments. From this visualization it is apparent that many of the established cadhesome components are among the most highly interconnected proteins in the network. Only 12% of the 273 proteins in the outermost circle are known components, compared to 50% of the 69 components in the three inner most circles. Importantly, the dense core of the network, which we defined as proteins making twenty or more interactions, also contains a large number of novel components that, based on their position in the network, are likely to be true E-cadhesome components (Fig. 4a, insert). Many of these proteins are structural: membrane-bound and cytoskeleton-bound adaptors and other cytoskeletal proteins. To better appreciate these novel proteins in a structural context we created a subnetwork of structural elements as shown in Figure 4b.
Figure 4. PrePPI-predicted E-Cad-BirA*-based cadhesome network.
(a) Depicted is the network of 2,288 PrePPI-predicted(Zhang, 2013 #30) interactions with a probability ≥0.5 for 435 proteins identified by E-Cad-BirA* MS. Proteins are grouped based on how many interactions they make (1-9, 10-19, 20-29, 30-39, 40-80), and arranged in concentric circles, organized from innermost circle (40-80 interactions) to outermost (1–9). Magenta lines represent interactions with database support, black lines represent interactions with none. Proteins that appear in the literature-based cadhesome(Zaidel-Bar, 2013 #9) are colored blue (except for E-cadherin, colored yellow), novel proteins are colored green; colored green with a blue outline are the novel proteins for which junctional localization was confirmed in this paper. (b) The depicted subnetwork is derived from part “a”. Included are structural proteins: receptor and transmembrane proteins, adaptors, actin dynamics regulators, and cytoskeletal proteins. Except for membrane-binding adaptors, all adaptors are organized based on the number of interactions (degree) separating them from E-cadherin (CDH1). Proteins and interactions are colored the same as in part “a”, with literature-based cadhesome interactions(Zaidel-Bar, 2013 #9) added and depicted in cyan.
The unbiased isolation of E-cadherin associated proteins described here identified many proteins from functional groups not previously considered integral to the cadhesome, such as trafficking-, transcription- and translation-related proteins. We used the PrePPI-based network of predicted interactions to examine the connectivity of these functional groups with known cadhesome components (Supplementary Fig. 3). Not surprisingly, we found 374 predicted interactions between novel adaptors and the 89 known cadhesome components in the BirA* E-cadhesome. Importantly, we also predicted that the 89 known cadhesome components make 62 interactions with novel trafficking/golgi/ER proteins, 44 interactions with novel DNA/transcription and RNA/translation proteins, and 17 interactions with novel metabolic enzymes (Supplementary Fig. 3), suggesting that proteins from these groups not only reside in close proximity to E-cadherin but also functionally interact with bona fide cadhesome components, although these interactions may not necessarily take place at cell-cell junctions.
The construction of an interaction network allowed us to separate proteins into groups according to their “distance” from E-cadherin, i.e., whether they bind E-cadherin directly, are 2nd degree neighbors, and so forth. We then plotted the iBAQ intensity of a protein in the MS as a function of its “distance” from E-cadherin (Supplementary Fig. 2b). We found the median iBAQ value for 1st degree neighbors of E-cadherin was 3-fold higher than the median iBAQ value for 2nd degree neighbors and there was little difference between 2nd and 3rd or 4th degree neighbors. However, the distribution of iBAQ values within each group is wide and thus iBAQ value alone cannot predict the “distance” from E-cadherin.
Response of E-cad-BirA* cadhesome to loss of cell-cell junctions
With the tools to quantitatively investigate the E-cadhesome in hand we next asked the following question: to what extent does the E-cadhesome depend on the formation of cadherin-mediated cell-cell junctions? Since the adhesive trans-dimers of E-cadherin depends on the presence of calcium we incubated MKN28 E-cad-BirA* cells with the calcium chelator EGTA for 24 hours and compared the intensity of E-cadhesome proteins derived from these cells with the control E-cadhesome. After 3h in EGTA cell-cell junctions unraveled and for the remaining 21h incubation in EGTA MKN28 cells remained detached from their neighbors, gradually adopting a rounded conformation (Fig. 5a). Cells remained viable during this period of time, as they readily spread and reformed cell-cell junctions upon replenishment of calcium to the media.
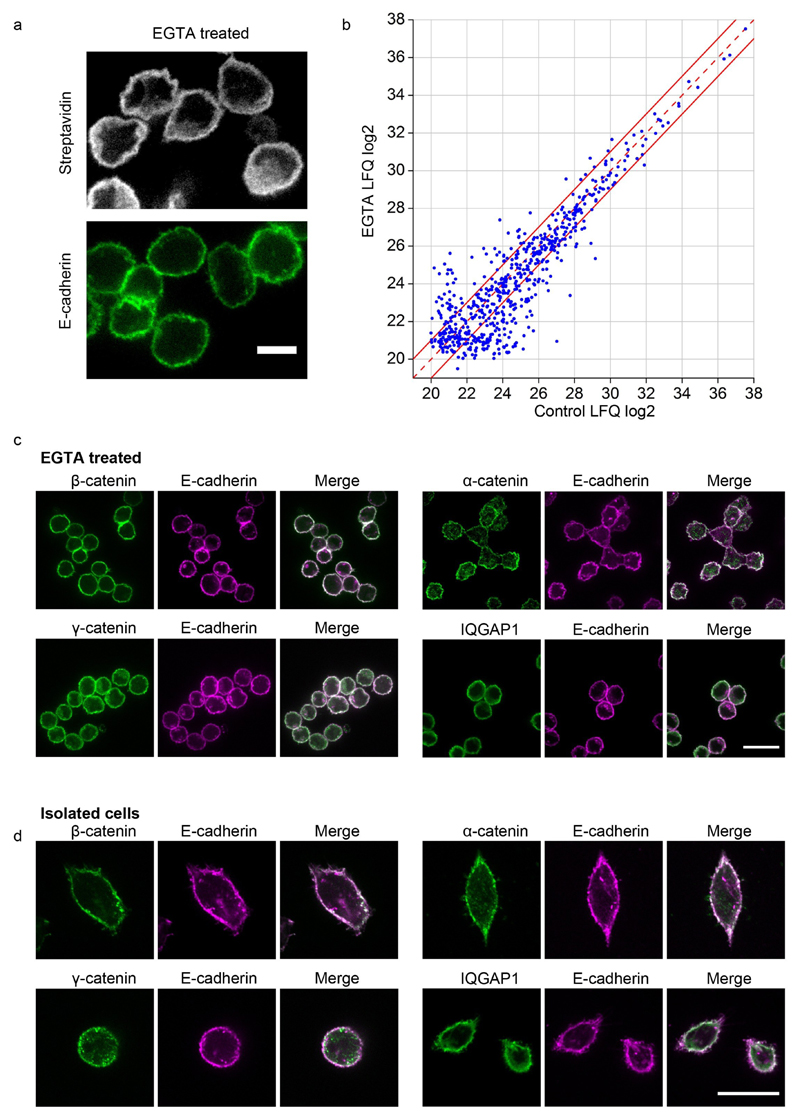
Figure 5. Effect of EGTA treatment on the composition of E-cad-BirA* MS cadhesome.
(a) MKN28 cells expressing E-cad-BirA* incubated with EGTA for 24h stained for E-cadherin and biotinylated proteins (streptavidin). Scale bar 10µm (b) Plot of the average label-free calculated summed peptide intensities (n=4 independent biological replicates) of the E-cad-BirA* cadhesome proteins (n=612) in control versus 24h EGTA treatment. Dashed red line indicates no difference between control and treatment and points between the two solid red lines demarcate the region within which proteins show less than a 2-fold difference. (c) After 24h incubation in EGTA MKN28 cells were doubly immunolabeled for E-cadherin along with β-catenin, α-catenin, γ-catenin, or IQGAP1. (d) Single MKN28 cells adhering to glass were doubly immunolabeled for E-cadherin along with β-catenin, α-catenin, γ-catenin, or IQGAP1. Scale bar 25µm.
Surprisingly, when comparing the intensity of the same protein under control and EGTA conditions, using the label-free quantification algorithm available in MaxQuant (33), we found very little change. As shown in figure 5b, the majority (60%) of proteins showed less than two-fold change, and the highest abundance proteins, including E-cadherin and the catenins, showed the least change. Among the low abundance proteins that showed more than a two-fold change we observed an enrichment of PDZ and SH3 domain proteins in the control E-cadhesome compared to the EGTA condition (Supplementary Fig. 4).
The apparent independence of the E-cadhesome on the formation of cell-cell junction was unexpected. To corroborate these finding we used immunofluorescence to examine the co-localization of several E-cadhesome components with E-cadherin in cells that have been incubated with EGTA for 24h. As shown in figure 5c we found β-catenin, α-catenin, γ-catenin and IQGAP to co-localize with E-cadherin at the periphery of the cells under these conditions. To exclude the possibility that calcium chelation interferes with normal E-cadherin endocytosis we also examined the localization of the same E-cadhesome components in sparsely seeded MKN28 cells that were allowed to adhere for 24h in normal media. As shown in figure 5d, single cells display cortical localization of all examined E-cadhesome proteins similar to EGTA-treated cells, confirming that, at least for the four adaptors we examined, co-localization with E-cadherin is independent of cell-cell junctions.
Response of E-cad-BirA* cadhesome to loss of tension
The composition of the integrin adhesome has been shown to be modulated by treatment of cells with the myosin inhibitor blebbistatin (13, 14). Cadherin adhesion complexes have also been shown to be mechanoresponsive (26–28), but so far only a few cadhesome proteins (e.g. vinculin) have been shown to be tension-sensitive (29, 30). Therefore, we took advantage of E-cad-BirA* to examine how the composition of the cadhesome changes upon myosin inhibition. We found that treating MKN28 cells with blebbistatin for one hour was sufficient to dramatically reduce tension at the junctions, as evident by their wavy and splayed appearance, primarily at apical regions (Fig. 6a). We confirmed that tension at junctions was reduced by laser microdissection (Fig. 6b). While laser ablation lead to mean recoil distance of 3.7±0.27 microns 30 seconds after ablation in control junctions, in cells treated with blebbistatin junctions recoiled significantly (P<0.0005, student’s t-test) less at 1.6±0.32 microns. Importantly, after one hour of blebbistatin treatment E-cad-BirA* was still localized at the cell-cell junction, as indicated by the streptavidin staining (Fig. 6a, lower panel).
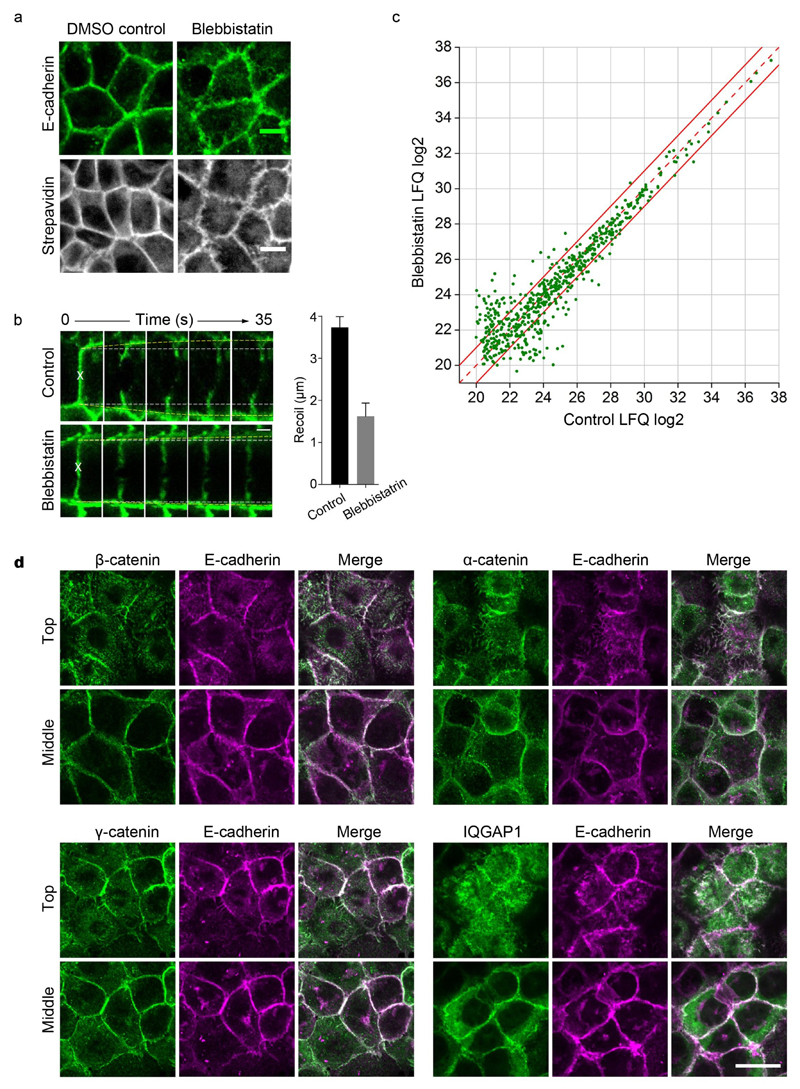
Figure 6. Effect of blebbistatin treatment on the composition of E-cad-BirA* MS cadhesome.
(a) Confluent MKN28 E-cad-BirA* cells incubated with biotin for 24h stained for E-cadherin or biotinylated proteins after a one hour treatment with either DMSO (negative control) or 50µM blebbistatin. Scale bar, 10 μm. (b) Measurement of recoil in blebbistatin-treated MKN28 cells following laser ablation. Representative images from an ablation experiment in control and blebbistatin-treated (50µM) cells are shown; white dashed lines represent the initial junction length while yellow lines indicate vertex separation in ablated samples. Data represent mean±s.e.m., ***p<0.0005; calculated from n=8 junctions in each experimental condition. Scale bar 5µm. (c) Plot of the average label-free calculated summed peptide intensities (n=4 independent biological replicates) of the E-cad-BirA* cadhesome proteins (n=612) in control versus 1h blebbistatin treatment. Dashed red line indicates no difference between control and treatment and points between the two solid red lines demarcate the region within which proteins show less than a 2-fold difference. (d) MKN28 cells treated with blebbistatin for 1h were fixed and co-stained for E-cadherin along with β-catenin, α-catenin, γ-catenin, or IQGAP1. Two confocal planes are shown: an upper Z-slice and a middle Z-slice. Scale bar 25µm.
When comparing the intensity of the biotinylated proteins isolated from control DMSO-treated E-cad-BirA* cells with the biotinylated proteins isolated from E-cad-BirA* cells treated with blebbistatin for one hour, we found surprisingly very little change. As shown in figure 6c, the majority of the proteins (73%) exhibited a difference of less than two-fold between control and blebbistatin conditions. Among the primarily low abundance proteins exhibiting more than a two-fold change we found PDZ and SH3 domain proteins to be enriched in control conditions, similar to what we found with EGTA treatment (Supplementary Fig. 4). Plotting the change from control in blebbistatin versus EGTA treatment demonstrates a positive correlation (Pearson correlation coefficient 0.42, P=1.3E-27) between the two treatments (Supplementary Fig. 4), indicating that both perturbations have similar effects on the E-cadhesome.
We used immunoflourescence to corroborate the MS results, labeling E-cadherin along with the adaptors β-catenin, α-catenin, γ-catenin and IQGAP in control MKN28 cells and in cells treated with blebbistatin for one hour. As shown in figure 6d, the effect of blebbistatin on junctions is most evident in the upper most focal planes. Even where junctions are frayed we continue to observe co-localization of E-cadherin with the tested adaptors. Moreover, the co-localization in the middle focal planes was indistinguishable from control non-treated cells (compare with Fig. 1b).
Discussion
High resolution MS is capable of accurately identifying large mixtures of proteins over a wide range of abundance. Therefore, the challenge in characterizing the protein composition of a given cellular structure lies in purifying its components for MS analysis. We used BioID, a technique in which a promiscuous biotin ligase, BirA*, is fused to a protein of interest, in this case E-cadherin, enabling biotinylation of vicinal proteins (32). This method was first used to identify proteins in close proximity to nuclear lamin-A and subsequently was applied to the tight junction protein ZO-1 (43). A similar method for spatially restricted biotinylation, employing a different enzyme, was used for proteomic mapping of the mitochondia (44). Recently, Van Itallie et. al. also fused BirA* to the cytoplasmic tail of E-cadherin (45). They identified 303 proteins associated with E-cadherin in MDCK cells, 127 of which overlap with the 612 proteins found in this paper (Supplementary Table 4). A notable difference between our studies is the choice of negative control. We used cells not expressing E-cad-BirA* as a negative control, whereas Van Itallie et. al. used cells expressing a cytoplasmic BirA*.
The exact radius within which vicinal proteins are biotinylated by BirA* is not established yet. However, Roux et. al. estimated that 50% of the proteins they identified using bioID reside within 20-30nm of BirA* (32). Van Itallie et. al. identified a different subset of proteins depending on which end of ZO-1 they fused BirA*, strengthening the notion that the radius of biotinylation is not much larger than the size of a single protein. Comparing the 408 proteins identified by the fusion of BirA* to either end of ZO-1(43) with the 612 proteins we identified by E-cad-BirA* MS we find an overlap of only 84 proteins (Supplementary Table 4), further supporting the specificity of the technique and the unique composition of tight versus adherens junctions.
A good indication of the efficacy of the E-cad-BirA* approach was the identification of 89 out of 173 literature-based cadhesome proteins. Of the 84 previously described components that were not identified by E-cad-BirA* most were cytoplasmic proteins, primarily kinases and phosphatases, as well as GTPases. Thus, it should be noted that the brief permeabilization step preceding lysis of cells for MS most likely introduced a bias towards identifying insoluble proteins. However, it may also be that some of the missed components are not expressed in MKN28 or that their localization is dependent on certain conditions. Future proteomic analysis, using different cell types and under various conditions will help to distinguish between these possibilities and refine the E-cadhesome.
Our proteomic analysis as well as localization analysis provided a wealth of candidate cadhesome proteins that can now be tested further. Importantly, in addition to the expansion of existing classes of cadhesome components our unbiased approach also identified proteins with functions that have not previously been thought of as integral to cadherin adhesions. These include, for example, proteins involved in endocytosis, translation and transcription. While there are some examples of transcription factors playing a second role in cell-cell junctions, most notably β-catenin (46), and while there is some anecdotal evidence for junctional localization of mRNA and translation machinery (47), our GFP-tagging of candidate proteins from these functional groups suggests that most of them do not localize to cell-cell junctions. Nevertheless, the true extent of intertwining between these cellular functions and cadherin adhesion warrants further study.
Although our protocol was aimed to purify proteins associated with E-cadherin at adherens junctions, the expression of GFP-tagged candidates clearly showed that many of the E-cad-BirA* interactome proteins are found in other locations in the cell. It is reasonable to believe E-cadherin interacts with these proteins at various stages of its lifetime and these interactions may have profound implications for cell adhesion. A recent genome wide RNAi screen in drosophila S2 cells uncovered over 800 genes required for cadherin-mediated cell adhesion, and only a fraction of them localized to cell-cell junctions (48). Between our list of E-cad-BirA* proteins and the list of genes from Toret et. al. (48) we found 64 overlapping genes, 39 of which are involved in trafficking, translation or transcription (Supplementary Table 4). 56 of the overlapping genes are novel and we consider them particularly promising candidates for further investigation since they have now been shown independently to be essential for cell adhesion and closely associated with E-cadherin. Examples of such proteins are the receptor SCARB1, the RhoGAP ARHGAP1, and the actin binding adaptors ANLN and LASP1.
Proximal biotinylation combined with quantitative mass spectrometry is a strong tool to address the molecular composition of adherens junctions. To understand how cell adhesion is regulated we need to have a comprehensive map of the cadhesome, and the resource we provide here goes a long way in that direction. Surprisingly, we found the E-cadhesome composition to be mostly independent of cell-cell junctions, suggesting that adherens junctions are unique by virtue of their structural organization, and/or posttranslational modification, which will need to be probed by other means.
Materials and Methods
Establishing E-cad-BirA* stable cell line and isolation of cadherin-proximal biotinylated proteins
To generate the E-cad-BirA* fusion construct, we replaced GFP on E-cadherin-GFP(49) (gift from James Nelson, Stanford University, USA) with BirA*(32) (gift from Brian Burke, IMB-A*, Singapore, also available from Addgene: http://www.addgene.org/35700/), using XhoI and MfeI restriction sites. For establishing a stable cell line, E-cad-BirA* was cloned into pJTI™ Fast DEST vector with a CMV promoter and integrated into MKN28 cell line using Jump-In™ Fast Gateway Kit (Life Technologies). Colonies stably expressing E-cad-BirA* were selected by 200µg/ml Hygromycin B, and screened by streptavidin staining.
For the purification of biotinylated proteins, E-cad-BirA* MKN28 cells were cultured with biotin-starving media (RPMI + dialyzed fetal bovine serum) until 24 h before the experiment, when their media was exchanged to DMEM containing biotin. Confluent cells (140mm dish) were treated with 50µM Blebbistatin or DMSO control for 1h, or with 1µM EGTA for 24h followed by permeabilization with ice cold 1% Tween-20 PBS. Cells were then lysed to collect biotinylated proteins with 300µl streptavidin beads (MyOne Steptavadin C1; Life Techonologies) as previously described (32). MKN28 wild type was used as a control for endogenous biotinylated protein and beads-non-specific binding. Samples of each condition were prepared in quadruplicate. For western blotting, one third of the amount above was loaded and the membrane was probed with Streptavdin-HRP (Life Technologies S911).
Mass spectrometry and bioinformatic analysis
Proteins were separated on 4-12% gradient NuPAGE Novex Bis-Tris gel (Invitrogen), in-gel digested with trypsin (50), and peptides concentrated and desalted on StageTips (51). Peptides were analyzed by EASY-nLC system (Thermo Fisher Scientific) coupled on line to a linear trap quadrupole (LTQ)-Orbitrap Elite via a nanoelectrospray ion source (Thermo Fisher Scientific). Chromatographic peptide separation was done in a 20 cm fused silica emitter (Thermo Fisher Scientific) packed in house with reversed-phase ReproSil-Pur C18-AQ, 1.9 µm resin (Dr. Maisch GmbH) and eluted with a flow of 200 nl/min from 5% to 60% solvent (80% ACN, in water, 0.5% acetic acid) over 110 min. The full scan MS spectra were acquired with a resolution of 120,000 at m/z 400 Th and target value of 1,000,000 charges in the Orbitrap. The top 10 most intense ions were sequentially isolated for fragmentation using high-energy collision dissociation (HCD) at the MSn target value 40,000 charges and recorded in the Orbitrap with a resolution of 15,000 at m/z 400 Th. All data were acquired with Xcalibur software (Thermo Fisher Scientific).
The MS .RAW files were processed with the MaxQuant software(33) version 1.3.8.2 and searched with Andromeda search engine(52) against the human UniProt database(34) (release-2013 05, 88,847 entries). To search parent mass and fragment ions, an initial mass deviation of 4 and 20 ppm, respectively, was required. Trypsin enzyme specificity and only peptides with a minimum length of 7 amino acids were selected. A maximum of two missed cleavages were allowed. Carbamidomethylation (Cys) was set as fixed modification. Oxidation (Met) and N-acetylation were considered as variable modifications. For identification of protein and peptide we required a maximum of 1% FDR. Scores were calculated in MaxQuant as described previously(33).
The reverse and common contaminants hits were removed from MaxQuant output. Proteins were quantified according to the MaxQuant label-free quantification (LFQ) algorithm(53). In order to perform Welsh test analysis between samples, the missing LFQ intensity values were replaced using the “Imputation, replace missing values from normal distribution” feature available in the MaxQuant module Perseus(54). Unique and razor (= most likely belonging to the protein group) peptides were used for protein quantification and we required proteins being quantified with a minimum of two ratio counts. Proteins for which different isoforms were quantified, only the isoform with the highest number of assigned peptides was included in the final list of the cadhesome proteins.
The three BirA* E-cadhesome (Junctional, Blebbistatin and EGTA) contain following proteins:
-
i)
Proteins not quantified at all in the cells without BirA* E-cadherin and quantified in minimum three of the four replicate experiments of BirA* expressing cells treated or not with Blebbistatin or EGTA.
-
ii)
Proteins identified and quantified in at least three replicate experiments of the BirA* non-treated cells or BirA* blebbistatin- or EGTA-treated cells, and that were differentially enriched compared to non-BirA* cells. Enriched proteins were selected by applying a single tailed Welch test controlled for multiple hypothesis testing by permutation-based FDR (5%).
For the estimated total abundance of the E-cadhesome proteins, the iBAQ intensity value implemented in MaxQuant software (=Intensity/number of theoretical tryptic peptides) was used. The iBAQ intensity of each protein was calculated as the sum of all measured iBAQ intensities in the analyzed samples (excluding samples from non-BirA* cells). For each protein the estimated abundance was calculated as (iBAQprotein/sumiBAQE-cadhesome)*100.
Validation of novel component by GFP fusion protein
For fusing GFP to the C-terminal of 122 candidate proteins, entry vectors containing open reading frames (gift from T.C. Cornvik, NTU, Singapore) were recombined with destination vector of Vivid Colors™ pcDNA™6.2/C-EmGFP-DEST (Life Technologies) using gateway cloning methods (Life Technologies). The resulting cDNA-GFP fusion constructs were transfected into MKN28 cells by Lipofectamine 2000, and observed by confocal microscopy.
Immunostaining
In staining of monolayer adherens junctions, the samples were fixed with 4% PFA for 30 min, permeabilized with 0.1% Triton X-100 for 20 min, and incubated with primary and secondary antibodies, or other staining reagents. Primary antibodies and reagents used: Anti-Ecadherin (BD Biosciences 610181, Sigma U3254), anti-β-catenin (Sigma C7082), anti-α-catenin (Cell Signalling 3134), anti-γ-catenin (Abcam 15153), anti-IQGAP1 (Santa Cruz 10792), streptavidin-Alexa-647 (Life technologies S32357), and TRITC-phalloidin (Sigma 77418). Species-specific secondary antibodies used were: anti-Rat (Life technologies A11077), anti-Mouse (Life technologies A31571, Abnova PAB 10733), anti-Rabbit (Abcam 6799, Abcam ab6717).
Microscopy
Confocal images were acquired using a Nikon Eclipse Ti inverted microscope with a Spinning-Disk confocal head (CSU-X1, Yokogawa Corporation), a laser launch unit (iLas2, Roper Scientific), an EMCCD camera (Evolve Rapid-Cal, Photometrics), a 60X and a 100X Plan Apo objectives (Nikon). MetaMorph software (Molecular Devices) was used for image acquisitions.
Comparison of junctional tension by laser microdissection
Laser ablation experiments were performed on wild-type and blebbistatin-treated (50µM, 30min) MKN28 cells stably expressing GFP-tagged E-Cadherin. Ablation experiments were carried out on a Nikon A1R confocal microscope equipped with an ultraviolet laser (355nm, PowerChip, Teem Photonics, France). Images were acquired at 1s time intervals using a 60X Plan-Apo 1.4NA objective (Nikon) with 2X digital magnification. The tightly focused UV laser was targeted on cell apical junctions for 300 ms with an average power of 450nW at the back aperture of the objective, while image acquisition was carried out at 488nm.
Network analysis
Each of the 612 E-cad-BirA* MS proteins was queried in the human PrePPI database (36) (http://bhapp.c2b2.columbia.edu/PrePPI) and all interactions with high confidence (probability ≥0.5) were retrieved. Only interactions between the 612 were retained and the resulting network was drawn in Cytoscape (39) version 3.0.2. We removed proteins that made either no interactions or only self interactions. Network topology was calculated using the Cytoscape plugIn NetworkAnalyzer (38). To generate a control set of proteins/interactions, we combined the 89 literature proteins identified in the BirA* cadhesome with 523 proteins randomly selected from the human proteome and repeated the same procedure.
Supplementary Material
One Sentence Summary.
Identifying novel network interacting with E-cadherin independently of cell adhesion.
Acknowledgments
We thank Brian Burke (IMB, A-STAR, Singapore) for the BirA* plasmid, Tobias Carl Cornvik (Nanyang Technological University, Singapore) for access to the ORFeome and MGC libraries, Yusuke Toyama (MBI, Singapore) for help with laser ablation, Ann Hedley (Cancer Research UK Beatson Institute) for informatics assistance, and Barry Honig (Columbia Univeristy, NY) for his support of PSM.
Funding: This work was supported by the National Research Foundation Singapore under its NRF fellowship (NRF-RF2009-RF001-074) awarded to RZB, and Cancer Research UK.
Footnotes
Author contributions: To be added later.
Competing interests: None.
Data and materials availability: The .raw MS files and search/identification files obtained with MaxQuant have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset) via the PRIDE partner repository. Data can be accessed using Accession: PXD000448; Username: review17262; Password: 5akYMTJ4.
References
- 1.Harrison OJ, et al. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19(2):244–56. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong S, Troyanovsky RB, Troyanovsky SM. Binding to F-actin guides cadherin cluster assembly, stability, and movement. J Cell Biol. 2013;201(1):131–43. doi: 10.1083/jcb.201211054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lecuit T. Adhesion remodeling underlying tissue morphogenesis. Trends Cell Biol. 2005;15(1):34–42. doi: 10.1016/j.tcb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Gumbiner BM, McCrea PD. Catenins as mediators of the cytoplasmic functions of cadherins. J Cell Sci Suppl. 1993;17:155–8. doi: 10.1242/jcs.1993.supplement_17.22. [DOI] [PubMed] [Google Scholar]
- 5.Yonemura S. Cadherin-actin interactions at adherens junctions. Curr Opin Cell Biol. 2011;23(5):515–22. doi: 10.1016/j.ceb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Bertocchi C, Vaman Rao M, Zaidel-Bar R. Regulation of adherens junction dynamics by phosphorylation switches. J Signal Transduct. 2012;2012:125295. doi: 10.1155/2012/125295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratheesh A, Yap AS. A bigger picture: classical cadherins and the dynamic actin cytoskeleton. Nat Rev Mol Cell Biol. 2012;13(10):673–9. doi: 10.1038/nrm3431. [DOI] [PubMed] [Google Scholar]
- 8.Delva E, Kowalczyk AP. Regulation of cadherin trafficking. Traffic. 2009;10(3):259–67. doi: 10.1111/j.1600-0854.2008.00862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaidel-Bar R. Cadherin adhesome at a glance. J Cell Sci. 2013;126(Pt 2):373–8. doi: 10.1242/jcs.111559. [DOI] [PubMed] [Google Scholar]
- 10.Hinck L, et al. Dynamics of cadherin/catenin complex formation: novel protein interactions and pathways of complex assembly. J Cell Biol. 1994;125(6):1327–40. doi: 10.1083/jcb.125.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaidel-Bar R, Geiger B. The switchable integrin adhesome. J Cell Sci. 2010;123(Pt 9):1385–8. doi: 10.1242/jcs.066183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humphries JD, et al. Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci Signal. 2009;2(87):ra51. doi: 10.1126/scisignal.2000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiller HB, et al. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 2011;12(3):259–66. doi: 10.1038/embor.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo JC, et al. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13(4):383–93. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeichi M, et al. Selective adhesion of embryonal carcinoma cells and differentiated cells by Ca2+-dependent sites. Dev Biol. 1981;87(2):340–50. doi: 10.1016/0012-1606(81)90157-3. [DOI] [PubMed] [Google Scholar]
- 16.Nagar B, et al. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380(6572):360–4. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- 17.Pokutta S, et al. Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur J Biochem. 1994;223(3):1019–26. doi: 10.1111/j.1432-1033.1994.tb19080.x. [DOI] [PubMed] [Google Scholar]
- 18.Pertz O, et al. A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J. 1999;18(7):1738–47. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haussinger D, et al. Calcium-dependent homoassociation of E-cadherin by NMR spectroscopy: changes in mobility, conformation and mapping of contact regions. J Mol Biol. 2002;324(4):823–39. doi: 10.1016/s0022-2836(02)01137-3. [DOI] [PubMed] [Google Scholar]
- 20.Vendome J, et al. Molecular design principles underlying beta-strand swapping in the adhesive dimerization of cadherins. Nat Struct Mol Biol. 2011;18(6):693–700. doi: 10.1038/nsmb.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Troyanovsky RB, Sokolov E, Troyanovsky SM. Adhesive and lateral E-cadherin dimers are mediated by the same interface. Mol Cell Biol. 2003;23(22):7965–72. doi: 10.1128/MCB.23.22.7965-7972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kartenbeck J, et al. Endocytosis of junctional cadherins in bovine kidney epithelial (MDBK) cells cultured in low Ca2+ ion medium. J Cell Biol. 1991;113(4):881–92. doi: 10.1083/jcb.113.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov AI, Nusrat A, Parkos CA. Endocytosis of epithelial apical junctional proteins by a clathrin-mediated pathway into a unique storage compartment. Mol Biol Cell. 2004;15(1):176–88. doi: 10.1091/mbc.E03-05-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borghi N, et al. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci U S A. 2012;109(31):12568–73. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smutny M, et al. Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat Cell Biol. 2010;12(7):696–702. doi: 10.1038/ncb2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyake Y, et al. Actomyosin tension is required for correct recruitment of adherens junction components and zonula occludens formation. Exp Cell Res. 2006;312(9):1637–50. doi: 10.1016/j.yexcr.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 27.Ladoux B, et al. Strength dependence of cadherin-mediated adhesions. Biophys J. 2010;98(4):534–42. doi: 10.1016/j.bpj.2009.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, et al. Mechanical tugging force regulates the size of cell-cell junctions. Proc Natl Acad Sci U S A. 2010;107(22):9944–9. doi: 10.1073/pnas.0914547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.le Duc Q, et al. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189(7):1107–15. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yonemura S, et al. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12(6):533–42. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 31.Taguchi K, Ishiuchi T, Takeichi M. Mechanosensitive EPLIN-dependent remodeling of adherens junctions regulates epithelial reshaping. J Cell Biol. 2011;194(4):643–56. doi: 10.1083/jcb.201104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roux KJ, et al. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196(6):801–10. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 34.The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38(Database issue):D142–8. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maglott D, et al. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33(Database issue):D54–8. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang QC, et al. PrePPI: a structure-informed database of protein-protein interactions. Nucleic Acids Res. 2013;41(Database issue):D828–33. doi: 10.1093/nar/gks1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang QC, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490(7421):556–60. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Assenov Y, et al. Computing topological parameters of biological networks. Bioinformatics. 2008;24(2):282–4. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 39.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu X, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007;21(9):1010–24. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
- 41.Delprato A. Topological and functional properties of the small GTPases protein interaction network. PLoS One. 2012;7(9):e44882. doi: 10.1371/journal.pone.0044882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reja R, et al. MitoInteractome: mitochondrial protein interactome database, and its application in 'aging network' analysis. BMC Genomics. 2009;10(Suppl 3):S20. doi: 10.1186/1471-2164-10-S3-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Itallie CM, et al. The N and C termini of ZO-1 are surrounded by distinct proteins and functional protein networks. J Biol Chem. 2013;288(19):13775–88. doi: 10.1074/jbc.M113.466193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhee HW, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339(6125):1328–31. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Itallie CM, et al. Biotin ligase tagging identifies proteins proximal to E-cadherin, including lipoma preferred partner, a regulator of epithelial cell-cell and cell-substrate adhesion. J Cell Sci. 2014;127(Pt 4):885–95. doi: 10.1242/jcs.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bienz M. beta-Catenin: a pivot between cell adhesion and Wnt signalling. Curr Biol. 2005;15(2):R64–7. doi: 10.1016/j.cub.2004.12.058. [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez AJ, et al. Visualization of mRNA translation in living cells. J Cell Biol. 2006;175(1):67–76. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toret CP, et al. A genome-wide screen identifies conserved protein hubs required for cadherin-mediated cell-cell adhesion. J Cell Biol. 2014;204(2):265–79. doi: 10.1083/jcb.201306082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams CL, et al. Mechanisms of epithelial cell-cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J Cell Biol. 1998;142(4):1105–19. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shevchenko A, et al. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–60. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 51.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–70. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 52.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 53.Luber CA, et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32(2):279–89. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 54.Cox J, Mann M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics. 2012;13(Suppl 16):S12. doi: 10.1186/1471-2105-13-S16-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.