Abstract
The ability to resolve glycans while attached to tryptic peptides would greatly facilitate glycoproteomics, as this would enable site-specific glycan characterization. Peptide/glycopeptide separations are typically performed using reversed-phase liquid chromatography (RPLC), where retention is driven by hydrophobic interaction. As the hydrophilic glycans do not interact significantly with the RPLC stationary phase, it is difficult to resolve glycopeptides that differ only in their glycan structure, even when these differences are large. Alternatively, glycans interact extensively with the stationary phases used in hydrophilic interaction chromatography (HILIC), and consequently, differences in glycan structure have profound chromatographic shifts in this chromatographic mode. Here, we evaluate HILIC for the separation of isomeric glycopeptide mixtures that have the same peptide backbone but isomeric glycans. Hydrophilic functional groups on both the peptide and the glycan interact with the HILIC stationary phase, and thus, changes to either of these moieties can alter the chromatographic behavior of a glycopeptide. The interactive processes permit glycopeptides to be resolved from each other based on differences in their amino acid sequences and/or their attached glycans. The separations of glycans in HILIC are sufficient to permit resolution of isomeric N-glycan structures, such as sialylated N-glycan isomers differing in α2-3 and α2-6 linkages, while these glycans remain attached to peptides.
Keywords: Isomeric Separation, HILIC, Fetuin, IgGs
INTRODUCTION
Protein glycosylation is a common post-translational modification that is known to affect many cellular processes, such as cell adhesion, receptor activation, and signal transduction.1–5 Alteration of glycosylation is associated with various human pathologies, such as cancers,6 Alzheimer’s disease,7 and rheumatoid arthritis.8 Consequently, the analysis of protein glycosylation is necessary to improve our understanding of various biologic processes, as well as to facilitate correlation of glycan structures with healthy and disease states.
Liquid chromatography (LC) coupled with mass spectrometry (MS) has been an enabling technology for the analysis of protein glycosylation. MS has developed into a valuable tool for glycan analysis because of its high sensitivity, selectivity, and throughput.9 LC complements MS analysis, as it can separate isomeric glycans/glycopeptides, and thus, the combination of LC/MS offers the ability to characterize and quantitate individual glycoforms in complex mixtures.
There are 2 general approaches to characterize glycans using LC/MS. In the first of these, the glycans are released from the glycoprotein enzymatically or chemically before LC/MS analysis.10 Various LC strategies have demonstrated the ability to analyze either native or derivatized glycans, including porous graphitized carbon,11 reversed-phase (RP),12 and HILIC.13 Such techniques have been used to accomplish separation, identification, quantification, and occasionally isomeric structure analysis of glycans.13, 14 For instance, α2-3/2-6 sialic acid (SA) linkage isomers of N-glycans can be resolved by HILIC.13 The solvents/buffers used in each of these separation approaches are MS compatible, and thus, all of these have been used for LC/MS analysis of glycans.
The liberation of the glycans before analysis simplifies the analysis; however, it causes the loss of information. Specifically, information on the sites of glycan attachment and the quantities of each glycan at individual glycosylation sites are lost when the glycans are released when >1 glycosylation site is present in the analyte. A second approach is to analyze the glycans present at each location by resolution of the intact glycopeptides produced by proteolysis of the glycoprotein, without release of the glycans.15–17 When examining glycopeptides, the peptide sequence analysis can enable identification of the site of glycan attachment. Often site occupancy by the relevant glycan is critical for biologic activity. For instance, with therapeutic antibodies, the glycans located in the variable domain region influence the serum clearance rate, whereas glycans in the constant domain region affect the activity.18 Consequently, the comprehensive characterization of protein glycosylation microheterogeneity—the identification and quantitation of isomeric glycan occupancy at potentially all glycosylation sites on a protein—is a significantly more difficult challenge than glycomic profiling and requires the development of new approaches to overcome these analytical challenges.
RPLC/MS is one of the most widely used methods to separate and analyze glycopeptides.19–21 With RPLC, retention of glycopeptides is predominantly driven by the hydrophobic character of the peptide portion, and as a result, glycans attached to peptides with different sequences typically elute at different times. However, because the glycan does not interact with the RPLC stationary phase, the glycans have a very limited effect on the separation, and thus, all glycopeptides with the same peptide backbone are poorly resolved. Consequently, RPLC does not separate isomeric glycans attached to the same glycosylation sites and offers limited selectivity, even for significant differences in glycan composition.
HILIC is a commonly used approach for the analysis of released glycans and is capable of resolving released glycans. This ability led us to investigate if HILIC would be capable of resolving the glycoforms of a glycopeptide. In this manuscript, we demonstrate that HILIC is capable of resolving isomeric glycoforms of glycopeptides and that the separations obtained on glycopeptides are similar to those obtained by HILIC of released glycans, with high-separation selectivity differences based on composition and even on branch position for complex glycans. Additionally, because the hydrophilic amino acid side chains of the glycopeptide also interact with the stationary phase, glycopeptides with different peptide sequences can also be separated. The ability to separate glycopeptides based on both the glycan and the peptide makes HILIC a useful tool in the glycoproteomic toolbox.
MATERIALS AND METHODS
Materials
Fetuin, human serum, trypsin [l-(tosylamido-2-phenyl)ethylchloromethylketone-treated], dl-DTT, idoacetamide (IDA), ammonium bicarbonate, ammonium formate, and formic acid (for LC-MS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Acetonitrile (ACN; HPLC grade) was purchased from Thermo Fisher Scientific Life Sciences (Waltham, MA, USA). Octadecyl (C18) disposable extraction columns were purchased from J.T. Baker (Avantor Performance Materials, Center Valley, PA, USA). All reagents were analytical grade.
Trypsin Digestion
Bovine fetuin (200 μg) was dissolved in 100 μl 50 mM ammonium bicarbonate. For human serum, a 50 μl aliquot (70 μg/μl protein) was mixed with 50 μl 100 mM ammonium bicarbonate. DTT (5 μl 200 mM) was added to both sample solutions to reduce the disulfide bonds. Sulfhydryl alkylation was carried out by adding 4 μl 1 M IDA to the sample, and the excess IDA was neutralized by adding 20 μl 200 mM DTT solution. Trypsin digestion was carried out at 37°C overnight, with the enzyme amount adjusted to establish the ratio of trypsin to sample protein at 1:20 (w/w). The trypsin-digested peptide and glycopeptide mixture were separated from the undigested protein, trypsin, and other impurities (DTT and IDA) by an RP C18 solid-phase extraction (SPE) column. The trypsin digests were loaded onto a C18 SPE column, which had been pre-equilibrated in 5% acetic acid. The peptides and glycopeptides were eluted with 5 ml 65% ACN in 5% acetic acid, which was collected and dried in the SpeedVac.
C18 Separation of N-Glycopeptides of Fetuin
The study used a Nexera Ultra-Fast LC (UFLC; Shimadzu, Kyoto, Japan) LC system and Agilent C18 columns (Eclipse XDB-C18; Agilent Technologies, Santa Clara, CA, USA; 4.6 mm × 15 cm, 5 μm particle size). The separation was carried out at a flow rate of 0.4 ml/min at room temperature, with a mobile phase A consisting of 99.9% H2O with 0.1% formic acid and mobile phase B of 99.9% ACN with 0.1% formic acid. A linear gradient of 5% mobile phase B to 40% mobile phase B in 60 min was used. Samples were dispersed in 0.1% formic acid in water and maintained at room temperature in the autosampler until analysis.
HILIC Separation of N-Glycopeptides
The study used a Nexera UFLC (Shimadzu) LC system and Halo Penta-HILIC columns (Advanced Materials Technology, Wilmington, DE, USA; 2.1 mm × 15 cm, 2.7 μm particle size). The separation was carried out at a flow rate of 0.4 ml/min at 60°C, with a mobile phase A consisting of 95% H2O/ACN with 50 mM ammonium formate (adjusted to pH 4.4 with formic acid) and mobile phase B as pure ACN.
For glycopeptides of fetuin, a linear gradient of 85% mobile phase B to 48% mobile phase B in 75 min was used. For glycopeptides of IgGs in human serum, the following segmented linear gradients were used: 1) 62% mobile phase B to 61.2% mobile phase B in 9 min, 2) 61.2% mobile phase B to 60.2% mobile phase B in 10 min, and 3) 60.2% mobile phase B to 58% mobile phase B in 11 min. In each case, the column was flushed at 25% mobile phase for 5 min before returning to the starting mobile phase composition.
SRM Detection
MS analysis was performed using the selected reaction monitoring (SRM) mode on a 4000 QTRAP (AB SCIEX, Concord, ON, Canada) mass spectrometer. In SRM mode, 2 stages of mass filtering are used in the triple quadrupole mass spectrometer. In the first stage, ions of interest (precursor ions) are selected in quadrupole 1 (Q1) and induced to fragment by collisional excitation with neutral gas in a collision cell (Q2). In the second stage, instead of obtaining full-scan tandem MS (MS/MS), where all the possible fragment ions derived from the precursor are mass analyzed in Q3, only a small number of specific fragment ions (transition ions) are analyzed in Q3. This targeted MS analysis using SRM enhances the lower detection limit for target analysts. The masses of glycopeptides in fetuin and the human serum IgGs (precursor masses) were predicted by adding the masses of targeted N-glycans to the masses of targeted tryptic peptides containing the N-glycosylation sites of interest. The mass-to-charge ratio (m/z) values used as the precursor ions used in the SRM experiments for the fetuin glycopeptides are listed in Supplemental Table 1 of the supporting information. For example, the m/z value of 1218.8 corresponds to the triply charged glycopeptide with a peptide backbone of LCPDCPLLAPLNDSR (GP15) and biantennary N-glycan with 2 SAs (Bi-2SA), which is abbreviated as GP15-Bi-2SA. The m/z values used as precursors in the SRM experiments performed on the glycopeptide of the human serum IgGs are listed in Supplemental Table 2. MS/MS experiments performed on the various glycopeptides revealed that each produced 2 intense fragment ions. The common fragment at m/z 365.7 corresponds to the oxonium ion of hexose-N-acetylhexoseamine (Hex-HexNAc). The other intense fragment ion corresponds to the complete peptide backbone for the selected precursor, combined with a single N-acetylglucosamine (GlcNAc) attached. These 2 fragments ions were used in the SRM experiment for all of the glycopeptides as Q3 transition ions and significantly reduced the possibility of false positives. A collision energy of 70 V and declustering potential of 40 V were selected as an appropriate compromise between selected ion intensity and background current. The dwell time for data collection was set at 100 ms, and unit resolution was used in both Q1 and Q3.
RESULTS AND DISCUSSION
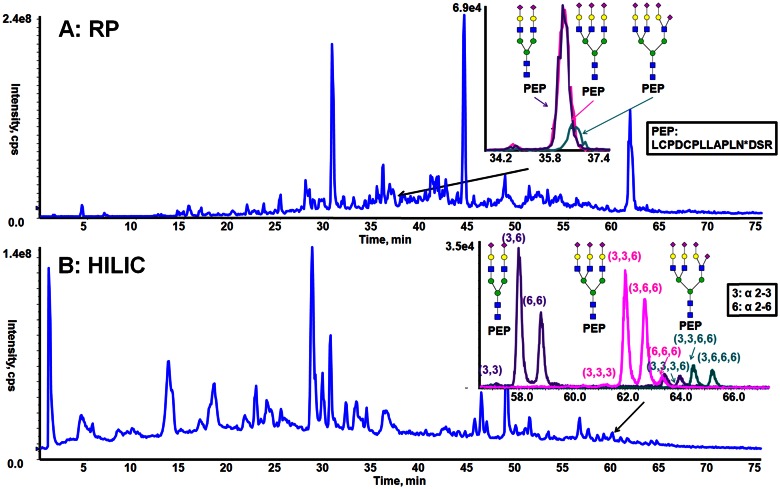
Both RP and HILIC were used to analyze N-linked glycopeptides from fetuin, and the results were compared to demonstrate the features of these 2 separation modes for resolving N-glycopeptide mixtures. Fetuin is a standard glycoprotein that has 3 well-characterized N-glycosylation sites, occupied by N-glycans consisting predominantly of bi-, tri-, and tetra-antennary complex structures, possessing variable degrees of sialylation.22–24 For this paper, the fetuin glycans are named using the following common nomenclature: the number of antenna (bi, tri, and tetra), followed by the number of SA residues (1-SA). The RP separation of N-glycopeptides of fetuin is shown in Fig. 1A. The extracted ion chromatograms (EICs) from full MS scans obtained for the glycoforms from 1 of the fetuin glycopeptides (Fig. 1A, inset) illustrate that GP15-Bi-2SA and GP15-Tri-3SA have a virtually identical elution profile. GP15-Tri-4SA is partially resolved from GP15-Bi-2SA and GP15-Tri-3SA. These ion traces show that the glycoforms attached to this peptide essentially coelute by RPLC and are consistent with the RP separation mechanism being driven by the hydrophobic interactions of the stationary phase with the peptide backbone. The hydrophilic glycan has minimal interaction with the stationary phase and thus, has little effect on the RP chromatographic behavior. Hence, even dramatic changes in glycan structures (e.g., bi- vs. triantennary N-glycans) only lead to minimal shifts in retention of glycopeptide glycoforms. A consequence caused by all of a glycopeptide’s glycoforms coeluting is the inability to differentiate between glycoforms that are authentically present in the sample, from ions corresponding to the m/z value of smaller glycoforms that result from in-source fragmentation of a larger glycoform(s). The presence of in-source-generated fragment ions can also cause an error in glycoform quantitation, as a signal can correspond to both an authentic glycoform and coincident fragments from a larger glycoform. Thus, whereas RP separations offer some benefits for glycopeptide analysis, development of alternative strategies could lead to greater certainty for accurate identification or quantitation of isomeric glycopeptide glycoforms.
FIGURE 1.
Behavior of tryptic glycopeptides of fetuin during RP chromatography and HILIC. A) The total ion chromatogram (TIC; blue trace) and EIC (inset graph: purple trace of GP15-Bi-2SA; pink trace of GP15-Tri-3SA; green trace of GP15-Tri-4SA) from the RP separation and (B) the TIC (blue trace) and EIC (inset graph: purple trace of GP15-Bi-2SA; pink trace of GP15-Tri-3SA; green trace of GP15-Tri-4SA) of HILIC separation of trypsin-digested fetuin.
The HILIC separation of targeted glycopeptides is shown in Fig. B. The EIC of the same glycopeptide glycoforms discussed above (Fig. 1B, inset) demonstrates that the glycoforms of GP15 (GP15-Bi-2SA, GP15-Tri-3SA, and GP15-Tri-4SA) are baseline resolved from each other. Comparison of N-glycopeptides separation with RP chromatography and HILIC confirms that the attached glycans have a significant contribution to HILIC retention compared with RP and that the retention processes generate sufficient selectivity to enable HILIC to resolve different glycopeptide glycoforms.
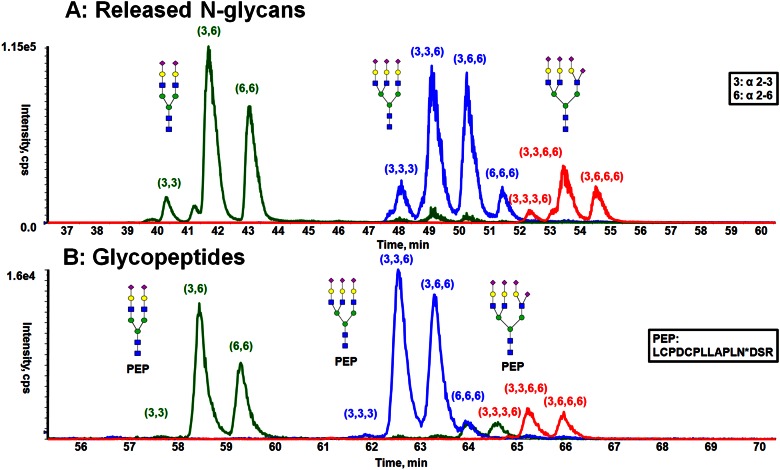
The multiple peaks observed in the HILIC for fetuin glycopeptides with the same mass suggest that these multiple peaks result from chromatographic resolution of isomeric glycoforms. Such elution profiles are similar to the HILIC separation of isomeric-released glycans, wherein the SA α2-3/6 linkage isomers can be resolved. The potential ability to separate isomeric glycans while still attached to peptides led us to compare the HILIC separation of released N-glycans of fetuin with the glycopeptides of fetuin. The HILIC separation of the procainamide (ProA)-labeled N-glycans from fetuin has been studied by exoglycosidase digestion and revealed that baseline separation was achieved for N-glycan isomers with α2-3/α2-6 SA linkages.13 As illustrated in Fig. 2A, isomeric glycans are being resolved based on the ratio of α2-3-to-α2-6 SA linkages present in the glycoform, with the α2-3-linked isomers eluting before the α2-6 glycoforms. In these experiments, the same LC gradient was used to analyze the ProA-labeled N-glycans and glycopeptides of fetuin. By comparing the retention time of glycopeptides (Fig. 2B) and ProA-glycans (Fig. 2A), it is clear that the presence of peptide backbone caused the glycopeptide to be retained longer in the HILIC separation than the ProA glycans. However, the presence of the peptide did not significantly alter the relative retention time differences between the glycoforms, as shown by the similarity of the chromatographic profiles of the glycopeptides and the ProA glycans. Analogous HILIC separation qualities were observed with the other N-linked glycopeptides of fetuin (GP30 and GP27). The chromatographic resolution between adjacent α2-3/α2-6 SA linkage isomers was determined for several of the glycoforms using Equation 1. The results from these calculations (Table 1) demonstrates that the HILIC resolution for glycopeptides is similar to that obtained for the released and derivatized N-glycans (Fig. 2A), while using the same gradient conditions. Therefore, HILIC appears to be equally capable of resolving N-linked glycopeptide glycoforms as well, as this approach resolves the tagged N-glycans.
FIGURE 2.
Comparison of the separation obtained from released N-glycans and glycopeptides of fetuin by HILIC. A) HILIC separation of ProA-labeled, released N-glycans. B) HILIC separation of glycopeptides with the same peptide backbone.
TABLE 1.
The chromatographic resolution between adjacent α2-3/α2-6 SA linkage isomers obtained from the analysis of glycopeptides and released glycans using the identical LC gradient conditions
| Peak 1 | Peak 2 | Resolution |
|
|---|---|---|---|
| Glycans | Glycopeptides | ||
| Bi-2SA(3,3) | Bi-2SA(3,6) | 4.56 | 3.80 |
| Bi-2SA(3,6) | Bi-2SA(6,6) | 4.09 | 3.67 |
| Tri-3SA(3,3,3) | Tri-3SA(3,3,6) | 3.41 | 2.30 |
| Tri-3SA(3,3,6) | Tri-3SA(3,6,6) | 3.53 | 2.79 |
| Tri-3SA(3,6,6) | Tri-3SA(6,6,6) | 4.02 | 2.75 |
| Tri-4SA(3,3,3) | Tri-4SA(3,3,6,6) | 4.09 | 2.56 |
| Tri-4SA(3,3,6,6) | Tri-4SA(3,6,6,6) | 3.49 | 2.12 |
| (1) |
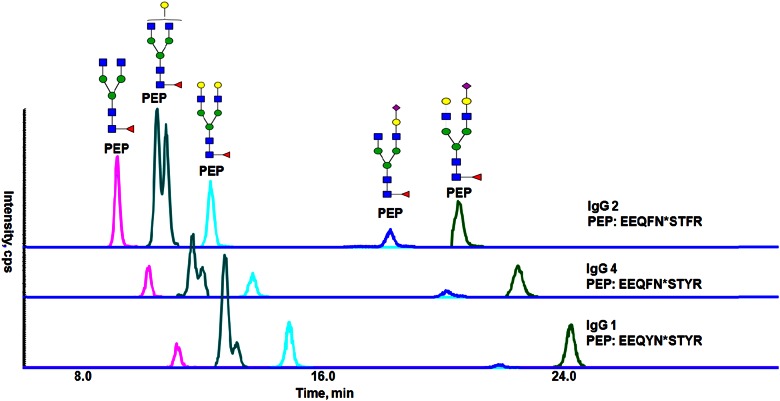
The ability of HILIC to resolve linkage glycoforms of glycopeptides led us to investigate the HILIC separation of a more complex glycopeptide mixture—those obtained by tryptic digestion of human serum IgGs, which occur in 4 subclasses (IgG1, IgG2, IgG3, and IgG4), all of which carry a single N-glycosylation site at asparagine (Asn)-297 in the conserved domain of the crystallizable fragment (Fc) region. After trypsin digestion, the amino acid sequences of glycopeptides from IgG1, IgG2/3, and IgG4 are relatively similar (Supplemental Table 2, supporting information). The N-glycans attached to Asn-297 are a predominantly complex biantennary structure with core fucosylation, and there are small populations of glycans carrying terminal α2-6 linked SAs or a bisecting GlcNAc. Consequently, this sample consists of glycopeptide mixtures with closely related peptide sequences in addition to glycan heterogeneity. The predicted m/z values of all of the known IgG glycopeptides are listed in Supplemental Table 2 (supporting information). The HILIC separation of several selected major N-glycopeptides from IgG1, IgG2/3, and IgG4 are displayed in Fig. 3. The retention order of the glycopeptides with different peptide sequences can be rationalized by the hydrophilicity of the peptide backbones. The amino acid sequences of IgG2/3 (EEQFNSTFR), IgG4 (EEQFNSTYR), and IgG1 (EEQYNSTYR) differ by a phenylalanine (F) to tyrosine (Y) substitution at 1 or 2 locations. As Y is more hydrophilic than F, IgG2/3 has the more hydrophobic amino acid sequence and glycopeptides with this backbone elute earliest. IgG4 has the intermediate hydrophobicity, and IgG1 is the most hydrophilic; all of these are consistent with their elution order (Fig. 3). The varying of glycans on the same peptide sequence exhibits greater HILIC retention for the species with larger glycans. Within these assemblies of glycans, separation of isomeric glycoforms is observed, as is shown by the 2 peaks detected for each glycopeptide possessing the N-glycan with a composition of Hex4-HexNAc4Fuc1 (G1F). This glycan structure has 2 isomers, resulting from attachment of the galactose to each of the 2 antennae.
FIGURE 3.
The HILIC separation of major N-glycopeptides in IgG1, IgG2, and IgG4.
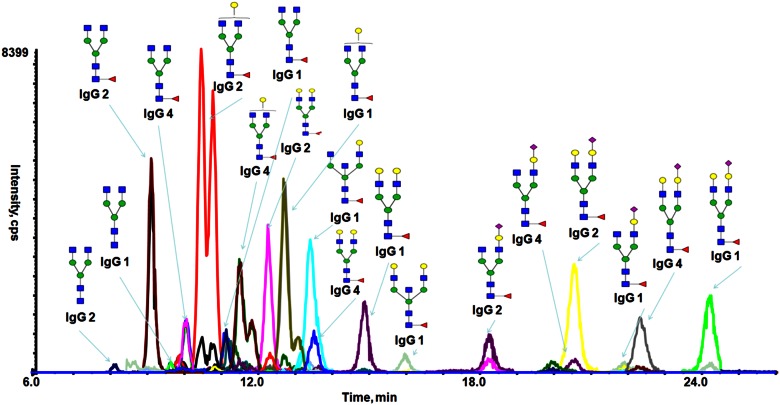
A scheduled SRM approach was developed to enable detection of the lower abundance glycoforms and thus, better evaluate the HILIC retention behavior from each of the expected glycoforms of the IgG glycopeptides. The glycan and peptide sequence heterogeneity of the IgG glycopeptides combines to yield a total 78 distinct precursor masses. Given this sample complexity and thus, the high sensitivity and a high dynamic range required to identify the potential glycopeptide structures that could be resolved by the 30 min-long LC/MS acquisition, parameters for SRM scheduling required knowledge of estimated retention times. A retention time was found experimentally for each glycopeptide glycoform, and these are listed in Supplemental Table 3. The sd of retention times from 10 LC runs ranged from 0.104 to 0.383 min, indicating good reproducibility of the HILIC separation. The detection window set for each transition is 3 min, selected for peak widths observed to be ∼50 s. Under these conditions, numerous glycopeptides of human serum IgGs are detected and are well resolved, as illustrated in Fig. 4.
FIGURE 4.
HILIC SRM analysis of human serum IgGs demonstrating the ability to resolve isomeric glycopeptide glycoforms.
CONCLUSION
Hydrophilic functional groups on both the peptide and the glycan interact with the HILIC stationary phase, and thus, both peptide and glycan structure determine retention of a glycopeptide. This characteristic enables the resolution of mixtures of closely related glycopeptides. The combination of glycopeptide amino acid sequence and glycan composition permits the resolution of glycoforms, which can extend to even resolving isomeric and positional glycan variants. Thus, LC/MS/MS can provide the site-specific glycan profiles at individual sequence sites from complex mixtures of glycoproteins.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the U.S. National Institutes of Health National Institute of General Medical Sciences Grants GM093747 and GM113666.
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Compton SJ, Renaux B, Wijesuriya SJ, Hollenberg MD. Glycosylation and the activation of proteinase-activated receptor 2 (PAR(2)) by human mast cell tryptase. Br J Pharmacol 2001;134:705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell 2006;126:855–867. [DOI] [PubMed] [Google Scholar]
- 3.Ernst B, Magnani JL. From carbohydrate leads to glycomimetic drugs. Nat Rev Drug Discov 2009;8:661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dwek RA, Lellouch AC, Wormald MR. Glycobiology: ‘the function of sugar in the IgG molecule’. J Anat 1995;187:279–292. [PMC free article] [PubMed] [Google Scholar]
- 5.Ray K, Clapp P, Goldsmith PK, Spiegel AM. Identification of the sites of N-linked glycosylation on the human calcium receptor and assessment of their role in cell surface expression and signal transduction. J Biol Chem 1998;273:34558–34567. [DOI] [PubMed] [Google Scholar]
- 6.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 2015;15:540–555. [DOI] [PubMed] [Google Scholar]
- 7.Charlwood J, Dingwall C, Matico R, et al. Characterization of the glycosylation profiles of Alzheimer’s β -secretase protein Asp-2 expressed in a variety of cell lines. J Biol Chem 2001;276:16739–16748. [DOI] [PubMed] [Google Scholar]
- 8.Malhotra R, Wormald MR, Rudd PM, Fischer PB, Dwek RA, Sim RB. Glycosylation changes of IgG associated with rheumatoid arthritis can activate complement via the mannose-binding protein. Nat Med 1995;1:237–243. [DOI] [PubMed] [Google Scholar]
- 9.Zaia J. Mass spectrometry and the emerging field of glycomics. Chem Biol 2008;15:881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raman R, Raguram S, Venkataraman G, Paulson JC, Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nat Methods 2005;2:817–824. [DOI] [PubMed] [Google Scholar]
- 11.Alley WR Jr, Madera M, Mechref Y, Novotny MV. Chip-based reversed-phase liquid chromatography-mass spectrometry of permethylated N-linked glycans: a potential methodology for cancer-biomarker discovery. Anal Chem 2010;82:5095–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Flynn GC. Analysis of N-glycans from recombinant immunoglobulin G by on-line reversed-phase high-performance liquid chromatography/mass spectrometry. Anal Biochem 2007;370:147–161. [DOI] [PubMed] [Google Scholar]
- 13.Tao S, Huang Y, Boyes BE, Orlando R. Liquid chromatography-selected reaction monitoring (LC-SRM) approach for the separation and quantitation of sialylated N-glycans linkage isomers. Anal Chem 2014;86:10584–10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hua S, An HJ, Ozcan S, et al. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst (Lond) 2011;136:3663–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalpathado DS, Irungu J, Go EP, etc. Comparative glycomics of the glycoprotein follicle stimulating hormone: glycopeptide analysis of isolates from two mammalian species. Biochemistry 2006;45:8665–8673. [DOI] [PubMed] [Google Scholar]
- 16.Conboy JJ, Henion JD. The determination of glycopeptides by liquid chromatography/mass spectrometry with collision-induced dissociation. J Am Soc Mass Spectrom 1992;3:804–814. [DOI] [PubMed] [Google Scholar]
- 17.Irungu J, Dalpathado DS, Go EP, et al. Method for characterizing sulfated glycoproteins in a glycosylation site-specific fashion, using ion pairing and tandem mass spectrometry. Anal Chem 2006;78:1181–1190. [DOI] [PubMed] [Google Scholar]
- 18.Zauner G, Selman MH, Bondt A, et al. Glycoproteomic analysis of antibodies. Mol Cell Proteomics 2013;12:856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huddleston MJ, Bean MF, Carr SA. Collisional fragmentation of glycopeptides by electrospray ionization LC/MS and LC/MS/MS: methods for selective detection of glycopeptides in protein digests. Anal Chem 1993;65:877–884. [DOI] [PubMed] [Google Scholar]
- 20.Peterman SM, Mulholland JJ. A novel approach for identification and characterization of glycoproteins using a hybrid linear ion trap/FT-ICR mass spectrometer. J Am Soc Mass Spectrom 2006;17:168–179. [DOI] [PubMed] [Google Scholar]
- 21.Harmon BJ, Gu X, Wang DI. Rapid monitoring of site-specific glycosylation microheterogeneity of recombinant human interferon-γ. Anal Chem 1996;68:1465–1473. [DOI] [PubMed] [Google Scholar]
- 22.Dziegielewska KM, Brown WM, Casey SJ, et al. The complete cDNA and amino acid sequence of bovine fetuin. Its homology with alpha 2HS glycoprotein and relation to other members of the cystatin superfamily. J Biol Chem 1990;265:4354–4357. [PubMed] [Google Scholar]
- 23.Küster B, Wheeler SF, Hunter AP, Dwek RA, Harvey DJ. Sequencing of N-linked oligosaccharides directly from protein gels: in-gel deglycosylation followed by matrix-assisted laser desorption/ionization mass spectrometry and normal-phase high-performance liquid chromatography. Anal Biochem 1997;250:82–101. [DOI] [PubMed] [Google Scholar]
- 24.Sheeley DM, Reinhold VN. Structural characterization of carbohydrate sequence, linkage, and branching in a quadrupole Ion trap mass spectrometer: neutral oligosaccharides and N-linked glycans. Anal Chem 1998;70:3053–3059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.