Abstract
Melanopus is a morphological group of Polyporus which contains species with a black cuticle on the stipe. In this article, taxonomic and phylogenetic studies on Melanopus group were carried out on the basis of morphological characters and phylogenetic evidence of DNA sequences of multiple loci including the internal transcribed spacer (ITS) regions, the large subunit nuclear ribosomal RNA gene (nLSU), the small subunit nuclear ribosomal RNA gene (nSSU), the small subunit mitochondrial rRNA gene sequences (mtSSU), the translation elongation factor 1-α gene (EF1-α), the largest subunit of RNA polymerase II (RPB1), the second largest subunit of RNA polymerase II (RPB2), and β-tubulin gene sequences (β-tubulin). The phylogenetic result confirmed that the previously so-called Melanopus group is not a monophyletic assemblage, and species in this group distribute into two distinct clades: the Picipes clade and the Squamosus clade. Four new species of Picipes are described, and nine new combinations are proposed. A key to species of Picipes is provided.
Introduction
Melanopus Pat. was established by Patouillard [1] as a specific genus which including stipitate polypores with black stipes. Then it was accepted as a synonym of Polyporus P. Micheli ex Adans. [2]. Núñez and Ryvarden [3] treated genus Melanopus as an infrageneric group of Polyporus. They defined this group with following characters: basidiocarps coriaceous, tough when dry, context thin, stipe with a black cuticle, skeleto-binding hyphae mostly solid and narrow when mature, basidiospores medium size to large (6–12 × 2–4 μm). Eleven species of Polyporus were accepted as members of group Melanopus by Núñez and Ryvarden [3]. Among these species, P. mikawai Lloyd was transferred into Neofavolus Sotome & T. Hatt. as N. mikawai (Lloyd) Sotome & T. Hatt. based on morphological and phylogenetic analyses [4].
Phylogenetically, P. badius (Pers.) Schwein., P. melanopus (Pers.) Fr. and P. tubaeformis (P. Karst.) Ryvarden & Gilb. grouped together and formed a clade with higher supports while others dispersed with lower supports [5,6]. Thus, Krüger et al. [6] indicated that “Melanopus” appeared to be a non-monophyletic assemblage of dark-stiped polypores. Sotome et al. [7] revealed that P. badius, P. dictyopus Mont. and P. tubaeformis could cluster together in a single clade with high supports while P. leprieurii Mont., P. varius (Pers.) Fr., P. squamosus (Huds.) Fr. and Datronia mollis (Sommerf.) Donk, D. scutellata (Schwein.) Domański, Pseudofavolus cucullatus (Mont.) Pat. gathered into another clade. In the latest study of Melanopus, a well supported Melanopus clade including eleven species was reported [8].
Recently, when studying the lentinoid and polyporoid fungi, Zmitrovich & Kovalenko [9] described a new genus Picipes Zmitr. et Kovalenko typified by Pi. badius (Pers.) Zmitr. et Kovalenko (= Polyporus badius) according to analyses of nLSU, ITS and EF1-α datasets. This genus is characterized by the following features: basidiomata annual, stipitate, morphotype polyporoid; pilei infundibuliform, covered with hard cuticle, without scales, smoke gray to castaneous or deeply brown; stipe covered with brownish to black cuticle; pores small (more than 5 per mm); hyphal system dimitic with uninflated both corioiloid and fibrous skeletals; clamps present or absent; basidiospores cylindrical, smooth, hyaline; on wood of frondose and coniferous trees causing a white rot. Polyporus badius, P. melanopus and P. tubaeformis were segregated from Polyporus and transferred into Picipes as Pi. badius, Pi. melanopus (Pers.) Zmitr. et Kovalenko and Pi. tubaeformis (P. Karst.) Zmitr. et Kovalenko [9].
In the current study, species diversity of group Melanopus in China was investigated, phylogenetic analysis based on 8 genes (including ITS, nLSU, EF1-α, mtSSU, β-tubulin, RPB1, RPB2 and nSSU) was carried out, and four new species matching the concept of Picipes are described and illustrated.
Materials and Methods
All thirty-five specimens examined in this study are publicly deposited in the herbaria of the Institute of Microbiology, Beijing Forestry University (BJFC, Beijing, China) and the Institute of Applied Ecology, Chinese Academy of Sciences (IFP, Shenyang, China). No permits were required for the described study because Chinese legislations do not forbid the access to studying fungi in Nature Reserves and National Parks. We confirm that the studies involve only fungi samples and these samples do not involve endangered or protected species.
Morphology
Macro-morphological descriptions were based on field notes. Special color terms followed Petersen [10]. Micro-morphological data were obtained from dried specimens, and observed under a light microscope following methods in Li et al. [11]. Microscopic features and measurements were made from slide preparations stained with 5% KOH solution, 1% Congo Red solution, Cotton Blue and Melzer’s reagent. Basidiospores were measured from sections cut from the tubes. To represent variation in the size of basidiospores, 5% of measurements were excluded from each end of the range, and are given in parentheses. The following abbreviations are used: IKI– = neither amyloid nor dextrinoid, KOH = 5% potassium hydroxide, CB+ = cyanophilous, CB– = acyanophilous, L = mean spore length (arithmetic average of all basidiospores), W = mean spore width (arithmetic average of all basidiospores), Q = variation in the L/W ratios between the specimens studied, Qm = mean Q, n (a/b) = number of basidiospores (a) measured from given number (b) of specimens. Microscopic features, such as basidiospores, basidia, hyphae and cystidioles, were observed and photographed at a magnification of up to ×1000 by Nikon Digital Sight DS-Fi1 microscope (Nikon Corporation, Tokyo, Japan), and quantified by the Image-Pro Plus 6.0 software (Media Cybernetics, Silver Spring, USA).
Molecular phylogeny
CTAB rapid plant genome extraction kit-DN14 (Aidlab Biotechnologies Co., Ltd., Beijing, China) and FH plant DNA kit II (Demeter Biotech Co., Ltd., Beijing, China) were used to extract total genomic DNA from dried specimens and to perform the polymerase chain reaction (PCR), according to the manufacturer’s instructions with some modifications [12,13]. ITS region was amplified with the primer pair ITS4 and ITS5 [14] while nLSU with LR0R and LR7 [15], EF1-α with EF1-983F and EF1-1567R [16], β-tubulin with Bt-1a and Bt-1b [17], nSSU with PNS1 and NS41 [18], mtSSU with MS1 and MS2 [14], RPB1 with RPB1-Af and RPB1-Cr [19], RPB2 with fRPB2-5F and bRPB2-7.1R [20,21]. The primer RPB1-2.2f (GAGTGTCCGGGGCATTTYGG) [22] sometimes was used as an alternative to RPB1-Af and bRPB2-6F [21] was used as an alternative to fRPB2-5F. The final PCR volume is 50 μl each tube which contains 1.5 μl primer (10 pM), 2 μl DNA extract, 20 μl ddH2O and 25 μl 2×EasyTaq PCR Supermix (TransGen Biotech Co., Ltd., Beijing, China). PCRs were performed on S1000™ Thermal Cycler (Bio-Rad Laboratories, California, USA).
PCR procedures for mtSSU, ITS, nSSU, β-tubulin and EF1-α were as following: (1) initial denaturation 94°C for 2 min, (2) denaturation at 94°C for 45 s, (3) annealing at 52°C (for mtSSU)/53°C (for ITS, nSSU and β-tubulin)/54°C (for EF1-α) for 45 s, (4) extension at 72°C for 1 min, (5) repeat for 36 cycles of last three steps, (6) final extension at 72°C for 10 min.
PCR procedure for nLSU was as following: (1) initial denaturation 94°C for 5 min, (2) denaturation at 94°C for 1 min, (3) annealing at 50°C for 1min 20 s, (4) extension at 72°C for 1 min 30 s, (5) repeat for 36 cycles of last three steps, (6) final extension at 72°C for 10 min.
For RPB1 and RPB2, the PCR procedure was used as following: initial denaturation 94°C for 2 min, followed by 9 cycles at 94°C for 45 s, 60°C for 45 s (minus 1°C per cycle) and 72°C for 1min 30 s, then followed by 36 cycles at 94°C for 45 s, 53°C for 1 min and 72°C for 1min 30 s, finally with a final extension at 72°C for 10 min.
All PCR products were purified and sequenced in the BGI (Beijing Genomics Institute), China, with the same primers. All newly generated sequences were deposited at GenBank and listed in Table 1. Trametes conchifer (Schwein.) Pilát was selected as outgroup.
Table 1. Species list of Polyporus and related genera used in this study and GenBank entries.
| No. | Species | Specimen No. | Country | GenBank accession No. | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS | nLSU | EF1-α | mtSSU | β-tubulin | RPB1 | RPB2 | nSSU | ||||
| 1 | Favolus acervatus | Cui 11053 | China | KU189774 a | KU189805 a | KU189920 a | KU189956 a | KU189864 a | KU189889 a | KU189994 a | KU189835 a |
| 2 | F. emerici | Cui 10926 | China | KU189776 a | KU189807 a | KU189922 a | - | KU189866 a | KU189890 a | KU189995 a | KU189837 a |
| 3 | F. roseus | PEN 33 | Malaysia | AB735975 | AB368099 | - | - | - | - | AB368156 | - |
| 4 | F. spathulatus | Dai 13615A | China | KU189775 a | KU189806 a | KU189921 a | KU189957 a | KU189865 a | - | - | KU189836 a |
| 5 | Neofavolus alveolaris | Dai 11290 | China | KU189768 a | KU189799 a | KU189913 a | KU189949 a | KU189859 a | KU189885 a | KU189982 a | KU189828 a |
| 6 | N. mikawai | Cui 11152 | China | KU189773 a | KU189804 a | KU189919 a | KU189955 a | KU189863 a | KU189888 a | KU189986 a | KU189834 a |
| 7 | Picipes admirabilis | Dai 1127 | China | KC572001 | - | - | - | - | - | - | - |
| 8 | Pi. americanus | JV 0509–149 (T) | USA | KC572002 | KC572041 | - | - | - | - | - | - |
| 9 | Pi. austroandinus | MR 10701 | Argentina | AF516569 | - | - | - | - | - | - | - |
| 10 | Pi. badius | Cui 10853 | China | KU189780 a | KU189811 a | KU189929 a | - | KU189871 a | KU189894 a | - | KU189844 a |
| 11 | Pi. badius | Cui 11136 | China | KU189781 a | KU189812 a | KU189930 a | KU189964 a | KU189872 a | KU189895 a | KU189990 a | KU189845 a |
| 12 | Pi. badius | Cui 10501 | China | KC572015 | KC572053 | KU189927 a | KU189962 a | KU189869 a | - | KU189989 a | KU189842 a |
| 13 | Pi. badius | Cui 10484 | China | KC572014 | KC572052 | KU189928 a | KU189963 a | KU189870 a | KU189893 a | - | KU189843 a |
| 14 | Pi. baishanzuensis | Dai 13418 (T) | China | KU189762 a | KU189793 a | KU189907 a | KU189945 a | KU189855 a | KU189882 a | KU189977 a | KU189823 a |
| 15 | Pi. baishanzuensis | Cui 11395 | China | KU189763 a | KU189794 a | KU189908 a | KU189946 a | KU189856 a | - | KU189978 a | KU189824 a |
| 16 | Pi. conifericola | Cui 9950 | China | KU189783 a | KU189814 a | KU189934 a | KU189968 a | KU189875 a | KU189897 a | KU189993 a | KU189848 a |
| 17 | Pi. conifericola | Dai 11114 (T) | China | JX473244 | KC572061 | KU189935 a | KU189969 a | - | - | - | KU189849 a |
| 18 | Pi. fraxinicola | Dai 2494 | China | KC572023 | KC572062 | KU189932 a | KU189966 a | - | - | - | - |
| 19 | Pi. melanopus | TENN 59326 | Austria | AF518759 | - | - | - | - | - | - | - |
| 20 | Pi. melanopus | MJ 400–93 | Czech | KC572027 | - | - | - | - | - | - | - |
| 21 | Pi. rhizophilus | Dai 11599 | China | KC572028 | KC572067 | KU189933 a | KU189967 a | KU189874 a | KU189896 a | KU189992 a | KU189847 a |
| 22 | Pi. submelanopus | Dai 13294 | China | KU189770 a | KU189801 a | KU189915 a | KU189951 a | KU189860 a | KU189886 a | KU189984 a | KU189830 a |
| 23 | Pi. submelanopus | Dai 13296 | China | KU189771 a | KU189802 a | KU189916 a | KU189952 a | KU189861 a | - | - | KU189831 a |
| 24 | Pi. subtropicus | Li 1928 | China | KU189758 a | KU189790 a | KU189904 a | KU189942 a | KU189854 a | KU189881 a | KU189976 a | KU189820 a |
| 25 | Pi. subtropicus | Cui 2662 (T) | China | KU189759 a | KU189791 a | KU189905 a | KU189943 a | - | - | - | KU189821 a |
| 26 | Pi. subtubaeformis | Dai 11870 (T) | China | KU189752 a | KU189784 a | KU189899 a | KU189937 a | KU189850 a | KU189876 a | KU189972 a | KU189815 a |
| 27 | Pi. subtubaeformis | Cui 10793 | China | KU189753 a | KU189785 a | KU189900 a | KU189938 a | KU189851 a | KU189877 a | KU189973 a | KU189816 a |
| 28 | Pi. taibaiensis | Dai 5746 (T) | China | KX196783 a | KX196784 a | KX196785 a | KX196786 a | - | - | - | KX196787 a |
| 29 | Pi. tibeticus | Cui 12215 (T) | China | KU189755 a | KU189787 a | KU189902 a | KU189940 a | KU189853 a | KU189879 a | KU189975 a | KU189818 a |
| 30 | Pi. tibeticus | Cui 12225 | China | KU189756 a | KU189788 a | KU189903 a | KU189941 a | - | KU189880 a | - | KU189819 a |
| 31 | Pi. tubaeformis | Niemela 6855 | Finland | KC572036 | KC572073 | - | - | - | - | - | - |
| 32 | Pi. tubaeformis | JV 0309–1 | USA | KC572034 | KC572072 | - | - | - | - | - | - |
| 33 | Pi. virgatus | CulTENN 11406 | Argentina | AF516582 | AJ488123 | ||||||
| 34 | Pi. virgatus | CulTENN 11219 | Argentina | AF516581 | AJ488122 | ||||||
| 35 | Polyporus arcularius | Cui 11398 | China | KU189766 a | KU189797 a | KU189911 a | KU189947 a | - | KU189884 a | KU189980 a | KU189826 a |
| 36 | P. brumalis | Cui 10750 | China | KU189765 a | KU189796 a | KU189910 a | - | KU189857 a | KU189883 a | KU189979 a | KU189825 a |
| 37 | P. ciliatus | Wei 1582 | China | KU189767 a | KU189798 a | KU189912 a | KU189948 a | KU189858 a | - | KU189981 a | KU189827 a |
| 38 | P. dictyopus | TENN 59385 | Belize | AF516561 | AJ487945 | - | - | - | - | - | - |
| 39 | P. guianensis | TENN 59093 | Argentina | AF516564 | AJ487947 | - | - | - | - | - | - |
| 40 | P. guianensis | TENN 58404 | Venezuela | AF516566 | AJ487948 | - | - | - | - | - | - |
| 41 | P. hapalopus | Yuan 5809 (T) | China | KC297219 | KC297220 | KU189918 a | KU189954 a | - | - | - | KU189833 a |
| 42 | P. leprieurii | TENN 58579 | Costa Rica | AF516567 | - | - | - | - | - | - | - |
| 43 | P. squamosus | Wang 555 | China | KU189779 a | KU189810 a | KU189926 a | KU189961 a | - | - | - | KU189841 a |
| 44 | P. squamosus | Cui 10595 | China | KU189778 a | KU189809 a | KU189925 a | KU189960 a | KU189868 a | KU189892 a | KU189988 a | KU189840 a |
| 45 | P. subvarius | Yu 2 (T) | China | AB587632 | AB587621 | KU189924 a | KU189959 a | - | - | - | KU189839 a |
| 46 | P. tuberaster | Dai 12462 | China | KU507580 a | KU507582 a | KU507590 a | KU507584 a | KU507588 a | - | - | KU507586 a |
| 47 | P. tuberaster | Dai 11271 | China | KU189769 a | KU189800 a | KU189914 a | KU189950 a | - | - | KU189983 a | KU189829 a |
| 48 | P. umbellatus | Pen 13513 | China | KU189772 a | KU189803 a | KU189917 a | KU189953 a | KU189862 a | KU189887 a | KU189985 a | KU189832 a |
| 49 | P. varius | Cui 12249 | China | KU507581 a | KU507583 a | KU507591 a | KU507585 a | - | KU507589 a | KU507592 a | KU507587 a |
| 50 | P. varius | Dai 13874 | China | KU189777 a | KU189808 a | KU189923 a | KU189958 a | KU189867 a | KU189891 a | KU189987 a | KU189838 a |
| 51 | Trametes conchifer | FP 106793sp | USA | JN164924 | JN164797 | JN164887 | - | - | JN164823 | JN164849 | - |
a indicates accession numbers for newly generated sequences; (T) indicates holotype specimen.
Phylogenetic analyses were done as in Zhao et al. [23–25]. All gene sequences were initially aligned separately using Clustal Omega [26] and then manually adjusted. One thousand partition homogeneity test (PHT) replicates of the combined dataset were tested by PAUP 4.0 beta 10 [27] to determine whether the partitions were homogeneous. The best-fit evolutionary model was selected by hierarchical likelihood ratio tests (hLRT) and Akaike information criterion (AIC) in MrModeltest 2.2 [28] after scoring 24 models of evolution by PAUP 4.0 beta 10. The best maximum likelihood (ML) phylogenetic bootstrap values (ML-BS) obtained from 1000 replicates were performed using RAxmlGUI [29], while the ML topology, maximum parsimony (MP) tree and bootstrap values (MP-BS) were performed using PAUP 4.0 beta 10. Bayesian phylogenetic inference and Bayesian posterior probabilities (BPPs) were performed with MrBayes v3.2 [30]. Four Markov chains were run for 5,000,000 generations until the split deviation frequency value <0.01, and sampled every 100th generation. All trees were viewed in FigTree 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Nomenclature Acts
The electronic version of this article in Portable Document Format (PDF) in a work with an ISSN or ISBN will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants, and hence the new names contained in the electronic publication of a PLOS article are effectively published under that Code from the electronic edition alone, so there is no longer any need to provide printed copies.
In addition, new names contained in this work have been submitted to MycoBank from where they will be made available to the Global Names Index. The unique MycoBank number can be resolved and the associated information viewed through any standard web browser by appending the MycoBank number contained in this publication to the prefix http://www.mycobank.org/MB/. The online version of this work is archived and available from the following digital repositories: PubMed Central and LOCKSS.
Results
232 new sequences (Table 1), which including 29 ITS, 29 nLSU, 36 EF1-α, 33 mtSSU, 25 β-tubulin, 22 RPB1, 23 RPB2 and 35 nSSU, were generated for this study. Other 45 related sequences (including 23 ITS, 18 nLSU, 2 RPB2, 1 EF1-α and 1 RPB1) used in phylogenetic analysis were downloaded from GenBank and listed in Table 1. The partition homogeneity test indicated all the eight different DNA sequences display a congruent phylogenetic signal (P value = 0.999). Thus, an 8-gene concatenated dataset resulted in an alignment with 7270 total characters (including 682 ITS + 1364 nLSU + 600 EF1-α + 738 mtSSU + 482 β-tubulin + 1245 RPB1 + 1053 RPB2 + 1106 nSSU nucleotides) was carried out. Among these characters, 4876 of them were constant, 488 variable characters were parsimony-uninformative and 1906 characters were parsimony- informative. In the MP analysis, 63,166,858 rearrangements were tried and two equally most parsimonious trees (length = 8184, CI = 0.481, RI = 0.622, RC = 0.299, HI = 0.519) were retained. While in the ML analysis, 33,295 rearrangements were tried and the best obtained tree scored 46291.85109. ML analysis used the GTR+I+G model and had an identical topology with the Bayesian trees. Topology from ML analysis was presented along with ML-BS (above 50%), MP-BS (above 50%) and BPPs (above 0.95) values (Fig 1, TreeBase submission ID: 18703).
Fig 1. Phylogeny of Polyporus and related genera inferred from ITS+nLSU+EF1-α+mtSSU+ β-tubulin+RPB1+ RPB2+nSSU data.
Topology is from ML analysis with maximum likelihood bootstrap support values (≥50, former), parsimony bootstrap support values (≥50, middle) and Bayesian posterior probability values (≥0.95, latter). The bold species are new from China.
The phylogenetic tree (Fig 1) shows that all sampled species within Polyporus group Melanopus were divided into two distinct clades:
The picipes clade: Besides the three species of Picipes, Pi. badius, Pi. melanopus and Pi. tubaeformis, nine Polyporus spp. (including P. admirabilis Peck, P. americanus Vlasák & Y.C. Dai, P. austroandinus Rajchenb. & Y.C. Dai, P. conifericola H.J. Xue & L.W. Zhou, P. fraxinicola L.W. Zhou & Y.C. Dai, P. rhizophilus Pat., P. submelanopus H.J. Xue & L.W. Zhou, P. taibaiensis Y.C. Dai and P. virgatus Berk. & M.A. Curtis) and four undescribed Picipes species are contained in the well supported picipes clade (100/98/1.00). Among these species, P. rhizophilus, which was morphologically treated as a member of group Polyporellus, is closely related to P. austroandinus (85/65/1.00), while P. admirabilis of group Admirabilis strongly clusters with P. fraxinicola (100/100/1.00). The four undescribed species are well supported as new species of Picipes.
The squamosus clade: Polyporus squamosus strongly clusters with P. dictyopus, P. guianensis Mont., P. leprieurii, P. subvarius C.J. Yu & Y.C. Dai and P. varius in the squamosus clade (96/79/1.00). Species in this clade usually produce stipes with black cuticle at the lower or entire part, although P. squamosus was morphologically treated as a member of group Polyporus.
The phylogenetic topology also shows four other clades with high supports:
The well supported core polyporus clade (94/77/1.00) contains P. hapalopus H.J. Xue & L.W. Zhou, P. tuberaster (Jacq. ex Pers.) Fr. and P. umbellatus (Pers.) Fr. It shows that Polyporus spp. in the core polyporus clade have closer relationships with species in squamosus clade. Polyporus tuberaster was selected as the lectotype species of Polyporus by Donk [31] and accepted by most succeeding researchers [3,4,7,8,32]. Morphologically, P. tuberaster was treated as a members of group Polyporus along with P. squamosus, however P. umbellatus was put into group Dendropolyporus [6].
Favolus clade and neofavolus clade are well supported as monophyletic lineages respectively (both 100/100/100 for Favolus and Neofavolus). These two clades are separately composed of species from genera Favolus and Neofavolus.
Polyporellus clade, which is composed of P. arcularius (Batsch) Fr., P. brumalis (Pers.) Fr. and P. ciliatus Fr. is well supported as a distinct group phylogenetically separated from other Polyporus (100/100/100).
Taxonomy
Picipes Zmitr. et Kovalenko
Basidiocarps annual, stipitate, of polyporoid morphotype; pilei fan-shaped to circular or infundibuliform, covered with hard cuticle, glabrous; corky to coriaceous when fresh and hard when dry; stipe usually covered with brownish to black cuticle when mature; pores round or angular; hyphal system dimitic with generative and skeleto-binding hyphae, uninflated; generative hyphae with clamp connections or simple septa; skeleto-binding hyphae strongly branched in trama; hyphae in cuticle bearing clamp connections or not, thick-walled with a wide lumen, usually unbranched; basidiospores oblong to cylindrical or fusiform, smooth, hyaline, less than 13 μm long and 5 μm wide; mainly growing on woods and causing a white rot, occasionally on ground or grass roots.
1 Picipes baishanzuensis J.L. Zhou & B.K. Cui, sp. nov. Figs 2A and 3
Fig 2. Basidiocarps of the four new Picipes species.
(A): Pi. baishanzuensis (Dai 13418); (B): Pi. subtropicus (Li 1611); (C): Pi. subtubaeformis (Dai 11870); (D): Pi. tibeticus (Cui 12215). Bars: A = 2 cm, B, C, D = 1 cm.
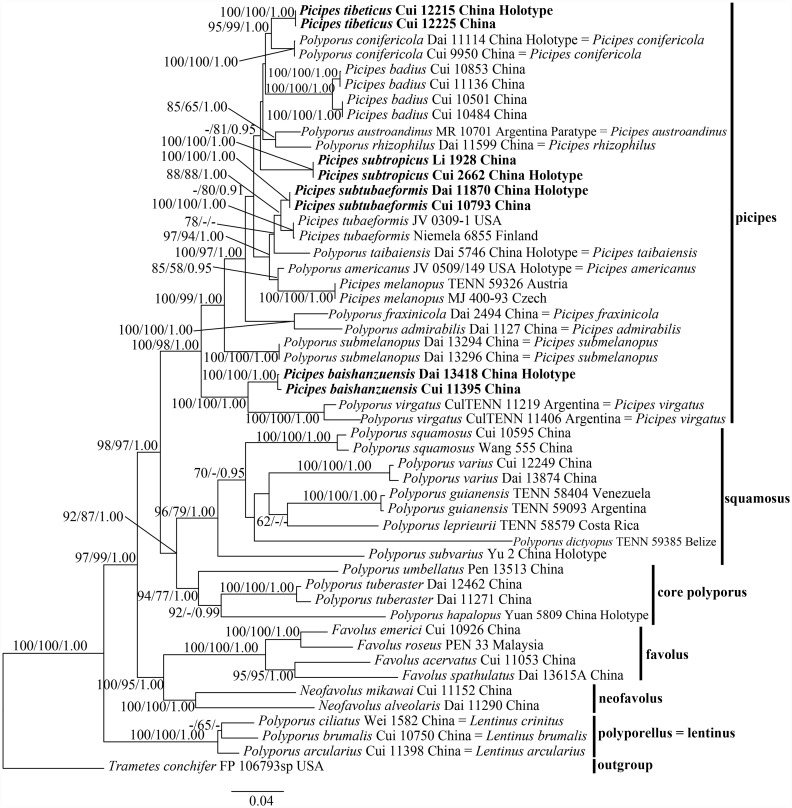
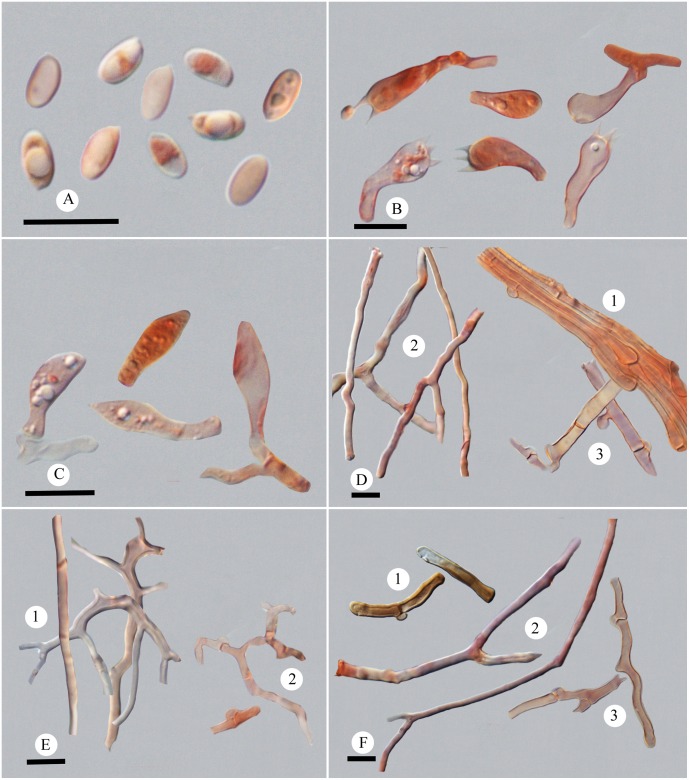
Fig 3. Microscopic structures of Picipes baishanzuensis.
(A): Basidiospores; (B): Basidia and basidioles; (C): Cystidioles; (D): Hyphae from context, 1 hyphae in cuticle, 2 generative hyphae, 3 skeleto-binding hyphae; (E): Hyphae from trama, 1 generative hyphae, 2 skeleto-binding hyphae; (F): Hyphae from stipe, 1 hyphae in cuticle, 2 generative hyphae, 3 skeleto-binding hyphae. Bars = 10 μm.
MycoBank NO.: MB 815517
Basidiocarps annual, centrally stipitate, solitary. Pilei infundibuliform. Pileal surface glabrous, reddish-brown to black in the center and becoming light ivory to pale-brown towards the edge, with radially aligned stripes. Pore surface white; pores round to angular, 3–6 per mm. Stipe black. Hyphal system dimitic; generative hyphae with clamp connections. Basidiospores cylindrical, 6.6–7.9 × 2.5–3.1 μm. On dead angiosperm tree, causing a white-rot.
Type: China. Zhejiang Prov., Qingyuan County, Baishanzu Nature Reserve, on dead angiosperm tree, 14 August 2013, Dai 13418 (holotype in BJFC).
Etymology: baishanzuensis (Lat.): referring to the locality of type specimens in Baishanzu Nature Reserve.
Fruitbody: Basidiocarps annual, centrally stipitate, solitary, coriaceous when fresh and tough when dry. Pilei infundibuliform, up to 5.5 cm wide and 2.5 mm thick. Pileal surface glabrous, reddish-brown to black in the center and becoming light ivory to pale-brown towards the edge in young specimens, black upon the whole pileus upon the old ones, with radially aligned stripes; margin straight when fresh and involute upon drying. Pore surface white when fresh, cream to buff upon drying; pores round to angular, 3–6 per mm; dissepiments thin, entire to slightly lacerate. Context white to buff, becoming woody hard upon drying, up to 1 mm thick. Tubes concolorous with pore surface, decurrent on the stipe, less than 1.5 mm thick. Stipe slender, bearing a black cuticle, wrinkled, 2.2 cm long and 5 mm in diam.
Hyphal structure: Hyphal system dimitic; generative hyphae bearing clamp connections, colorless, thin-walled; skeleto-binding hyphae colorless, thick-walled, with arboriform branches and tapering ends, IKI–, CB+; tissue unchanged in KOH.
Context: Generative hyphae frequent, colorless, thin-walled, frequently branched from clamp connections, 2–5.5 μm in diam., usually inflating at the branching area; skeleto-binding hyphae dominant, colorless, thick-walled with a wide lumen, moderately branched, interwoven, 1.7–6.8 μm in diam. Hyphae in cuticle bearing clamp connections, thin-walled with a wide lumen, with buff inclusion inside, parallel arranged into a palisade, 2.7–6 μm in diam.
Tubes: Generative hyphae frequent, usually present near hymenium, colorless, thin-walled, occasionally branched, 2–3.8 μm in diam.; skeleto-binding hyphae dominant, colorless, thick-walled with a wide lumen, frequently with dendroid branching, strongly interwoven, 0.9–3.3 μm in diam. Cystidia absent; cystidioles infrequent, subulate, 16–21 × 3.2–5.3 μm; basidia clavate, with a basal clamp and four sterigmata, 13.4–27 × 4.6–6.5 μm; basidioles in shape similar to basidia, smaller than basidia.
Stipe: Generative hyphae frequent, colorless, thin-walled, occasionally branched, 1.9–5.5 μm in diam.; skeleto-binding hyphae colorless, thick-walled with a wide to narrow lumen, moderately branched, interwoven, 1.9–4.3 μm in diam. Hyphae in cuticle bearing clamp connections, thick-walled with a wide lumen, with buff to brown inclusion inside and arranged in a palisade, 3.1–6 μm in diam.
Basidiospores: Basidiospores cylindrical, thin-walled, colorless, smooth, usually bearing one to three guttules, IKI–, CB–, (5.8–)6.6–7.9(−8) × (2.4–)2.5–3.1(−3.3) μm, L = 7.04 μm, W = 2.82 μm, Q = 2.14–2.84, Qm = 2.5 (n = 90/3).
Rot type: A white rot.
Additional specimens (paratypes) examined: China. Zhejiang Prov., Qingyuan County, Baishanzu Nature Reserve, on dead angiosperm tree, 14 September 2012, Cui 11392 (BJFC). Zhejiang Prov., Qingyuan County, Baishanzu Nature Reserve, on dead angiosperm tree, 24 July 2013, Cui 11395 (BJFC).
2 Picipes subtropicus J.L. Zhou & B.K. Cui, sp. nov. Figs 2B and 4
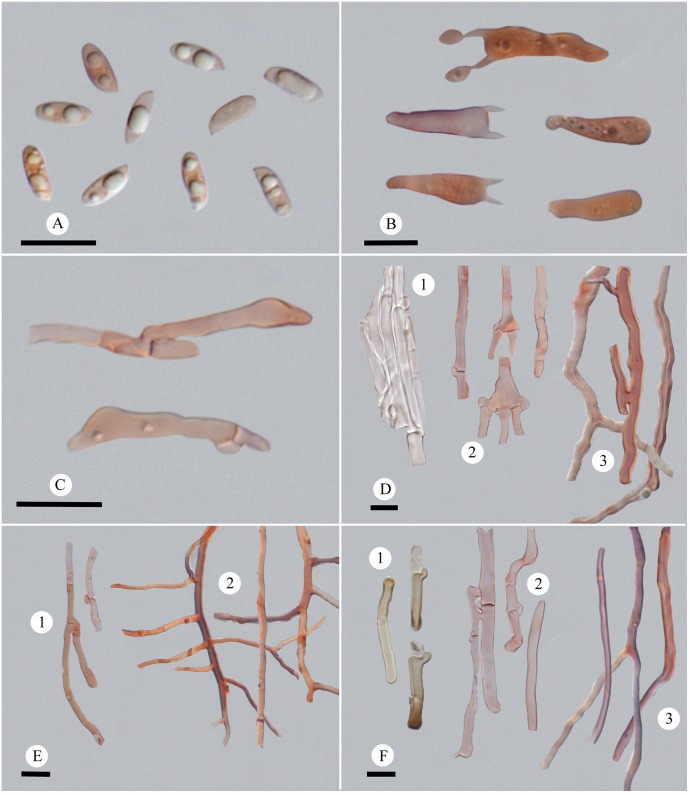
Fig 4. Microscopic structures of Picipes subtropicus.
(A): Basidiospores; (B): Basidia and basidioles; (C): Cystidioles; (D): Hyphae from context, 1 skeleto-binding hyphae, 2 generative hyphae, 3; (E): Hyphae from trama, 1 skeleto-binding hyphae, 2 generative hyphae; (F): Hyphae from stipe, 1 hyphae in cuticle, 2 skeleto-binding hyphae, 3 generative hyphae. Bars = 10 μm.
MycoBank NO.: MB 815518
Basidiocarps annual, laterally stipitate, gregarious. Pilei fan-shaped to semicircular. Pileal surface glabrous, always black towards the base and becoming reddish-brown to orange-brown towards the edge. Pore surface white; pores angular to subcircular, 8–9 per mm when young and becoming 5–7 per mm when aged. Stipe short or with a flattened base, with a black cuticle. Hyphal system dimitic; generative hyphae with clamp connections. Basidiospores cylindrical, 5.1–6.2 × 2.2–2.7 μm. On fallen angiosperm branch, causing a white-rot.
Type: China. Zhejiang Prov., Lin’an, Tianmushan Nature Reserve, on fallen angiosperm branch, 10 October 2005, Cui 2662 (holotype in BJFC).
Etymology: subtropicus (Lat.): referring to the geographic distribution in subtropical regions.
Fruitbody: Basidiocarps annual, laterally stipitate, gregarious, coriaceous when fresh and tough when dry. Pilei fan-shaped to semicircular, up to 4.8 cm wide and 2.5 mm thick. Pileal surface glabrous, black towards the base and becoming reddish-brown to orange-brown towards the edge when fresh, frequently becoming black to chestnut upon the whole pileus, sometimes brown-beige or pastel-yellow towards the edge when dry; margin straight when fresh and straight or slightly involute upon drying. Pore surface white when fresh, white to brown-beige when dry; pores angular to subcircular, 8–9 per mm when young and becoming 5–7 per mm when aged; dissepiments thin, entire to slightly lacerate. Context white to buff and woody hard upon drying, up to 2 mm thick. Tubes white when fresh and white to brown-beige upon drying, less than 1 mm thick, decurrent on one side of the stipe. Stipe very short or forming a flattened base, bearing a black cuticle, up to 5 mm long and 5 mm in diam.
Hyphal structure: Hyphal system dimitic; generative hyphae bearing clamp connections, colorless, thin-walled; skeleto-binding hyphae colorless, thick-walled, with arboriform branches and tapering ends, IKI–, CB+; tissue unchanged in KOH.
Context: Generative hyphae infrequent, colorless, thin-walled, rarely branched, 1.9–4.7 μm in diam.; skeleto-binding hyphae dominant, colorless, thick-walled with a wide to narrow lumen, moderately branched, interwoven, 1.7–4.4 μm in diam. Hyphae in cuticle bearing clamp connections, thin-walled with a wide lumen, with buff to yellowish-brown inclusion inside, parallel arranged into a palisade, 1.6–3.2 μm in diam.
Tubes: Generative hyphae frequent, usually present near hymenium, colorless, thin-walled, 1.5–3.6 μm in diam.; skeleto-binding hyphae dominant, colorless, thick-walled with a wide to narrow lumen, frequently with dendroid branching, strongly interwoven, 1.2–4.2 μm in diam. Cystidia absent; cystidioles frequent, subulate, 14.5–22.8 × 3.2–5.1 μm; basidia clavate, with a basal clamp and four sterigmata, 12.5–27 × 4.8–6.4 μm; basidioles in shape similar to basidia, smaller than basidia.
Stipe: Generative hyphae infrequent, colorless, thin-walled, 2–4.5 μm in diam.; skeleto- binding hyphae colorless, thick-walled with a narrow lumen to solid, moderately branched, interwoven, 0.9–5.3 μm in diam. Hyphae in cuticle bearing clamp connections, thick-walled, with a narrow lumen, with dark brown inclusion inside and arranged in a palisade, 2.5–5.6 μm in diam.
Basidiospores: Basidiospores cylindrical, thin-walled, colorless, smooth, occasionally bearing one or two guttules, IKI–, CB–, (4.7–)5.1–6.2(–6.6) × 2.2–2.7(−2.9) μm, L = 5.6 μm, W = 2.5 μm, Q = 1.92–2.96, Qm = 2.24 (n = 120/4).
Rot type. A white rot.
Additional specimens (paratypes) examined: China. Zhejiang Prov., Qingyuan County, Baishanzu Nature Reserve, on fallen angiosperm branch, 14 September 2013, Cui 11393 (BJFC). Guangdong Prov., Fengkai County, Heishiding Nature Reserve, on fallen angiosperm branch, 3 April 2014, Li 1611 (BJFC). Guangdong Prov., Fengkai County, Heishiding Nature Reserve, on fallen angiosperm branch, 3 April 2014, Li 1928 (BJFC).
3 Picipes subtubaeformis J.L. Zhou & B.K. Cui, sp. nov. Figs 2C and 5
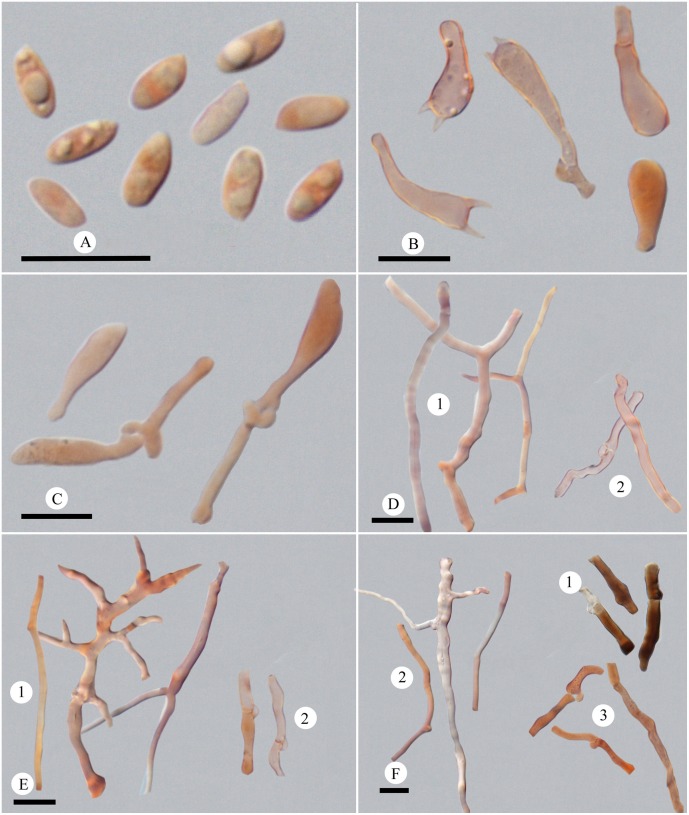
Fig 5. Microscopic structures of Picipes subtubaeformis.
(A): Basidiospores; (B): Basidia and basidioles; (C): Cystidioles; (D): Hyphae from context, 1 Hyphae in cuticle, 2 generative hyphae, 3 skeleto-binding hyphae; (E): Hyphae from trama, 1 generative hyphae, 2 skeleto-binding hyphae; (F): Hyphae from stipe, 1 hyphae in cuticle, 2 generative hyphae, 3 skeleto-binding hyphae. Bars = 10 μm.
MycoBank NO.: MB 815519
Basidiocarps annual, centrally to laterally stipitate, solitary. Pilei irregularly semicircular or elliptical, with shallow central depression. Pileal surface reddish-brown to chestnut in the center or towards the stipe and changing to signal-orange to clay-brown towards the edge. Pore surface buff to festucine, shining; pores angular, 4–6 per mm. Stipe with a terra-brown to black cuticle. Hyphal system dimitic; generative hyphae with clamp connections. Basidiospores oblong to cylindrical, 5.7–6.8 × 2.7–3.1 μm. On angiosperm wood, causing a white-rot.
Type: China. Anhui Prov., Huangshan, Huangshan Mountain, on fallen angiosperm branch, 20 October 2010, Dai 11870 (holotype in BJFC).
Etymology: subtubaeformis (Lat.): referring to the morphological similarity to Picipes tubaeformis.
Fruitbody: Basidiocarps annual, centrally to laterally stipitate, solitary, coriaceous when fresh and tough when dry. Pilei irregularly semicircular or ellipsoidal, with shallow central depression, up to 7.8 cm wide and 2 mm thick. Pileal surface glabrous, reddish-brown to chestnut in the center or towards the stipe and changing to signal-orange to clay-brown towards the edge when dry in young specimens, becoming reddish-brown to chestnut upon whole surface in mature ones, with radially aligned stripes; margin involute upon drying. Pore surface buff to festucine when dry, shining; pores round to angular, 4–6 per mm; dissepiments thin, entire to lacerate. Context white to buff, becoming woody hard upon drying, up to 1 mm thick. Tubes concolorous with pore surface, less than 1.5 mm thick, sometimes decurrent on one side of stipe. Stipe bearing a terra-brown to black cuticle, up to 1.2 cm long and 3.5 mm in diam.
Hyphal structure: Hyphal system dimitic; generative hyphae bearing clamp connections, colorless, thin-walled; skeleto-binding hyphae colorless, thick-walled, with arboriform branches and tapering ends, IKI–, CB+; tissue unchanged in KOH.
Context: Generative hyphae infrequent, colorless, thin-walled, frequently branched, 1.6–4.3 μm in diam.; skeleto-binding hyphae dominant, colorless, thick-walled with a narrow lumen to solid, moderately branched, interwoven, 1.4–4.8 μm in diam. Hyphae in cuticle bearing clamp connections, thick-walled with a wide lumen, with buff inclusion inside, parallel arranged into a palisade, 1–3.5 μm in diam.
Tubes: Generative hyphae frequent, usually present near hymenium, colorless, thin-walled, 1.5–3.4 μm in diam.; skeleto-binding hyphae dominant, colorless, thick-walled with a wide to narrow lumen, frequently with dendroid branching, strongly interwoven, 1.2–3.6 μm in diam. Cystidia absent; cystidioles frequent, subulate, 16.7–25 × 3.5–5.5 μm; basidia clavate, with a basal clamp and four sterigmata, 15.7–29 × 5.1–6.2 μm; basidioles in shape similar to basidia, smaller than basidia.
Stipe: Generative hyphae frequent, colorless, thin-walled, 1.8–6.5 μm in diam.; skeleto-binding hyphae colorless, thick-walled with a narrow lumen, 1–6.4 μm in diam. Hyphae in cuticle bearing clamp connections, thick-walled with a wide lumen, with brown to dark brown inclusion inside and arranged in a palisade, 1.5–4.3 μm in diam.
Basidiospores: Basidiospores oblong to cylindrical, thin-walled, colorless, smooth, usually bearing one or two guttules, (5.3–)5.7–6.8(–7.1) × (2.4–)2.7–3.1(−3.4) μm, L = 6.18 μm, W = 2.91 μm, Q = 1.88–2.5, Qm = 2.13 (n = 60/2).
Rot type. A white rot.
Additional specimen (paratype) examined: China. Sichuan Prov., Luding County, Hailuogou Forest Park, on dead angiosperm tree, 20 September 2012, Cui 10793 (BJFC).
4 Picipes tibeticus J.L. Zhou & B.K. Cui, sp. nov. Figs 2D and 6
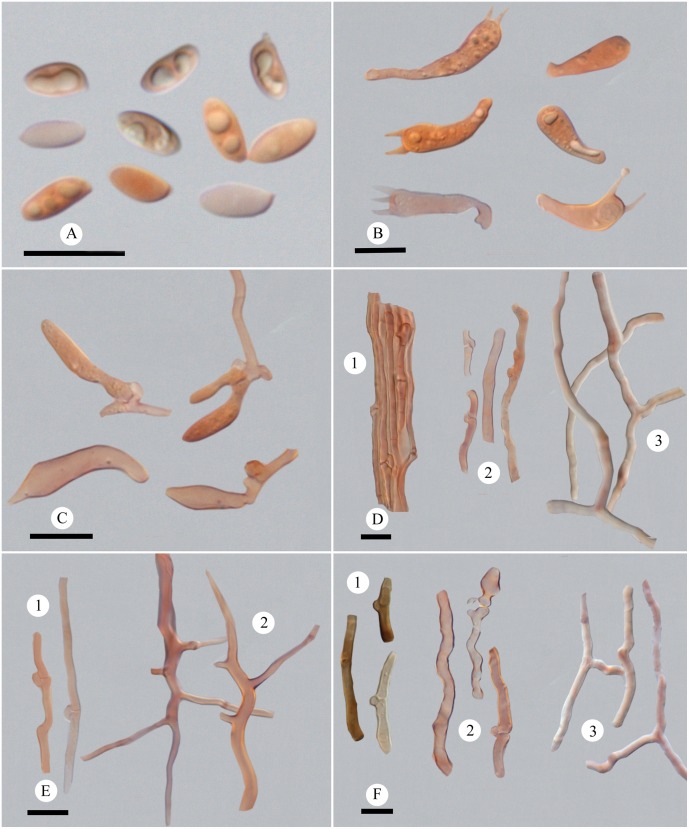
Fig 6. Microscopic structures of Picipes tibeticus.
(A): Basidiospores; (B): Basidia and basidioles; (C): Cystidioles; (D): Hyphae from context, 1 hyphae in cuticle, 2 skeleto-binding hyphae, 3 generative hyphae; (E): Hyphae from trama, 1 skeleto-binding hyphae, 2 generative hyphae; (F): Hyphae from stipe, 1 hyphae in cuticle, 2 skeleto-binding hyphae, 3 generative hyphae. Bars = 10 μm.
MycoBank NO.: MB 815520
Basidiocarps annual, centrally to laterally stipitate, solitary or scattered. Pilei irregularly fan-shaped or semicircular, usually shallow towards the stipe. Pileal surface orange-brown to brown, more or less radially wrinkled when dry. Pore surface white; pores angular to subcircular, 6–9 per mm. Stipe with a black cuticle. Hyphal system dimitic; generative hyphae with clamp connections. Basidiospores oblong, 5–5.9 × 2.8–3.3 μm. On fallen branch of Abies or Picea, causing a white-rot.
Type: China. Xizang Autonomous Region (Tibet), Mêdog County, on fallen branch of Abies, 20 September 2014, Cui 12215 (holotype in BJFC).
Etymology: tibeticus (Lat.): referring to the locality of the type specimens in Tibet.
Fruitbody: Basidiocarps annual, centrally to laterally stipitate, solitary or scattered, coriaceous when fresh and tough when dry. Pilei irregular fan-shaped or semicircular, usually shallow towards the stipe, up to 10.5 cm wide and 1 mm thick. Pileal surface glabrous, orange-brown to brown when fresh, becoming orange-brown to reddish-brown or blackish-brown upon drying, more or less radially wrinkled when dry; margin straight or slightly involute when fresh and involute upon drying. Pore surface white when fresh, becoming buff to yellow-orange when dry; pores angular to subcircular, 6–9 per mm; dissepiments thin, entire to slightly lacerate. Context white when fresh, becoming woody hard upon drying, up to 0.5 mm thick. Tubes concolorous with pore surface, less than 0.9 mm thick, decurrent. Stipe bearing a black cuticle, wrinkled, up to 4.5 cm long and 9 mm in diam.
Hyphal structure: Hyphal system dimitic; generative hyphae bearing clamp connections, colorless, thin-walled; skeleto-binding hyphae colorless, thick-walled, with arboriform branches and tapering ends; IKI–, CB+; tissue unchanged in KOH.
Context: Generative hyphae infrequent, colorless, thin-walled, 2.4–7 μm in diam.; skeleto-binding hyphae dominant, colorless, thick-walled with a narrow lumen to solid, moderately branched, interwoven, 1.6–4.4 μm in diam. Hyphae in cuticle bearing clamp connections, thin-walled with a wide lumen, with buff inclusion inside, parallel arranged into a palisade, 1.6–10 μm in diam.
Tubes: Generative hyphae frequent, usually present near hymenium, colorless, thin-walled, occasionally branched, 1.8–3.2 μm in diam.; skeleto-binding hyphae dominant, colorless, thick-walled with a narrow lumen to solid, frequently with dendroid branching, strongly interwoven, 0.9–4.5 μm in diam. Cystidia absent; cystidioles infrequent, subulate, 14.5–21 × 4–5.3 μm; basidia clavate, with a basal clamp and four sterigmata, 15.3–20 × 5.7–6.8 μm; basidioles in shape similar to basidia, smaller than basidia.
Stipe: Generative hyphae infrequent, colorless, thin-walled, occasionally branched, 2.2–5.1 μm in diam.; skeleto-binding hyphae colorless, thick-walled with a narrow lumen to subsolid, moderately branched, interwoven, 1.9–6.6 μm in diam. Hyphae in cuticle bearing clamp connections, thick-walled with a wide lumen, with buff to brown inclusion inside and arranged in a palisade, 2.5–5.4 μm in diam.
Basidiospores: Basidiospores oblong, thin-walled, colorless, smooth, usually bearing one or two guttules, (4.8–)5–5.9(−6.2) × (2.5–)2.8–3.3(−3.5) μm, L = 5.48 μm, W = 3.01 μm, Q = 1.6–2.07, Qm = 1.82 (n = 90/3).
Rot type. A white rot.
Additional specimens (paratypes) examined: China. Xizang Autonomous Region (Tibet), Nyingchi County, Lulang, on fallen branch of Picea, 24 September 2010, Cui 9651 (BJFC). Xizang Autonomous Region (Tibet), Mêdog County, on fallen branch of Abies, 20 September 2014, Cui 12225 (BJFC).
5 New combinations of Picipes
Picipes admirabilis (Peck) J.L. Zhou & B.K. Cui, comb. nov. [MB 817136]
Basionym: Polyporus admirabilis Peck, Bulletin of the Torrey Botanical Club 26: 69 (1899). [MB 157762]
≡ Cerioporus admirabilis (Peck) Zmitr. et Kovalenko, International Journal of Medicinal Mushrooms 18 (1): 33 (2016). [MB 812034]
Picipes americanus (Vlasák & Y.C. Dai) J.L. Zhou & B.K. Cui, comb. nov. [MB 817137]
Basionym: Polyporus americanus Vlasák & Y.C. Dai, Fungal Diversity 64: 136 (2014). [MB 803796]
Picipes austroandinus (Rajchenb. & Y.C. Dai) J.L. Zhou & B.K. Cui, comb. nov. [MB 817138]
Basionym: Polyporus austroandinus Rajchenb. & Y.C. Dai, Fungal Diversity 64: 138 (2014). [MB 803797]
Picipes conifericola (H.J. Xue & L.W. Zhou) J.L. Zhou & B.K. Cui, comb. nov. [MB 817139]
Basionym: Polyporus conifericola H.J. Xue & L.W. Zhou, Mycological Progress 13(1): 139 (2014). [MB 801216]
Picipes fraxinicola (L.W. Zhou & Y.C. Dai) J.L. Zhou & B.K. Cui, comb. nov. [MB 817140]
Basionym: Piptoporus fraxineus Bondartsev & Ljub., Novosti Sistematiki Nizshikh Rastenii 2: 135 (1965). [MB 337052]
≡ Polyporus fraxinicola L.W. Zhou & Y.C. Dai, Fungal Diversity 64: 141 (2014). [MB 803799]
Picipes rhizophilus (Pat.) J.L. Zhou & B.K. Cui, comb. nov. [MB 817141]
Basionym: Polyporus rhizophilus Pat., Journal de Botanique 8: 219 (1894). [MB 150169]
≡ Cerioporus rhizophilus (Pat.) Zmitr. et Kovalenko, International Journal of Medicinal Mushrooms 18 (1): 33 (2016). [MB 812040]
Picipes submelanopus (H.J. Xue & L.W. Zhou) J.L. Zhou & B.K. Cui, comb. nov. [MB 817142]
Basionym: Polyporus submelanopus H.J. Xue & L.W. Zhou, Mycotaxon 122: 436 (2013). [MB 800237]
Picipes taibaiensis (Y.C. Dai) J.L. Zhou & B.K. Cui, comb. nov. [MB 817143]
Basionym: Polyporus rhododendri Y.C. Dai & H.S. Yuan, Annales Botanici Fennici 46 (1): 58 (2009). [MB 540894]
≡ Polyporus taibaiensis Y.C. Dai, Fungal Diversity 64: 142 (2014). [MB 803798]
Picipes virgatus (Berk. & M.A. Curtis,) J.L. Zhou & B.K. Cui comb. nov. [MB 817144]
Basionym: Polyporus virgatus Berk. & M.A. Curtis, Botanical Journal of the Linnean Society 10: 304 (1869). [MB 202513]
≡ Leucoporus virgatus (Berk. & M.A. Curtis) Pat., Énumération Méthodique des Champignons Recueillis à la Guadeloupe et à la Martinique: 25 (1903). [MB 102236]
Discussion
Picipes baishanzuensis was collected from subtropical area of China. It is characterized by its radially striped infundibuliform pileus with a slender black stipe. In phylogenetic analysis (Fig 1), it strongly clustered with Pi. virgatus (100/100/1.00). Morphologically, Pi. virgatus and Pi. baishanzuensis share infundibuliform pileus, similar pore size, decurrent tubes and wrinkled dark stipe; however, the basidiospores of Pi. virgatus are much larger (9–12.5 × 4–5 μm for Pi. virgatus and 6.6–7.9 × 2.5–3.1 μm for Pi. baishanzuensis) [3]. Polyporus tuberaster also has depressed pileus and decurrent pores, but its pileus is covered with dark brown flecks, pores (0.5–2 per mm) and basidiospores (12–14.5 × 4.8–6 μm) [33] are much larger than Pi. baishanzuensis; besides, P. tuberaster usually grows on the ground, arising from a black underground sclerotium [3,34].
Picipes subtropicus was found in subtropical areas of China. It can be identified by a continuous variation in pore size, bright pileal surface color, short black stipe-like base and medium cylindrical basidiospores (5.1–6.2 × 2.2–2.7 μm). In phylogenetic analysis (Fig 1), it did not cluster with any other species in our study set. Polyporus dictyopus also has chestnut upper surface when aged, in addition, its pore size and pore surface are similar to Pi. subtropicus; but P. dictyopus has longer and thicker stipe (up to 3 cm long and 1 cm thick), larger basidiospores (7–8.5 × 2.5–4 μm) and pantropical distribution [3]. Picipes badius share similar basidiocarps and pore size with P. subtropicus; but it differs in its larger basidiospores (7.5–9.5 × 3–3.5 μm), simple-septate generative hyphae and absence of cystidioles [3]. Picipes baishanzuensis was also found in subtropical areas of China, but its infundibuliform pilei, slender stipe and lager basidiospores (6.6–7.9 × 2.5–3.1 μm) are quite different from Pi. subtropicus.
Picipes subtubaeformis was described from temperate zone of China. It can be distinguished by the irregularly semicircular or elliptical pileus, terra-brown to black stipe, and oblong to cylindrical basidiospores (5.7–6.8 × 2.7–3.1 μm). In the phylogenetic analysis (Fig 1), Pi. subtubaeformis grouped together with Pi. tubaeformis (88/88/1.00); morphologically, both of them have orange to reddish-brown pileus and dark stipe, but Pi. tubaeformis differs in its slender stipe and basidiospores (6–7.8 × 2.3–3.2 μm, L = 6.49 μm, W = 2.75 μm) [35]. Both Pi. virgatus and Pi. subtubaeformis have reddish-brown or chestnut basidiocarps with centrally to laterally dark stipe, but the former one has both larger pores (3–4 per mm) and basidiospores [3]; moreover, Pi. virgatus is absence of cystidioles [3]. Picipes taibaiensis is another temperate species described from China. It has similar upper pileal surface color with Pi. subtubaeformis, but the flabelliform or spathulate pileus, larger basidiospores (7.5–10.5 × 3.2–3.8 μm) and fusoid cystidioles make it different from Pi. subtubaeformis [36].
Picipes tibeticus is a special species found from eastern Tibetan Plateau. it can be identified by its reddish-brown to blackish-brown fan-shaped or semicircular basidiocarps, small angular pores (6–9 per mm), oblong basidiospores (5–5.9 × 2.8–3.3 μm) and growth on coniferous trees. Phylogenetically, it grouped together with Pi. conifericola (95/99/1.00; Fig 1). Morphologically, Pi. conifericola and Pi. badius have similar basidiocarps and substrates as Pi. tibeticus, but the former two have larger basidiospores (6–8 × 2.3–3.1 μm for Pi. conifericola [8]; 7.5–9 × 3–3.5 μm for P. badius [3]). Picipes submelanopus resembles Pi. tibeticus in having dark pileal surface, black-stipitate basidiocarps and buff pore surface, but it differs from Pi. tibeticus in terrestrial habit, larger pores (2–3 per mm) and basidiospores (8–10 × 3–3.9 μm). In addition, Pi. submelanopus has both simple septate and clamped generative hyphae [37].
Picipes admirabilis was was initially collected on wood of apple trees in northeastern United States [38]. Lloyd considered that Pi. admirabilis is a variety of P. varius or belongs to group Melanopus for its black stipe [39,40]. Núñez and Ryvarden treated Pi. admirabilis, P. gayanus Lév. and P. pseudobetulinus (Murashk. ex Pilát) Thorn, Kotir. & Niemelä as members of Polyporus group Admirabilis [3]. Among these three species, P. pseudobetulinus has recently been combined into Favolus as F. pseudobetulinus (Murashk. ex Pilát) Sotome & T. Hatt. [4]. Recently, Zmitrovich & Kovalenko [9] regarded Pi. admirabilis as a member of genus Cerioporus Quél. But according to our phylogenetic analysis, Pi. admirabilis strongly clusters in the picipes clade (Fig 1). Morphologically, Pi. admirabilis has a long (up to 8 cm) and pale buff to black stipe, firm-corky basidiocarps and uninflated hyphae [3]. Based on our specimen collected from northeast of China, the balck stem, corky basidiocarps, uninflated hyphae and strongly branched skeleto-binding hyphae in trama are more similar to species of Picipes.
Picipes rhizophilus was treated as a member of group Polyporellus, and it is a special polypore merely grows on the grass roots [3,7,41,42]. But, recently Zmitrovich & Kovalenko [9] transferred Pi. rhizophilus into Cerioporus as C. rhizophilus (Pat.) Zmitr. et Kovalenko. In our current phylogenetic analysis, Pi. rhizophilus strongly groups into the picipes clade (Fig 1). According to our collections, Pi. rhizophilus has dark brown to black stipe, soft-corky basidiocarps, uninflated hyphae, strongly branched skeleto-binding hyphae in trama and cylindrical basidiospores (8.2–10.1 ×3.2–4.1 μm, L = 9.15 μm, W = 3.63 μm, Qm = 2.52). These above-mentioned features fit Picipes well, so we consider this species as a member of Picipes.
Picipes is showed to be a monophyletic group based on the 8-gen-squences data analysis, and sixteen species are included in this clade (Fig 1). Among these species, Pi. badius was previously put into Royoporus A.B. De by De [43] for its simple septa. But our analysis strongly supported Pi. badius as a member of Picipes. Corner [44] considered that Pi. badius, P. dictyopus and Pi. melanopus were conspecific, but our analysis showed that they are three different species. Zmitrovich & Kovalenko [9] considered that Picipes only includes the species with small pores (more than 5 per mm), cylindrical basidiospores and lignicolous habit. According to our study with more samples, we find several species with large pores, oblong basidiospores and terrestrial habit are also members of Picipes.
Polyporus umbellatus is a particular species that merely grows on the ground from a sclerotium close to stumps of hardwoods, and characterized by numerous stipitate pilei arising from a common, strongly branched stipe [3]. It was morphologically treated as a member of Polyporus group Dendropolyporus [3]. Phylogenetically, P. umbellatus was reported as a distinctive species that could not cluster with any other species [7,9]. Zmitrovich & Kovalenko [9] transferred P. umbellatus into genus Cladomeris Quél. But in our analysis, P. umbellatus strongly clusters with P. tuberaster and P. hapalopus in the core polyporus clade (Fig 1). Morphologically, basidiomata of the three species are fleshy when fresh and brittle upon drying [3,45]. Besides, all of them usually have inflated skeleton-binding hyphae up to 17 μm wide, generative hyphae dominant to almost monomitic in trama, cylindrical basidiospores and light-colored stipes. Thus, the core polyporus clade is treated as a natural group of Polyporus.
Species in the squamosus clade together with several species of Datronia Donk were transferred into Cerioporus [9]. Zmitrovich & Kovalenko [9] considered that Cerioporus spp. have inflated skeleto-binding hyphae, but the skeleto-binding hyphae of P. guianensis and P. leprieurii are uninflated and up to 5 μm wide [3]. So we prefer to maintain their previously taxonomic names here.
Previous phylogenetic analyses showed that favolus clade and neofavolus clade did not gather together and favolus clade has closer relationships with P. tuberaster [4,8]. But our phylogenetic result (Fig 1) shows that Favolus spp. closely related to Neofavolus spp. rather than other Polyporus spp. Seelan et al. [46] estimated that the ancestral state for Neofavolus is angular pores. They transferred Lentinus suavissimus Fr., a gilled species with sub-poroid lamellae, into genus Neofavolus. Zmitrovich & Kovalenko [9] also treated Lentinus suavissimus as a member of Neofavolus and named it N. suavissimus (Fr.) Zmitr. et Kovalenko, but this name is illegitimate because of its earlier homonym N. suavissimus (Fr.) J.S. Seelan.
The relationships of Lentinus Fr. and Polyporus have been suspected for a long time. Both Pegler [47] and Singer [48] believed that Lentinus divides from polypores, and this assumption had been evidenced by Hibbett & Vilgalys [49] and Hibbett & Donoghue [50]. Molecular phylogenetic studies also showed that species of group Polyporellus has a much closer relationship with Lentinus compared with other Polyporus spp. [4,7,9,46,51–56]. Krüger [5] and Krüger & Gargas [57] proposed a Lentinus–Polyporellus clade alliance to unite species of Lentinus s. str. and group Polyporellus which have the inflated generative hyphae. Seelan et al. [46] considered the ancestral hymenophoral configuration for species of Lentinus and Polyporellus group is circular pores, with independent transitions to angular pores and lamellae. Zmitrovich [58] combined species in polyporellus clade into Lentinus as L. arcularius (Batsch) Zmitr., L. brumalis (Pers.) Zmitr., L. crinitus (L.) Fr. and L. tricholoma (Mont.) Zmitr. But the last name is illegitimate because of its earlier homonym L. tricholoma Berk. & Cooke. Then Zmitrovich & Kovalenko [9] renamed P. tricholoma to L. flexipes (Fr.) Zmitr. et Kovalenko. In this article, we prefer to treat species in polyporellus clade as members of Lentinus.
Our phylogenetic analysis based on multiple gene sequences data of ITS, nLSU, EF1-α, mtSSU, β-tubulin, RPB1, RPB2 and nSSU suggested that species of group Melanopus distribute into two different clades: picipes clade and squamosus clade. This conclusion verified the view that Melanopus group is not a monophyletic assemblage of dark-stiped Polyporus species, and whether having black cuticle on stipe or not, is not a sufficient feature to define the natural Melanopus group. In our study, sixteen species including four new species of Picipes are recognized. A key to species of Picipes is provided.
Key to species of Picipes
-
1a
Growing on woods or ground………………………………………… 2
-
1b
Growing on grass roots…………………………………………………. Pi. rhizophilus
-
2a
Generative hyphae bearing simple septa……………………………… 3
-
2b
Generative hyphae only with clamps……………………………………….. 4
-
3a
Pores 2–3 per mm, generative hyphae bearing both simple septa and clamp connections, growing on ground or hardwoods, basidiospores 8–10 × 3–3.9 μm……………………………………………….. Pi. submelanopus
-
3b
Pores 5–6 per mm, generative hyphae only with simple septa, growing on hardwoods, basidiospores 6.5–8 × 3–3.8 μm……………………………………………………………………. Pi. badius
-
4a
Stipe very short or attach to the substrate with a flattened base…………………………………… 5
-
4b
Stipe usually more than 1 cm long……………………………………………………………………….. 6
-
5a
Pores 8–9 per mm when young and becoming 5–7 per mm when mature, basidiospores 5.1–6.2 × 2.2–2.7 μm……………………………………………………………………………………… Pi. subtropicus
-
5b
Pores 3–5 per mm, growing on Rhododendron, basidiospores 7.5–10.5×3.2–3.8 μm……………………………………………………………………………………………Pi. taibaiensis
-
6a
Basidiospores more than 9 μm long…………………………… 7
-
6b
Basidiospores less than 9 μm long……………………………. 8
-
7a
Growing on Austrocedrus or Lomatia woods, pores 4–5 per mm, basidiospores 9–11.5 × 3–3.8 μm………………………………………………………………………………………………………………………………….Pi. austroandinus
-
7b
Growing on hardwoods, pores 3–4 per mm, basidiospores 9–12.5 × 4–5 μm…………………………………………. Pi. virgatus
-
8a
Cystidioles present……………………………………………………..9
-
8b
Cystidioles absent……………………………………………………. Pi. fraxinicola
-
9a
Pores 3–6 per mm…………………………………………….. 10
-
9b
Pores 6–10 per mm………………………………………………. 14
-
10a
Basidiospores usually more than 3 μm wide…………………………………………. 11
-
10b
Basidiospores usually less than 3 μm wide…………………………………………. 12
-
11a
Cystidioles subulate……………………………………….. Pi. admirabilis
-
11b
Cystidioles fusoid……………………………………….. Pi. melanopus
-
12a
Pilei not infundibuliform in shape……………………………………. 13
-
12b
Pilei infundibuliform in shape……………………………………… Pi. baishanzuensis
-
13a
Pilei nearly circular, basidiospores 7–9 × 2.5–3.1 μm…………………………………….. Pi. americanus
-
13b
Pilei irregularly semicircular or elliptical, basidiospores 5.7–6.8 × 2.7–3.1 μm……………………………….Pi. subtubaeformis
-
14a
Basidiospores more than 6 μm in length………………………………….. 15
-
14b
Basidiospores 5–5.9 × 2.8–3.3 μm………………………………………….. Pi. tibeticus
-
15a
Growing on coniferous trees………………………………… Pi. conifericola
-
15b
Growing on hardwoods………………………………………. Pi. tubaeformis
Acknowledgments
The authors would like to thank Dr. Wen-Min Qin (Institute of Applied Ecology, Chinese Academy of Sciences, China) and Dr. Fang Li (Sun Yat-Sen University, China) for loan of specimens. Prof. Yu-Cheng Dai (Beijing Forestry University, China) is grateful for help in field collections.
Data Availability
All sequences files are available from the GenBank database (accession number(s) are listed in Table 1).
Funding Statement
The research was financed by the National Natural Science Foundation of China (Project No. 31422001).
References
- 1.Patouillard N. Les Hyménomycètes d'Europe. Paris: Paul Klincksieck; 1887. [Google Scholar]
- 2.Donk MA. The generic names proposed for Polyporaceae. Persoonia. 1960; 1: 173–302. [Google Scholar]
- 3.Núñez M, Ryvarden L. Polyporus (Basidiomycotina) and related genera. Oslo: Fungiflora; 1995. [Google Scholar]
- 4.Sotome K, Akagi Y, Lee SS, Ishikawa NK, Hattori T. Taxonomic study of Favolus and Neofavolus gen. nov. segregated from Polyporus (Basidiomycota, Polyporales). Fungal Divers. 2013; 58: 245–266. 10.1007/s13225-012-0213-6 [DOI] [Google Scholar]
- 5.Krüger D. Monographic studies in the genus Polyporus (Basidiomycotina). Ph.D. diss., University of Tennessee. 2002. Available: http://trace.tennessee.edu/utk_graddiss/2135
- 6.Krüger D, Petersen RH, Hughes KW. Molecular phylogenies and mating study data in Polyporus with special emphasis on group ‘‘Melanopus” (Basidiomycota). Mycol Prog. 2006; 5: 185–206. 10.1007/s11557-006-0512-y [DOI] [Google Scholar]
- 7.Sotome K, Hattori T, Ota Y, To-anun C, Salleh B, Kakishima M. Phylogenetic relationships of Polyporus and morphologically allied genera. Mycologia. 2008; 100: 603–615. 10.3852/07-191r [DOI] [PubMed] [Google Scholar]
- 8.Dai YC, Xue HJ, Vlasák J, Rajchenberg M, Wang B, Zhou LW. Phylogeny and global diversity of Polyporus group Melanopus (Polyporales, Basidiomycota). Fungal Divers. 2014; 64: 133–144. 10.1007/s13225-013-0248-3 [DOI] [Google Scholar]
- 9.Zmitrovich IV, Kovalenko AE. Lentinoid and polyporoid fungi, two generic conglomerates containing important medicinal mushrooms in molecular perspective. Int J Med Mushrooms. 2016; 18: 23–38. 10.1615/intjmedmushrooms.v18.i1.40 [DOI] [PubMed] [Google Scholar]
- 10.Petersen JH. Farvekort. The Danish Mycological Society´s colour-chart. Greve: Foreningen til Svampekundskabens Fremme; 1996.
- 11.Li HJ, Cui BK, Dai YC. Taxonomy and multi-gene phylogeny of Datronia (Polyporales, Basidiomycota). Persoonia. 2014; 32: 170–182. 10.3767/003158514x681828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen JJ, Cui BK, Zhou LW, Korhonen K, Dai YC. Phylogeny, divergence time estimation, and biogeography of the genus Heterobasidion (Basidiomycota, Russulales). Fungal Divers. 2015; 71: 185–200. 10.1007/s13225-014-0317-2 [DOI] [Google Scholar]
- 13.Han ML, Chen YY, Shen LL, Song J, Vlasák J, Dai YC, et al. Taxonomy and phylogeny of the brown-rot fungi: Fomitopsis and its related genera. Fungal Divers. 2016; [Google Scholar]
- 14.White TJ, Bruns TD, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky J, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic; 1990. pp. 315–322. 10.1016/b978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- 15.Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several species of Cryptococcus. J Bacteriol. 1990; 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005; 97: 84–98. 10.3852/mycologia.97.1.84 [DOI] [PubMed] [Google Scholar]
- 17.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microb. 1995; 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hibbett DS. Phylogenetic evidence for horizontal transmission of group I introns in the nuclear ribosomal DNA of mushroom-forming fungi. Mol Biol Evol. 1996; 13: 903–917. 10.1093/oxfordjournals.molbev.a025658 [DOI] [PubMed] [Google Scholar]
- 19.Matheny PB, Liu YJ, Ammirati JF, Hall BD. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am J Bot. 2002; 89: 688–698. 10.3732/ajb.89.4.688 [DOI] [PubMed] [Google Scholar]
- 20.Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerse II subunit. Mol Bio Evol. 1999; 16: 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- 21.Matheny PB. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol Phylogenet Evol. 2005; 35: 1–20. 10.1016/j.ympev.2004.11.014 [DOI] [PubMed] [Google Scholar]
- 22.Binder M, Larsson KH, Matheny PB, Hibbett DS. Amylocorticiales ord. nov. and Jaapiales ord. nov.: Early diverging clades of Agaricomycetidae dominated by corticioid forms. Mycologia. 2010; 102: 865–880. 10.3852/09-288 [DOI] [PubMed] [Google Scholar]
- 23.Zhao CL, Cui BK, Dai YC. New species and phylogeny of Perenniporia based on morphological and molecular characters. Fungal Divers. 2013; 58: 47–60. 10.1007/s13225-012-0177-6 [DOI] [Google Scholar]
- 24.Zhao CL, He XS, Wanghe KY, Cui BK, Dai YC. Flammeopellis bambusicola gen. et. sp. nov. (Polyporales, Basidiomycota) evidenced by morphological characters and phylogenetic analysis. Mycol Prog. 2014; 13: 771–780. 10.1007/s11557-014-0960-8 [DOI] [Google Scholar]
- 25.Zhao CL, Chen H, Song J, Cui BK. Phylogeny and taxonomy of the genus Abundisporus (Polyporales, Basidiomycota). Mycol Prog. 2015; 14: 38 10.1007/s11557-015-1062-y [DOI] [Google Scholar]
- 26.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li WZ, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011; 7: 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and other methods). Version 4.0b10 Sunderland: Sinauer; 2002. [Google Scholar]
- 28.Nylander J. MrModeltest2.2. Computer software distributed by the University of Uppsala. 2004.
- 29.Michalak S. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012; 12: 335–337. 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- 30.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2 efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012; 61: 539–42. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donk MA. Revision de Niederlandischen Homobasidiomycetes. Aphyllophoraceae II. Medd Bot Mus Herb Rijhs Universit Utrecht. 1933; 9: 1–278. [Google Scholar]
- 32.Singer R. The Agaricales in modern taxonomy, 4th ed Koenigstein: Koeltz Scientific Books; 1986. [Google Scholar]
- 33.Dai YC. Changbai wood-rotting fungi 11. Species of Polyporus sensu stricto. Fungal Sci. 1999; 14: 67–77. [Google Scholar]
- 34.Jahn H. Der sklerotien porling P. tuberaster (Pers. ex Fr.) Fr. (P. lentus Berk.). Westfälische Pilzbriefe. 1980; 11: 125–144. [Google Scholar]
- 35.Dai YC. Changbai wood-rotting fungi 5. Study on Polyporus mongolicus and P. tubaeformis. Ann Bot Fennici. 1996; 33: 153–163. [Google Scholar]
- 36.Dai YC, Yuan HS, Wang HC, Yang F, Wei YL. Polypores (Basidiomycota) from Qin Mts. in Shaanxi Province, Central China. Ann Bot Fenn. 2009; 46: 54–61. 10.5735/085.046.0105 [DOI] [Google Scholar]
- 37.Xue HJ, Zhou LW. Polyporus submelanopus sp. nov. (Polyporales, Basidiomycota) from Northwest China. Mycotaxon. 2012; 122: 433–441. 10.5248/122.433 [DOI] [Google Scholar]
- 38.Peck CH. New species of fungi. Bulletin of the Torrey Botanical Club. 1899; 26: 66–71. [Google Scholar]
- 39.Burnham SH. The admirable popyporus in the flora of the Lake George region. Torreya. 1916; 16: 139–142. [Google Scholar]
- 40.Dodge BO. Fungi producing heart-rot of apple trees. Mycologia. 1916; 8: 5–15. 10.2307/3753323 [DOI] [Google Scholar]
- 41.Ryvarden L, Gilbertson RL. European polypores 2. Syn Fungorum 1994; 7: 394–743. [Google Scholar]
- 42.Sotome K, Hattori T, Kakishima M. Polyporus phyllostachydis sp. nov. with notes on other rhizophilic species of Polyporus. Mycoscience. 2007; 48: 42–46. 10.1007/s10267-006-0328-2 [DOI] [Google Scholar]
- 43.De AB. Royoporus–a new genus for Favolus spathulatus. Mycotaxon. 1996; 60: 143–148. [Google Scholar]
- 44.Corner EJH. Ad Polyporaceas II. Polyporus, Mycobonia, and Echinochaete. Beih Nova Hedw. 1984; 78: 1–129. [Google Scholar]
- 45.Xue HJ, Zhou LW. Polyporus hapalopus sp. nov. (Polyporales, Basidiomycota) from China based on morphological and molecular data. Mycol Progress. 2014; 13: 811–817. 10.1007/s11557-014-0964-4 [DOI] [Google Scholar]
- 46.Seelan JS, Justo A, Nagy LG, Grand EA, Redhead SA, Hibbett D. Phylogenetic relationships and morphological evolution in Lentinus, Polyporellus and Neofavolus, emphasizing southeastern Asian taxa. Mycologia. 2015; 107: 460–74. 10.3852/14-084 25661717 [DOI] [PubMed] [Google Scholar]
- 47.Pegler DN. The genus Lentinus: a world monograph Kew Bull Add Ser 10 London: Her Majesty’s Stationary Office; 1983. [Google Scholar]
- 48.Singer R. The Agaricales in modern taxonomy. Königstein: Koeltz Scientific Books; 1986. [Google Scholar]
- 49.Hibbett DS, Vilgalys R. Phylogenetic relationships of Lentinus (Basidiomycotina) inferred from molecular and morphological characters. Syst Bot. 1993; 18: 409–433. 10.2307/2419417 [DOI] [Google Scholar]
- 50.Hibbett DS, Donoghue MJ. Progress toward a phylogenetic classification of the Polyporaceae through parsimony analysis of mitochondrial ribosomal DNA sequences. Can J Bot. 1995; 73 (Suppl 1): S853–S861. 10.1139/b95-331 [DOI] [Google Scholar]
- 51.Hibbett DS, Donoghue MJ. Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in Homobasidiomycetes. Syst Biol. 2001; 50: 215–241. 10.1080/10635150151125879 [DOI] [PubMed] [Google Scholar]
- 52.Hibbett DS, Thorn RG. Basidiomycota: Homobasidiomycetes In: McLaughlin DJ, McLaughlin EG, Lemke PA, editors. The mycota VII part B systematics and evolution. Berlin & Heidelberg: Springer-Verlag; 2001. [Google Scholar]
- 53.Ko KS, Jung HS. Phylogenetic evaluation of Polyporus s. str. based on molecular sequences. Mycotaxon. 2002; 82: 315–322. [Google Scholar]
- 54.Binder M, Hibbett DS, Larsson KH, Larsson E, Langer E, Langer G. The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes). Syst Biodivers. 2005; 3(2): 1–45. 10.1017/s1477200005001623 [DOI] [Google Scholar]
- 55.Binder M, Justo A, Riley R, Salamov A, Lopez-Giraldez F, Sjökvist E, et al. Phylogenetic and phylogenomic overview of the Polyporales. Mycologia. 2013; 105: 1350–1373. 10.3852/13-003 [DOI] [PubMed] [Google Scholar]
- 56.Sotome K, Hattori T, Ota Y, Kakishima M. Second report of Polyporus longiporus and its phylogenetic position. Mycoscience. 2009; 50: 415–420. 10.1007/s10267-009-0506-0 [DOI] [Google Scholar]
- 57.Krüger D, Gargas A. The basidiomycete genus Polyporus—an emendation based on phylogeny and putative secondary structure of ribosomal RNA molecules. Feddes Repert. 2004; 115: 530–546. 10.1002/fedr.200311052 [DOI] [Google Scholar]
- 58.Zmitrovich IV. The Taxonomical and nomenclatural characteristics of medicinal mushrooms in some genera of Polyporaceae. Int J Med Mushrooms, 2010, 12(1). 87–89. 10.1615/intjmedmushr.v12.i1.80 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequences files are available from the GenBank database (accession number(s) are listed in Table 1).