Abstract
Thymic T-cell development is initiated from bone marrow-derived multi-potent thymus seeding progenitors (TSPs). During the early stages of thymocyte differentiation progenitors become T-cell restricted. However, the cellular environments supporting these critical initial stages of T-cell development within the thymic cortex are not known. We here use the dependence of early, c-Kit–expressing thymic progenitors on Kit ligand (KitL) to show that CD4–CD8–c-Kit+CD25– DN1-stage progenitors associate with, and depend on the membrane-bound form of KitL (mKitL) provided by, a cortex-specific KitL-expressing vascular endothelial cell (VEC) population. In contrast, the subsequent CD4–CD8–c-Kit+CD25+ DN2 stage progenitors associate selectively with cortical thymic epithelial cells (cTECs) and depend on cTEC-presented mKitL. These results show that the dynamic process of early thymic progenitor differentiation is paralleled by migration-dependent changes to the supporting niche, and identify VECs as a thymic niche cell, with mKitL as a critical ligand.
The niches that maintain tissue stem cells have been extensively characterized over the past 3 decades, leading to a much improved understanding of their constituent cell types and extracellular matrix components, and the signals these provide to dynamically regulate stem cell behavior1,2. In contrast, little is known about the physical environments dedicated to supporting the progenitor cells derived from tissue stem cells. This is due to several factors, including their transient nature and changing phenotype during the differentiation process, contrasting with the relative stability and phenotypic homogeneity of stem cell populations. That specific progenitor niches exist was first suggested by the identification of erythroid islands, where central macrophages provide support for developing erythroblasts3. More recently, a Cxcl12-dependent, bone-associated lymphoid progenitor niche was proposed4,5, the latter study emphasizing the usefulness of critical ligands in the identification of essential niche cell types.
T-cell development is initiated in the thymic cortex, where multi-potent thymus seeding progenitors (TSPs) enter through P-selectin–mediated extravasation at the cortico-medullary junction (CMJ)6. As they migrate through the thymic cortex they progress through the CD4/CD8 double negative stages 1-4 (DN1-4) of thymocyte differentiation to form CD4+CD8+ thymocytes, which then migrate to the medulla to undergo negative selection where self-reactive T-cells are eliminated7. DN3 thymocytes are the first fully T-cell restricted progenitors, whereas DN1 and DN2 cells undergo expansion and gradual lineage restriction. This process is supported by Dll4 expressed on cortical thymic epithelial cells (cTECs) as a critical Notch ligand for DN1/DN2 thymocytes8,9. Other regulators of thymic progenitor pool size and progression include interleukin (IL-)7, Ccl19, Ccl25 and Cxcl1210–14, whereas BMP4 and Wnt4 are involved in thymocyte differentiation15–17. However, while these factors are expressed in the thymic stroma18, their cellular source(s), and hence the physical niches in which thymic progenitors develop, are yet to be identified.
The c-Kit receptor is selectively expressed on early thymic progenitors (DN1/DN2). A thymic Kit ligand (KitL) source is critical for early thymic progenitor development, as KitL-deficient thymi transplanted into wild type recipient mice show defective T-cell development19, but the cell type(s) providing the ligand remain unknown. Moreover, KitL exists both as a membrane-associated (mKitL) and a secreted (sKitL) form, and little is known about the specific physiological roles of these two KitL molecules20, a question particularly relevant to the identification of cellular niches supporting defined progenitors through direct cell-cell interaction.
We here set out to define the cellular source(s) and molecular form of KitL involved in supporting the earliest stages of c-Kit+ multi-potent thymocyte progenitor development. We observed that, in addition to TECs, a distinct subset of vascular endothelial cells (VECs), selectively located in the thymic cortex, expressed high levels of KitL. DN1 thymocytes were closely associated with mKitL expressing VECs, and VEC-specific loss of mKitL resulted in a strong depletion of DN1 thymocytes, including ETPs. DN2 thymocytes did not associate closely with VECs, and were instead principally dependent on mKitL presented by TECs for their maintenance. Overall, these results identify thymic VECs as a novel and critical component of the developing thymocyte niche, and mKitL as a critical niche-presented ligand, demonstrating that thymic progenitor niches are dynamic structures to which distinct stromal cell populations contribute in a progenitor differentiation stage-dependent manner.
Results
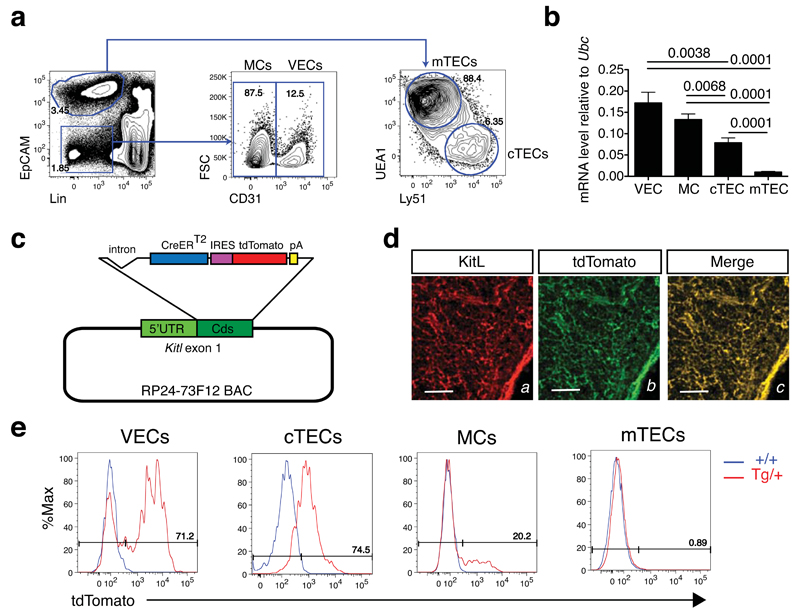
To identify the thymic stromal cells with the potential to support ETP differentiation through KitL production we first fractionated the thymic stroma into its major components: vascular endothelial cells (VEC), mesenchymal cells (MC) and thymic epithelial cells (TEC) by cell sorting. TECs were further subdivided into cortical (cTEC) and medullary (mTEC) subtypes21 (Figure 1a and Supplementary Figure 7). We next determined the expression of Kitl in these cell types. We observed that Kitl mRNA was expressed in VECs, MCs and cTECs, with VECs expressing the highest levels, but barely detectable in mTECs (Figure 1b). To quantify Kitl expression at the single cell level, and to visualize and prospectively isolate of Kitl expressing cells, we generated a BAC transgenic reporter mouse line expressing tdTomato from the Kitl promoter (Figure 1c). The reporter was validated by double immunofluorescence of KitL and tdTomato, showing co-localization of tdTomato and KitL in thymic sections (Figure 1d). Flow cytometric analysis of Kitl-tdTomatotg/+ thymi showed that, consistent with the gene expression analysis, expression of Kitl-tdTomato-derived tdTomato (henceforth KT) in TECs was confined to cTECs, with no expression detectable in mTECs (Figure 1e). Significantly, VECs contained distinct KT+ and KT– populations, the former comprising approximately 70% of the VEC population. Finally, a minor fraction (ca. 20%) of MCs were KT+. The abundance of a distinct KT+ VECs, combined with their high Kitl expression, was consistent with a role for this cell population in c-Kit+ thymocyte differentiation.
Figure 1. Distinct patterns of Kit ligand expression in thymic stromal cell types.
(a) Representative flow cytometry plots and gating strategy for the detection of the indicated thymic stromal cell populations. VEC: vascular endothelial cells; MC: mesenchymal cells; mTEC: medullary thymic epithelial cells; cTEC: cortical thymic epithelial cells.
(b) Real-time qPCR analysis of Kitl mRNA expression in sorted VECs, MCs, cTECs and mTECs. Cell populations were sorted from 4-week old C57Bl/6 mice and Kitl mRNA quantified by real-time qPCR. Values represent averages after normalization to Ubc mRNA. N=3 biological replicates, each measured in technical triplicate. Significance of differences in expression level were determined using Student’s t-test; P-values are shown. Error bars show the SEM. For source data see Supplementary Table 5.
(c) Schematic representation of Kitl-CreERT2-IRES-tdTomato BAC transgene. IRES: internal ribosomal entry site; pA: polyadenylation site; Cds: coding sequence.
(d) Double immunofluorescence analysis of thymic frozen section from of Kitl-tdTomatotg/+ mouse using antibodies against KitL (left panel; red) and tdTomato (center panel; green). KitL/tdTomato co-localization is shown (right panel, merge). Scale bars: 100μm.
(e) Representative flow cytometric analysis of tdTomato expression in the indictaed thymic stromal cell populations from Kitl-tdTomatotg/+ mice and wild type controls. The fraction of tdTomato+ cells is shown for each population.
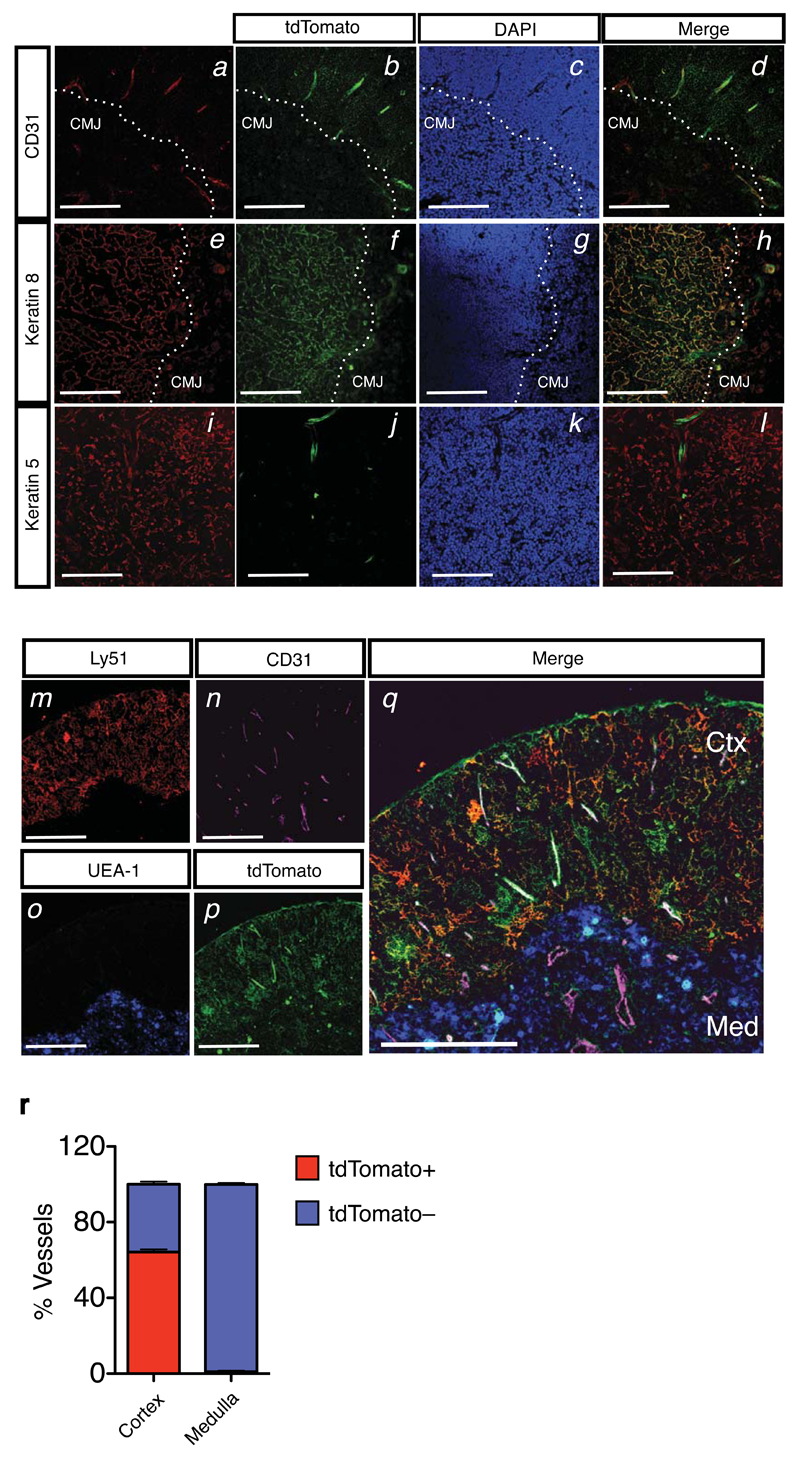
To localize thymic KT+ cells we performed immunohistochemistry using markers specific for VEC (CD31), cTEC (keratin 8 (K8)), and mTECs (K5). KT expression was confined to the cortical side of the CMJ, and co-localized with both CD31 and K8, but not K5, consistent with the flow cytometric analysis (Figure 2a-l). Simultaneous labeling of tdTomato, cTECs (Ly51), mTECs (UEA-1) and VECs (CD31) confirmed the selective expression of KT in cortically located VECs and cTECs (Figure 2m-q). Strikingly, whereas around 60% of the cortical vascular structures were KT+, no KT+ VECs were observed in the medulla (Figure 2r). The KT+ VEC population therefore defines a distinct VEC subset exclusive to the thymic cortex, the area of ETP immigration.
Figure 2. Localization of Kit ligand expressing thymic stromal subsets to the cortico-medullary junction and cortex.
(a-d) Immunofluorescence analysis of Kitl-tdTomatotg/+ thymic section using antibodies against CD31 (panel a, red), and tdTomato (panel b, green). DAPI staining was used to identify the cortico-medullary junction (CMJ). Colocalization of CD31 and tdTomato is shown (panel d). Scale bars: 300μm.
(e-h) Analysis as in (a-d), using anti-K8 (panel e, red) and anti-tdTomato (panel f, green) antibodies. Scale bars: 300μm.
(i-l) Analysis as in (a-d), using anti-K5 (panel i, red) and anti-tdTomato (panel j, green) antibodies. Scale bars: 300μm.
(m-q) Immunofluorescence analysis of Kitl-tdTomatotg/+ thymic section using antibodies against Ly51 (panel m, red), CD31 (panel n, pink), UEA-1 (panel o, blue), tdTomato (panel p, green). Colocalization is shown (panel q). Note the preferential location of tdTomato+CD31+ VECs to the cortex (Ctx), whereas medullary (Med) VECs (CD31+) are tdTomato–. Scale bars: 300μm.
(r) Quantification of the tdTomato+CD31+ and tdTomato–CD31+ VECs across the cortical and medullary regions. Result shown as average percentage of the total vascular structures counted in the cortical region (n=1475) and medulla (n=341) from 3 biological replicates and 2 separate experiments. Bars show the SEM.
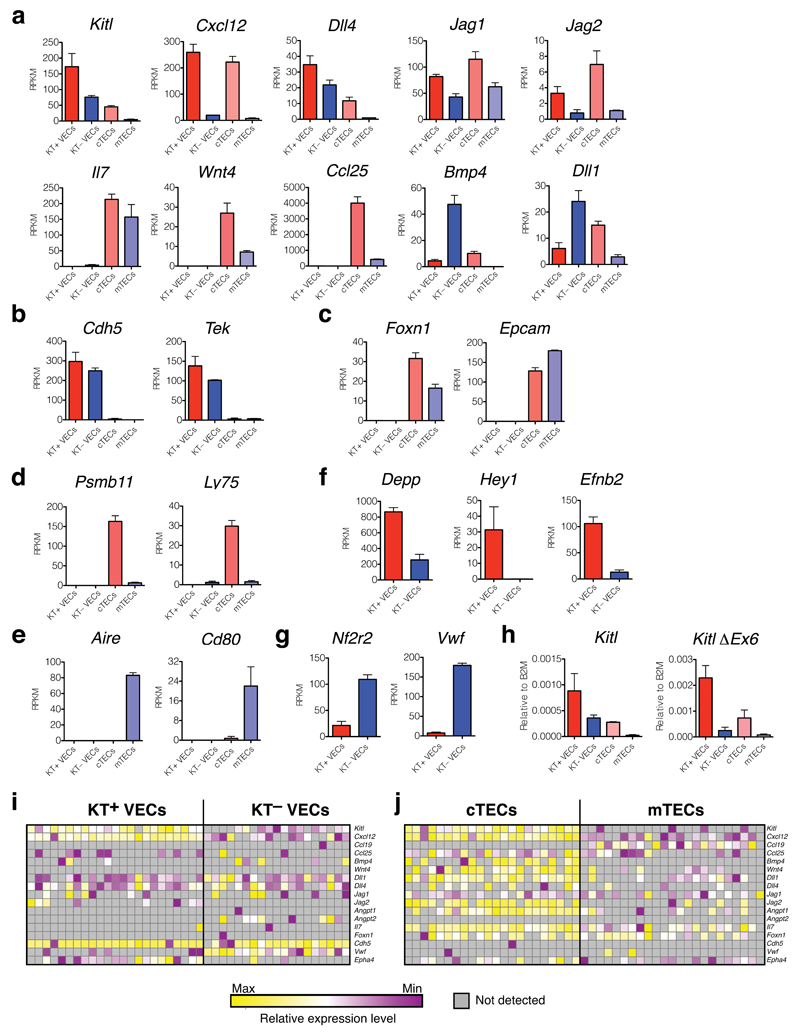
To explore the molecular differences between KT+ and KT– stromal cell populations, we performed RNA sequencing-based global gene expression profiling, comparing KT+ and KT– VECs, as well as cTECs and mTECs. This analysis showed that Kitl, Notch ligands (Dll1, Dll4, Jag1, Jag2), Bmp4, Wnt4, Ccl25 and Cxcl12 were expressed more highly by cTECs compared to mTECs, consistent with their importance for DN1/DN2 differentiation. Notably, several ligands (Kitl, Cxcl12, Dll4, Jag1, Jag2) were expressed at higher or comparable levels by KT+ VECs, supporting a niche role for this distinct cortical VEC population (Figure 3a). To confirm the identity of the sorted populations we examined the expression of Cdh5 and Tek (encoding vascular endothelial (VE)-Cadherin and Angiopoietin receptor; Figure 3b), and Foxn1 and Epcam (epithelial/TEC-specific markers; Figure 3c), and found these to be selectively expressed in the VEC and TEC populations, respectively. To validate the TEC sub-fractionation we examined expression of Psmb11 and Ly75 (encoding the cTEC-specific beta5T proteasome subunit and the CD205 cTEC marker, respectively; Figure 3d), and Aire and Cd80 (both mTEC-specific genes; Figure 3e) and found the expected selective expression in cTECs and mTECs populations. Comparison of KT+ and KT– VEC populations showed higher expression of arterial markers (Depp, Hey1, Efnb2; Figure 3f) in KT+ VECs, and of the venous marker Nf2r222 and Vwf in KT– VECs (Figure 3g). We also confirmed expression of the mKitL encoding Kitl splice variant lacking exon 6 in KT+ VECs and cTECs, with low expression in KT– VECs and mTECs (Figure 3h, expression data summarized in Supplementary Figure 1a). Finally, we measured the expression of Kitl in single TECs and VECs, confirming high and consistent expression of Kitl mRNA in KT+ VECs compared to KT– VECs, and cTECs compared to mTECs. This analysis also showed that Kitl and Cxcl12 are co-expressed by virtually by both cTECs and KT+ VECs (Fig. 3i,j and Supplementary Figure 1b).
Figure 3. Thymic stromal cell subsets show distinct ligand expression profiles.
(a) mRNA expression levels for ligands involved in T-cell development in KT+ VECs, KT– VECs, cTECs and mTECs, as indicated, measured by RNA sequencing. Bars show the average reads per kilobase transcript per million reads (RPKM) from 3 biological replicates; error bars represent standard errors.
(b,c) Analysis as in (a) of VEC-specific (b) and TEC-specific (c) gene expression.
(d,e) Analysis as in (a) of cTEC-specific (d) and mTEC-specific (e) gene expression.
(f,g)Analysis as in (a) of genes associated with arterial (f) and venous endothelium (g).
(h) Real-time qPCR analysis of Kitl mRNA and ΔEx6 Kitl mRNA expression in sorted KT+ and KT– VECs, cTECs and mTECs from (a). Values represent averages after normalization to B2m mRNA. N=3 biological replicates, each measured in technical triplicate. For source data see Supplementary Table 5.
(I,j) mRNA expression in single KT+ VECs, KT– VECs (h) and cTECs and mTECs (i) using microfluidics-based qPCR. Expression values are normalized to B2m for each gene, and subsequently to the average value for each individual gene. Normalization was done separately for VECs and TECs. Each column represents a single cell.
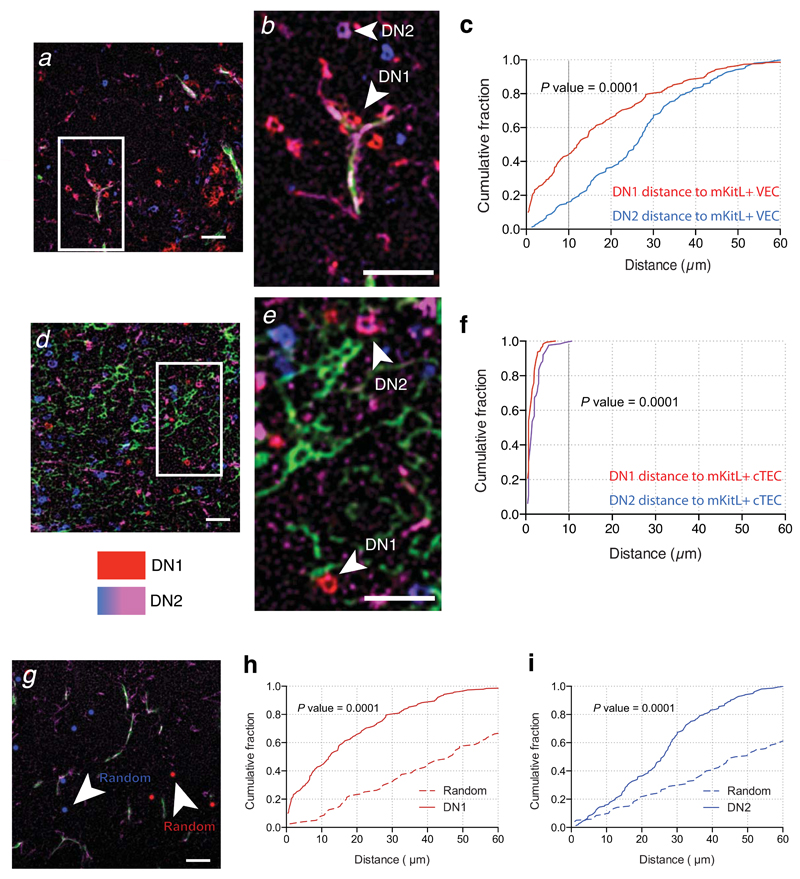
These findings identified molecularly distinct thymic VEC subpopulations. In particular, the KT+ VEC subset displayed arterial identity, expressed many ligands necessary for thymocyte development, including Kitl and Cxcl12. Together with their selective cortical localization this suggested that KT+ VECs could play a role in promoting thymic progenitor development. We therefore examined the location of DN1 (defined as c-Kit+CD25–) and DN2 (c-Kit+CD25+) cells relative to VECs and cTECs, using ICKitL-specific antibody to specifically label mKitL-expressing cells. Quantification of the distance between c-Kit+ thymocytes and mKitL+ VECs showed that the majority of DN1 cells were located in close proximity (<10μm) to mKitL+ VECs (56%; median distance 7μm), whereas only a small fraction of DN2 cells were located <10μm from mKitL+ VECs (12%; median distance 28μm) (Figure 4a-c). In contrast, all c-Kit+ thymocytes were located <10μm of a mKitL+ cTEC (Figure 4d-f), consistent with the dense network of cTECs within the cortex, and their ubiquitous KitL expression. Comparison of DN1 and DN2 localization to that obtained by randomly distributed cells confirmed that the proximity of DN1 and DN2 cells to vascular endothelium was statistically significant (Figure 4g-i).
Figure 4. c-Kit+ thymocyte progenitors home to mKitL expressing cortical VECs and TECs.
(a) Immunofluorescence analysis thymic sections using antibodies against CD31 (green), intracellular KitL+ (ICKitL; pink), c-Kit (red) and CD25 (blue) to identify mKitL+ VECs (CD31+ICKitL+) and c-Kit+ thymocyte progenitors (DN1: c-Kit+CD25–; DN2: c-Kit+CD25+). Scale bar: 100μm
(b) Insert from (a) showing mKitL+ VEC-associated DN1 thymocyte (arrowhead) and DN2 thymocyte (arrow) indicated. Scale bar: 100μm
(c) Distribution of the distances between ICKitL+ VECs and DN1 and DN2 thymocytes measured from sections stained as in (a). The plot shows the cumulative fraction of cells closer than the indicated distance. The significance of the difference in distance distribution between DN1 and DN2 thymocytes was calculated using the Kolmogorov-Smirnov test. DN1: N=189; DN2: N=153 from 3 biological replicates.
(d,e) Analysis as in (a,b) using antibodies against against Ly51 (green), ICKitL (pink), c-Kit (red) and CD25 (blue) to identify mKitL+ cTECs (Ly51+ICKitL+) and c-Kit+ thymocyte progenitors (DN1: c-Kit+CD25–; DN2: c-Kit+CD25+). Scale bars: 100μm
(f) Analysis as in (c) of the distances between ICKitL+ VECs and DN1 and DN2 thymocytes measured from sections stained as in (d). DN1: N=158; DN2: N=127 from 3 biological replicates.
(g) Immunofluorescence analysis of thymic sections with antibodies against CD31 (green), intracellular KitL (pink) (used to generate data in (a)) was used as a platform to generate a random distribution of DN1 and DN2 cells. The original DN1 and DN2 cell staining was removed and replaced with randomly distributed red (DN1) and blue (DN2) dots that resembled the size and the in vivo frequency for these cells.
(h) Distribution of the distances between ICKitL+ VECs/DN1 vs ICKitL+ VECs/Random. The significance of the difference in the distance distribution was calculated using the Kolmogorov-Smirnov test. P value is shown. DN1: N=189 (from panel c); Random: N=80.
(i) Distribution of the distances between ICKitL+ VECs/DN2 vs ICKitL+ VECs/Random. The significance of the difference in the distance distribution was calculated using the Kolmogorov Smirnov test. P value is shown. DN2: N=153 (from panel c); Random: N=120.
Their proximity to c-Kit+ DN1 cells raised the possibility of mKitL+ VECs acting as part of an ETP niche. We therefore examined the role of mKitL expressed on VECs, since its membrane association should confine its action to directly adjacent cells. The physiological role of mKitL has previously been examined using the Steel-Dickie (Sld) mutation, which generates a frame shift mutation eliminating the transmembrane domain of KitL23,24. It is assumed that sKitL is expressed normally by the Sld allele. However, the KitL serum level in Sld/+ was comparable to that seen in Sl/+ mice, which are heterozygous for a Steel null mutation, and in compound Sl/Sld mice serum KitL was barely detectable (Supplementary Figure 2a). The described effects of the Sld mutation25–27, in particular in combination with a Steel null allele, therefore cannot be attributed solely to the loss of mKitL. To analyze mKitL function in specific cell types we therefore generated a mouse line where exon 7 of the Kitl gene, encoding the transmembrane domain, is flanked by loxP sites (KitlLEx7; Supplementary Figure 2b). Germline deletion of this exon (generating the KitlΔEx7 allele) still allows production of sKitL with the same biological activity as that produced by the Sld allele (Supplementary Figure 2c,d). Mice homozygous for the KitlΔLEX7 allele showed the perinatal lethality, and severe developmental and hematopoietic abnormalities normally associated with Steel alleles28, including loss of thymic cellularity. Serum KitL in KitlΔEx7/ΔEx7 mice (henceforth ΔEx7 mice) was reduced (Supplementary Figure 2e), but only to a level similar to that of Sl/+ mice (Supplementary Figure 2a) (P=0.65), a level sufficient to sustain normal bone marrow lymphopoiesis and thymopoiesis in the presence of mKitL (Supplementary Figure 2f-i). While the strong phenotype of ΔEx7 mice, compared to Sl/+ mice, supported a critical physiological role of mKitL, their severe and pleiotropic phenotype precluded conclusions about the cellular origin of the thymic defect.
To determine the contribution of VEC-associated mKitL to early thymic progenitor development we deleted Kitl exon 7 using two different VEC-specific Cre drivers, Tie2-Cre29 and Pdgfb-CreERT230. To confirm their selective expression in thymic VECs we performed lineage tracing (Tie2-Cre) using the Rosa26-EYFP reporter31 and direct readout of EGFP expression (using the IRES-EGFP moiety of the Pdgfb-CreERT2 allele), respectively (Supplementary Figure 3a-d). In both cases fluorescence was observed in 80-90% of VECs, but not in other stromal cells.
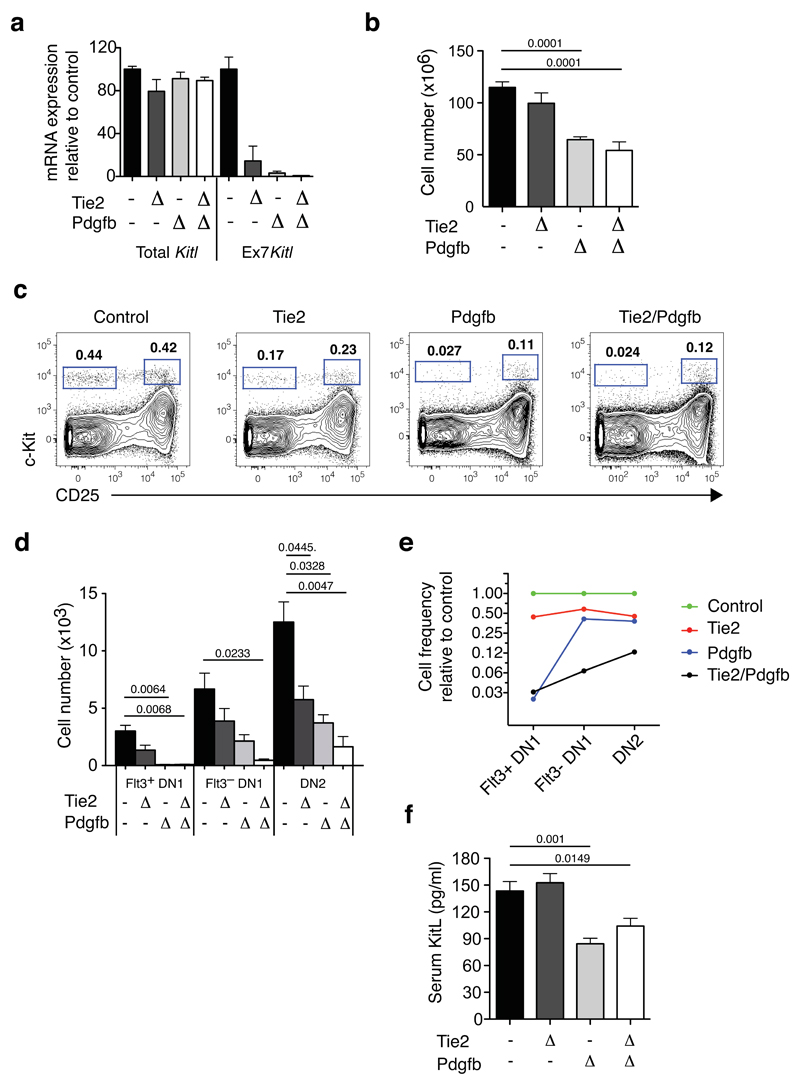
We induced VEC-specific Kitl Ex7 deletion using the Tie2-Cre and Pdgfb-CreERT2 drivers, in the latter case inducing recombination with tamoxifen, generating Tie2ΔEx7 and PdgfbΔEx7 mice, respectively. Analysis 7 days after the last tamoxifen injection showed no significant effect of tamoxifen on thymopoiesis, nor did Pdgfb-CreERT2 activation significantly affect thymopoiesis or bone marrow lymphoid progenitor populations (Supplementary Figure 3g-k). Quantitative PCR using an amplicon spanning exons 7 and 8 confirmed loss of Kitl exon 7, without any significant depletion of total Kitl mRNA (measured using an exon 2-3 spanning amplicon) (Figure 5a). The deletion efficiencies obtained were high for both Tie2-Cre (86%) and Pdgfb-CreERT2 (97%). Combined Tie2-Cre and Pdgfb-CreERT2 (generating Tie2/PdgfbΔEx7 mice) achieved near-complete (>99%) exon 7 deletion.
Figure 5. Cortical vascular expression of mKitL is critical for thymocyte progenitor homeostasis.
(a). qPCR analysis of KItl mRNA levels using Kitl exon 2-3 spanning (Total Kitl) and exon7-8 (Ex7 Kitl) spanning amplicons was performed on sorted thymic VECs from Control (KitlLEx7/LEx7; n=10), Tie2ΔEx7 (n=4), PdgfbΔEx7 (n=5) and Tie2/PdgfbΔEx7 (n=6) mice, collected from 5 independent experiments. To induce Pdgfb-CreERT2–mediated deletion, mice were injected twice with 3mg tamoxifen with a 2-day interval, and analyzed 7 days after the last injection, at 4-5 weeks of age. Values shown are average expression as percentage of the control value; error bars indicate standard errors. For source data see Supplementary Table 5.
(b) The number of total thymocytes in 4-5 weeks old Control (KitlLEx7/LEx7; n=20), Tie2ΔEx7 (n=7), Pdgf ΔEx7 (n=5) and Tie2/Pdgf ΔEx7 (n=4) mice, from 8 independent experiments and tamoxifen treated as in (a). The Cre drivers used are indicated on the x-axis. Values are group averages; error bars show standard errors. Student’s t-test P-values relative to the control group are shown. For source data see Supplementary Table 5.
(c) Representative flow cytometric analysis of c-Kit+ DN thymocyte progenitors from the mice in (a). The gates used to identify DN1 (c-Kit+CD25–) and DN2 cells (c-Kit+CD25+) are indicated.
(d) Absolute number of phenotypic Flt3+DN1, Flt3–DN1 and DN2 cells mice from (a). Values are group averages; error bars show standard errors. Student’s t-test P-values relative to the control group are shown. For source data see Supplementary Table 5.
(e) Graph showing the average abundance of Flt3+DN1, Flt3-DN1 and DN2 cells in Tie2ΔEx7, Pdgf ΔEx7 and Tie2/Pdgf ΔEx7 mice relative to Cre- Control mice.
(f) KitL concentration in the serum of the mice in (a). Values are group averages; error bars show standard errors. Student’s t-test P-values relative to the control group are shown. For source data see Supplementary Table 5.
VEC-specific mKitL loss resulted in a 50% decrease in thymocyte numbers (Figure 5b). This was accompanied by a selective and much more extensive loss of the earliest c-Kit+ thymic progenitors, as Flt3+DN1 cells, and downstream Flt3–DN1 and DN2 thymocyte progenitors were all reduced following recombination with either driver, and in particular by their combined effect (Figure 5c,d). Correcting for the diminished size of the thymus, depletion was most evident for the earliest progenitors: When the VEC-specific Cre lines were combined, Flt3+DN1 cells were decreased 30 fold, Flt3–DN1 cells 16 fold, and DN2 cells 8 fold (Figure 5e), showing that VEC-associated mKitL is most critically required in the DN1 compartment, consistent with the close association of this progenitor population with mKitL+ vascular structures. Importantly, the severity of the phenotype correlated with the efficiency of Ex7 deletion, being most profound in the Tie2/PdgfbΔEx7 genotype, supporting that the phenotype observed was a direct effect of mKitL depletion. The decrease in thymic progenitors did not result from disrupted thymic architecture (Supplementary Figure 4), and proportional numbers of stromal cell types were maintained (Supplementary Figure 5a). Finally, there was no detectable impairment of subsequent thymocyte differentiation stages, but rather a partial recovery at the CD4 and CD8 single positive stages (SP4 and SP8, respectively). (Supplementary Figure 5b). The observed progenitor phenotype and decreased thymus cellularity was therefore due to a direct and selective effect on the earliest DN1 and DN2 thymocyte progenitors, rather than indirect effects on stromal cells or subsequent thymocyte differentiation stages. Since recombination was induced systemically in VECs, potentially decreasing availability of circulating KitL, we examined serum KitL levels, observing a 35% decrease in Tie2/PdgfbΔEx7 mice (Figure 5f), clearly above the level of circulating KitL compatible with normal bone marrow lymphopoiesis and thymopoiesis in Sl/+ mice (Supplementary Figure 2a), and we observed no changes to bone marrow lymphopoiesis upon VEC-specific mKitL deletion (Supplementary Figure 6a,b).
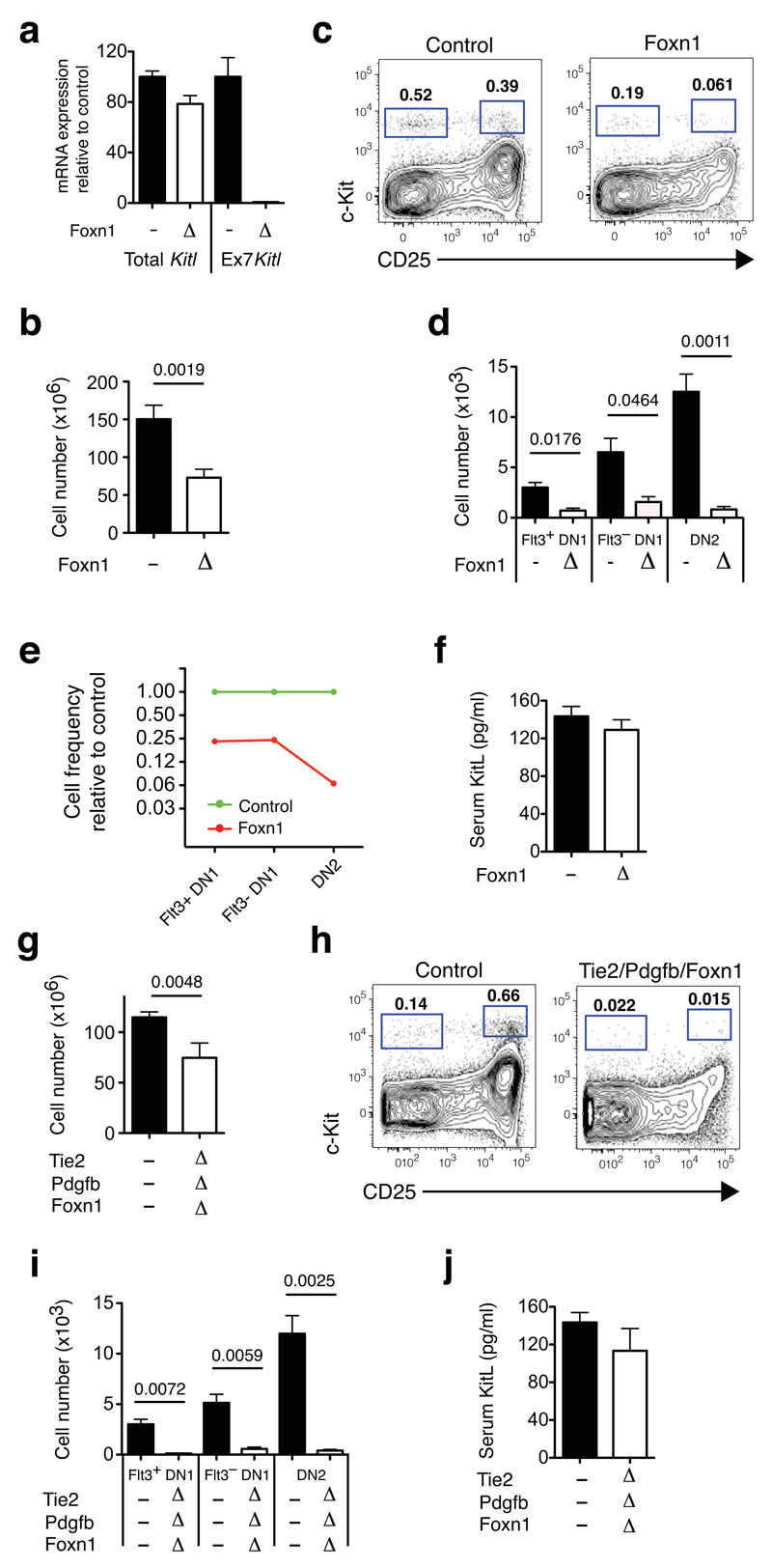
To investigate the role of mKitL in cTECs we used the Foxn1-Cre driver32. We confirmed its TEC-selective and efficient recombination by lineage tracing, (>98% of TECs YFP labeled; Supplementary Figure 3e,f). Crossing the Foxn1-Cre32 driver to the KitlLEx7 allele generating Foxn1ΔEx7 mice. By qPCR we observed near-complete deletion of Kitl exon 7 in Foxn1ΔEx7 TECs (>99%), with only a mild (20%) decrease in the total Kitl mRNA level (Figure 6a). Also here thymus cellularity was decreased (Figure 6b), with a significant depletion of Flt3+DN1 (4-fold) and Flt3–DN1 cells (4 fold). However, in contrast to VEC specific mKitL loss, an even more profound loss of DN2 cells (15 fold) was observed (Figure 6c-e), consistent with the DN2 population lacking proximity to mKitL+ VECs and being principally supported by mKitL+ TECs. Consistent with the restricted Foxn1-Cre recombination pattern we saw no significant decrease of circulating KitL (Figure 6f), changes to bone marrow lymphopoiesis (Supplementary Figure 6c,d), thymic morphology, (Supplementary Figure 4) proportions of thymic stromal cells (Supplementary Figure 5a) or downstream thymic progenitor subsets (Supplementary Figure 5c) in Foxn1ΔEx7 mice. Finally, combining VEC and TEC deletion we observed the expected decrease in thymic cellularity (Figure 6g), and achieved a synergistic depletion of both DN1 and DN2 cells (>20 fold relative decrease) (Figure 6h,i), with serum sKitL maintained above the level compatible with normal lymphopoiesis (Figure 6j). Also in these mice bone marrow lymphoid progenitor numbers, thymic morphology, as well as frequencies of stromal cells and partial recovery of subsequent thymocyte differentiation stages were preserved (Supplementary Figure 4, 5c, 6e,f).
Figure 6. Effect of thymic epithelial mKitL deletion on thymocyte number and thymic progenitors.
(a) qPCR analysis as in (Fig. 5e) of KItl gene expression in sorted cTECs from KitlLEx7/LEx7 (n=8) and Foxn1ΔEx7 mice (n=6) collected from 2 independent experiments.
(b) Total thymic cellularities in 4-5 weeks old Control (KitlLEx7/LEx7; n=18) and Foxn1ΔEx7 mice (n=6) collected over 4 independent experiments. The Cre driver used is indicated on the x-axis. Values are group averages; error bars show standard errors. The Student’s t-test P-value relative to the control group is shown.
(c) Representative flow cytometric analysis of c-Kit+ DN thymocyte progenitors from the mice in (a). The gates used to identify DN1 (c-Kit+CD25–) and DN2 progenitors (c-Kit+CD25+) are indicated.
(d) Absolute number of phenotypic Flt3+DN1, Flt3–DN1 and DN2 thymocyte progenitors in mice from (a). The Cre drivers used are indicated on the x-axis. Values are group averages; error bars show standard errors. Student’s t-test P-values relative to the control group are shown.
(e) Graph showing the average abundance of Flt3+DN1, Flt3–DN1 and DN2 cells in Foxn1ΔEx7 mice relative to Cre– Control mice.
(f) KitL concentration in the serum of the mice in (a). Values are group averages; error bars show standard errors.
(g) Total thymic cells in 4-5 weeks old Control (KitlLEx7/LEx7; n=18), Tie2/Pdgfb/Foxn1ΔEx7 (n=5) mice analysed over 4 independent experiments.. The Cre drivers used are indicated on the x-axis. Mice were tamoxifen treated as in Fig. 5a. Values are group averages; error bars show standard errors. Student’s t-test P-values relative to the control group are shown.
(h) Representative flow cytometric analysis of c-Kit+ DN thymocyte progenitors from the mice in (g). The gates used to identify DN1 (c-Kit+CD25–) and DN2 cells (c-Kit+CD25+) are indicated.
(i) Absolute number of phenotypic Flt3+DN1, Flt3–DN1 and DN2 thymocyte progenitors in mice from (g). The Cre drivers used are indicated on the x-axis. Values are group averages; error bars show standard errors. Student’s t-test P-values relative to the control group are shown.
(j) KitL concentration in the serum of the mice in (g). Values are group averages; error bars show standard errors. P-values relative to the control group are shown.
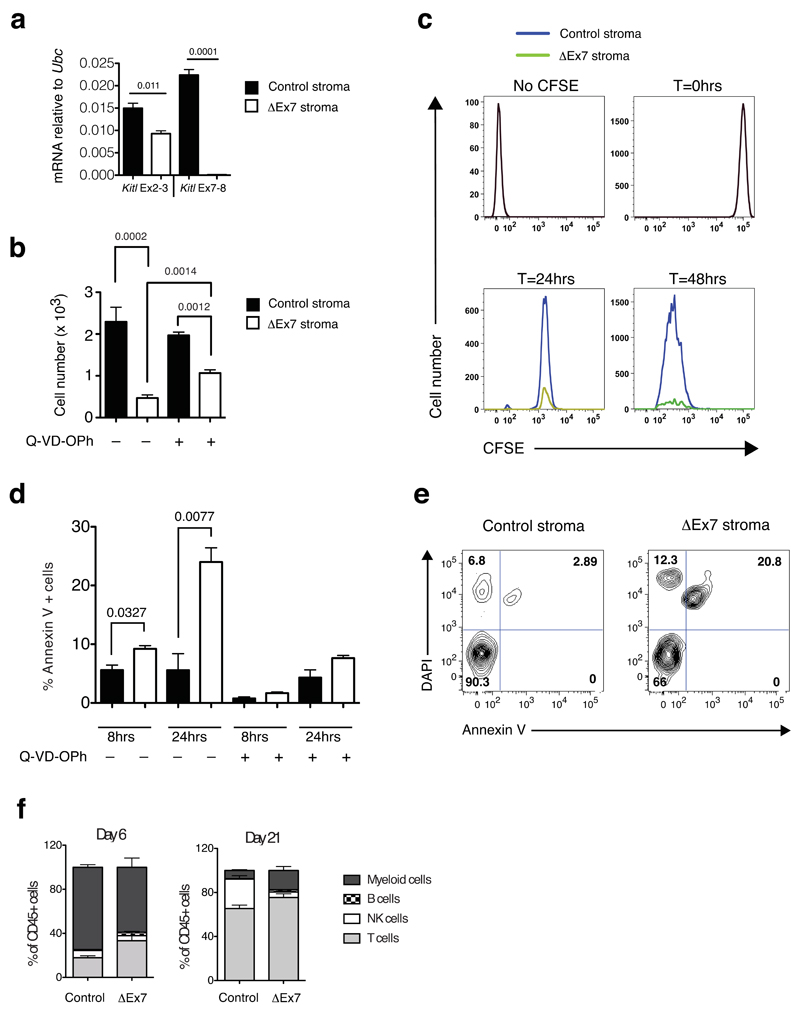
To address how mKitL promotes thymic progenitor differentiation we performed Rosa26-CreERT2–mediated mKitL-depletion in primary thymic stromal cultures (Figure 7a). Co-culture of wild type DN1 cells with ΔEx7 stroma resulted in significantly decreased thymic progenitor expansion compared to wild-type control cultures (Figure 7b). We therefore labeled input cells with carboxyfluorescein-succinimidyl-ester (CFSE) and measured their proliferative history by dye dilution, and observed no difference in CFSE dilution between ΔEx7 and control stromal cultures (Figure 7c). In contrast, Annexin V-staining revealed a significant increase in thymic progenitor apoptosis in the absence of mKitL, which could be reversed by the addition of the Q-VD-OPh pan-caspase inhibitor (Figure 7d,e), partially rescuing progenitor expansion (Figure 7b). Finally, to assess mKitL requirement for multi-potency, DN1 cells were plated on wild type or mKitL-deficient stroma. Both generated, in addition to T-cells, myeloid and other lymphoid lineages (Figure 7f). A key function of mKitL is therefore to promote the survival of early thymic progenitors by preventing caspase-dependent apoptosis, whereas it is dispensable for their proliferation and multi-lineage differentiation.
Figure 7. mKitL promotes early thymocyte progenitor survival.
(a) qPCR analysis as in Fig, 5a of cultured thymic stroma from 4-week old KitlLEx7/LEx7;Rosa26-CreERT2 (ΔEx7 stroma) and Kitl+/+;Rosa26-CreERT2 (control (Con) stroma) treated with tamoxifen (4OHT). Each value is the mean, normalized to Ubc mRNA, of 3 biological replicates, each measured in technical triplicate. Error bars show standard errors. Significance of difference in expression level was determined using Student’s t-test; P-values are shown. For source data see Supplementary Table 5.
(b) Total live thymocyte cell number from at 48 after plating DN1 cells on Con and ΔEx7 stroma, in the presence or absence of Q-VD-OPh as indicated (–Q-VD-OPh: N=7; +Q-VD-OPh: n=3 biological replicates pooled over 2 independent experiments). Error bars show standard errors. Significance of difference in expression level was determined using Student’s t-test; P-values are shown.
(c) Representative histogram plots of CFSE labeled DN1 cells 24hrs and 48hrs after plating onto Con or ΔEx7 stroma. Profiles of unstained cells (No CFSE) and CFSE labeled cells prior to plating (T=0hrs) are shown. n at 24 hours=6; n at 48 hours=7. n represents biological replicates pooled over 3 different experiments
(d) Percentage of Annexin V+ cells at 8hrs and 24hrs of culture in control and ΔEx7 cocultures, in the presence or absence of Q-VD-OPh as indicated. n=3 biological replicated for all conditions, pooled over 2 independent experiments. Error bars show standard errors. Significance of difference in expression level was determined using Student’s t-test; P-values are shown. For source data see Supplementary Table 5.
(e) Representative FACS plot at the 24 hour time point of the data shown in (d).
(f) Lineage readout at days 6 and 21 from 3x103 sorted ETPs plated onto control and ΔEx7 primary thymic stroma. n=3 biological replicates for all conditions, from 2 independent experiments. Error bars show standard errors. For source data see Supplementary Table 5.
Discussion
According to the current paradigm early stages of thymocyte development are supported by cTECs which express ligands required for T-cell lineage commitment and early T-cell progenitor expansion18,33,34, whereas a niche role for thymic VECs was not previously explored. We here show that several of these ligands (including Kitl, Cxcl12, Dll4, Jag1, Jag2) are highly expressed by a distinct, cortex-restricted subpopulation of thymic VECs. KitL+ VECs are closely and selectively associated with DN1 thymocytes, and in agreement with this, loss of mKitL from thymic VECs results in a profound and selective decline in DN1 thymocyte numbers. Furthermore, as membrane-bound KitL is required for VECs to support c-Kit+ thymic progenitors their niche function is mediated through direct cell-cell contact. Together, these results identify KitL+ VECs as a critical niche cell type for the earliest thymic progenitors in the cortex. We also show that cTECs, but not mTECs, express KitL, and that TEC-specific loss of mKitL results in depletion of both DN1 and DN2 thymocytes. cTECs therefore collaborate with mKitL+ VECs to generate niches for the earliest multi-potent c-Kit+ ETPs by providing mKitL to support DN thymocyte development through dynamic, stage-specific cell-cell interaction.
The Kit receptor is exclusively expressed on DN1 and DN2 thymic progenitors, and in line with this, these progenitors were selectively reduced in mKitL VEC- and TEC-deleted mice, in agreement with previous studies of Sl/Sl mice19, whereas no effect on the differentiation of downstream DP, SP4 and SP8 thymocytes was observed, supporting that the decrease in thymic cellularity is due to depletion of the c-Kit+ thymocyte progenitors in the absence of VEC- and TEC-expressed mKitL. In vitro mKitL was dispensable for proliferation and multi-lineage differentiation of c-Kit+ ETPs, but prevented their capsase-dependent apoptosis. While additional signals may regulate ETP survival in vivo, this indicates that promoting ETP survival is a key mKitL function, distinct from that provided by Notch signaling, which acts on the same populations to mediate T-cell lineage restriction8. This complementarity of function is consistent with the observation that Notch ligands and KitL promote T-lymphopoiesis in a non-redundant manner in vivo when expressed in Foxn1 null TECs that otherwise fail to support T-cell differentiation33.
The Sld allele has been used to assign physiological function to mKitL in erythropoiesis, smooth muscle pacemaking and reproduction25–27, assuming that the Sld allele does not significantly alter systemic KitL levels. This assumption was not previously tested, and when doing so we detected only trace levels of sKitL in compound Sl/Sld mice. A significant component of the phenotypes associated with Sld mutation could therefore be due to loss of systemic sKitL. Soluble KitL generated by the Sld allele contains a C-terminal peptide sequence additional to that generated through cleavage within the exon 6-encoded sequence23,24. This does not affect its biological activity when produced recombinantly23, but clearly does affect in vivo availability. The soluble KitL generated by exon 7 deletion in the present study contains a distinct C-terminal extension peptide, and is both biologically active and physiologically stable. The KitLΔEx7 allele will therefore be important for studies of the physiological role of mKitL.
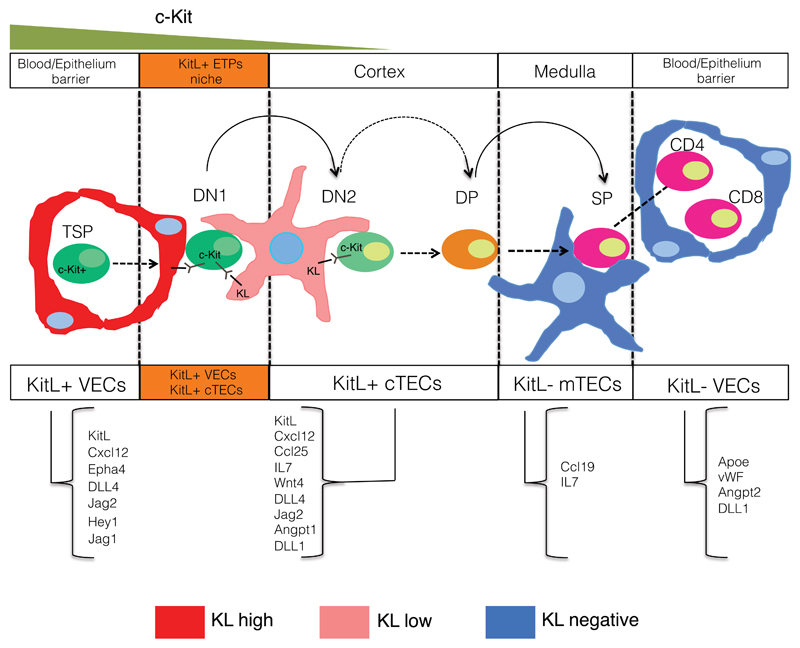
VECs play important roles in tissue morphogenesis, regeneration and tissue stem cell maintenance35. In particular, KitL produced by VECs participates in the generation of embryonic hematopoietic clusters36 and maintenance of hematopoietic stem cells37. However, as both sKitL and mKitL were depleted it remains unclear if the observed effects were mediated systemically or in a niche context. In the case of the earliest thymocyte progenitors we now demonstrate that distinct VEC and cTEC mKitL expressing niche cell types provide stage-specific support of early thymocyte progenitors: while mKitL produced by VECs is particularly critical for DN1 maintenance, cTEC-presented mKitL is required for maintaining DN2 cells. This requirement is reflected in the local organization of c-Kit+ thymocyte progenitors, where DN1 thymocytes are selectivly associated with KitL+ VECs. TSPs are believed to enter the thymus at the cortico-medullary junction, where KitL+ VECs are abundant on the cortical side. The initial dependence of c-Kit+ thymocyte progenitors on KitL+ VECs may therefore reflect their point of entry. They subsequently migrate away from the vasculature, becoming dependent on TEC-presented mKitL at the DN2 stage. Since mKitL levels are lower in cTECs compared to KT+ VECs, other factors likely control the migration process. This model for c-Kit+ thymocyte niche cell interactions and dynamics is illustrated in Figure 8. It will be of interest to examine if other differentiating progenitors display similar dynamic interactions with supporting niche cells.
Figure 8. Model of the cellular niches occupied by c-Kit+ thymocyte progenitors.
Schematic diagram of the sequential interaction of c-Kit thymocyte progenitors with KitL+ VEC and cTEC stromal cells. The ligands found to be expressed by the different stromal cell subsets are indicated.
Online methods
Mouse lines
Pdgfb-iCreERT2 transgenic mice were kindly provided by Dr M. Fruttiger30. Foxn1-Cre transgenic mice were kindly provide by Dr. C Blackburn32. Rosa26-CreERT2 mice were kindly provided by Dr. J. Nathans38. Sl/+ (stock number 000693) Sld/+ (stock number 000160), Rosa26-EYFP reporter mice31 (stock number 006148) and Tie2-Cre29 (stock number 008863) transgenic mice were purchased from the Jackson Laboratory. Sl/Sld mice were generated by intercrossing Sl/+ and Sld/+ mice. The Sld allele was re-sequenced to ensure that it retained the described genomic alteration23,24.
To generate the Kitl-tdTomato transgene the RP24-73F12 BAC backbone (BacPac Resources, CHORI) was engineered by recombineering to replace the endogenous Kitl ATG and the remainder of exon 1 with a cassette composed of a hybrid intron (to allow for a controlled splicing event), a Kozak-ATG-CreERT2 moiety, an IRES followed by a myristoylated tdTomato dimer, and a SV40 polyA site. Insertion of the cassette was screened for with beta-galactosidase selection marker flanked by FRT sites. After successful clonal isolation and verification by sequencing, the marker gene was removed by bacterially expressed Flp-recombinase. No other modifications were applied to the BAC. DNA was prepared for injection using the Large Construct Kit (Qiagen, Hilden) and mice generated by ICSI as described39.
The Kitl floxed exon 7 targeting vector was generated through recombinase-mediated BAC shaving and insertion of a LoxP site into intron 6 and an FRT-neo-FRT-LoxP cassette into intron 7. Targeting was done by homologous recombination in IB10/C ES cells. After generation of chimeras and germline transmission the FRT-Neo-FRT recombined using the Rosa26-Flpe strain40, to generate the KitlLEx7 allele.
All recombineered junctions and coding sequences were sequenced before transgenic lines were generated. Complete construct sequences are available upon request.
Mouse lines were backcrossed to C57Bl/6 (>5 generations) where required, and maintained on this background. 4-6 weeks old male mice have been used in all the experiments. All mice were bred and maintained in accordance with UK Home Office regulations. Experimental protocols were approved by the Edinburgh University School of Biological Sciences Ethical Review Committee, and the Oxford University Clinical Medicine Ethical Review Committee. Mice were genotyped by PCR using the primers listed in Supplementary Table 1.
Tamoxifen treatment
Tamoxifen (Sigma) was dissolved in ethanol at 60mg/ml. The equivalent of 3mg was mixed at a 1:1:2 ratio with Cremophor (Sigma) and PBS and injected subcutaneously (SC) twice, with a 2 day interval. Mice were analyzed one week after the last injection, at an age of 4-5 weeks. We did not observe any toxicity of the inducible Cre, as no difference in thymic cellularity, DN1/DN2 cell number, bone marrow cellularity or lymphoid progenitor numbers were observed in Tamoxifen-induced Pdgfb-CreERT2 mice compared to non-transgenic controls (Supplementary Figure 3g-j). Likewise, no dramatic effect was observed of the Tamoxifen treatment alone on thymocyte subsets with this setting (Supplementary Figure 3k).
Thymic stromal cell preparation
Dissected thymi were finely chopped and the obtained fragments digested in a mix of Collagenases I and 3 (Worthington) (3mg/ml), dispase II (Roche) (7mg/ml), DNase I (Invitrogen) (2U/ml) at 37°C with gentle agitation for 20min. The dissociated cells were centrifuged at 1300rpm for 5 min and resuspended in ammonium chloride solution (Gey solution) for 1 min to lyse the erythrocytes and the reaction stopped by adding PBS, 5% FBS, 25mM EDTA. To facilitate the thymic stromal cell sorting the samples were depleted of CD45+ cells by using the anti mouse CD45 microbeads (Miltenyi). The FACS gating strategy is shown in Supplementary Figure 7. In the case of mesencymal cells (MCs), the lack of CD45+ cells in this population was verified by establishing the absence of detectable Ptprc mRNA.
Primary thymic stromal cell-DN1 co-culture
Unfractionated thymic cells were prepared as described above (see Thymic stromal cell preparation). After the complete organ disaggregation the cells were re-suspended in IMDM (Life Technologies/Gibco), 10% heat inactivated FBS (Life Technologies/Gibco), 1% Penicillin (Life Technologies/Gibco) and plated in collagen treated plates. The collagen (Stem Cell Technologies) was diluted in PBS at 70μg/ml final concentration and used to coat tissue culture treated plated for 45’ at RT. In vitro recombination was achieved by adding 1μM 4-Hydroxytamoxifen (SIGMA) to the culture every 2 days, by removing 50% of the medium volume and adding fresh medium back, for a total of 8 days. Before the DN1 seeding, the wells were extensively washed to remove all non-adherent cells and tamoxifen, and fresh medium was added.
CD4 and CD8 depletion
To prepare CD4/CD8-depleted thymocytes, thymi were disaggregated as above and re-suspended in 500μl volume. Biotinylated anti-CD4 (eBioscience 10-0043) and anti-CD8 (eBioscience 13-0081) (0.5μg/ml) were added to the cells and left for 15’ at 4C. The cells were washed and anti-biotin microbeads (Miltenyi-10μl x 108) added for 10’at 4C. The samples were then passed onto the AutoMacs machine by using the “depletes” programme.
CFSE labeling
The thymic cell suspension was re-suspended at 10x106 cells/ml of PBS and . CFSE was added to a final concentration of 1μM, and cells incubated for 10’ at room temperature in the dark. The labeling procedure was stopped by adding 5 volumes of FBS, followed by washing to remove excess dye.
Thymic section preparation and immunofluorescence
Freshly isolated thymi were washed in PBS and included in OCT for snap freezing. The OCT blocks were sliced at 7μm thickness and sections stored at -80°C. The sections were dried at RT for 5 before fixing in acetone for 5’ at 4°C and left at RT for 5’ to ensure the acetone evaporation. The sections were then re-hydrated for 30’ in PBS at RT and blocked in 5% BSA in PBS for 10’ RT. Fluorochrome conjugated or unconjugated primary antibodies were diluted in PBS and sections stained for 1h and than washed several times in PBS before adding the secondary antibody for 45’ at RT. Sections were than counterstained with DAPI (0.2 μg/ml) and mounted with DXP mounting medium (Fisher Scientific). Images were acquired with a Zeiss 780 confocal inverted microscope and processed using FiJi software.
To produce the randomized DN1 and DN2 simulation, 2D images were generated using Fiji analysis software41. In each image, Gaussian blobs (sigma=3) of equal size to the DN1 and DN2 positive cells were placed randomly by sampling a uniform random distribution for the x and y coordinate position. In the original analysed images, the DN1 and DN2 staining was then removed and the staining replaced with the simulated images which contained 4 randomly positioned red cells (DN1) and 6 randomly distributed blue cells (DN2). The recombined images were then analysed by assessing the distance between the simulated DN1 (random, R-DN1) and DN2 (random, R-DN2) cells and the original CD31+mKitL+ VEC staining. Antibodies used for the immunofluorescence are listed in Supplementary Table 2.
Flow cytometry of hematopoietic cells
Mouse T cells were isolated by meshing the thymi on 70 μm cell strainers and resuspended in PBS, 5% FBS, 2mM EDTA. 25x106 cells were used for staining and analysis of the ETPs (gating strategy shown in Supplementary Fig. 8a). Mouse bone marrow cells were isolated by crushing the bones in PBS, 5% FBS, 2mM EDTA and the recovered cells filtered through 70 μm cell strainers. 30x106 cells were used to stain and analyse the bone marrow lymphoid progenitor populations (Supplementary Fig. 8b). Data were acquired using LSR Fortessa (BD biosciences) and analysed using FlowJo software. Sorting was performed by using a FACSAria III (BD Biosciences). The antibodies used are listed in Supplementary Table 3.
RNA-sequencing
Samples for RNA sequencing were prepared using the SMARTer™ Ultra Low RNA kit for Illumina Sequencing (Clontech) according to manufacturers instructions as previously described42. Briefly, cDNA libraries were prepared from 200 (VECs, cTECs and mTECs) cells, with 4-5 biological replicates from each genotype. Cells were sorted directly into lysis buffer supplemented with RNase Inhibitor (Clontech). cDNA library was prepared according to manufacturer instructions with 15 cycles of amplification. Amplified cDNA libraries validated with a distinct peak spanning 400bp to 9000 bp in size as measured by High Sensitivity DNA kit (Agilent) on Agilent 2100 Bioanalyzer were further processed for RNA sequencing. Libraries were prepared for Illumina sequencing using the NexteraXT DNA Sample Preparation Kit (Illumina) using the Manufacturers recommended protocol. The size of the amplified fragments was confirmed by Agilent High Sensitivity DNA kit (Agilent). 4-5 samples with different indices were pooled per lane and sequenced on a HiSeq2000 (Illumina) generating single–end, 51 bp reads. 51-bp reads were aligned against the mouse genome obtained from the build 37 assembly by NCBI (mm9) using Tophat243. Non-uniquely mapped reads were discarded. Uniquely mapped reads were counted per exon of each RefSeq gene model and reads per kilobase of a concatenated exon length per million mapped reads (RPKMs) were calculated and assigned to each gene locus using custom R scripts.
Multiplex quantitative real time PCR analysis of single cells
For cDNA synthesis and preamplification of target genes CellsDirect One-Step qRT-PCR kit (Invitrogen) was used. Cells were sorted directly into PCR plates or 0.2 mL PCR tubes containing 2.5μL gene specific 0.2x TaqMan gene expression assays (Applied Biosystems), 5μL of CellsDirect 2x Reaction mix (Invitrogen), 1.2μL CellsDirect RT/Taq mix, 1.2μL TE buffer and 0.1μL SUPERase-In RNase Inhibitor (Ambion) to a total volume of 10μL. Conditions for reverse transcription and target gene amplification were: 15 min at 50°C; 2min at 95°C; 22 cycles of 95°C for 15s and 60°C for 4min. Preamplified products were diluted 1:5 in TE buffer and analyzed on a 48x48 Dynamic Array (Fluidigm) using the following PCR cycling condition: 95°C for 10min; 40 cycles of 95°C for 15s and 60°C for 60s. Data was analyzed using the ΔΔCt method as described44; results were normalized to B2m. Single cells not expressing Hprt or Actinb or B2m were excluded from the analysis. Taqman assays used for multiplexed quantitative real-time PCR are listed in Supplementary Table 4.
Serum KitL measurements
Mouse serum samples were collected from peripheral blood clotted for 2 hr at room temperature by centrifuging for 20min at 2000g and assessed by murine Kitl ELISA kits (R&D systems) according to manufacturer’s instructions. Optical density measurements were determined using a micro plate reader (Molecular Devices-SpectraMaxM2e) set to 450nm with wavelength correction set to 570 nm.
Recombinant KitL activity measurements
KitlΔex7/+ RNA was used for direct RT-STA PCR. Conditions for reverse transcription and target gene amplification were: 50min at 50°C; 2min at 95°C; 42 cycles of 95°C for 30s, gradient of 55-65°C and 72°C for 3min. Primers (Life Technologies): (F): 5’-TCC TCC TCC TCC TCT TCC TC-3’; (R): AAG CAG GGC TTT CTG TTC AA). Sld/+ RNA was converted to cDNA by mixing 1μl random hexamer primers (50ng/μl) (Fermentas), 1μl dNTPs (10mM) (Fermentas) and 10μl RNA incubated for 10min at 65°C, centrifuged briefly then put immediately on ice. 4 μl 5x First Strand buffer (MMLV-RT kit), 2μl 0.1 M DTT (MMLV-RT kit), 1μl RNase-out RNAse inhibitor and 1μl Superscript II Reverse Transcriptase (SSII) (all Life Technologies) were added then incubated for 10min at 25°C, followed by 50min at 42°C then 15min at 70°C. Gene amplification was then carried out using two rounds of PCR. Conditions for target gene amplification were 30s at 98°C; 35 cycles of 98°C for 10s, gradient of 55-65°C for 30s, 72°C for 10s then final 10min at 72°C using primers (F): 5’-AGA GGC CAG AAA CTA GAT CC-3’; (R): 5’-GGT AGC AAG AAC AGG TAA GG-3’).
KitL PCR products were cloned using TOPO Cloning®Kit PCR®II-TOPO®vector and then transformed into One Shot®TOP10 Chemically Competent Cells (both Life Technologies). sKitl cDNA was inserted into a pMSCV-iresGFP plasmid by digesting with Hpa I/Xho I and Pvu I/ Xho I (Fermentas / NEB) ligated using T4 DNA ligase and transformed into TOP10 E. coli cells (both Life Technologies) according to manufacturer’s protocol. Sld Kitl was subcloned into mKitl- pMSCV-iresGFP using BamH I, Bgl II and Nsi I (Fermentas). Correct ligation of sKitl and Sld Kitl into pMSCV-iresGFP vectors was confirmed by digestion with Spe I (NEB). Samples were sequenced using M13R and KitLR primers (M13R: 5’-CAG GAA ACA GCT ATG ACC-3’; KitL cDNA R: 5’- GCC ACT GTG CGA AGG TAA C-3’).
Human embryonic kidney (HEK) 293T/17 cells (293T, ATCC Number: CRL-11268) cells were transfected with KitL-pMSCV-iGFP vectors in DMEM/10%FCS/1%PS using FuGene (Promega) according to the manufacturers’ instructions. Transfected 293T cells were incubated in a humidified atmosphere in 5% CO2 at 37°C for 48hr. The supernatants from the transfected cells were collected and then evaluated for the level of sKitl by ELISA as described above. MC/9 cells (ATCC Number: CRL- 8306) were plated at 250,000 cells/well in 96-well plate, cultured in 100µl supernatant and incubated in a humidified atmosphere, 5% CO2 at 37°C for 48 hr (Sanyo CO2 incubator). CellTiter96r AQueous Non-Radioactive Cell Proliferation Assay MTS reagent (Promega) was then added to cells, incubated for 4hr (according to manufacturer’s instructions) and absorbance detected at 490nm (Molecular Devices -SpectraMax M2e). Absorbance values were then used to calculate percentage of maximum proliferation of cells (in relation to proliferation of cells in the recommended optimal medium: DMEM high glucose/10%FCS/10% rat T-STIM/0.05mM β-mercaptoethanol). Cell lines were regularly screened for mycoplasma contamination.
Data reproducibility
For Figures where representative images are shown, the number of times the experiments were repeated are as follows: Figure 1a,d,e: 4 times; Figure 2a-l: 6 times; Figure 2m-q: 3 times; Figure 4a,b,d,e,g: 6 times. Supplementary Figure 3b: 3 times; 3d: 5 times; 3f: 7 times.
Statistical analysis
Data sets were analyzed using Student’s t-test after testing for normality (F-test), except for comparison of cell distributions, where the Kolmogorov-Smirnov test was used. No statistical method was used to predetermine sample size, and experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Accession numbers
RNA sequencing data files have been deposited in the NCBI GEO database (accession number GSE62741).
Supplementary Material
Acknowledgements
We thank Dr. C. Blackburn, Dr. J. Nathans and Dr. M. Fruttiger for providing Foxn1-Cre, Rosa26-CreERT2 and Pdgfb-CreERT2 mice, respectively, Ajoy Samraj for screening of targeted ES cells, and the Wolfson Imaging Centre for image analysis. This work was supported by an MRC Strategic Award and MRC Program Grant to CN, by an MRC Program Grant to SEJ, by Cancer Research UK and by Leukemia and Lymphoma Research. AM is the recipient of an MRC Senior Clinical fellowship.
Footnotes
Author Contributions.
MB carried out mouse genetic experiments and imaging, and associated data analysis; RF and SM generated Sl/Sld and ΔEx7 mice and measured KitL levels and activity; AS screened Kitl targeted ES cells; ST analyzed RNASeq data; TL provided protocols for single cell profiling; AG generated RNAseq libraries; AM provided protocols for single cell profiling; SEJ and CN conceived the project and wrote the manuscript with MB.
References
- 1.Scadden DT. Nice neighborhood: emerging concepts of the stem cell niche. Cell. 2014;157:41–50. doi: 10.1016/j.cell.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 3.Chasis JA, Mohandas N. Erythroblastic islands: niches for erythropoiesis. Blood. 2008;112:470–478. doi: 10.1182/blood-2008-03-077883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, et al. Osteoblasts support B-lymphocyte commitment and differentiation from hematopoietic stem cells. Blood. 2007;109:3706–3712. doi: 10.1182/blood-2006-08-041384. [DOI] [PubMed] [Google Scholar]
- 5.Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunting MD, Comerford I, McColl SR. Finding their niche: chemokines directing cell migration in the thymus. Immunol Cell Biol. 2011;89:185–196. doi: 10.1038/icb.2010.142. [DOI] [PubMed] [Google Scholar]
- 7.Love PE, Bhandoola A. Signal integration and crosstalk during thymocyte migration and emigration. Nat Rev Immunol. 2011;11:469–477. doi: 10.1038/nri2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 9.Koch U, et al. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205:2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Freeden-Jeffry U, Solvason N, Howard M, Murray R. The earliest T lineage-committed cells depend on IL-7 for Bcl-2 expression and normal cell cycle progression. Immunity. 1997;7:147–154. doi: 10.1016/s1074-7613(00)80517-8. [DOI] [PubMed] [Google Scholar]
- 11.Sitnicka E, et al. Critical role of FLT3 ligand in IL-7 receptor independent T lymphopoiesis and regulation of lymphoid-primed multipotent progenitors. Blood. 2007;110:2955–2964. doi: 10.1182/blood-2006-10-054726. [DOI] [PubMed] [Google Scholar]
- 12.Kwan J, Killeen N. CCR7 directs the migration of thymocytes into the thymic medulla. J Immunol. 2004;172:3999–4007. doi: 10.4049/jimmunol.172.7.3999. [DOI] [PubMed] [Google Scholar]
- 13.Trampont PC, et al. CXCR4 acts as a costimulator during thymic beta-selection. Nat Immunol. 2010;11:162–170. doi: 10.1038/ni.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah DK, Zuniga-Pflucker JC. An overview of the intrathymic intricacies of T cell development. J Immunol. 2014;192:4017–4023. doi: 10.4049/jimmunol.1302259. [DOI] [PubMed] [Google Scholar]
- 15.Tsai PT, Lee RA, Wu H. BMP4 acts upstream of FGF in modulating thymic stroma and regulating thymopoiesis. Blood. 2003;102:3947–3953. doi: 10.1182/blood-2003-05-1657. [DOI] [PubMed] [Google Scholar]
- 16.Heinonen KM, et al. Wnt4 regulates thymic cellularity through the expansion of thymic epithelial cells and early thymic progenitors. Blood. 2011;118:5163–5173. doi: 10.1182/blood-2011-04-350553. [DOI] [PubMed] [Google Scholar]
- 17.Hager-Theodorides AL, et al. Bone morphogenetic protein 2/4 signaling regulates early thymocyte differentiation. J Immunol. 2002;169:5496–5504. doi: 10.4049/jimmunol.169.10.5496. [DOI] [PubMed] [Google Scholar]
- 18.Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat Rev Immunol. 2006;6:127–135. doi: 10.1038/nri1781. [DOI] [PubMed] [Google Scholar]
- 19.Rodewald HR, Kretzschmar K, Swat W, Takeda S. Intrathymically expressed c-kit ligand (stem cell factor) is a major factor driving expansion of very immature thymocytes in vivo. Immunity. 1995;3:313–319. doi: 10.1016/1074-7613(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 20.Lennartsson J, Ronnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev. 2012;92:1619–1649. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- 21.Gray DH, Chidgey AP, Boyd RL. Analysis of thymic stromal cell populations using flow cytometry. J Immunol Methods. 2002;260:15–28. doi: 10.1016/s0022-1759(01)00493-8. [DOI] [PubMed] [Google Scholar]
- 22.Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by developmental reprogramming of venous cells. Nature. 2010;464:549–553. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brannan CI, et al. Steel-Dickie mutation encodes a c-kit ligand lacking transmembrane and cytoplasmic domains. Proc Natl Acad Sci U S A. 1991;88:4671–4674. doi: 10.1073/pnas.88.11.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flanagan JG, Chan DC, Leder P. Transmembrane form of the kit ligand growth factor is determined by alternative splicing and is missing in the Sld mutant. Cell. 1991;64:1025–1035. doi: 10.1016/0092-8674(91)90326-t. [DOI] [PubMed] [Google Scholar]
- 25.Gu Y, Runyan C, Shoemaker A, Surani MA, Wylie C. Membrane-bound steel factor maintains a high local concentration for mouse primordial germ cell motility, and defines the region of their migration. PLoS One. 2011;6:e25984. doi: 10.1371/journal.pone.0025984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikkelsen HB, Malysz J, Huizinga JD, Thuneberg L. Action potential generation, Kit receptor immunohistochemistry and morphology of steel-Dickie (Sl/Sld) mutant mouse small intestine. Neurogastroenterol Motil. 1998;10:11–26. doi: 10.1046/j.1365-2982.1998.00082.x. [DOI] [PubMed] [Google Scholar]
- 27.Kapur R, et al. Signaling through the interaction of membrane-restricted stem cell factor and c-kit receptor tyrosine kinase: genetic evidence for a differential role in erythropoiesis. Blood. 1998;91:879–889. [PubMed] [Google Scholar]
- 28.Russell ES. Hereditary anemias of the mouse: a review for geneticists. Adv Genet. 1979;20:357–459. [PubMed] [Google Scholar]
- 29.Kisanuki YY, et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 30.Claxton S, et al. Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis. 2008;46:74–80. doi: 10.1002/dvg.20367. [DOI] [PubMed] [Google Scholar]
- 31.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordon J, et al. Specific expression of lacZ and cre recombinase in fetal thymic epithelial cells by multiplex gene targeting at the Foxn1 locus. BMC Dev Biol. 2007;7:69. doi: 10.1186/1471-213X-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calderon L, Boehm T. Synergistic, context-dependent, and hierarchical functions of epithelial components in thymic microenvironments. Cell. 2012;149:159–172. doi: 10.1016/j.cell.2012.01.049. [DOI] [PubMed] [Google Scholar]
- 34.Krause DS, Scadden DT. Deconstructing the complexity of a microenvironmental niche. Cell. 2012;149:16–17. doi: 10.1016/j.cell.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramasamy SK, Kusumbe AP, Adams RH. Regulation of tissue morphogenesis by endothelial cell-derived signals. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasaki T, et al. Regulation of hematopoietic cell clusters in the placental niche through SCF/Kit signaling in embryonic mouse. Development. 2010;137:3941–3952. doi: 10.1242/dev.051359. [DOI] [PubMed] [Google Scholar]
- 37.Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23:2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreira PN, Pozueta J, Giraldo P, Gutierrez-Adan A, Montoliu L. Generation of yeast artificial chromosome transgenic mice by intracytoplasmic sperm injection. Methods Mol Biol. 2006;349:151–161. doi: 10.1385/1-59745-158-4:151. [DOI] [PubMed] [Google Scholar]
- 40.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- 41.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramskold D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grover A, et al. Erythropoietin guides multipotent hematopoietic progenitor cells toward an erythroid fate. J Exp Med. 2014;211:181–188. doi: 10.1084/jem.20131189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.