Abstract
Leptospirosis is a zoonotic disease usually acquired by contact with water contaminated with urine of infected animals. However, few molecular methods have been used to monitor or quantify pathogenic Leptospira in environmental water samples. Here we optimized a DNA extraction method for the quantification of leptospires using a previously described Taqman-based qPCR method targeting lipL32, a gene unique to and highly conserved in pathogenic Leptospira. QIAamp DNA mini, MO BIO PowerWater DNA and PowerSoil DNA Isolation kits were evaluated to extract DNA from sewage, pond, river and ultrapure water samples spiked with leptospires. Performance of each kit varied with sample type. Sample processing methods were further evaluated and optimized using the PowerSoil DNA kit due to its performance on turbid water samples and reproducibility. Centrifugation speeds, water volumes and use of Escherichia coli as a carrier were compared to improve DNA recovery. All matrices showed a strong linearity in a range of concentrations from 106 to 10° leptospires/mL and lower limits of detection ranging from <1 cell /ml for river water to 36 cells/mL for ultrapure water with E. coli as a carrier. In conclusion, we optimized a method to quantify pathogenic Leptospira in environmental waters (river, pond and sewage) which consists of the concentration of 40 mL samples by centrifugation at 15,000×g for 20 minutes at 4°C, followed by DNA extraction with the PowerSoil DNA Isolation kit. Although the method described herein needs to be validated in environmental studies, it potentially provides the opportunity for effective, timely and sensitive assessment of environmental leptospiral burden.
Introduction
Leptospirosis is a zoonotic disease caused by pathogenic spirochetes of the genus Leptospira [1,2]. Clinical symptoms range from mild flu-like infections to life-threatening manifestations such as Weil’s disease and pulmonary hemorrhage, the latter showing case fatality rates as high as 50% [3,4]. Leptospirosis has become particularly prevalent in poor urban and peri-urban communities from tropical developing countries [5–7]. In such locations, deficient sewer, drainage and refuse collection systems provide optimal ecological conditions for the thriving of synanthropic rodents such as Rattus rattus and R. norvergicus, which are natural reservoirs of pathogenic leptospires [8,9]. Human infection occurs predominantly by contact of abrasions or cuts in the skin or mucous membranes with water or soil contaminated with urine of rodents chronically infected with leptospires [2,10,11]. Large leptospirosis outbreaks are commonly associated to heavy rainfall and flooding events [12–17] and outdoor recreational activities involving water contact [18–22], highlighting its waterborne transmission.
Transmission dynamics and clinical progression are modulated by virulence characteristics of the infecting strain, by the host immune status and inherent susceptibility factors; and by the Leptospira inoculum size [2,23]. Thereby, the assessment and quantification of the leptospiral burden in environmental water samples would be valuable to guide public health interventions, and help expand the current knowledge about the leptospirosis zoonotic cycle and the spatial and temporal distribution of leptospires in the environment. Specifically, the availability of quantitative methods would allow the evaluation of the impact of interventions through the observation of decreasing environmental bacterial loads. Furthermore, the determination of the environmental burden may help to inform health authorities before adopting preventive measures such as closing recreational areas or access to rivers or other water sources during heavy rainfall events.
To date, culture isolation and animal inoculation have been the most used methods to detect leptospires in the environment. These methods are laborious and time-consuming and their lack of sensitivity may lead to false-negative results [24]. Inhibitors potentially present in complex environmental samples can hinder in vitro growth of leptospires and impair their isolation. Additionally, pathogenic leptospires are easily outgrown in culture by contaminating bacteria and saprophytic leptospires [1,11]. The use of molecular methods might overcome the limitations inherent to culture- and animal-based methods and provide quantitative information about the concentration of leptospires in contaminated water. A number of studies have successfully quantified pathogenic Leptospira in environmental waters (i.e. wells, streams, rivers, spring and standing water) using qPCR assays targeting the 16S rRNA [23,25] or lipL32 genes [26–28].
Different centrifugation or filtration methods and commercial DNA extraction kits were used, but only Vein et al., 2012 [27] compared the efficiency of the DNA extraction procedures. However, the comparison was based exclusively on the total quantity of DNA extracted and the linearity and lower limit of detection were not determined. In this study we aimed to optimize a method to concentrate and extract leptospiral DNA from environmental water samples to be quantified with a previously described Taqman-based qPCR method targeting the lipL32 gene of pathogenic Leptospira species [29]. This qPCR has been successfully applied in clinical samples [15] and animal tissues [30–32] showing high specificities. This procedure provides the opportunity for an effective and timely assessment of environmental leptospiral risk.
Materials and Methods
Water samples
Three water matrices were collected within the metropolitan area of Atlanta, GA, USA in March, 2010 and tested in this study. Raw sewage was provided by the staff of Snapfinger Creek Advanced Wastewater Treatment Facility (Decatur, GA, USA). Pond water was collected with the permission of the land owner from a pond located in Decatur, GA (N33°48’07” W84°18’26”) and river shoreline water was collected in the Chattahoochee River National Recreation Area in Roswell, GA (N33°59’17” W84°17’38”), with the permission of the Park Research Coordinator. The water collection procedures did not involve or affect any endangered and protected species. In all cases, an eight-liter batch was collected in a clean container and stored at 4°C until use. Ultrapure, distilled, nuclease-free water (Gibco BRL, Gaithersburg, USA) was used in all the experiments as an inhibitor-free control.
Bacterial strains and culture
L. interrogans serovar Copenhageni strain Fiocruz L1-130 (ATCC® BAA-1198) and Escherichia coli strain F2747 (provided by the Special Bacteriology Reference Laboratory, CDC) were used in the spiking experiments. L. interrogans was grown in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) [33,34] medium at 29°C for five days and counted using a Petroff-Hauser chamber under dark-field microscopy. E. coli was cultured in blood agar plates at 37°C for 24 hours and the concentration was estimated by suspending isolated colonies in sterile PBS (0.01 M, pH = 7.2) and correcting the density to 0.5 McFarland’s scale.
Selection of DNA extraction kit
Four aliquots of 40 mL of each water matrix were spiked with 1 × 105 leptospires/mL and centrifuged at 3,000 × g for 20 minutes at room temperature. The resuspended pellets were submitted to DNA extraction using either the QIAamp DNA mini kit (QIAGEN, Valencia, CA) or the PowerSoil DNA Isolation kit (MO BIO, Carlsbad, CA), following the manufacturers’ instructions. PowerWater DNA Isolation kit (MO BIO, Carlsbad, CA) was used to extract DNA from two additional, non-centrifuged aliquots of each water matrix. Samples were filtered through 11μm filter papers to remove large particulate debris and filtrates were passed through 0.22 μm membranes, which were aseptically removed and used for DNA extraction as per manufacturer’s recommendation. For consistency, the experiment was repeated three independent times in duplicates. For further experiments, DNA extraction was performed using PowerSoil DNA Isolation kit (MO BIO).
Optimization of the concentration protocol
To evaluate the efficiency of two centrifugation protocols, aliquots of 40 mL of each water matrix were spiked with 1 × 105 leptospires/mL and centrifuged using either protocol A (3,000 × g for 20 minutes at room temperature), or protocol B (15,000 × g for 20 minutes at 4°C). After careful removal of supernatants, the resuspended pellets were submitted to DNA extraction. As a control for loss of deposited matter due to supernatant removal, DNA was extracted from 200 μl of each spiked sample without a previous centrifugation step. To improve cell pelleting, 40 mL aliquots of ultrapure water spiked with 1 × 105 leptospires/mL were additionally spiked with varying amounts of E. coli (none, 1 × 105, 1 × 106 and 1 × 107/mL) as a carrier [35]. An ultrapure water sample spiked only with 1 × 107/mL E. coli was used as a negative control. All experiments were repeated three independent times in duplicates. Subsequent experiments were performed using the centrifugation procedure B.
Selection of the optimal sample volume
To determine the optimal sample volume, aliquots of 40, 80, 200 and 400 mL of each water matrix were spiked with 1 × 105 leptospires/mL and 1 × 107 E. coli/mL (except for sewage), centrifuged using the centrifugation procedure B, and pellets were submitted to DNA extraction. The experiment was repeated three independent times in duplicates. Subsequent experiments were conducted using 40 mL of water.
Linearity of extraction and determination of lower limit of detection (LLOD)
To compare the extraction linearity over a range of concentrations and determine the LLOD for each water matrix, 40 mL of each sample were spiked with a range of concentrations from 1 × 106 leptospires/mL to 1 × 10° leptospires/mL. All samples were also spiked with 1 × 107 E. coli/mL, with the exception of sewage. For comparison purposes, an additional group of ultrapure water samples was spiked with Leptospira but no E. coli. All samples were centrifuged and submitted to DNA extraction as described above. All experiments were repeated three independent times in duplicates.
Quantitative real-time PCR
DNA extracts were tested in triplicate by qPCR using previously described oligonucleotides [29]. The reaction mix consisted of 12.5 μl of Platinum Quantitative PCR SuperMix-UDG (Invitrogen, Carlsbad, CA), 500 nM of forward and reverse primers, 100 nM of probe, 5 μl of DNA extract and ultrapure water to a final volume of 25 μl. Amplification was performed on a ABI 7500 Real-Time PCR System (Applied Biosystems, Foster, CA) using standard conditions for 45 cycles. All samples with quantification cycles lower than 40 were considered positive. Genomic DNA obtained from L. interrogans serovar Copenhageni strain Fiocruz L1-130 was used to construct a standard curve with concentrations ranging from 1 × 107 to 1 × 10° GEq/5 μl, based on its genome size of 4,627 Mb [36]. Non-template controls were included after every five samples. All water matrices were proven negative for the presence of pathogenic Leptospira by qPCR before being used in the experiments. No amplification signal was detected when water samples spiked exclusively with E. coli were tested by qPCR.
Statistical analysis
To facilitate the comparison among experiments performed with different sample volumes, the recovery of leptospiral DNA was expressed in GEq/mL in all cases. All results were log10-transformed prior to further statistical analysis. Student’s unpaired t test was used to assess the difference between the means when two groups were compared and Tukey-Kramer post hoc test was used to correct for multiple comparisons. A linear regression model was used to evaluate the DNA extraction linearity over a range of concentrations and the slopes and y-intercepts of best-fit lines were compared by analysis of covariance. Probit regression analysis was used to estimate the LLOD targeting a 95% hit-rate (IBM SPSS software v.19, SPSS Inc., Chicago, IL). All the other statistical analysis were performed using GraphPad Prism 6.02 (GraphPad Software, San Diego California).
Results and Discussion
In this study we have optimized a method for the sample concentration and DNA extraction of environmental water matrices (river water, pond water and sewage) that may be used to monitor the leptospiral burden using a previously described qPCR method [29]. First, we evaluated the performance of three commercial kits: PowerWater DNA Isolation kit (MO BIO), QIAamp DNA Mini kit (QIAGEN) and PowerSoil DNA Isolation kit (MO BIO). PowerWater kit recovered more DNA than the others for river and ultrapure water samples (Table 1), but PowerSoil kit showed the highest efficiency when DNA was extracted from turbid samples (sewage and pond water) (p<0.0001). In addition, it presented the lowest DNA recovery variability among the four water types, being thus selected for further experiments. The underperformance of PowerWater kit for pond and sewage samples might be due to the clogging of the filter at the filtration step, which limits its ability to process large volumes of water. Furthermore, previous studies have reported the outperformance of MO BIO DNA extraction kits over QIAamp DNA mini kit for particulate-rich water samples [37–39]. The two MO BIO kits, specifically optimized to extract DNA from environmental samples, include a bead beating step that improves both bacterial desorption from sediments and cell disruption. DNA shearing caused by the bead beating process may lead to weaker interactions with sample particles than those that would be observed with intact DNA. Shorter, fragmented DNA would therefore be easier to desorb during DNA extraction [40]. In addition, MO BIO kits include a step for cationic flocculation of humic substances, which are removed prior to column loading. As a result, these chemical complexes do not compete with DNA for the silica binding sites in the column, leading to a higher final DNA recovery [41].
Table 1. Comparison of the efficiency of DNA extraction for three commercial kits from four water types spiked at 1 × 105 Leptospira/mL.
| mean log10 Leptospira GEq/mL ± SD† | |||
|---|---|---|---|
| Water matrix | PowerWater DNA Isolation Kit‡ | QIAamp DNA Mini Kit§ | PowerSoil DNA Isolation Kit§ |
| Ultrapure | 4.62 ± 0.30 | 3.27 ± 0.15 | 3.33 ± 0.02 |
| River | 4.65 ± 0.01 | 4.56 ± 0.09 | 3.71 ± 0.18 |
| Pond | 3.91 ± 0.13 | Not detected | 4.17 ± 0.09 |
| Sewage | 1.90 ± 0.62 | 0.68 ± 0.59 | 3.37 ± 0.02 |
†Data represent the mean results obtained from three independent experiments. Statistical analysis revealed that the difference in DNA extraction efficiency upon comparison of the three kits tested was significant and was observed for all the four water matrices (P<0.0001).
‡DNA was extracted from 40 mL aliquots, without a centrifugation step.
§Forty mL aliquots were centrifuged at 3,000×g for 20 minutes at room temperature. The respective pellets were used for DNA extraction.
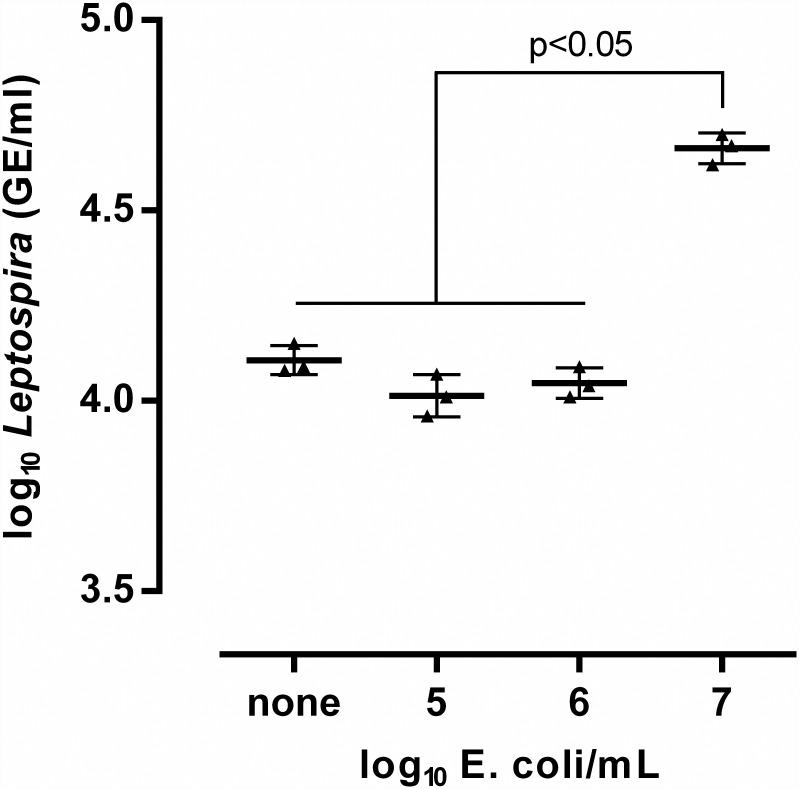
Two different centrifugation protocols (A: 3,000 ×g for 20 minutes at room temperature, and B: 15,000×g for 20 minutes at 4°C) were compared using the MO BIO PowerSoil DNA Isolation kit. These two centrifugation speeds have been used in previous studies [23,27,26], although the specific effect of the speed in the recovery of DNA has not been determined. The difference between mean leptospiral DNA quantities obtained with both centrifugation protocols was not statistically significant, except for river water in which more DNA was recovered using protocol B (p = 0.0005). Both protocols yielded lower recoveries than those obtained from the aliquots which were not centrifuged (p < 0.05) (Table 2). Despite these lower recoveries, the centrifugation protocol B was chosen for further tests because it allowed the concentration of at least 200 times more sample volume than the direct extraction (40 ml and 200 μL, respectively). Since the concentration of pathogenic leptospires in environmental water is presumably low (~103 leptospires/ml even in high-risk endemic areas [23]), larger volumes are required to provide adequate load estimations. In parallel, we also evaluated whether the addition of different amounts of E. coli as a carrier would improve leptospiral DNA recovery from ultrapure water. The addition of 1 × 107 E. coli/mL to ultrapure water significantly improved the recovery of leptospiral DNA (p < 0.05), but had no effect with smaller concentrations (Fig 1). Thus, E. coli can act as a carrier in clear water and co-sediment with leptospires to generate larger and more visible pellets less prone to be washed away upon supernatant removal. Consequently, 1 × 107 E. coli/mL was added to water aliquots used in the subsequent tests, except for sewage. We did not add E. coli to sewage because it naturally contains large amounts E. coli and other enteric bacteria, which already produce large visible pellets.
Table 2. Effect of different centrifugation protocols on the recovery of leptospiral DNA from four water samples spiked at 1 × 105 Leptospira/mL.
| mean log10 Leptospira GEq/mL ± SD | ||||
|---|---|---|---|---|
| Water matrix | No centrifugation† | Centrifugation Protocol | ||
| A‡ | B§ | P value | ||
| Ultrapure | 4.92±0.15 | 3.34±0.17 | 3.42±0.09 | 0.5360 |
| River | 4.39±0.02 | 3.35±0.01 | 3.77±0.07 | 0.0005 |
| Pond | 4.50±0.01 | 4.20±0.09 | 4.18±0.08 | 0.7950 |
| Sewage | 4.25±0.12 | 3.58±0.07 | 3.44±0.05 | 0.0512 |
†DNA was extracted from 200μL aliquots, without a centrifugation step.
‡Forty mL aliquots were centrifuged at 3,000×g for 20 minutes at room temperature. The respective pellets were used for DNA extraction.
§ Forty mL aliquots were centrifuged at 15,000×g for 20 minutes at 4°C. The respective pellets were used for DNA extraction
Fig 1. Effect of the addition of varying amounts of E. coli on Leptospira recovery from ultrapure water.
Aliquots were spiked with 1 × 105 Leptospira/mL and with 1 × 105, 1 × 106 and 1 × 107 E. coli/mL. Ultrapure water spiked with 1 × 105 Leptospira/mL was used as negative control. Error bars represent the geometric mean ± SD of the concentrations as determined by qPCR in three independent experiments.
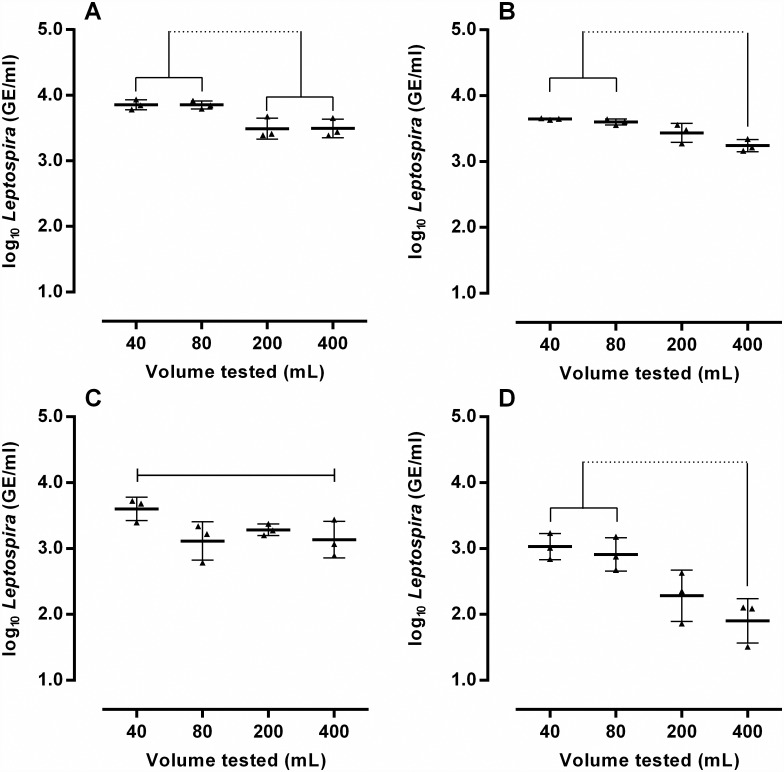
We then compared the efficiency of DNA extraction using different sample volumes (40, 80, 200 and 400 mL) to determine the best initial volume for Leptospira quantification in environmental waters. For all samples except pond water, qPCR quantification from 40 mL and 80 mL was not statistically different (Fig 2). However, leptospiral quantification obtained from 40 mL aliquots was significantly higher than those obtained from 200 mL and 400 mL aliquots for the four samples. For all the water matrices, an increase in sample volume led to a decrease in leptospiral DNA quantification (Fig 2). This observation may be due to a higher loss of bacteria during centrifugation with larger sample volumes or a saturation of the binding capacity of the DNA extraction kit. Alternatively, although the PowerSoil DNA Isolation kit had been designed to remove inhibitory substances from environmental samples, we cannot rule out the possibility that the amount of inhibitors contained in large water aliquots surpassed the kit’s removal capacity, particularly in sewage. A limitation of our study is the lack of an internal control to monitor the efficiency of DNA extraction and PCR amplification. Since it is not possible to precisely predict the amount of inhibiting compounds in environmental samples, an internal control would be useful to correlate the presence of inhibitors with DNA amplification efficiency [37].
Fig 2. Determination of the optimal volume to be tested by qPCR for different water types: ultrapure water (A), river (B); pond (C) and sewage (D).
Different volumes of each water type were spiked with 1 × 105 leptospires/mL to assess Leptospira recovery. Error bars represent the geometric mean ± SD of the concentrations as determined by qPCR in three independent experiments. Continuous lines connect groups whose average leptospiral DNA concentrations are not statistically significant (p>0.05). Groups whose averages are significantly different (p<0.05) are connected by a dashed line.
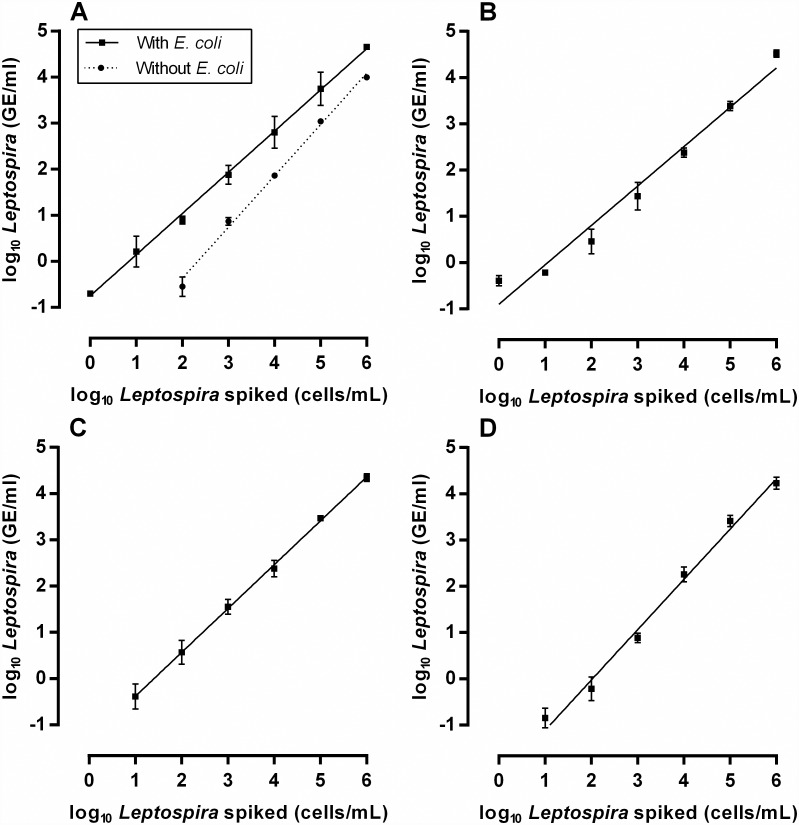
Finally, we determined the range of linearity and the LLOD of the method by spiking 40mL of each water matrix with 1 × 106 leptospires/mL to 1 × 10° leptospires/mL. As established in the previous experiment, 1 × 107 E. coli/mL were added to all samples, except for sewage. We observed a strong linear relationship between the concentration of Leptospira spiked and the concentration determined by qPCR after the extraction procedure (R2 > 0.97 in all cases) (Table 3 and Fig 3). Analysis of covariance of the slope and y-intercept of the best-fit lines calculated for each of the five samples tested showed that all the lines were significantly different between them (p < 0.0001). The 95% hit-rate LLODs were estimated at 36 cells/mL for ultrapure water with E. coli, 4,833 cells/mL for ultrapure water without E. coli, 11 cells/mL for pond water and 18 cells/mL for sewage water. It was not possible to calculate the 95% hit-rate LLOD for river water since all the replicates tested were positive, which indicated that it was lower than 1 cell/mL. Altogether, the strong linearity and low LLOD in all the water matrices indicate that the optimized DNA extraction method is suitable for the detection of a wide range of Leptospira concentrations in environmental waters.
Table 3. Lower limit of detection and best-fit lines calculated from four water types spiked with leptospires*.
| Best-fit line† | ||||
|---|---|---|---|---|
| Water matrix | LLOD (cells/mL)‡ | R2 | Slope ± SD | Y-intercept ± SD |
| Ultrapure (with E. coli) | 36 | 0.98 | 0.89 ± 0.027 | -0.75 ± 0.101 |
| Ultrapure (without E. coli) | 4,833 | 0.99 | 1.11 ± 0.026 | -2.60 ± 0.114 |
| River | <1 | 0.97 | 0.85 ± 0.036 | -0.90 ± 0.131 |
| Pond | 11 | 0.99 | 0.95 ± 0.023 | -1.33 ± 0.091 |
| Sewage | 18 | 0.99 | 1.09 ± 0.034 | -2.20 ± 0.137 |
* In three different experiments, water samples were spiked with 1 × 106 Leptospira/mL and 10-fold serially diluted down to 1 × 10° Leptospira/mL
†Calculated from linear regression of data obtained from 3 independent experiments.
‡As determined by Probit regression analysis.
Fig 3. Determination of the lower limit of detection for water samples obtained ultrapure water (A), river (B), pond (C) and sewage (D).
Ultrapure water (A) was tested with and without the addition of 1 × 107 E. coli/mL. All the samples were spiked with 106 Leptospira/mL and 10-fold serially diluted down to 1 Leptospira/mL. Error bars represent the geometric mean ± SD of three independent experiments.
In summary, we optimized a method to quantify pathogenic Leptospira in environmental waters (river, pond and sewage). The process includes concentration of 40 mL samples by centrifugation at 15,000×g for 20 minutes at 4°C, followed by extraction with the PowerSoil DNA Isolation kit (MO BIO). Although the optimized method needs to be further validated in environmental studies, it is a promising tool for the environmental monitoring of pathogenic Leptospira that may help to inform public health interventions aimed to reduce the burden of the disease.
Acknowledgments
The authors thank Jay Ash from the Snapfinger Creek Advanced Wastewater Treatment Facility and the Chattahoochee River National Recreation Area for providing water samples; Dr. David Cox of the Syphilis Laboratory Reference and Research Branch (CDC) for technical advice; and Jean Jordan and Dr. John McQuiston from the Special Bacterial Reference Laboratory (CDC) for providing the E. coli strain.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by funding from the National Institute of Allergy and Infectious Diseases (www.niaid.nih.gov) (award numbers R01 AI052473, U01 AI088752, R25 TW009338, R01 TW009504 and R01 AI121207 to AIK). INR was supported by a sandwich Ph.D. scholarship (BEX 066509-6) from CAPES (Brazilian Ministry of Education). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Faine S, Adler B, Bolin C, Perolat P. Leptospira and Leptospirosis. 1st ed Melbourne: MedScience; 1999. [Google Scholar]
- 2.Ko AI, Goarant C, Picardeau M (2009) Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol 7: 736–747. 10.1038/nrmicro2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, et al. (2003) Leptospirosis: A zoonotic disease of global importance. Lancet Infect Dis 3: 757–771. [DOI] [PubMed] [Google Scholar]
- 4.McBride AJ, Athanazio DA, Reis MG, Ko AI (2005) Leptospirosis. Curr Opin Infect Dis 18: 376–386. [DOI] [PubMed] [Google Scholar]
- 5.Ko AI, Reis MG, Dourado CMR, Johnson WD Jr, Riley LW (1999) Urban epidemic of severe leptospirosis in Brazil. Lancet 354: 820–825. [DOI] [PubMed] [Google Scholar]
- 6.Maciel EAP, de Carvalho ALF, Nascimento SF, de Matos RB, Gouveia EL, Reis MG, et al. (2008) Household transmission of Leptospira infection in urban slum communities. PLoS Negl Trop Dis 2: e154 10.1371/journal.pntd.0000154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reis RB, Ribeiro GS, Felzemburgh RDM, Santana FS, Mohr S, Melendez AXTO, et al. (2008) Impact of environment and social gradient on Leptospira infection in urban slums. PLoS Negl Trop Dis 2: e228 10.1371/journal.pntd.0000228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa F, Porter FH, Rodrigues G, Farias H, de Faria MT, Wunder EA, et al. (2014) Infections by Leptospira interrogans, Seoul virus, and Bartonella spp. among Norway rats (Rattus norvegicus) from the urban slum environment in Brazil. Vector Borne Zoonotic Dis 14: 33–40. 10.1089/vbz.2013.1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felzemburgh RDM, Ribeiro GS, Costa F, Reis RB, Hagan JE, Melendez AXTO, et al. (2014) Prospective study of leptospirosis transmission in an urban slum community: role of poor environment in repeated exposures to the leptospira agent. PLoS Negl Trop Dis 8: e2927 10.1371/journal.pntd.0002927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa F, Wunder EA, De Oliveira D, Bisht V, Rodrigues G, Reis MG, et al. (2015) Patterns in Leptospira Shedding in Norway Rats (Rattus norvegicus) from Brazilian Slum Communities at High Risk of Disease Transmission. PLoS Negl Trop Dis 9: e0003819 10.1371/journal.pntd.0003819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levett PN (2001) Leptospirosis. Clin Microbiol Rev 14: 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goarant C, Laumond-Barny S, Perez J, Vernel-Pauillac F, Chanteau S, Guigon A. (2009) Outbreak of leptospirosis in New Caledonia: diagnosis issues and burden of disease. Trop Med Int Health 14: 926–929. 10.1111/j.1365-3156.2009.02310.x [DOI] [PubMed] [Google Scholar]
- 13.Desvars A, Jégo S, Chiroleu F, Bourhy P, Cardinale E, Michault, A. (2011) Seasonality of human leptospirosis in Reunion Island (Indian Ocean) and its association with meteorological data. PLoS One 6: e20377 10.1371/journal.pone.0020377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tangkanakul W, Smits HL, Jatanasen S, Ashford DA (2005) Leptospirosis: an emerging health problem in Thailand. Southeast Asian J Trop Med Public Health 36: 281–288. [PubMed] [Google Scholar]
- 15.Weinberger D, Baroux N, Grangeon J-P, Ko AI, Goarant C (2014) El Niño Southern Oscillation and leptospirosis outbreaks in New Caledonia. PLoS Negl Trop Dis 8: e2798 10.1371/journal.pntd.0002798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amilasan AST, Ujiie M, Suzuki M, Salva E, Belo MCP, Koizumi N, et al. (2012) Outbreak of leptospirosis after flood, the Philippines, 2009. Emerg Infect Dis 18: 91–94. 10.3201/eid1801.101892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith JKG, Young MM, Wilson KL, Craig SB (2013) Leptospirosis following a major flood in Central Queensland, Australia. Epidemiol Infect 141: 585–590. 10.1017/S0950268812001021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monahan AM, Miller IS, Nally JE (2009) Leptospirosis: risks during recreational activities. J Appl Microbiol 107: 707–716. 10.1111/j.1365-2672.2009.04220.x [DOI] [PubMed] [Google Scholar]
- 19.Morgan J, Bornstein SL, Karpati AM, Bruce M, Bolin CA, Austin, CC, et al. (2002) Outbreak of leptospirosis among triathlon participants and community residents in Springfield, Illinois, 1998. Clin Infect Dis 34: 1593–1599. [DOI] [PubMed] [Google Scholar]
- 20.Sejvar JJ, Bancroft E, Winthrop K, Bettinger J, Bajani M, Bragg S, et al. (2003) Leptospirosis in “Eco-Challenge” athletes, Malaysian Borneo, 2000. Emerg Infect Dis 9: 702–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stern EJ, Galloway R, Shadomy SV, Wannemuehler K, Atrubin D, Blackmore C, et al. (2010) Outbreak of leptospirosis among adventure race participants in Florida, 2005. Clin Infect Dis 50: 843–849. 10.1086/650578 [DOI] [PubMed] [Google Scholar]
- 22.Agampodi SB, Karunarathna D, Jayathilala N, Rathnayaka H, Agampodi TC, Karunanayaka L. (2014) Outbreak of leptospirosis after white-water rafting: sign of a shift from rural to recreational leptospirosis in Sri Lanka? Epidemiol Infect 142: 843–846. 10.1017/S0950268813001465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganoza CA, Matthias MA, Collins-Richards D, Brouwer KC, Cunningham CB, Segura ER, et al. (2006) Determining risk for severe leptospirosis by molecular analysis of environmental surface waters for pathogenic Leptospira. PLoS Med 3: 1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wuthiekanun V, Chierakul W, Limmathurotsakul D, Smythe LD, Symonds ML, Dohnt MF, et al. (2007) Optimization of culture of Leptospira from humans with leptospirosis. J Clin Microbiol 45: 1363–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viau EJ, Boehm AB (2011) Quantitative PCR-based detection of pathogenic Leptospira in Hawai’ian coastal streams. J Water Health 9: 637–646. 10.2166/wh.2011.064 [DOI] [PubMed] [Google Scholar]
- 26.Rawlins J, Portanova A, Zuckerman I, Loftis A, Ceccato P, Willingham AL, et al. (2014) Molecular detection of leptospiral DNA in environmental water on St. Kitts. Int J Environ Res Public Health 11: 7953–7960. 10.3390/ijerph110807953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vein J., Perrin A., Berny PJ., Benoit E., Leblond A., Kodjo A. (2012) Adaptation of a real-time PCR method for the detection and quantification of pathogenic leptospires in environmental water. Can J Microbiol 58: 828–835. 10.1139/w2012-060 [DOI] [PubMed] [Google Scholar]
- 28.Muñoz-Zanzi C, Mason MR, Encina C, Astroza A, Romero A (2014) Leptospira contamination in household and environmental water in rural communities in southern Chile. Int J Environ Res Public Health 11: 6666–6680. 10.3390/ijerph110706666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR (2009) Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis 64: 247–255. 10.1016/j.diagmicrobio.2009.03.014 [DOI] [PubMed] [Google Scholar]
- 30.Muñoz-Zanzi C, Mason M, Encina C, Gonzalez M, Berg S (2014) Household characteristics associated with rodent presence and Leptospira infection in rural and urban communities from Southern Chile. Am J Trop Med Hyg 90: 497–506. 10.4269/ajtmh.13-0334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cosson JF, Picardeau M, Mielcarek M, Tatard C, Chaval Y, Suputtamongkol Y, et al. (2014) Epidemiology of Leptospira Transmitted by Rodents in Southeast Asia. PLoS Negl Trop Dis 8: e2902 10.1371/journal.pntd.0002902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer-Scholl A, Hammerl J, Schmidt S, Ulrich R, Pfeffer M, Woll D, et al. (2014) Leptospira spp. in Rodents and Shrews in Germany. Int J Environ Res Public Health 11: 7562–7574. 10.3390/ijerph110807562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson RC, Harris VG (1967) Differentiation of pathogenic and saprophytic letospires. I. Growth at low temperatures. J Bacteriol 94: 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellinghausen HC, McCullough WG (1965) Nutrition of Leptospira Pomona and growth of 13 other serotypes: Fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am J Vet Res 26: 45–51. [PubMed] [Google Scholar]
- 35.Elväng AM, Westerberg K, Jernberg C, Jansson JK (2001) Use of green fluorescent protein and luciferase biomarkers to monitor survival and activity of Arthrobacter chlorophenolicus A6 cells during degradation of 4-chlorophenol in soil. Environ Microbiol 3: 32–42. [DOI] [PubMed] [Google Scholar]
- 36.Nascimento ALTO, Ko AI, Martins EAL, Monteiro-Vitorello CB, Ho PL, Haake DA, et al. (2004) Comparative Genomics of Two Leptospira interrogans Serovars Reveals Novel Insights into Physiology and Pathogenesis. J Bacteriol 186: 2164–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behets J, Declerck P, Delaedt Y, Creemers B, Ollevier F (2007) Development and evaluation of a Taqman duplex real-time PCR quantification method for reliable enumeration of Legionella pneumophila in water samples. J Microbiol Methods 68: 137–144. [DOI] [PubMed] [Google Scholar]
- 38.Dauphin LA, Moser BD, Bowen MD (2009) Evaluation of five commercial nucleic acid extraction kits for their ability to inactivate Bacillus anthracis spores and comparison of DNA yields from spores and spiked environmental samples. J Microbiol Methods 76: 30–37. 10.1016/j.mimet.2008.09.004 [DOI] [PubMed] [Google Scholar]
- 39.Whitehouse CA, Hottel HE (2007) Comparison of five commercial DNA extraction kits for the recovery of Francisella tularensis DNA from spiked soil samples. Mol Cell Probes 21: 92–96. [DOI] [PubMed] [Google Scholar]
- 40.Bonot S, Courtois S, Block J-C, Merlin C (2010) Improving the recovery of qPCR-grade DNA from sludge and sediment. Appl Microbiol Biotechnol 87: 2303–2311. 10.1007/s00253-010-2686-0 [DOI] [PubMed] [Google Scholar]
- 41.Lloyd KG, Macgregor BJ, Teske A (2010) Quantitative PCR methods for RNA and DNA in marine sediments: maximizing yield while overcoming inhibition. FEMS Microbiol Ecol 72: 143–151. 10.1111/j.1574-6941.2009.00827.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.