Abstract
The abscisic acid (ABA)-, stress-, and ripening-induced (ASR) protein is a plant-specific hydrophilic transcriptional factor involved in fruit ripening and the abiotic stress response. To date, there have been no studies on the role of ASR genes in delayed flowering time. Here, we found that the ASR from banana, designated as MaASR, was preferentially expressed in the banana female flowers from the eighth, fourth, and first cluster of the inflorescence. MaASR transgenic lines (L14 and L38) had a clear delayed-flowering phenotype. The number of rosette leaves, sepals, and pedicel trichomes in L14 and L38 was greater than in the wild type (WT) under long day (LD) conditions. The period of buds, mid-flowers, and full bloom of L14 and L38 appeared later than the WT. cDNA microarray and quantitative real-time PCR (qRT-PCR) analyses revealed that overexpression of MaASR delays flowering through reduced expression of several genes, including photoperiod pathway genes, vernalization pathway genes, gibberellic acid pathway genes, and floral integrator genes, under short days (SD) for 28 d (from vegetative to reproductive transition stage); however, the expression of the autonomous pathway genes was not affected. This study provides the first evidence of a role for ASR genes in delayed flowering time in plants.
Introduction
Flowering time is crucial for pollination and reproductive success in higher plants [1, 2], which is regulated through four major pathways, the photoperiod-, vernalization-, autonomous-, and gibberellic acid (GA)-dependent pathways, in Arabidopsis [3–5]. Photoperiod, or the duration of light in a given day, is a critical cue that flowering plants utilize to effectively assess seasonal information and coordinate their reproductive development in synchrony with the external environment [6]; photoperiod thus controls flowering time by regulating the expression of a number of key genes, such as CONSTANS (CO), EARLY FLOWERING4 (ELF4), and EARLY FLOWERING3 (ELF3) [6, 7]. Flowering in some plants can be stimulated by exposure to long periods of low non-freezing temperatures, which is known as varnalization, and is regulated by the FRIGIDA (FRI) and FLOWERING LOCUS C (FLC) genes [4, 8]. However, the autonomous pathway associated with the GA pathway integrates developmental signals to regulate plant flowering time [9]. Recent studies have reported that floral regulatory pathways regulate the expression of floral integrator genes such as SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) and LEAFY (LFY) [10, 11]. In addition, a series of transcription factors including CO, FLC, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL), and CAPRICE (CPC) control flowering time by regulating the target genes expression of these pathways [6, 10, 12–14].
The ABA-, stress-, and ripening-induced (ASR) protein is a plant-specific hydrophilic transcriptional factor widely distributed to approximately 20 monocot, dicot, and gymnosperm plant species belonging to the group 7 of the LATE EMBRYONGENESIS ABUNDANT proteins [15, 16]. It is a small size protein (~13 kDa) localized to both the nucleus and the cytoplasm, and contains Zn2+-dependent DNA binding activity at the N-terminus and a nuclear localization signal at the C-terminus [17, 18]. The number of ASR orthologous genes varies from 1 to 9 in different plant species [18] but, surprisingly, orthologs have not been identified in Arabidopsis thaliana and crucifer Thlaspi caerulescens [16]. Several ASR orthologous and paralogous genes are involved in fruit ripening and in the response to various abiotic stresses, particulary salt and drought stress tolerance [15–18]. Increasing evidence has also indicated that ASRs are involved in the regulation of floral development [19, 20]. In lily, ASR orthologous proteins accumulate only at the later stage of pollen maturation and these levels remain steady in mature and vital pollen [21]. Tomato ASR1 and ASR4 are expressed in flower organs [19], and tobacco ASR in vivo binds to a transcription factor bZIP involved in floral development [20]. Overexpression of the ASR gene affects sugar trafficking, flower development, and fruit development [18, 20, 22]; however, the role of ASR in regulating plant flowering time has not been reported.
Banana (Musa spp.), the second ranking fruit crop in the world, has a large, dark purple-red inflorescence and produces female, male, and bisexual flowers. Bud differentiation and fruit yield are largely determined by female flowering time. Our previous studies showed that MaASR enhances drought stress tolerance [16]. In the present study, we found that the overexpression of MaASR in Arabidopsis could result in a clear delayed-flowering phenotype. Microarray and quantitative real-time PCR (qRT-PCR) results demonstrated that the expression of a number of key genes involved in the flowering regulator pathways, including photoperiod-, vernalization-, GA-pathways, and floral integrator, are down-regulated by MaASR overexpression to delay flowering time. This study has identified the role of ASR genes in delayed flowering time for the first time, and this finding may enable regulation of flowering time in plant breeding and a genetic improvement of plant yields.
Materials and Methods
Plant materials
Banana (M. acuminata L. AAA group, cv. ‘Dwarf Cavendish’) (ITC0002) inflorescence was obtained from a banana plantation (Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, Haikou, Hainan province, China). Roots, leaves, rhizomes, fruits, and female flowers from the tenth (F10), ninth (F9), eighth (F8), fourth (F4), and first (F1) cluster of the inflorescence were collected to analyze MaASR expression. All materials were separately frozen in liquid N2 and stored at -80°C until later analysis.
A. thaliana (Columbia ecotype) seeds were purchased from the Arabidopsis Biological Resource Center (Ohio University, Columbus, USA). DH5α Escherichia coli and LBA4404 Agrobacterium tumefaciens strains were provided by Professor Jiaming Zhang from the Chinese Academy of Tropical Agricultural Sciences. All Arabidopsis seeds were sown on a 1:1:8 mixture (by weight) of vermiculite, perlite, and peat moss, respectively, and were grown at 22°C with 70% humidity and short day condition (SD, 8 h light/16 h dark cycle) illuminated by Sylvania GRO LUX fluorescent lamps (Utrecht, Netherlands). When A. thaliana produced 12–14 rosette leaves, they were grown at 70% humidity and long day (LD) condition with 16 h light/8 h dark cycle to promote flowering.
Cloning, subcellular localization and expression analysis of MaASR
The full-length cDNA encoding MaASR was amplified with the primers MaASR-F and MaASR-R (S1 Table) based on the expressed sequence tag (EST) of MaASR isolated from a banana fruit cDNA library [23] with the adapter primers Ptr-F and Ptr-R (S1 Table). The MaASR full-length cDNA sequences were submitted to GenBank (http://www.ncbi.nlm.nih.gov/Banklt/index.html). Amino acid sequences were compared using the DNAMAN software package (Version 5.2.2, Canada).
The Open Reading Frame (ORF) of MaASR was inserted into a pCAMBIA1304-GFP expression vector to generate a MaASR-GFP fusion protein under the control of a cauliflower mosaic virus (CaMV) 35S promoter. The recombinant plasmid was transferred to the A. tumefaciens strain LBA4404 and introduced into Nicotiana benthamiana leaves as described previously by Goodin et al. [24]. After 48 h of incubation on MS at 25°C, fluorescence was examined using fluorescence microscopy (LSM700, Carl Zeiss, Germany).
MaASR expression was assayed by qRT-PCR in an iQ5 real-time PCR detection system (Bio-Rad, USA) using the SYBR ExScript RT-PCR kit (TaKaRa, Japan). A series of primer and template dilutions were performed to acquire the optimal primer and template concentrations for amplifying the target genes prior to quantification experiments. Primers that had high specificity and efficiency on the basis of melting curve analysis were used to conduct quantification analysis (S1 Table). Moreover, PCR products were sequenced to confirm the specificity of primer pairs. Amplification efficiencies of primer pairs ranged from 0.9 to 1.1. ACTIN (accession No. EF672732) and UBQ (accession No. XP009390884.1) that were verified to be constitutive expression and suitable to be used as internal controls were used as reference genes to normalize transcriptional levels of MaASR gene (S1 Table). The relative expression levels of MaASR gene were verified in triplicate and calculated using the 2−ΔΔCT method [25].
Plant transformation and blot analysis of transgenic plants
A MaASR coding region driven by a 35S promoter was inserted into the pBI121 vector by replacing the β-glucuronidase following digestion with BamHI and SacI. The pBI121-MaASR was transferred into an A. tumefaciens strain LBA4404. Transgenic Arabidopsis plants were generated using the floral dip-mediated infiltration method [26]. Homozygous T3 kanamycin-resistant lines L14 and L38 were used for blot analyses and functional investigation.
Southern blot was used to determine the integration of MaASR to the A. thaliana genome. Probes were prepared from the PCR product using primers (5′-ccgaggagaagcaccaccac-3′ and 5′-gccaccgctgcagcgatctcc tc-3′) and were labelled with DIG-dUTP according to the manufacturer’s instructions (Roche Applied Science, Germany). A Northern blot probe was labelled using a random primer labeling system (Roche Applied Science, Germany) and hybridized according to the manufacturer’s instructions (Roche Applied Science, Germany). Western blotting was performed using MaASR monoclonal antibodies (Abmart, China) diluted 1:500. After hybridization, the membrane was washed and exposed to X-ray film (Kodak BioMax MS system) according to Miao et al. [27].
Phenotype observation of MaASR transgenic plants
Rosette leaves number, bolting, and flowering time of MaASR transgenic lines L14 and L38 and WT were analyzed according to the methods of Diallo et al. [28]. The statistical analysis is listed in S2 Table. Vegetative growth, bolting, and flowering phenotype of the transgenic lines and WT were photographed. Floral organs phenotypic differences of early flowering between transgenic lines and WT under LD condition were observed using a dissecting microscope (OLMPUS-SZX12).
Total RNA extraction and cDNA synthesis from MaASR transgenic lines and WT
RNAs were isolated from MaASR transgenic plants L14 and L38 and WT at 14 d and 28 d under the SD, 14 d and 28 d under the LD condition, respectively, using a plant RNA Kit (QIAGEN, Germany). The first strand of cDNA was synthesized using a RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Ontario) according to the manufacturer’s instructions. RNA quality was assessed by the fractionation of total RNA on a 1.2% (w/v) agarose gel and imaged using the GelDox XR system (Bio-Rad, USA). All RNA samples prepared (A260/A280 ratio = 1.8~2.0; rRNA ratio (28S/18S)>0.9) were suitable for microarray and expression analysis.
Microarray profiling and data analysis
cDNAs prepared from MaASR transgenic lines L14 and WT were used for microarray analysis. Each sample included three biological replicates for L14 (designed as L14-1, L14-2, and L14-3) and WT (designed as WT1, WT2, and WT3). Labeling and hybridization was performed using the 29 k Arabidopsis Genome Array (Arabidopsis thaliana Genome Oligo Set Version3.0, http://www.operon.com) according to the procedure described by Patterson et al. [29]. Controls were also printed on glass slides using a SmartArray microarrayer (CapitalBio Corp.). The resulting images were analyzed with LuxScanTM3.0 software (CapitalBio Corp.) and identified using the methods of Miao et al. [30]. DEGs were identified using a P value <0.05, false discovery rate (FDR) <0.05, and a fold change ≥2.0.
qRT-PCR analysis of flowering-related pathway genes
Primers that had high specificity and efficiency on the basis of melting curve analysis and agarose gel electrophoresis were designed with Primer premier 5.0 software (http://www.premierbiosoft.com/) and used to conduct quantification analysis (S1 Fig). Primer sequences of AtFCA, AtFLK, AtFRI, AtGAI, AtLFY, AtRGL1, AtVRN1, AtFLC, AtFVE, AtSOC1, AtCol1, AtCol2, AtNAP, AtTCH2, AtSEP3, AtCol9, AtCO, AtELF3, AtELF4, AtNGA1, and AtMAF5 are listed in S1 Table. Amplification efficiencies of primer pairs ranged from 0.9 to 1.1. The AtACTIN and AtUBQ [31] that were verified to be constitutively expressed and suitable for use as internal controls were as reference genes to normalize transcriptional levels of target genes in this study (S1 Table). The relative expression of the tested genes with three replicates of each sample was assessed according to the 2−ΔΔCT method [25].
Statistical analysis
For all generated data, at least three biological replicates were performed for each sample. Then, one-way ANOVA and Duncan’s multiple range tests were performed at a 5% significance level (P values <0.05) using SPSS software (version 13.0). The statistical results were reported as mean±SD.
Results
Sequence analysis, subcellular localization and expression pattern of MaASR in banana female flowers
The cDNA of ASR from banana, MaASR, was 747 bp in length containing a 432 bp open reading frame (ORF), which encoded a protein of 143 amino acids with a 251 bp 5′ untranslated region (UTR) and a 64 bp 3′ UTR. The sequences of the cloned MaASR were registered in GenBank under the accession No. AY628102. Compared to the other ASR amino acid sequences, MaASR contained a small N-terminal DNA binding site (HHHRLFHH) and a longer putative nuclear C-terminal localization signal (KRDAKNEAEEASGKKHHHHL) (S2 Fig). MaASR protein was localized in the nucleus and plasma membrane (S3 Fig).
MaASR expression was detected in roots, leaves, rhizomes, female flowers (the first cluster from the upper inflorescence), and fruits. Female flowers showed the highest levels, along with roots; the lowest level was found in rhizomes. The level of MaASR expression in flowers was approximately 12-fold higher than that in rhizomes (S4 Fig). Significant differences in MaASR expression were detected in the female flowers from the tenth (F10), ninth (F9), eighth (F8), fourth (F4), and first (F1) cluster from the upper inflorescence (Fig 1A). At F10, the MaASR gene began to express and reached a maximum at F1, which was approximately 20-fold higher than that at F10 and F9 (Fig 1B), suggesting that expression of MaASR could play a role in banana female flower development.
Fig 1. Expression of MaASR gene in banana female flowers from the upper inflorescence.

(A) The female flowers from the tenth (F10), ninth (F9), eighth (F8), fourth (F4), and first (F1) cluster from the upper inflorescence. (B) Relative expression level in banana female flowers. The y-axis represents the relative fold-difference in mRNA level, which was calculated using the 2-ΔΔCT formula with ACTIN and UBQ as internal controls. The vertical bars represent the mean ± SD of three replicates.
MaASR overexpression caused a clear delayed flowering phenotype
Floral organs of two transgenic lines, L14 and L38, were significantly shorter (0.67-fold) than that of the WT (Fig 2A). The number of sepal and pedicel trichomes in the L14 line was greater than that of the L38 line. These trichomes are rarely present in the sepal and pedicel of WT, indicating that floral organ morphological changes in transgenic lines are relavant to the overexpression of MaASR. Southern blot showed that L14 and L38 harbored two and one copies of MaASR, respectively (S5 Fig). Northern and Western blots confirmed that MaASR transcripts were present in the two transgenic lines compared to WT in which expression was absent (S5 Fig).
Fig 2. Flowering phenotypes of MaASR transgenic plants.
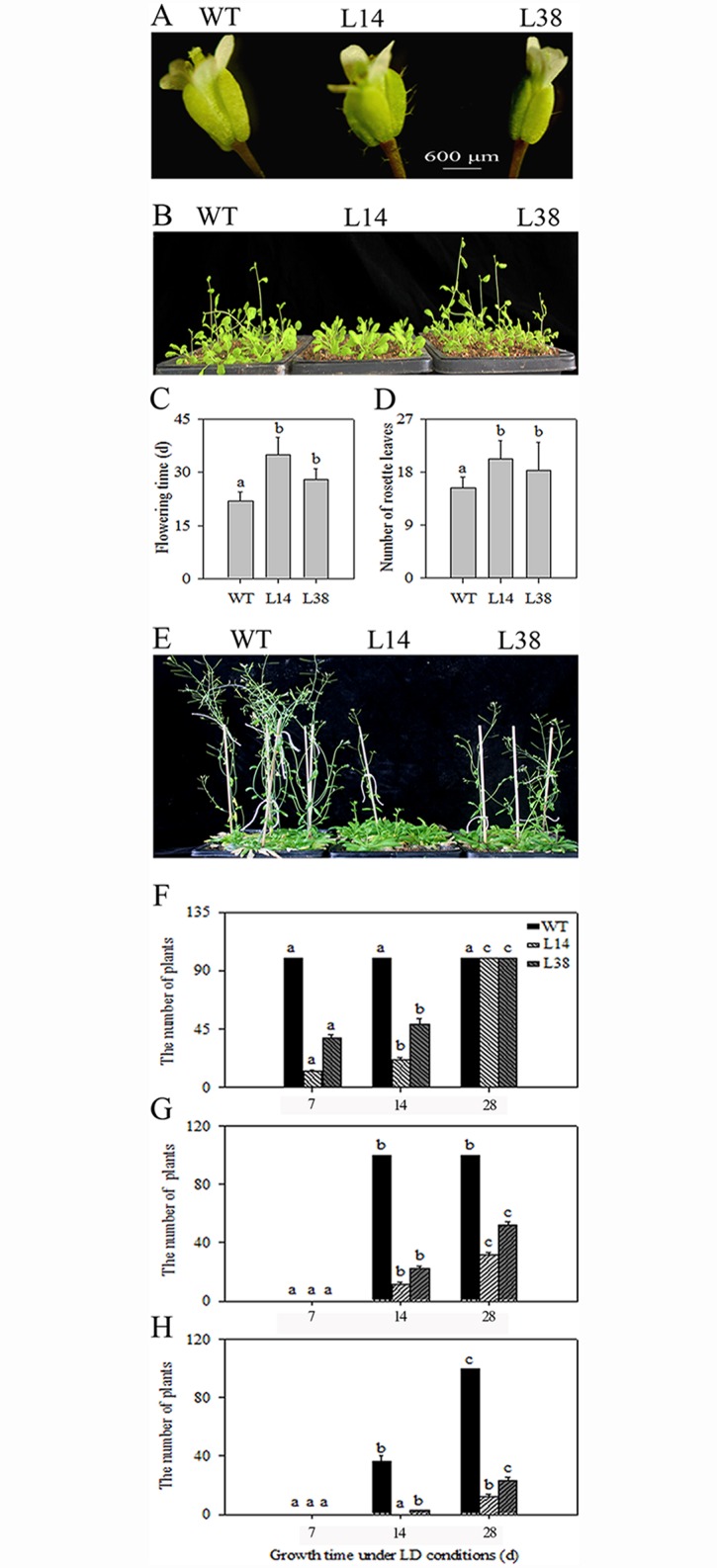
(A) Phenotype of floral organs detached from the same position and developmental stage of 14 d under LD in WT and transgenic plants. (B) Plants at 14 d under LD. (C) Flowering days of MaASR transgenic lines L14 and L38. (D) The number of rosette leaves of MaASR transgenic lines L14 and L38. (E) Plants at 28 d under LD. (F) The number of plants at flower buds stage. (G) The number of plants at mid-flower stage. (H) The number of plants at full-bloom stage.
Under 7 d of LD conditions, most WT plants, as well as the MaASR transgenic line L38, displayed bolting but the L14 line did not (Fig 2B). The number of days required for bolting was 22, 35, and 28 d in WT, L14, and L38, respectively (Fig 2C). The average number of rosette leaves in the WT, L14, and L38 lines was 15, 20, and 18, respectively (Fig 2D). These results showed that the number of rosette leaves produced by the MaASR transgenic lines was significantly greater than those produced by WT.
Under 28 d of LD conditions, WT reached the full-bloom stage and pods were observed; however, the bolting number of the L14 and L38 lines was significantly lower than that of WT (Fig 2E). Statistical analyses showed that flower buds were formed in WT under LD conditions for 7 d. The L14 and L38 lines only formed 12.76% and 38.33% of flower buds, respectively, until 28 d, at which point the flower buds of the transgenic lines were fully formed (Fig 2F). Under LD conditions for 14 d, all flower buds from WT were at mid-flower stage; the number of flower buds at the mid-flower stage in lines L14 and L38 was only 11.17% and 22.24%, respectively (Fig 2G). By 28 d, WT reached full bloom, but the flower number in lines L14 and L38 was only 12.21% and 23.50%, respectively (Fig 2H). These data demonstrate that MaASR transgenic lines have a significantly delayed flowering phenotype with respect to flower buds at mid-flower and full bloom stages.
Microarray analysis and screening of the differential expressed genes (DEGs) between MaASR transgenic plants and WT
Microarray analysis was used to determine the DEGs affected by MaASR overexpression compared to the WT. Each sample included three biological replicates for L14 (designed as L14-1, L14-2, and L14-3) and WT (designed as WT1, WT2, and WT3). TreeView representation of L14-1-vs-WT1, L14-2-vs-WT2, and L14-1-vs-WT3 libraries is shown in Fig 3A. Based on a fold change ≥2.0 and P value < 0.05, a total of 747 DEGs were identified, including 559 up-regulated genes (S3 Table) and 188 down-regulated (S4 Table) genes in L14 vs WT. All DEGs were mapped to the Gene Ontology (GO) database with respect to biological processes, molecular functions, and cellular components (S6 Fig).
Fig 3. TreeView representation of ESTs from microarray data (L14 vs WT) and functional classification of flowering-related candidate genes.
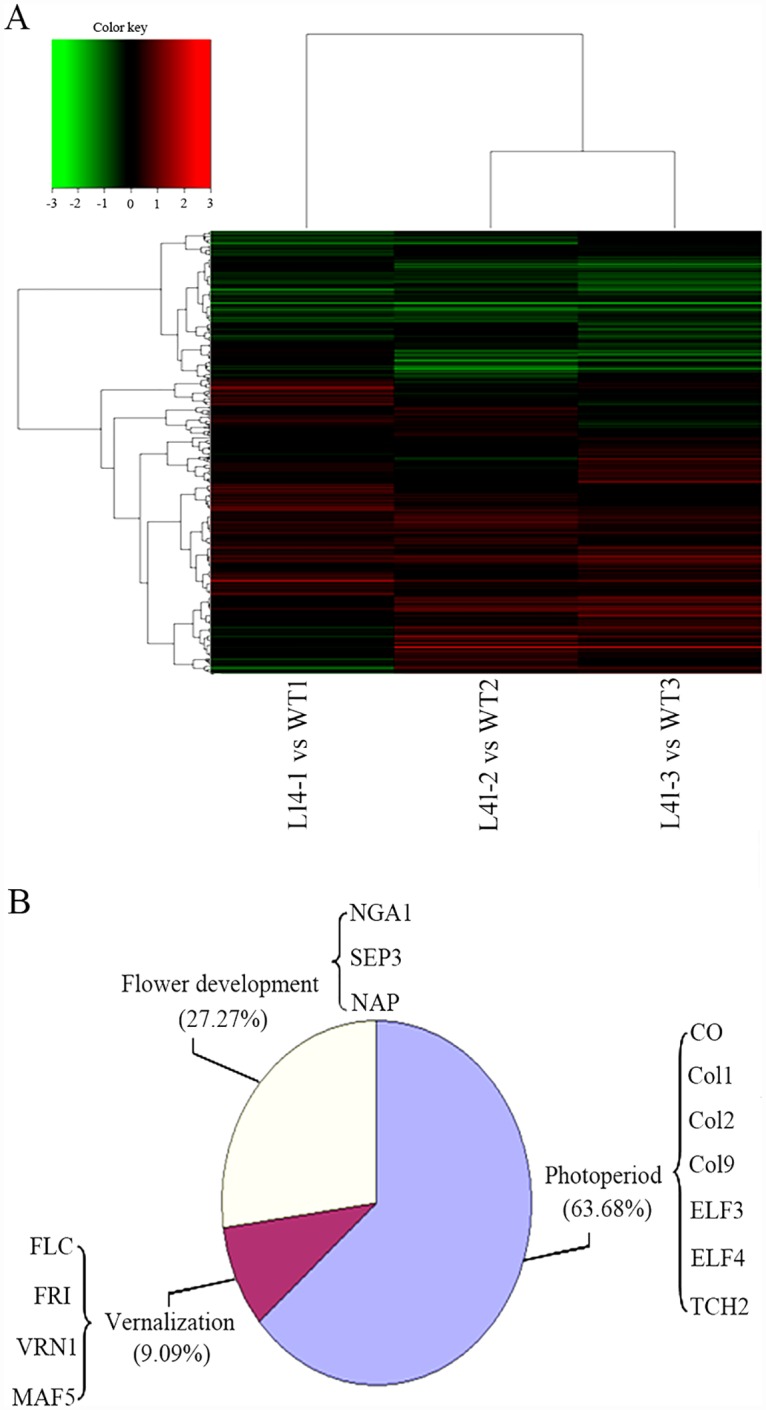
(A) Gene expression profile of transgenic plants L14 and WT. (B) Functional classification of candidate genes. Red: up-regulated genes; Green: down-regulated genes.
The 11 candidate genes involved in flowering included 6 up-regulated (Col9, ELF3, ELF4, TCH2, NGA1, and NAP) genes and 5 down-regulated (CO, Col1, Col2, MAF5, and SEP3) genes in L14 (Fig 3B). The 11 candidate genes were then divided into three pathways, the photoperiod pathways (CO, Col1, Col2, Col9, ELF3, ELF4, and TCH2), vernalization pathways (MAF5), and flowering development pathways (NGA1, SEP3, and NAP) (Fig 3B). These key pathway genes were subjected to further detailed expression analysis using qRT-PCR.
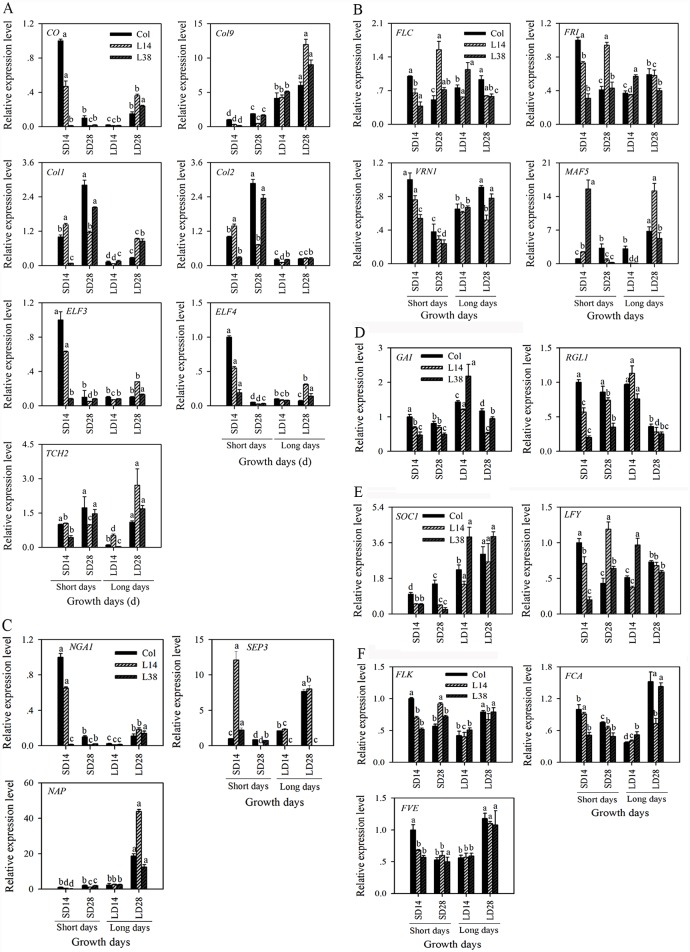
Overexpression of MaASR reduces the expression of photoperiod pathway genes under SD for 28 d
The expression of seven photoperiod pathway genes (CO, Col1, Col2, Col9, ELF3, ELF4, and TCH2) was examined between WT and the MaASR-overexpressing transgenic plants (L14 and L38) (Fig 4A). The expression pattern of CO in WT and MaASR transgenic lines was similar to Col1, Col2, ELF3, ELF4, and TCH2 but it was reversed with Col9 (Fig 4A). Compared to WT, the expression levels of CO, Col1, Col2, Col9, ELF3, ELF4, and TCH2 in MaASR transgenic lines were significantly lower than that of the WT under SD for 28 d (from vegetative to reproductive transition stage), specifically in the L14 line compared to the L38 line (Fig 4A), suggesting that different copy numbers of L14 and L38 can affect expression levels in transgenic plants. These results showed that overexpression of MaASR reduced the photoperiod pathway genes expression levels at 28 d under SD.
Fig 4. Expression analysis of photoperiod pathway genes, vernalization pathway genes, flower development related genes, GA pathway genes, floral integrator genes, and autonomous pathway genes in WT and MaASR transgenic plants.
(A) Photoperiod pathway genes. (B) Vernalization pathway genes. (C) Flower development related genes. (D) GA pathway genes. (E) Floral integrator genes. (F) Autonomous pathway genes. WT: Wild-type; L14, L38: MaASR transgenic lines. Data are represented as mean ± SD of biological replicates (n = 3). Means denoted by the same letter do not significantly differ when set at P<0.05 as determined by Duncan’s multiple range tests.
Overexpression of MaASR reduces the expression of vernalization pathway genes (VRN1and MAF5) under SD for 28 d
Four key genes involved in vernalization pathway (FLC, FRI, VRN1, and MAF5) were screened by microarray analysis based on previous studies of Arabidopsis [4, 32, 33]. FLC and FRI act as inhibitors of flowering in the vernalization pathway [4]. VRN1 and MAF5 could play an opposite role of FLC [32]. In Fig 4B, the expression of FLC and FRI was lower in the MaASR transgenic lines under SD for 14 d than that in WT. Transgenic plants exhibited enhanced expression of FLC and FRI compared to WT under SD for 28 d but the expression of VRN1 and MAF5 was lower in the transgenic lines under SD for 28 d, suggesting that MaASR overexpression increases FLC and FRI expression and decreases VRN1 and MAF5 expression under SD for 28 d to delay flowering time.
Overexpression of MaASR alters the expression pattern of flower development related genes
Three flowering development pathway genes (NGA1, SEP3, and NAP) were screened by microarray analysis (Fig 3B). The expression levels of NGA1, SEP3, and NAP were lower in the transgenic lines under SD for 28 d compared to WT (Fig 4C). SEP3 expression was significantly different between the WT and transgenic lines. SEP3 expression gradually increased in the WT from 14 d under SD to 28 d under LD but its expression in the MaASR transgenic lines declined rapidly from 14 d under SD to 28 d under SD and then increased gradually in L14 from 14 d under LD to 28 d under LD (Fig 4C). These results suggest that MaASR overexpression suppresses the expression of flowering development pathway genes (NGA1 and NAP), altering the expression pattern of SEP3.
Overexpression of MaASR reduces expression of GA pathway genes and floral integrator genes under SD for 28 d, but did not affect expression of autonomous pathway genes
Based on previous studies in Arabidopsis, several GA pathway genes (GAI and RGL1), floral integrator genes (SOC1 and LFY), and autonomous pathway genes (FLK, FCA, and FVE) have been identified by qRT-PCR [34, 35]. The expression levels of GAI and RGL1 in transgenic plants at 14 d under SD, 28 d under SD, and 28 d under LD were lower than in the WT (Fig 4D). SOC1 expression was lower in transgenic lines before flowering (from 14 d under SD to 28 d under SD) compared to WT; however, LFY expression was higher in the transgenic lines at 28 d under SD compared to WT (Fig 4E). FLK, FCA, and FVE promote flowering via negative regulation of FLC transcriptional levels in the autonomous pathway [36]. FLK, FCA, and FVE expression revealed similar trends from 14 d under SD to 28 d under LD between WT and transgenic plants (Fig 4F), indicating that MaASR overexpression reduces the expression of GA pathway genes (GAI and RGL1) and floral integrator gene (SOC1) at 28 d under SD but does not affect the expression of autonomous pathway genes.
A tentative model of the main genes involved in the flowering pathway in MaASR-overexpressed plants
A tentative model of the flowering regulatory network associated with MaASR overexpression was developed (Fig 5). Three genes, FLC, SOC1 and LFY, are in the core of the network. In the model, photoperiod-related genes (CO, ELF3, ELF4, Col1, Col2, Col9, and TCH2) inhibit flowering time by repressing SOC1 and LFY expression in MaASR transgenic plants at 28 d under SD condition. Vernalization pathways primarily access the network by inhibiting expression of FLC. GA pathway genes inhibit the expression of SOC1. Flower developmental pathway genes are directly regulated by LFY to affect flower organ formation.
Fig 5. A tentative model showing the main genes involved in the multiple flowering pathway in MaASR overexpressed plants.
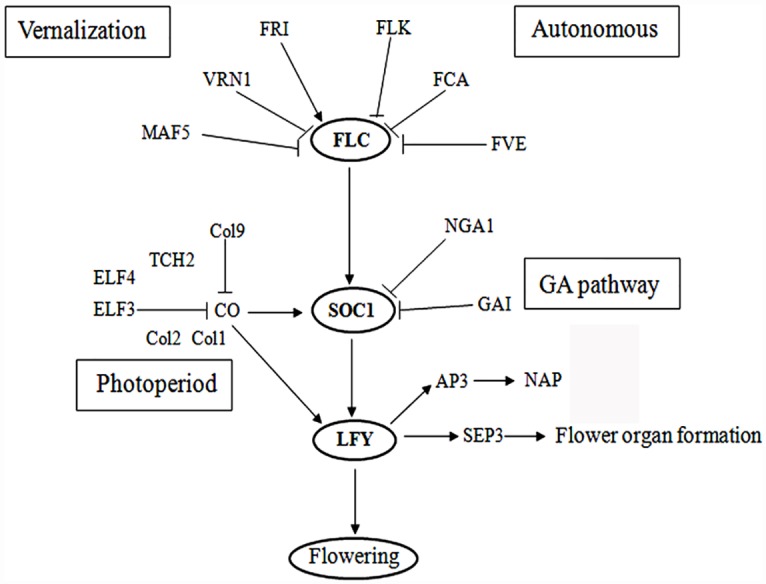
Autonomous pathway genes regulate flowering time by affecting FLC expression between WT and transgenic plants but MaASR overexpression does not affect expression of autonomous pathway genes. These findings indicate that MaASR delays flowering time by reducing expression of several photoperiod pathway genes, vernalization pathway genes, and GA pathway genes, while expression of several other flower development related genes (NGA1, SEP3, and NAP) and floral integrator genes (SOC1) are inhibited by MaASR overexpression.
Discussion
Despite extensive studies on the role of MaASR in fruit-ripening and in response to various abiotic stresses (mainly salt and drought stress tolerance) [16, 23], prior to this study, MaASR’s role in regulating flowering time in bananas was not studied. For the first time, herein we demonstrated that MaASR overexpression resulted in a clear delayed-flowering phenotype. The numbers of rosette leaves, sepal, and pedicel trichomes in transgenic Arabidopsis plants, L14 and L38, were significantly greater than those of WT under LD conditions. Similar observations have been made for other key flowering genes, such as FLC and AERIAL ROSETTE I (ART1), in causing enlarged basal rosette of leaves, developed adaxial trichome formation, and floral reversion for delayed flowering [37, 38]. Additional results showed that delaying in flowering time due to MaASR overexpression was caused mainly by the attenuated expression of several photoperiod pathway genes (CO, Col1, Col2, ELF3, and ELF4), vernalization pathway genes (VRN1 and MAF5), flowering development pathway genes (NGA1, SEP3, and NAP), GA pathway genes (GAI and RGL1), and floral integrator genes (SOC1) under SD for 28 d (from vegetative to reproductive transition stage). Interestingly, the expression of autonomous pathway genes was not affected.
CO is an important floral regulator in the photoperiod pathway, integrating the circadian and light signals to control flowering time in the early stage of Arabidopsis growth [39, 40]. Col9, Col1, and Col2 encode zinc finger proteins and are homologous genes of CO; Col9 delays flowering by reducing CO expression in Arabidopsis and over-expression of Col1 and Col2 can shorten the period of circadian rhythms [40, 41]. ELF3 and ELF4 negatively regulate CO transcription [42, 43]. In this study, Col9 expression showed a reverse pattern with CO expression, as in Arabidopsis [40]. While the expression levels of CO, Col1, Col2, ELF3, and ELF4 in MaASR transgenic plants were significantly lower than that of the WT under SD for 28 d (from vegetative to reproductive transition stage) (Fig 4A), overexpression of MaASR reduced several photoperiod pathway gene expression to prevent the switch from vegetative to reproductive growth, consequently delaying flowering time; this result was also supported by microarray analysis (Fig 3), indicating that photoperiod pathway may play a pivotal role in the regulating flowering time of MaASR. Further experiments will be required to determine the interaction mechanism between photoperiod pathway genes and MaASR.
FLC is an inhibitor of flowering in the vernalization pathway by binding the SOC1 promoter to regulate flowering time in Arabidopsis [4]. The FRI increases FLC levels and affects flowering time [4]. VRN1 is responsive to low temperature and could participate in the vernalization pathway to help regulate flowering time [33]. MAF5 could play an opposite role to FLC in the vernalization response [32]. In this study, the expression levels of FLC and FRI were higher in MaASR transgenic lines than in the WT at 28 d under SD conditions but VRN1 and MAF5 expression levels were lower in transgenic lines than in the WT at 28 d under SD conditions (Fig 4B), suggesting that MaASR overexpression could increase FLC and FRI transcription and reduce VRN1 and MAF5 expression levels at 28 d under SD conditions to delay flowering.
NGA1 belongs to the AP2 transcription factor family and inhibits stigma and style development via negative regulation of SOC1 expression in Arabidopsis [44]. In this study, the expression levels of NGA1 in MaASR transgenic lines were lower from 14 d to 28 d under SD compared to WT (Fig 4C), suggesting that MaASR overexpression may affect floral development by repressing NGA1 expression. SEP3 affects floral organ formation by controlling LFY expression [45]. Here, the expression pattern of SEP3 was significantly different between WT and the transgenic lines. SEP3 expression gradually increased in WT, but its expression in the transgenic lines declined rapidly from 14 under SD to 14 under LD (Fig 4C), suggesting that MaASR overexpression altered expression pattern of SEP3 to affect floral organ formation. Further studies are required in order to fully understand the interaction between the regulatory networks in MaASR overexpression and other flowering development-related pathway genes.
GAI and RGL1 belong to the DELLA subfamily and are negative regulators of GA in the flowering process [34]. Here, the expression levels of GAI and RGL1 in MaASR transgenic plants were found to be lower than that in WT at 28 d under SD conditions (Fig 4D), as in Arabidopsis [34]. SOC1, encodes a MADS box transcription factor and integrates multiple flowering signals derived from photoperiod, temperature, and hormone signals to prevent premature differentiation of the floral meristem [35]. In this study, SOC1 expression was lower in MaASR transgenic lines before flowering (from 14 d under SD to 28 d under SD) compared to the WT (Fig 4E). LFY is a master regulator of flowering and of flower development, and acts as part of a switch that mediates the transition from the vegetative to the reproductive phase of plant development [46]. Here, LFY expression levels were lower in MaASR transgenic lines under SD for 14 d before the transition from the vegetative to the reproductive stage (Fig 4E), suggesting early MaASR overexpression was repressed from the vegetative to the reproductive phase transition by reduced LFY expression.
FLK, FCA, and FVE are three members of an autonomous pathway that cause a late-flowering phenotype in Arabidopsis [36]; however, some mutations in FLK gave rise to phenotypes with only slightly delayed flowering [47]. FCA interacts with FY in regulating flowering time [48] and FVE participates in the regulation of flowering time by repressing FLC transcription [49]. In this study, the expression patterns of FLK, FCA, and FVE genes were similar between MaASR transgenic lines and WT (Fig 4F), suggesting that delayed flowering due to MaASR overexpression may not affect the expression of autonomous pathway genes.
Conclusions
MaASR gene is isolated and characterized from banana. Subcellular localization analysis showed that MaASR protein was localized in the nucleus and plasma membrane. Differences in the expression of MaASR gene were detected in different developmental stages of banana female flowers. MaASR transgenic lines showed a clear delayed-flowering phenotype. Overexpression of MaASR was able to delay flowering time by reducing the expression of several genes, including photoperiod pathway genes, vernalization pathway genes, GA pathway genes, and floral integrator genes, under SD for 28 d during the transition period from vegetative to reproductive phase, but without affecting the expression of autonomous pathway genes. This study provides a new insight into the regulatory mechanisms of flowering time and warrants further studies on MaASR that may lead to the development of strategies to regulate flowering time in banana and other flowering plants.
Supporting Information
(DOC)
(A) Domain: Zn2+-dependent DNA binding site in the N-terminal. (B) Domain: a conserved nuclear localization signal in the C-terminal.
(TIF)
(A) Green fluorescence in dark field. (B) Green fluorescence in bright field.
(TIF)
(TIFF)
(A) Southern blot analysis of MaASR transgenic lines L14 and L38. (B) Northern blot analysis of MaASR expression in transgenic lines L14 and L38. (C) Western blot analysis of MaASR expression in transgenic lines L14 and L38.
(TIFF)
(A) Cellular components. (B) Molecular functions. (C) Biological processes.
(TIFF)
(DOC)
(DOC)
(XLS)
(XLS)
Acknowledgments
The authors would like to acknowledge funding from the National Natural Science Foundation of China (NSFC, No. 31401843), the Modern Agro-industry Technology Research System (No. CARS-32), and the Natural Science Foundation of Hainan Province (No. 314100).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors would like to acknowledge funding from the National Natural Science Foundation of China (NSFC, No. 31401843), the Modern Agro-industry Technology Research System (No. CARS-32), and the Natural Science Foundation of Hainan Province (No. 314100).
References
- 1.Rosas U, Mei Y, Xie Q, Banta JA, Zhou RW, Seufferheld G, et al. Variation in Arabidopsis flowering time associated with cis-regulatory variation in CONSTANS. Nat Commun. 2014; 5: 3651 10.1038/ncomms4651 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castède S, Campoy JA, Le Dantec L, Quero-García J, Barreneche T, Wenden B, et al. Mapping of candidates genes involved in bud dormancy and flowering time in sweet cherry (Prunus avium). PLoS One. 2011; 10: e0143250 10.1371/journal.pone.0143250 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanano S, Goto K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell. 2011; 23: 3172–3184. 10.1105/tpc.111.088641 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2011; 13: 935–941. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shu K, Chen Q, Wu Y, Liu R, Zhang H, Wang S, et al. ABSCISIC ACID-INSENSITIVE 4 negatively regulates flowering through directly promoting Arabidopsis FLOWERING LOCUS C transcription. J Exp Bot. 2016; 67: 195–205. 10.1093/jxb/erv459 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golembeski GS, Kinmonth-Schultz HA, Song YH, Imaizumi T. Photoperiodic flowering regulation in Arabidopsis thaliana. Adv Bot Res. 2014; 72: 1–28. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson M, Staiger D. Time to flower: interplay between photoperiod and the circadian clock. J Exp Bot. 2015; 66: 719–730. 10.1093/jxb/eru441 . [DOI] [PubMed] [Google Scholar]
- 8.Henderson IR, Shindo C, Dean C. The need for winter in the switch to flowering. Annu Rev Genet. 2003; 37: 371–392. . [DOI] [PubMed] [Google Scholar]
- 9.Hu Q, Jin Y, Shi H, Yang W. GmFLD, a soybean homolog of the autonomous pathway gene FLOWERING LOCUS D, promotes flowering in Arabidopsis thaliana. BMC Plant Biol. 2014; 14: 263 10.1186/s12870-014-0263-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y, Yeom M, Kim H, Lim J, Koo HJ, Hwang D, et al. GIGANTEA and EARLY FLOWERING 4 in Arabidopsis exhibit differential phase-specific genetic influences over a diurnal cycle. Mol Plant. 2012; 5: 678–687. 10.1093/mp/sss005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moon J, Suh S, Lee H, Choi K, Hong CB, Paek NC, et al. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003; 35: 613–623. . [DOI] [PubMed] [Google Scholar]
- 12.Wada T, Tominaga-Wada R. CAPRICE family genes control flowering time through both promoting and repressing CONSTANS and FLOWERING LOCUS T expression. Plant Sci. 2015; 241: 260–265. 10.1016/j.plantsci.2015.10.015 . [DOI] [PubMed] [Google Scholar]
- 13.Ding F, Zhang S, Chen H, Su Z, Zhang R, Xiao Q, et al. Promoter difference of LcFT1 is a leading cause of natural variation of flowering timing in different litchi cultivars (Litchi chinensis Sonn.). Plant Sci. 2015; 241: 128–137. 10.1016/j.plantsci.2015.10.004 . [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, An L, Nquyen TH, Liang H, Wang R, Liu X, et al. The cloning and functional characterization of peach CONSTANS and FLOWERING LOCUS T homologous genes PpCO and PpFT. PLoS One. 2015; 23: e0124108 10.1371/journal.pone.0124108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ricardi MM, González RM, Zhong S, Domínguez PG, Duffy T, Turjanski PG, et al. Genome-wide data (ChIP-seq) enabled identification of cell wall-related and aquaporin genes as targets of tomato ASR1, a drought stress-responsive transcription factor. BMC Plant Biol. 2014; 14: 29 10.1186/1471-2229-14-29 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Hu W, Wang Y, Feng R, Zhang Y, Liu J, et al. The MaASR gene as a crucial component in multiple drought stress response pathways in Arabidopsis. Funct Integr Genomics. 2015; 15: 247–260. 10.1007/s10142-014-0415-y . [DOI] [PubMed] [Google Scholar]
- 17.Tiwari V, Chaturvedi AK, Mishra A, Jha B. Introgression of the SbASR-1 gene cloned from a halophyte Salicornia brachiata enhances salinity and drought endurance in transgenic groundnut (Arachis hypogaea) and acts as a transcription factor. PloS One. 2015; 10: e0135541 10.1371/journal.pone.0131567 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominguez PG, Frankel N, Mazuch J, Balbo I, Iusem N, Fernie AR, et al. ASR1 mediates glucose-hormone cross talk by affecting sugar trafficking in tobacco plants. Plant Physiol. 2013; 161: 1486–1500. 10.1104/pp.112.208199 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golan I, Dominguez PG, Konrad Z, Shkolnik-Inbar D, Carrari F, Bar-Zvi D. Tomato ABSCISIC ACID STRESS (ASR) gene family revisited. PLoS One. 2014; 9: e107117 10.1371/journal.pone.0107117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang CY, Wu CH, Jauh GY, Huang JC, Lin CC, Wang CS. The LLA23 protein translocates into nuclei shortly before desiccation in developing pollen grains and regulates gene expression in Arabidopsis. Protoplasma. 2008; 233: 241–254. 10.1007/s00709-008-0016-5 . [DOI] [PubMed] [Google Scholar]
- 21.Wang CS, Liau YE, Huang JC, Wu TD, Su CC, Lin CH. Characterization of a desiccation-related protein in lily pollen during development and stress. Plant Cell Physiol. 1998; 39: 1307–1314. . [DOI] [PubMed] [Google Scholar]
- 22.Frankel N, Nunes-Nesi A, Balbo I, Mazuch J, Centeno D, Iusem ND, et al. ci21A/Asr1 expression influences glucose accumulation in potato tubers. Plant Mol Biol. 2007; 63: 719–730. . [DOI] [PubMed] [Google Scholar]
- 23.Xu BY, Su W, Liu JH, Wang JB, Jin ZQ. Differentially expressed cDNAs at the early stage of banana ripening identified by suppression subtractive hybridization and cDNA microarray. Planta. 2007; 226: 529–539. . [DOI] [PubMed] [Google Scholar]
- 24.Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002; 31: 375–383. . [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schimittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods. 2001; 25: 402–408. . [DOI] [PubMed] [Google Scholar]
- 26.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998; 16: 735–743. . [DOI] [PubMed] [Google Scholar]
- 27.Miao HX, Sun PG, Liu WX, Xu BY, Jin ZQ. Identification of genes encoding granule-bound starch synthase involved in amylose metabolism in banana fruit. PLoS One. 2014; 9: e88077 10.1371/journal.pone.0088077 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diallo A, Kane N, Agharbaoui Z, Badawi M, Sarhan F. Heterologous expression of wheat VERNALIZATION 2 (TaVRN2) gene in Arabidopsis delays flowering and enhances freezing tolerance. PLoS One. 2010; 5: e8690 10.1371/journal.pone.0008690 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao W, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol. 2006; 24: 1140–1150. . [DOI] [PubMed] [Google Scholar]
- 30.Miao HX, Ye ZX, Hu GB, Qin YH. Comparative transcript profiling of gene expression between self-incompatible and self-compatible mandarins by suppression subtractive hybridization and cDNA microarray. Mol Breeding. 2015; 35: 47 10.1007/s11032-015-0204-x [DOI] [Google Scholar]
- 31.Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J. 2008; 6: 609–618. . [DOI] [PubMed] [Google Scholar]
- 32.Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell. 2003; 15: 1159–1169. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lü J, Suo H, Yi R, Ma Q, Nian H. Glyma11g13220, a homolog of the vernalization pathway gene VERNALIZATION 1 from soybean [Glycine max (L.) Merr.], promotes flowering in Arabidopsis thaliana. BMC Plant Biol. 2015; 15: 232 10.1186/s12870-015-0602-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen CK, Chang C. Arabidopsis RGL1 encodes a negative regulator of gibberellin response. Plant Cell. 2002; 14: 87–100. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J, Lee I. Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot. 2010; 61: 2247–2254. 10.1093/jxb/erq098 . [DOI] [PubMed] [Google Scholar]
- 36.Marquardt S, Boss PK, Hadfield J, Dean C. Additional targets of the Arabidopsis autonomous pathway members, FCA and FY. J Exp Bot. 2006; 57: 3379–3386. . [DOI] [PubMed] [Google Scholar]
- 37.Poduska B, Humphrey T, Redweik A, Grbić V. The synergistic activation of FLOWERING LOCUS C by FRIGIDA and a new flowering gene AERIAL ROSETTE 1 underlies a novel morphology in Arabidopsis. Genetics. 2003; 163: 1457–1465. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matías-Hernández L, Aguilar-Jaramillo AE, Cigliano RA, Sanseverion W, Pelaz S. Flowering and trichome development share hormonal and transcription factor regulation. J Exp Bot. 2016; 67: 1209–1219. 10.1093/jxb/erv534 . [DOI] [PubMed] [Google Scholar]
- 39.Ayre BG, Turgeon R. Graft transmission of a floral stimulant derived from CONSTANS. Plant Physiol. 2004; 135: 2271–2278. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng XF, Wang ZY. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005; 43: 758–768. . [DOI] [PubMed] [Google Scholar]
- 41.Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 2001; 26: 15–22. . [DOI] [PubMed] [Google Scholar]
- 42.Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell. 2001; 13: 1305–1315. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011; 475: 398–402. 10.1038/nature10182 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fourquin C, Ferrándiz C. The essential role of NGATHA genes in style and stigma specification is widely conserved across eudicots. New Phytol. 2014; 202:1001–1013. 10.1111/nph.12703 . [DOI] [PubMed] [Google Scholar]
- 45.Castillejo C, Romera-Branchat M, Pelaz S. A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J. 2005; 43: 586–596. . [DOI] [PubMed] [Google Scholar]
- 46.Li W, Zhou Y, Liu X, Yu P, Cohen JD, Meyerowitz EM. LEAFY controls auxin response pathways in floral primordium formation. Sci Signal. 2013; 6: ra23 10.1126/scisignal.2003937 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mockler TC, Yu X, Shalitin D, Parikh D, Michael TP, Liou J, et al. Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci U S A. 2014; 101: 12759–12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jang YH, Park HY, Kim SK, Lee JH, Suh MC, Chung YS, et al. Survey of rice proteins interacting with OsFCA and OsFY proteins which are homologous to the Arabidopsis flowering time proteins, FCA and FY. Plant Cell Physiol. 2009; 50: 1479–1492. 10.1093/pcp/pcp093 . [DOI] [PubMed] [Google Scholar]
- 49.Ausín I, Alonso-Blanco C, Jarillo JA, Ruiz-García L, Martínez-Zapater JM. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet. 2004; 36: 162–166. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(A) Domain: Zn2+-dependent DNA binding site in the N-terminal. (B) Domain: a conserved nuclear localization signal in the C-terminal.
(TIF)
(A) Green fluorescence in dark field. (B) Green fluorescence in bright field.
(TIF)
(TIFF)
(A) Southern blot analysis of MaASR transgenic lines L14 and L38. (B) Northern blot analysis of MaASR expression in transgenic lines L14 and L38. (C) Western blot analysis of MaASR expression in transgenic lines L14 and L38.
(TIFF)
(A) Cellular components. (B) Molecular functions. (C) Biological processes.
(TIFF)
(DOC)
(DOC)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.