Abstract
Callerya speciosa (Champ.) ScHot is a woody perennial plant in Fabaceae, the roots of which are used medicinally. The storage roots of C. speciosa are derived from fibrous roots, but not all fibrous roots can develop into storage roots. To detect key genes involved in storage roots formation, we performed Illumina sequencing of the C. speciosa storage roots and fibrous roots. De novo assembly resulted in 161,926 unigenes, which were subsequently annotated by BLAST, GO and KEGG analyses. After expression profiling, 4538 differentially expressed genes were identified. The KEGG pathway enrichment analysis revealed changes in the biosynthesis of cytokinin, phenylpropanoid, starch, sucrose, flavone and other secondary metabolites. Transcription factor-related differentially expressed genes (DEGs) were also identified, including such gene families as GRAS, COL, MIKC, ERF, LBD, and NAC. The DEGs related to light signaling, starch, sugar, photohormones and cell wall-loosening might be involved in the formation of storage roots. This study provides the first transcriptome profiling of C. speciosa roots, data that will facilitate future research of root development and metabolites with medicinal value as well as the breeding of C. speciosa.
Introduction
Callerya speciosa (Champ.) ScHot is a woody perennial plant in Fabaceae and distributed over South China and Southeast Asia. Its roots have long been used for food and as a traditional medicinal herb, with properties of toxin removal, heat clearance from the lungs to relieve cough, liver purging and kidney invigoration. More recently, C. speciosa has been widely planted because the wild plants are on the brink of extinction, and rapid propagation has been applied for the manufacture-scale production of seedlings. In previous studies of C. speciosa, we have developed SSR markers to analyze population differences and diversity among natural populations, screened superior variety, and investigated the application of cryopreservation [1, 2]. The thickness and starch content are the main factors of quality of the storage roots. The storage roots (SRs) are string- or spindle-shaped and may be lignified and stop expanding under uncertain conditions. The FRs of C. speciosa possess the potential to form SRs, but limited FRs can transfer into SRs, the mechanism by which this occurs is poorly understood.
During evolution, some plants have acquired the ability to differentiate leaves, stems, or roots into storage organs in response to drought or freezing conditions [3]. The mechanisms of storage-organ formation have been investigated in root and tuber crops such as potato (Solanum tuberosum), sweet potato (Ipomoea batatas), cassava and yam. In particular, the tuberization mechanism has been most studied in potato. Homologs of the flowering locus T (FT) family are described as key components of signals inducing the formation of potato tuber and onion bulb [4, 5]. In potato, CONSTANS (CO) homologs repress the expression of mobile tuberizing signals under non-inductive long-day conditions (LD). StCOL2, which is expressed at higher levels in potato than other CO homologs, may repress tuberization by directly regulating the transcription of StSP5G, a putative tuberization repressor that may inhibits the expression of the mobile tuberizing signal StSP6A [4, 6]. In addition, potato homologs of CYCLING DOF FACTOR 1 (StCDF1), GIGANTEA (GI) and Flavin-binding kelch repeat F-box protein 1 (FKF1) form a complex that degrades StCDF1, thereby repressing the expression StCOL1 and StCOL2 and inducing tuber formation [7, 8]. Several plant hormones have been proven to be involved in tuberization in potato [9]. For instance, a decrease in the level of gibberellic acid (GA) is required for tuberization onset, indicating that GA plays an inhibitory role in tuberization [10, 11], and given the dramatic change in StARF and StPIN family gene expression during tuberization, it has been suggested that auxin functions as a promoter of tuber formation [12, 13]. By promoting cell division during tuberization onset and sink formation, cytokinins may also serve as universal regulators of storage-organ formation [14]. Some other mobile signals have been found to regulate tuberization in potato. miR156 and miR172, two major components of the flowering age pathway, were shown to travel to the stolon via the phloem and modulate tuber formation, with miR156 accumulating in stolons under LD and miR172 during tuberization onset [15, 16]. Overexpression of StBEL5 and POTATO HOMEOBOX1 (POTH1) in potato resulted in enhanced tuberization, and movement of the transcripts was demonstrated [17–19]. Furthermore, many tuberization-related transcription factors (TFs) such as NAC family genes, MADS-box family genes and knotted-like homeobox genes have also been found in sweet potato [20–25].
Several studies have applied transcriptome analysis to detect key genes involved in storage-organ formation of root and tuber crops. Firon et al. [26] performed transcriptome profiling of sweet potato roots, reporting up-regulated starch biosynthesis and down-regulated lignin biosynthesis in SRs compared to FRs. Sun et al. [27] studied tuberous root development in Rehmannia glutinosa using dynamic transcriptome profiling, and Shan et al. [28] used digital gene expression (DGE) tag profiling analysis to identify putative genes involved in photoperiodic tuberization in potato. Additionally, Sojikul et al. [29] revealed phytohormone action during cassava storage root initiation through genome-wide microarray analysis, and Yang et al. [30] employed expression profiling with microarray to demonstrate an active process of glycolysis/gluconeogenesis in cassava storage roots. As there is limited genomic and transcriptomic information for C. speciosa to date, in the present study, we applied Illumina RNA-seq to perform the first transcriptome profiling of C. speciosa roots to find candidate genes involved in SR formation and to provide sequence resources for further analysis.
Results
Sequencing and de novo assembly
To characterize transcriptome differences between SRs and FRs, total RNA was extracted in replicates to prepare cDNA libraries and then subjected to sequencing (S1 Fig). A total of 315,665,026 clean reads of 90 nt were generated (Table 1) and subsequently de novo assembled using Trinity, resulting in 161,926 unigenes with an N50 value of 2107 nt and a mean length of 1285 nt (Table 2). Using the CEGMA pipeline, we checked the completeness of the assembly by similarity searches of 248 conserved eukaryotic core genes [31]. The result indicated that 95.97% of the core genes were completely assembled and 3.81% were partially assembled, suggesting the completeness of the assembly.
Table 1. Statistics of reads.
| Libraries | Number of Clean Reads | Total Length of Reads (nt) | |
|---|---|---|---|
| Storage Roots | SR1 | 54,454,978 | 4,900,948,020 |
| SR2 | 51,348,702 | 4,621,383,180 | |
| SR3 | 51,218,046 | 4,609,624,140 | |
| Fibrous Roots | FR1 | 51,766,438 | 4,658,979,420 |
| FR2 | 54,840,008 | 4,935,600,720 | |
| FR3 | 52,036,854 | 4,683,316,860 |
Table 2. Statistics of de novo assembly.
| Libraries | Percentage of reads assigned to Unigenes | Number of Unigenes | Total Length | Mean Length | N50 | |
|---|---|---|---|---|---|---|
| Storage Roots | SR1 | 58.74% | 77,956 | 80,657,943 | 1039 | 1727 |
| SR2 | 56.74% | 90,238 | 98,402,901 | 1090 | 1807 | |
| SR3 | 54.34% | 84,060 | 87,889,002 | 1046 | 1724 | |
| Fibrous Roots | FR1 | 57.32% | 90,013 | 95,982,678 | 1066 | 1762 |
| FR2 | 53.95% | 117,282, | 119,457,513 | 1019 | 1750 | |
| FR3 | 56.61% | 92,268 | 99,420,748 | 1078 | 1774 | |
| All | 56.28% | 161,926 | 208,090,130 | 1285 | 2107 |
Functional annotation of the unigenes
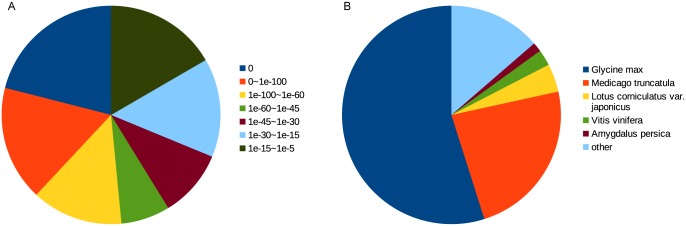
For functional annotations, the assembly unigenes were searched against the NCBI non-redundant protein (Nr), Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Cluster of Orthologous Groups (COG), NCBI Nucleotide (NT) and SwissProt databases using BLAST with an e-value cutoff of 1e-5. Of the 161,926 assembly unigenes, 125,967 matched to sequences in the databases. According to the BLAST results in the order of Nr, SwissProt, KEGG and COG, we predicted the coding sequences (CDSs) of 109,421 of the unigenes matching to sequences in the protein databases. Of those unigenes for which no sequence was matched, we predicted the CDSs of 8,159 using ESTScan. There are still 44,346 (27.39%) unigenes that have no CDS, some of which may function as non-coding RNAs. Based on the results of BLAST searches against the Nr database, 111,706 unigenes match to sequences of Glycine max (61,295), Medicago truncatula (26,330), Lotus corniculatus var. japonicus (4649), Vitis vinifera (2662), Amygdalus persica (1535), Ricinus communis (905) and other species (14,330) (Fig 1B). Of the 111,706 unigenes, 65,639 (58.76%) showed an e-value less than le-45, and 53,610 (47.99%) showed sequence similarity greater than 80% (Fig 1A).
Fig 1. Summary of BLASTx results for the NR database.
(A) E-value distribution of unigenes with BLAST hits in NR; (B) Species-based distribution of unigenes with BLAST hits in NR.
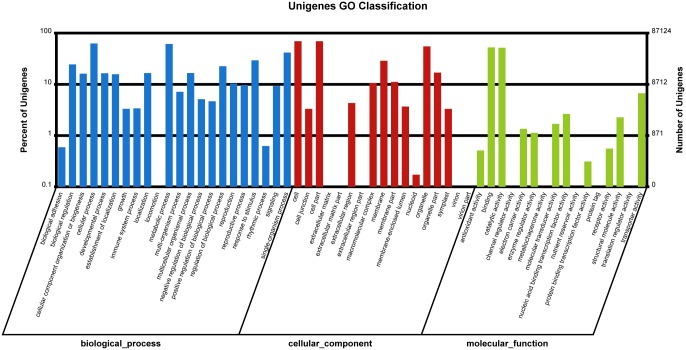
To further characterize the function of unigenes, we applied GO functional classification to unigenes based on the results of BLAST against the Nr database. In total, 87,124 unigenes were classified into 56 functional groups according to the categories of biological process, cellular component and molecular function (Fig 2). Among the 22 functional groups in biological process, cellular process (54,563) and metabolic process (53,339) are the most highly represented groups, followed by single-organism process (36,337), response to stimulus (25,626), biological regulation (21,305), regulation of biological process (19,606), multicellular organismal process (14,433), localization (14,424), and developmental process (14,305), among others. Similar to biological process, unigenes were grouped into 16 sub-groups of molecular function, among which binding (45,752), catalytic activity (45,038), transporter activity (5818), nucleic acid binding transcription factor activity (2299), and structural molecule activity were the top 5 represented groups.
Fig 2. GO classification of unigenes.
The number and percent of unigenes assigned to each GO category were provided in vertical axis.
To further classify and predict functions, we also matched unigenes to the COG database (S2 Fig). A total of 47,162 unigenes were assigned to 25 functional categories: general function prediction only (15,610), transcription (9090), replication, recombination and repair (8366), signal transduction mechanisms (7114), posttranslational modification, protein turnover, and chaperones (5983), translation, ribosomal structure and biogenesis (5377), carbohydrate transport and metabolism (4884), and function unknown (4627), among others.
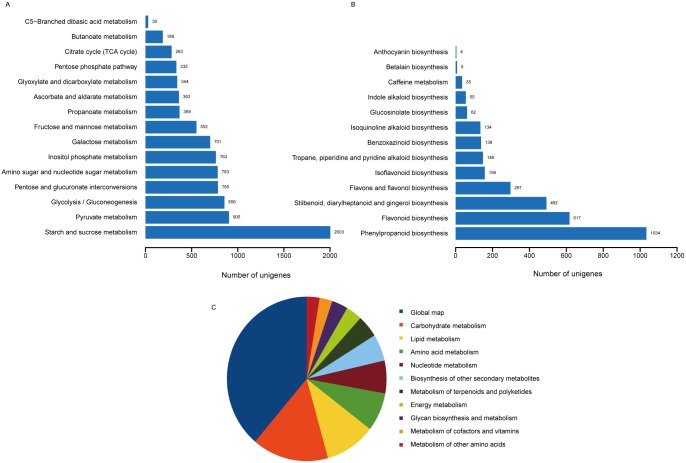
Based on a BLAST search against the KEGG database, 69,375 unigenes were assigned to KEGG pathways. Metabolic pathways and biosynthesis of secondary metabolites were the most represented pathways, followed by plant-pathogen interaction, plant hormone signal transduction, RNA transport, and spliceosome. Among metabolic sub-pathways, global map was the most represented, followed by carbohydrate metabolism, lipid metabolism, amino acid metabolism, nucleotide metabolism, and biosynthesis of other secondary metabolites (Fig 3C). For carbohydrate metabolism, sub-pathways of starch and sucrose metabolism, pyruvate metabolism and glycolysis/gluconeogenesis were the most highly represented (Fig 3A); phenylpropanoid biosynthesis, flavonoid biosynthesis, stilbenoid, diarylheptanoid and gingerol biosynthesis, flavone and flavonol biosynthesis and isoflavonoid biosynthesis were the top five represented sub-pathways of metabolism of other secondary metabolites (Fig 3B). Otherwise, some researches indicated that that many likely medical substances such as isoliquiritigenin, pterocapan, daucosterol, beta-Sitosterol, stigmasterol, chalcone, flavonoid, polysaccharide existed in roots of C. speciose, of which genes and pathways were identified in KEGG annotations [32, 33].
Fig 3. Summary of KEGG annotation of unigenes.
(A) Number of unigenes involved in sub-pathways of carbohydrate metabolism. (B) Number of unigenes involved in sub-pathways of metabolism of other metabolites. (C) Number of unigenes involved in sub-pathways of metabolic pathway.
Differential expression analysis
We first estimated the expression level of unigenes using fragments per kilobase of transcript per million fragments mapped (FPKM) values. The SR libraries and FR libraries contained 142,510, and 156,700 unigenes, with 19,416 specifically expressed in the SR libraries and 5,266 in the FR libraries. Using the NOIseq method proposed by Tarazona et al. [34] for differential expression analysis with |log2(SR/FR)|≥1 and probability≥0.8, we identified 4538 differentially expressed genes, among which 491 were up-regulated and 4047 down-regulated (S1 Table).
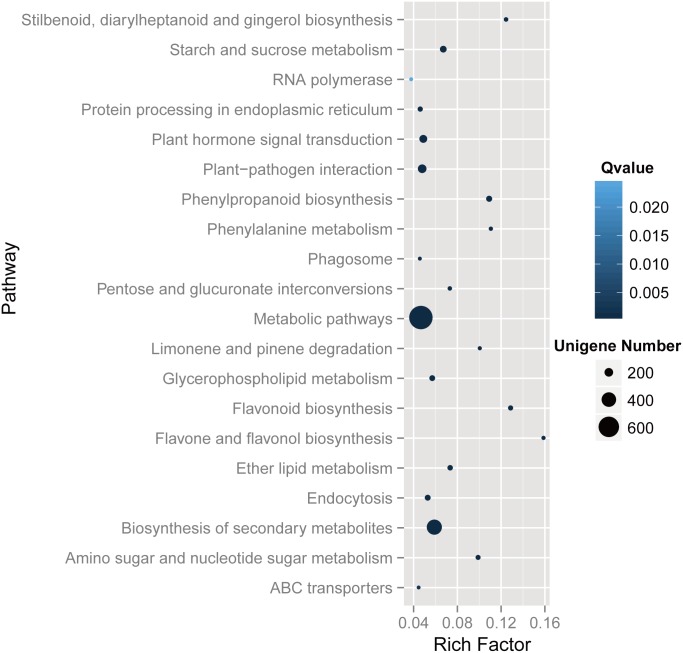
To explore the function of DEGs, 1579 of 4538 DEGs were assigned KEGG annotations and then subjected to KEGG pathway enrichment analysis. As a result, DEGs were classified to 118 pathways, 46 of which showed significant enrichment. Fig 4 illustrates the top 20 enrichment pathways. Metabolic pathways (699), Biosynthesis of secondary metabolites (420), and Plant-pathogen interaction (200) were the most common pathways, followed by Plant hormone signal transduction (172), Starch and sucrose metabolism (133), Phenylpropanoid biosynthesis (112), Endocytosis (104), Glycerophospholipid metabolism (102), Ether lipid metabolism (93), Protein processing in endoplasmic reticulum (84), and Flavonoid biosynthesis (79), among others.
Fig 4. KEGG enrichment analysis of DEGs.
The enriched kEGG pathways were with a q-value≤0.05. Enrichment factor: the ratio between the number of DEGs and all unigenes enriched in a particular pathway.
Identification of transcription factor genes among DEGs
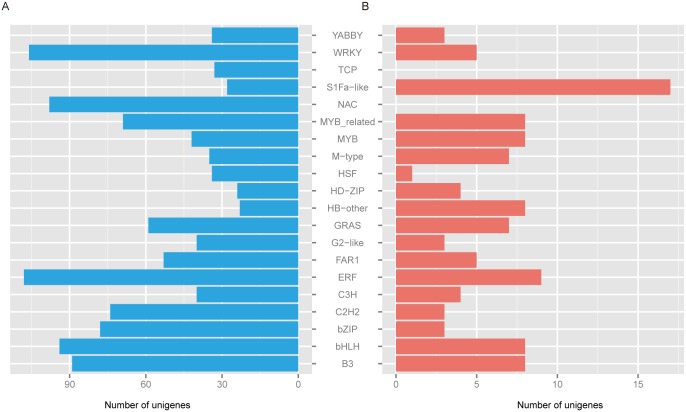
To identify transcription factor (TF)-associated unigenes, BLASTx was used to search PlantTFDB for DEG sequences [35], with 1597 DEGs classified into 54 transcription factor gene families. The ERF (117) gene family was the most represented, followed by WRKY (111), B3 (110), bHLH (102), NAC (98), bZIP (81), MYB_related (78), C2H2 (77), GRAS (66), FAR1 (58), MYB (50), S1Fa-like (45), C3H (44), G2-like (43), and M-type (42), among others (Fig 5). Interestingly, the majority of the DEGs classified into transcription factor gene families were down-regulated DEGs, with 19 gene families having no up-regulated DEGs.
Fig 5. Summary of DEGs related to transcription factors.
(A) Up-regulated unigenes related to transcription factors. (B) Down-regulated unigenes related to transcription factors.
Identification of DEGs involved in formation of storage roots
We identified DEGs might be involved in SR formation (Table 3, S2 Table). First, we examined unigenes might be involved in flowering timing, revealing homologs of CO, FT, TERMINAL FLOWER 1 (TFL1), and Flowering-promoting factor 1 (FDF1). Homologs of FT and TFL1, CONSTANS-LIKE 2 and CONSTANS-LIKE 5 were up-regulated, whereas homologs of FDF1 and EARLY FLOWERING 4 (ELF3) were down-regulated. Moreover, many TFs might be involved in flower and root development were identified. A large number of plant-specific transcription factors belong to NAC domain-containing protein families, some of which are involved in cell division and expansion, such as NAC21/22, NAC29 and NAC043; these homologs were down-regulated in SRs. LOB-domain (LBD) genes comprise a large family of plant-specific and DNA-binding transcription factors, the functions of which are largely unknown [36], and homologs of LBD4 and LBD5 were up-regulated but homologs of LBD13, LBD38 and LBD41 down-regulated. RAP1 belongs to the BHLH family of transcription factors, which are involved in anthocyanin biosynthesis and phytohormone signaling, and the homolog was found to be up-regulated in SRs. ARFs are transcription factors that response to auxin and function in promoting flowering, stamen development, and floral organ abscission as well as repressing cell division and organ growth [37]; homologs of ARF4, ARF5, ARF6, ARF9 and ARF18 were down-regulated. In the meanwhile, the homologs of auxin influx transporter LAX1, LAX2, LAX3 and LAX5 were down-regulated in SRs.
Table 3. Putative DEGs involved in storage root formation (Part of S2 Table).
| Unigene | Log2 FC | Probability | Homologue ID | E-value | Homolog name |
|---|---|---|---|---|---|
| CL13177.Contig1_All | 7.676971 | 0.969675 | sp|Q00081|GLGL1_SOLTU | 0 | Glucose-1-phosphate adenylyltransferase large subunit 1 |
| CL10402.Contig2_All | 6.264433 | 0.953126 | sp|P52417|GLGS2_VICFA | 0 | Glucose-1-phosphate adenylyltransferase small subunit 2 |
| CL14063.Contig6_All | 4.805002 | 0.939578 | sp|P53536|PHSL_VICFA | 0 | Alpha-1,4 glucan phosphorylase L isozyme, chloroplastic/amyloplastic |
| CL12657.Contig1_All | 3.97013 | 0.933003 | sp|Q43092|SSG1_PEA | 0 | Granule-bound starch synthase 1, chloroplastic/amyloplastic |
| CL10444.Contig2_All | 2.351379 | 0.886854 | sp|Q0WVX5|SSY4_ARATH | 3E-103 | Probable starch synthase 4, chloroplastic/amyloplastic |
| CL11429.Contig2_All | -1.28089 | 0.815526 | sp|Q8LCP6|GUN10_ARATH | 0 | Endoglucanase 10 |
| CL13908.Contig14_All | 4.335879 | 0.825065 | sp|Q9LZS3|GLGB2_ARATH | 0 | 1,4-Alpha-glucan-branching enzyme 2–2, chloroplastic/amyloplastic |
| CL886.Contig15_All | 7.343399 | 0.928711 | sp|P55231|GLGL3_ARATH | 4E-89 | Glucose-1-phosphate adenylyltransferase large subunit 3, chloroplastic |
| CL14396.Contig5_All | 4.665488 | 0.934589 | sp|Q94AZ2|STP13_ARATH | 0 | Sugar transport protein 13 |
| CL1456.Contig8_All | 1.190382 | 0.807632 | sp|Q9SEK3|HXK1_SPIOL | 0 | Hexokinase-1 |
| CL6463.Contig3_All | 3.836333 | 0.928029 | sp|Q9LPS1|HXK3_ARATH | 0 | Hexokinase-3 |
| CL9768.Contig4_All | 3.855858 | 0.917584 | sp|Q38997|KIN10_ARATH | 1E-133 | SNF1-related protein kinase catalytic subunit alpha KIN10 |
| CL9768.Contig3_All | 2.961163 | 0.889714 | sp|Q38997|KIN10_ARATH | 2E-156 | SNF1-related protein kinase catalytic subunit alpha KIN10 |
| Unigene59772_All | -6.52946 | 0.831999 | sp|P92958|KIN11_ARATH | 5E-66 | SNF1-related protein kinase catalytic subunit alpha KIN11 |
| Unigene59262_All | -8.0805 | 0.940236 | sp|P92958|KIN11_ARATH | 9E-31 | SNF1-related protein kinase catalytic subunit alpha KIN11 |
| CL4742.Contig10_All | 2.805577 | 0.854161 | sp|Q9SYM4|TPS1_ARATH | 0 | Alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1 |
| CL4742.Contig7_All | 1.85385 | 0.805582 | sp|Q9SYM4|TPS1_ARATH | 0 | Alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1 |
| Unigene25815_All | -1.20994 | 0.813003 | sp|Q9LMI0|TPS7_ARATH | 9E-20 | Probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7 |
| CL9903.Contig2_All | 1.880861 | 0.86867 | sp|Q9FUD3|BZIP9_ARATH | 8E-52 | Basic leucine zipper 9 |
| Unigene33406_All | -3.26232 | 0.920375 | sp|B9DGI8|BZP63_ARATH | 1E-10 | Basic leucine zipper 63 |
| Unigene1547_All | -3.65527 | 0.924065 | sp|B9DGI8|BZP63_ARATH | 4E-12 | Basic leucine zipper 63 |
| CL6174.Contig7_All | 1.380675 | 0.831663 | sp|Q9LXL5|SUS4_ARATH | 0 | Sucrose synthase 4 |
| CL6174.Contig5_All | -1.8308 | 0.861009 | sp|O24301|SUS2_PEA | 0 | Sucrose synthase 2 |
| CL11300.Contig1_All | 1.980318 | 0.874639 | sp|Q43876|SPSA_VICFA | 0 | Probable sucrose-phosphate synthase |
| CL11300.Contig2_All | 1.668228 | 0.852002 | sp|Q43876|SPSA_VICFA | 0 | Probable sucrose-phosphate synthase |
| CL12981.Contig1_All | -10.8547 | 0.993212 | sp|O23547|EXLB1_ARATH | 3.00E-89 | Expansin-like B1 |
| CL14227.Contig1_All | -4.78227 | 0.932096 | sp|Q38865|EXPA6_ARATH | 1.00E-130 | Expansin-A6 |
| Unigene25102_All | 4.717632 | 0.938405 | sp|O80622|EXP15_ARATH | 2.00E-116 | Expansin-A15 |
| CL3091.Contig7_All | 2.983223 | 0.909582 | sp|O80622|EXP15_ARATH | 1.00E-70 | Expansin-A15 |
| CL3091.Contig8_All | 2.833029 | 0.90457 | sp|Q9LDR9|EXP10_ARATH | 7.00E-49 | Expansin-A10 |
| CL3091.Contig6_All | 3.0807 | 0.916087 | sp|Q9C554|EXPA1_ARATH | 6.00E-71 | Expansin-A1 |
| Unigene41911_All | -2.86906 | 0.893997 | sp|Q8LDW9|XTH9_ARATH | 3.00E-57 | Xyloglucan endotransglucosylase/hydrolase protein 9 |
| Unigene40780_All | -3.84624 | 0.922292 | sp|Q38857|XTH22_ARATH | 4.00E-85 | Xyloglucan endotransglucosylase/hydrolase protein 22 |
| CL13489.Contig1_All | -4.17811 | 0.923414 | sp|Q9ZSU4|XTH14_ARATH | 9.00E-38 | Xyloglucan endotransglucosylase/hydrolase protein 14 |
| CL1947.Contig6_All | -1.50402 | 0.827194 | sp|Q38909|XTH28_ARATH | 6.00E-134 | Probable xyloglucan endotransglucosylase/hydrolase protein 28 |
| CL7466.Contig3_All | -2.06881 | 0.875615 | sp|Q9SJL9|XTH32_ARATH | 3.00E-123 | Probable xyloglucan endotransglucosylase/hydrolase protein 32 |
| Unigene37972_All | -1.24476 | 0.807754 | sp|Q9C9Q8|PMTT_ARATH | 5.00E-45 | Probable pectin methyltransferase QUA2 |
| Unigene21277_All | -1.42031 | 0.821245 | sp|Q9LXK7|PME32_ARATH | 2.00E-139 | Probable pectinesterase/pectinesterase inhibitor 32 |
| CL11485.Contig2_All | -1.70682 | 0.855194 | sp|O81301|PME40_ARATH | 8.00E-161 | Probable pectinesterase/pectinesterase inhibitor 40 |
| CL9924.Contig2_All | -1.77975 | 0.85495 | sp|Q43111|PME3_PHAVU | 0 | Pectinesterase 3 |
| Unigene17922_All | -1.8481 | 0.863072 | sp|Q9FK05|PME61_ARATH | 8.00E-71 | Probable pectinesterase/pectinesterase inhibitor 61 |
| Unigene36964_All | -1.91615 | 0.856509 | sp|O04887|PME2_CITSI | 0 | Pectinesterase 2 |
| CL9508.Contig4_All | -3.24505 | 0.921022 | sp|Q1JPL7|PME18_ARATH | 7.00E-172 | Pectinesterase/pectinesterase inhibitor 18 |
| CL11485.Contig3_All | -4.46623 | 0.926086 | sp|O81301|PME40_ARATH | 2.00E-106 | Probable pectinesterase/pectinesterase inhibitor 40 |
| Unigene66068_All | -3.75972 | 0.903985 | sp|Q38890|GUN25_ARATH | 8.00E-123 | Endoglucanase 25 |
| CL9596.Contig1_All | 3.275602 | 0.922434 | sp|Q9FS16|EXTN3_ARATH | 1.00E-32 | Extensin-3 |
| CL9470.Contig2_All | -1.51796 | 0.821949 | sp|Q9T0K5|LRX3_ARATH | 0 | Leucine-rich repeat extensin-like protein 3 |
| CL13561.Contig2_All | -2.72166 | 0.896394 | sp|O65375|LRX1_ARATH | 4.00E-32 | Leucine-rich repeat extensin-like protein 1 |
| Unigene33730_All | -2.89631 | 0.913111 | sp|Q9LUI1|LRX6_ARATH | 7.00E-15 | Leucine-rich repeat extensin-like protein 6 |
| Unigene16895_All | -2.91198 | 0.898041 | sp|Q9LJ64|PLRX1_ARATH | 1.00E-111 | Pollen-specific leucine-rich repeat extensin-like protein 1 |
| CL13561.Contig1_All | -3.59081 | 0.926325 | sp|Q9M1G9|EXTN2_ARATH | 2.00E-103 | Extensin-2 |
| Unigene1149_All | -5.75528 | 0.948599 | sp|Q9SN46|LRX5_ARATH | 2.00E-31 | Leucine-rich repeat extensin-like protein 5 |
| Unigene26568_All | -1.662 | 0.845571 | sp|O81081|LAC2_ARATH | 9.00E-23 | Laccase-2 |
| Unigene47867_All | -9.01966 | 0.975263 | sp|Q9FLB5|LAC12_ARATH | 2.00E-64 | Laccase-12 |
| CL3996.Contig1_All | -3.93481 | 0.921552 | sp|Q9SIY8|LAC5_ARATH | 3.00E-145 | Laccase-5 |
| Unigene13230_All | -2.55428 | 0.898325 | sp|Q9SR40|LAC7_ARATH | 2.00E-104 | Laccase-7 |
| Unigene58553_All | -2.56409 | 0.895477 | sp|Q9ZRF1|MTDH_FRAAN | 1.00E-96 | Probable mannitol dehydrogenase |
| Unigene37208_All | -2.15881 | 0.872666 | sp|O82515|MTDH_MEDSA | 0 | Probable mannitol dehydrogenase |
Many unigenes related to cell wall-loosening were found, such as homologs of expansins, extensins, pectin-related genes, xyloglucan endotransglucosylase/hydrolases and cellulase. However, only homologs of Expansin-A15 (EXPA15), Expansin-A10 (EXPA10), Expansin-A1 (EXPA1) and extensin-3 were up-regulated, whereas other genes related to cell wall-loosening proteins were down-regulated.
Furthermore, the expression levels of phytohormone biosynthetic genes were determined. Auxin, gibberellin, and abscisic acid biosynthesis-related genes, including homologs of Abscisic-aldehyde oxidase, Flavin-containing monooxygenase YUCCA8, Indole-3-acetic acid-amido synthetase GH3.6, Jasmonic acid-amido synthetase JAR1, 1-Aminocyclopropane-1-carboxylate synthase, and Gibberellin 20 oxidase 2, were down-regulated. In contrast, a cytokinin biosynthesis-related unigene, a homolog of Adenylate isopentenyltransferase 5, was up-regulated.
Starch synthesis-related unigenes, such as homologs of Starch-branching enzyme, Granule-bound starch synthase, and ADP-glucose synthase, were also up-regulated. Additionally, sucrose-related unigenes homologous to Sucrose synthase 4 (SUS4) and Sucrose-phosphate synthase (SPS) were up-regulated, whereas a homolog of Sucrose synthase 2 (SUS2) was down-regulated. Moreover, unigenes linking sugar and development, including homologs of Hexokinase-1 (HXK1), SNF1-related protein kinase catalytic subunit alpha KIN10 (SNRK1.1), Basic leucine zipper 9 (BZIP9), and Alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1 (TPS1), were up-regulated.
qRT-PCR validation of DEGs
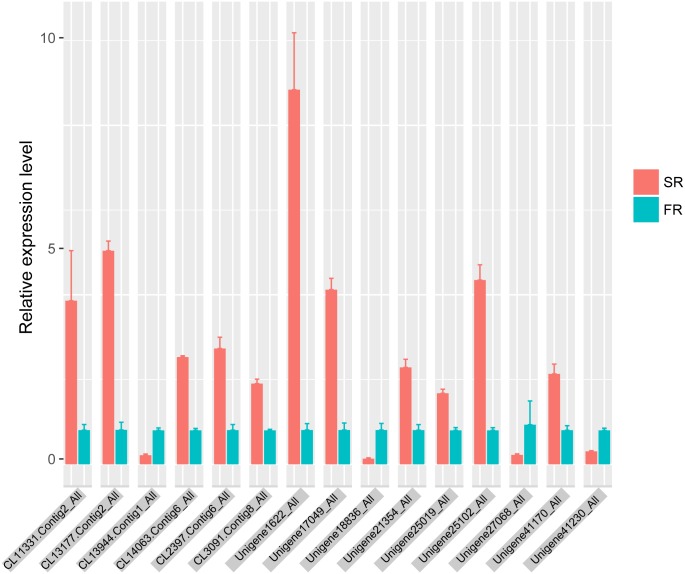
Fifteen DEGs were selected for qRT-PCR, all of which were found to be up-regulated or down-regulated in agreement with the differential expression analysis by RNA-seq (Fig 6). CL11331.Contig2_All, CL13177.Contig2_All, CL14063.Contig6_All and Unigene25102_All were up-regulated more than 25-fold in SRs, and the relative was up-regulated expression level less than 7-fold; the qRT-PCR-based results for Unigene41230_All, Unigene27068_All and Unigene18836_All also revealed a lower degree of relative down-regulation. This result suggests that high amounts of the unigene are not completely attributable to a high rate of gene expression.
Fig 6. qRT-PCR validation of selected DEGs.
SR, relative expression level of SR DEGs; FR, relative expression level of FR DEGs.
Discussion
This study provides a comparative transcriptome analysis between C. speciosa SRs and FRs. A total of 4538 DEGs were identified. KEGG pathway enrichment analysis revealed that DEGs were enriched in pathways such as starch and sucrose metabolism, plant hormone signal transduction, and phenylpropanoid biosynthesis, indicating changes in carbon flow and phytohormone signaling might happened during the formation of SRs of C. speciosa. DEGs were also enriched into the pathways Flavonoid biosynthesis, Flavone and flavonol biosynthesis, Isoflavonoid biosynthesis, Benzoxazinoid biosynthesis, Isoquinoline alkaloid biosynthesis, Indole alkaloid biosynthesis, and Glucosinolate biosynthesis, demonstrating the different secondary metabolite biosynthesis between SRs and FRs. Furthermore, DEGs related to transcription factors were predicted and involved many transcription factor gene families including bZIP, GRAS, COL, MIKC, ERF, LBD, NAC and HB-other families, which may participate in sugar and phytohormone signaling, flowering regulation, root development and cell expansion, indicating a wide range of transcription factors might be involved in the formation of SRs.
Unigenes involved in light signaling and flowering
Appropriate photoperiod is acquired for initiation of storage organ in potato, lotus and onion [38–40]. And light is necessary for initiation of tuberous roots of R. glutinosa, with the intonation of tuberous roots were completely blocked when shaded with sunshade net [41]. Light signaling related genes COL2, COL5, Early flowering 4 (ELF4) and serine/threonine protein kinase WNK8 differently expressed between SRs and FRs. COL5 affects the expression FT and induce flowering in short-days [42], and COL2 responds to light stimulus but not influence flowering in A. thaliana [43]. ELF4 and WNK8 control flowering time in A. thaliana, with ELF4 involved in circadian rhythms and WNK8 modulating photoperiod response [44, 45]. FT and TFL1 both function in flowering time control and tuberization in potato, were up-regulated in SRs of C. speciose [4, 46]. Therefore, light signaling might be involved in the formation of SRs of C. speciose, and some flowering related genes expressed in roots may also have distinct functions in root development. Although the storage organs of C. speciose, R. glutinosa, potato, lotus and onion derive from different organs, similar regulation mechanisms may exist, on which light signaling might play a key role.
Large populations of mRNAs were found to be able to move between shoots and roots in Arabidapsis thaliana [47]. The mobility of mRNAs related to transcript abundance and half-life [48]. The mRNA of FT is involved in flowering induction by long distance movement in A. thaliana [49]. Some researches indicate that the expression of FT in underground stolons is activated by mobile FT protein in potato [4], which has not been found in other plants. However, there is no evidence that FT mRNA can transport from leaves into roots, although many mRNAs may able to move between shoots and roots in C. speciose. Therefore, further research is needed to determine whether the mRNA of FT in roots of C. speciose is transported from leaves or expresses locally.
Unigenes related to sucrose and starch
A surplus of sugar in plants may be one of the main causes of storage root formation. Indeed, sugars can function as signaling molecular and gene regulators, regulating processes such as growth, flowering transition and tuber formation [50]. Sucrose has been found to promote tuber formation in potato, yam and R. glutinosa [51–53]. Sucrose synthase (SUS) converts sucrose into fructose and UDP-glucose, functioning as a sucrose-clearing enzyme [54]. Conversely, Sucrose-phosphate synthase (SPS), the key enzyme catalyzing the synthesis of sucrose, drives sucrose accumulation. The sucrose synthase alleles up-regulated in overall, for that homolog of SUS4 showed an expression level 10-fold than homolog of SUS2, whereas SUS4 up-regulated and SUS2 down-regulated in SRs. The up-regulated sucrose synthase may also play a key role in the formation of SR of C. speciose.
The main form of carbohydrate stored by SRs of C. speciose is starch. The starch content of dried SRs is 49.3%, while dry matter content of the SRs is about 67% [55]. The storage organ of C. speciose and sweet potato both are modified roots that accumulated starch. The homologs of ADP-glucose pyrophosphorylase and granule-bound starch synthase, the key enzyme for starch synthesis, and sucrose synthase functioned in both sucrose synthesis and cleavage, up-regulated during SRs formation both in sweet potato and C. speciose, with similar expression pattern during SRs formation [26, 56]. The homolog of beta-fructofuranosidase, which functions as sucrose cleavage enzyme, showed a very low expression level, with a mean FPKM value of 0.16 and 0.07 in SRs and FRs of C. speciose respectively. The expression level of beta-fructofuranosidase of sweet potao were very low and decreased during SRs development [26, 56], whereas the expression level was higher in SRs than FRs in C. speciose, indicating that beta-fructofuranosidase may play different roles during SRs development between C. speciose and sweet potato. The transcriptome data also suggested that sucrose synthase was predominant sucrose cleavage enzyme for starch accumulation during SRs development of C. speciose, which is similar with sweet potato. Furtherly, study on proteomic of cassava during formation of tuberous roots indicated that 14-3-3 protein up-regulated and play important roles in starch accumulation [57]. But homologs of 14-3-3 like protein C and 14-3-3 protein 7 down-regulated in SRs, showing a possible and different role of 14-3-3 protein during SRs formation and starch accumulation.
Laccases, which are involved in the lignin catabolic process, are required for secondary cell wall lignification [58, 59], and Cinnamyl alcohol dehydrogenase (CAD) also participates in lignin biosynthesis by catalyzing the final step of lignin monomer production. Homologs of laccases and CAD were down-regulated, indicating that lignin biosynthesis and lignification might decreased during SRs formation. The homolog of Anthocyanin regulatory C1, the key trans-acting factor required for anthocyanin biosynthesis, and the homolog of Root-specific chalcone synthase were both up-regulated in SRs, suggesting that changes in sugar metabolism may accompany changes in anthocyanin biosynthesis.
Crosstalk between phytohormones, sugars and calcium signaling
Phytohormones play key roles in tuber or tuberous root formation. In potato, GA inhibits tuber formation and promotes stolon elongation [60]; in sweet potato, high cytokinin and auxin levels promote storage root initiation and growth [61, 62], and ABA levels positively correlate with the thickening of sweet potato storage roots [63]. GA-, ABA-, ethylene-, auxin-related biosynthesis unigenes were down-regulated, whereas cytokinin-related unigenes were up-regulated, indicating that cytokinin may participle in the formation of SRs in C. speciosa.
Plants possess systems that link carbon status and development, in wich the Hexokinase (HXK) play a key role. HXK-based signaling interacts negatively with auxin but positively with cytokinin [64, 65]. And HXK1 signaling involves extensive crosstalk with plant hormone signaling via interaction with F-actin [66]. The up-regulation of homologs of HXK1, suggested that sugars may affect the formation of SRs partly via the regulation of complex networks of sugar-sensing systems in C. speciosa.
Sugar-based signaling pathways also interact with the calcium signaling pathway. CIPKs function together with calcium-sensing CBLs, comprising a CPIK-CBL network involved in plant signal transduction in response to abiotic stress [67]. AtCIPK4 was found to be induced by salt stress, whereas OsPK4, the rice CIPK4 homolog, was induced by illumination, cytokinin treatment and nutrient deprivation [68, 69]. AtCIPK7 is induced by cold and sugars and may function in sugar metabolism through the phosphorylation of sucrose synthase [67, 69]. In our study, homologs of CIPK4 and CIPK7 were up-regulated in SRs, indicating that CIPKs may also be involved in regulating the formation of SRs response environment and endogenous cues.
Unigenes involved in cell-loosening and root development
Growth in cell volume begins with selective cell wall-loosening, in which cell wall stress is relaxed, resulting in consequential water uptake and cell enlargement [70]. The growing plant cell wall consists of one or more layers of cellulose microfibrils, with each microfibril directly contacting another or interacting with matrix polymers embedded in a hydrophilic matrix [71]. Expansins meditate acid-induced growth through non-lytic cell wall loosing. Based on sequence based phylogeny, plant expansins can be classed into two large families, EXPA and EXPB, and two small families, EXPLA and EXPLB [71]. In C. speciose, homologs of EXPA1, EXPA10 and EXPA15 are up-regulated and homologs of EXPA6 and EXPLB1 are down-regulated, of which the function need to character in the further research. Several transcription factors have been found to regulate expansins genes. Arabidopsis thaliana homeobox 12 (ATHB12) increases the expression of AtEXP10, promoting cell expansion and leaf growth [72]. In SRs of C. speciose, ATHB12 is down-regulated and the differential expression of expansins may attributable to the homologs of ATHB12.
It has been proposed that the root architecture influences the formation of tuberous root, with the development of lateral roots preventing lignification of the stele in the main axis of the adventitious root, which described in the swelling of sweet potato [26, 73]. LBD family genes are involved in root formation and are up-regulated in sweet potato tuberous roots [26, 74]. In addition, LBD3 and LBD4 are induced by cytokinin and required for a flat and asymmetrical leaf lamina in Arabidopsis [36]. LBD4 may also be involved in SRs formation of C. speciose, with up-regulation of homologs of LBD4 in SRs. NAC21/NAC22 may function as a transcription activator mediating auxin signaling to promote lateral root development [75], and NAC29 may function in the transition between active cell division and cell expansion [76]. NAC043, also called NAC SECONDERY CELL WALL THICKENING PROMOTING FACTOR 1 (NST1), functions together with NST2 and NST3 in cell wall thickening and cell wall lignification [77–79]. The differences of root architecture between C. speciose SRs and FRs might partly resulted from the down-regulation of homologs of NAC21/ NAC22, NAC29 and NAC043 in SRs, which is need further research.
Conclusion
We performed comparative analysis of C. speciose SRs and FRs, discovering a series of genes associates with light signaling, phytohormones, starch, sugar and cell wall-loosening differentially expressed between SRs and FRs, which may be involved in the formation of SRs. However, further research is required to determine the DEGs responsible for SR formation, which will shed lights on the mechanism of SR formation and help in the breeding of C. speciose.
Methods
Plant material
Three in vitro-grown seedlings transplanted in a greenhouse for 5 months were selected for the present study. The experimental site is the Institute of Tropical Crop Genetic Resources, Chinese Academy of Tropical Agricultural Sciences (CATAS). The greenhouse are on the conditions of temperature of 20–35°C, relative humidities of 85%-95%, transmittance of 20% and natural photoperiod. The three plants were planted in the nursery bed and watered and fertilized weekly. The potting soil was a mixture of equal parts coconut fibre, sand and turfy soil. The roots were sampled before fertilizing and watering at 9 am, when the soil was dry and the seedlings were exposed to light 2 h. The materials collected from each plant were grounded to fine powder using liquid nitrogen and mixed, and send to BGI Life Tech Co., Ltd (Shenzhen, China) for Illumina sequencing.
RNA extraction, library preparation and sequencing
Total RNA was extracted using TRIzol® Reagent (Invitrogen) following the manufacturer’s instructions. The integrity of the RNA samples was analyzed by agarose gel electrophoresis, and the purity and concentration were evaluated using an Agilent 2100 Bioanalyzer and NanoDrop (Thermo Scientific, USA). All RNA samples had an RNA integrity number (RIN) higher than 8. Libraries were prepared using Illumina TruSeq RNA Sample Preparation Kit (Illumina, USA) according to the manufacturer’s instructions. After checking quality and quantity, mRNA was isolated from total RNA using poly-(T) oligo-attached magnetic beads. The isolated mRNA was then sheared into short fragments by mixing with fragmentation buffer and used as templates for cDNA synthesis. cDNA was purified with a QIAquick PCR Purification Kit (QIAGEN), followed by end repair and acetylation of the 3’ ends. The Illumina pair-end adapters were then connected with the cDNAs. Finally, the cDNA libraries were checked using a Agilent 2100 Bioanalyzer and ABI StepOnePlus Real-time PCR system and sequenced using an Illumina HiSeqTM 2000 (Illumina Inc., San Diego, CA, USA). The sequence reads were submitted into the NCBI Short Read Archive (SRA) database and included in NCBI BioProject database with accession number PRJNA309919 (http://www.ncbi.nlm.nih.gov/bioproject/309919).
De novo assembly and gene annotation
After the filtering of low-quality reads and removing adaptors, clean reads were assembled using Trinity de novo transcriptome assembly [80]. The unigenes in each sample were processed for sequence splicing and redundancy removal to generate a non-redundant unigene dataset. The core eukaryotic gene-mapping approach (CEGMA) was used to assess the completeness of the de novo assembly. The unigenes were then searched against protein and nucleotide databases, including SwissProt, NCBI's non-redundant protein database (NR), NCBI's non-redundant nucleotide database (NT), Kyoto Encyclopedia of Genes and Genomes (KEGG) and COG (Cluster of Orthologous Groups) using BLASTX with an e-value cutoff of 1e-5. The unigenes were annotated by GO with Blast2GO sing the BLAST results for NR [81]. The unigenes were assigned with KEGG annotations using the BLAST results for KEGG [82]. Based on the results of protein database searches, the sequence direction and coding region were determined for unigenes with BLAST hits; ESTscan was used to predict coding regions for those without BLAST hits [83].
Unigene expression analysis
FPKM was calculated to assess unigene expression. NOISeq, a nonparametric approach, was used to determine the DEGs of samples with replicates, with a fold change≥2 and probability≥0.8 [34]. To characterize the function of DEGs, KEGG pathway enrichment analysis was performed using KOBAS 2.0 (q-value≤0.05) [83]. DEGs were also searched in PlantTFDB to determine DEGs related to TFs [35].
Validation of qRT-PCR
Fifteen DEGs were selected for qRT-PCR validation of gene expression. Gene-specific primers were designed with PRIMER 6.0 software (University of Plymouth); the primers are listed in S3 Table. Total RNA was extracted from SRs and FRs described as above. The total RNA were treated with DNaseI (Takara).With validation of the expression level of several reference genes using qRT-PCR and expression stability analysis the reference genes by Genorm (https://genorm.cmgg.be) [84], the Actin-2 gene was used as an internal control (S3 Fig). The standard curve of each selected DEG was obtained by qRT-PCR with a series of cDNA dilutions. The 10 μl reaction mixture for qRT-PCR consisted of 5 μl of 2× SYBR Green Master Mix Reagent (Applied Biosystems), 0.2 μM of gene-specific primers and 50 ng of cDNA sample. The amplification reactions were performed with following program: 95°C for 10 min and 45 cycles of 95°C for 5 s and 60°C for 30 s. The relative expression levels of DEGs were calculated with the 2−△△Ct method. qRT-PCR was performed with 3 biological replicates and 3 technical replicates for each experiment.
Supporting Information
(JPG)
(TIF)
ACT2: Actin-2; ACT7: Actin-7; REFA1: Elongation factor 1-alpha; TUBA1: Tubulin alpha-1 chain; UBQ10: Polyubiquitin 10.
(TIF)
(XLS)
(DOC)
(DOC)
Data Availability
All RNA-seq files are available from the NCBI BioProject database (accession number PRJNA309919).
Funding Statement
This work was funded by The Ministry of Agriculture Species Resource Production Project (15RZZY-29), The Fundamental Scientific Research Funds for CATAS-TCGRI (1630032013012), and The Special Fund for AGRO-Scientific Research in The Public Interest (201303117). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Li Z, Li T, Xu L, Panis B. Cryopreservation of Callerya speciosa (Champ.) Schot through droplet-vitrification. Propagation of Ornamental Plants. 2013;13(4):189–95. [Google Scholar]
- 2.Li L, Li Z, Li K, Huang B, Xu L. Development and characterization of EST-SSR markers in the Chinese medicinal plant Callerya Speciosa (Fabaceae). Applications in Plant Sciences. 2013;1(6):1200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navarro C, Cruz-Oró E, Prat S. Conserved function of FLOWERING LOCUS T (FT) homologues as signals for storage organ differentiation. Current opinion in plant biology. 2015;23:45–53. 10.1016/j.pbi.2014.10.008 [DOI] [PubMed] [Google Scholar]
- 4.Navarro C, Abelenda JA, Cruz-Oró E, Cuéllar CA, Tamaki S, Silva J, et al. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature. 2011;478(7367):119–22. 10.1038/nature10431 [DOI] [PubMed] [Google Scholar]
- 5.Lee R, Baldwin S, Kenel F, McCallum J, Macknight R. FLOWERING LOCUS T genes control onion bulb formation and flowering. Nature communications. 2013;4. [DOI] [PubMed] [Google Scholar]
- 6.González-Schain ND, Díaz-Mendoza M, Żurczak M, Suárez-López P. Potato CONSTANS is involved in photoperiodic tuberization in a graft—transmissible manner. The Plant Journal. 2012;70(4):678–90. 10.1111/j.1365-313X.2012.04909.x [DOI] [PubMed] [Google Scholar]
- 7.Kloosterman B, Abelenda JA, Gomez MdMC, Oortwijn M, de Boer JM, Kowitwanich K, et al. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature. 2013;495(7440):246–50. 10.1038/nature11912 [DOI] [PubMed] [Google Scholar]
- 8.Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Developmental cell. 2009;17(1):75–86. 10.1016/j.devcel.2009.06.015 [DOI] [PubMed] [Google Scholar]
- 9.Rodríguez-Falcón M, Bou J, Prat S. Seasonal control of tuberization in potato: conserved elements with the flowering response. Annu Rev Plant Biol. 2006;57:151–80. [DOI] [PubMed] [Google Scholar]
- 10.Xu X, van Lammeren AA, Vermeer E, Vreugdenhil D. The role of gibberellin, abscisic acid, and sucrose in the regulation of potato tuber formation in vitro. Plant Physiology. 1998;117(2):575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kloosterman B, Navarro C, Bijsterbosch G, Lange T, Prat S, Visser RG, et al. StGA2ox1 is induced prior to stolon swelling and controls GA levels during potato tuber development. The Plant Journal. 2007;52(2):362–73. [DOI] [PubMed] [Google Scholar]
- 12.Roumeliotis E, Kloosterman B, Oortwijn M, Kohlen W, Bouwmeester HJ, Visser RG, et al. The effects of auxin and strigolactones on tuber initiation and stolon architecture in potato. Journal of experimental botany. 2012;63(12):4539–47. 10.1093/jxb/ers132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faivre-Rampant O, Cardle L, Marshall D, Viola R, Taylor MA. Changes in gene expression during meristem activation processes in Solanum tuberosum with a focus on the regulation of an auxin response factor gene*. Journal of Experimental Botany. 2004;55(397):613–22. [DOI] [PubMed] [Google Scholar]
- 14.Guivarc'h A, Rembur J, Goetz M, Roitsch T, Noin M, Schmulling T, et al. Local expression of the ipt gene in transgenic tobacco (Nicotiana tabacum L. cv. SR1) axillary buds establishes a role for cytokinins in tuberization and sink formation. Journal of experimental botany. 2002;53(369):621–9. Epub 2002/03/12. . [DOI] [PubMed] [Google Scholar]
- 15.Martin A, Adam H, Díaz-Mendoza M, Żurczak M, González-Schain ND, Suárez-López P. Graft-transmissible induction of potato tuberization by the microRNA miR172. Development. 2009;136(17):2873–81. 10.1242/dev.031658 [DOI] [PubMed] [Google Scholar]
- 16.Bhogale S, Mahajan AS, Natarajan B, Rajabhoj M, Thulasiram HV, Banerjee AK. MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant physiology. 2014;164(2):1011–27. 10.1104/pp.113.230714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee AK, Chatterjee M, Yu Y, Suh S-G, Miller WA, Hannapel DJ. Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. The Plant Cell. 2006;18(12):3443–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin T, Sharma P, Gonzalez DH, Viola IL, Hannapel DJ. The impact of the long-distance transport of a BEL1-like messenger RNA on development. Plant physiology. 2013;161(2):760–72. 10.1104/pp.112.209429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahajan A, Bhogale S, Kang IH, Hannapel DJ, Banerjee AK. The mRNA of a Knotted1-like transcription factor of potato is phloem mobile. Plant molecular biology. 2012;79(6):595–608. 10.1007/s11103-012-9931-0 [DOI] [PubMed] [Google Scholar]
- 20.You MK, Hur CG, Ahn YS, Suh MC, Jeong BC, Shin JS, et al. Identification of genes possibly related to storage root induction in sweetpotato. FEBS letters. 2003;536(1):101–5. [DOI] [PubMed] [Google Scholar]
- 21.Kim S-H, Mizuno K, Fujimura T. Isolation of MADS-box genes from sweet potato (Ipomoea batatas (L.) Lam.) expressed specifically in vegetative tissues. Plant and cell physiology. 2002;43(3):314–22. [DOI] [PubMed] [Google Scholar]
- 22.Kim S-H, Hamada T, Otani M, Shimada T. Isolation and characterization of MADS box genes possibly related to root development in sweetpotato (Ipomoea batatas L. Lam.). Journal of Plant Biology. 2005;48(4):387–93. [Google Scholar]
- 23.Ku AT, Huang Y-S, Wang Y-S, Ma D, Yeh K-W. IbMADS1 (Ipomoea batatas MADS-box 1 gene) is involved in tuberous root initiation in sweet potato (Ipomoea batatas). Annals of botany. 2008;102(1):57–67. 10.1093/aob/mcn067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noh SA, Lee H-S, Huh EJ, Huh GH, Paek K-H, Shin JS, et al. SRD1 is involved in the auxin-mediated initial thickening growth of storage root by enhancing proliferation of metaxylem and cambium cells in sweetpotato (Ipomoea batatas). Journal of experimental botany. 2010;61(5):1337–49. 10.1093/jxb/erp399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka M, Kato N, Nakayama H, Nakatani M, Takahata Y. Expression of class I knotted1-like homeobox genes in the storage roots of sweetpotato (Ipomoea batatas). Journal of plant physiology. 2008;165(16):1726–35. 10.1016/j.jplph.2007.11.009 [DOI] [PubMed] [Google Scholar]
- 26.Firon N, LaBonte D, Villordon A, Kfir Y, Solis J, Lapis E, et al. Transcriptional profiling of sweetpotato (Ipomoea batatas) roots indicates down-regulation of lignin biosynthesis and up-regulation of starch biosynthesis at an early stage of storage root formation. BMC genomics. 2013;14(1):460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun P, Xiao X, Duan L, Guo Y, Qi J, Liao D, et al. Dynamic transcriptional profiling provides insights into tuberous root development in Rehmannia glutinosa. Frontiers in Plant Science. 2015;6:396 10.3389/fpls.2015.00396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan J, Song W, Zhou J, Wang X, Xie C, Gao X, et al. Transcriptome analysis reveals novel genes potentially involved in photoperiodic tuberization in potato. Genomics. 2013;102(4):388–96. 10.1016/j.ygeno.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 29.Sojikul P, Saithong T, Kalapanulak S, Pisuttinusart N, Limsirichaikul S, Tanaka M, et al. Genome-wide analysis reveals phytohormone action during cassava storage root initiation. Plant molecular biology. 2015;88(6):531–43. 10.1007/s11103-015-0340-z [DOI] [PubMed] [Google Scholar]
- 30.Yang J, An D, Zhang P. Expression Profiling of Cassava Storage Roots Reveals an Active Process of Glycolysis/GluconeogenesisF. Journal of integrative plant biology. 2011;53(3):193–211. 10.1111/j.1744-7909.2010.01018.x [DOI] [PubMed] [Google Scholar]
- 31.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC bioinformatics. 2003;4(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zong X, Lai F, Wang Z, Wang J. Studies on chemical constituents of root of Millettia speciosa. Journal of Chinese Medicinal Materials. 2009;32(4):520–1. [PubMed] [Google Scholar]
- 33.Wang Z, Lai F, Wang M, Wang J. Chemical constituents of the roots of Millettia speciose. Chinese Journal of Tropical Crops. 2011;32(12):2378–80. [Google Scholar]
- 34.Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome research. 2011;21(12):2213–23. 10.1101/gr.124321.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin J, Zhang H, Kong L, Gao G, Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic acids research. 2013:gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Husbands AY. Molecular and biochemical analyses of the LOB DOMAIN genes, a DNA-binding family of transcription factors in plants: University of California, Riverside; 2007. [Google Scholar]
- 37.Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant molecular biology. 2002;49(3–4):373–85. [PubMed] [Google Scholar]
- 38.Ravi V, Naskar S, Makeshkumar T, Babu B, Krishnan BP. Molecular physiology of storage root formation and development in sweet potato (Ipomoea batatas (L.) Lam.). J Root Crops. 2009;35(1):1–27. [Google Scholar]
- 39.Aksenova NP, Sergeeva LI, Kolachevskaya OO, Romanov GA. Hormonal regulation of tuber formation in potato. Bulbous Plants: Biotechnology. 2013:1. [Google Scholar]
- 40.Masuda J-I, Urakawa T, Ozaki Y, Okubo H. Short photoperiod induces dormancy in Lotus (Nelumbo nucifera). Annals of botany. 2006;97(1):39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Yang Y, Li X, Gu L, Wang F, Feng F, et al. Analysis of integrated multiple ‘omics’ datasets reveals the initiation and determination mechanisms of tuberous root formation in R. glutinosa. Journal of experimental botany. 2015:erv288. [DOI] [PubMed] [Google Scholar]
- 42.Hassidim M, Harir Y, Yakir E, Kron I, Green RM. Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta. 2009;230(3):481–91. 10.1007/s00425-009-0958-7 [DOI] [PubMed] [Google Scholar]
- 43.Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. The Plant Journal. 2001;26(1):15–22. [DOI] [PubMed] [Google Scholar]
- 44.Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognár L, Nagy F, et al. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature. 2002;419(6902):74–7. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Liu K, Liao H, Zhuang C, Ma H, Yan X. The plant WNK gene family and regulation of flowering time in Arabidopsis. Plant Biology. 2008;10(5):548–62. 10.1111/j.1438-8677.2008.00072.x [DOI] [PubMed] [Google Scholar]
- 46.Guo J-L, Yu C-L, Fan C-Y, Lu Q-N, Yin J-M, Zhang Y-F, et al. Cloning and characterization of a potato TFL1 gene involved in tuberization regulation. Plant Cell, Tissue and Organ Culture (PCTOC). 2010;103(1):103–9. [Google Scholar]
- 47.Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nature Plants. 2015;1(4). [DOI] [PubMed] [Google Scholar]
- 48.Kopriva S, Morris RJ. Transcript abundance explains mRNA mobility data in Arabidopsis thaliana. The Plant Cell. 2016:tpc. 00956.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li C, Gu M, Shi N, Zhang H, Yang X, Osman T, et al. Mobile FT mRNA contributes to the systemic florigen signalling in floral induction. Scientific reports. 2011;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eveland AL, Jackson DP. Sugars, signalling, and plant development. Journal of Experimental Botany. 2012;63(9):3367–77. 10.1093/jxb/err379 [DOI] [PubMed] [Google Scholar]
- 51.Perl A, Aviv D, Willmitzer L, Galun E. In vitro tuberization in transgenic potatoes harboring ß-glucoronidase linked to a patatin promoter: effects of sucrose levels and photoperiods. Plant science. 1991;73(1):87–95. [Google Scholar]
- 52.Ovono PO, Kevers C, Dommes J. Effects of reducing sugar concentration on in vitro tuber formation and sprouting in yam (Dioscorea cayenensis–D. rotundata complex). Plant Cell, Tissue and Organ Culture (PCTOC). 2009;99(1):55–9. [Google Scholar]
- 53.Xue J-P, Shi L-Y, Zhang A-M. Microtuber induction in vitro from Rehmannia glutinosa. China journal of Chinese materia medica. 2002;27(11):824–7. [PubMed] [Google Scholar]
- 54.Wang F, Sanz A, Brenner ML, Smith A. Sucrose synthase, starch accumulation, and tomato fruit sink strength. Plant Physiology. 1993;101(1):321–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Comparisons of Nutrition and Medicinal Qualities of Millettia speciosa Champ Root from Hainan and Guangxi Provinces in China. Modern Chinese Medicine. 2015;17(8):808–11. [Google Scholar]
- 56.Li X-Q, Zhang D. Gene expression activity and pathway selection for sucrose metabolism in developing storage root of sweet potato. Plant and cell physiology. 2003;44(6):630–6. [DOI] [PubMed] [Google Scholar]
- 57.Wang X, Chang L, Tong Z, Wang D, Yin Q, Wang D, et al. Proteomics Profiling Reveals Carbohydrate Metabolic Enzymes and 14-3-3 Proteins Play Important Roles for Starch Accumulation during Cassava Root Tuberization. Scientific Reports. 2016;6:19643 10.1038/srep19643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. The Plant Cell. 2005;17(8):2281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. The Plant Cell. 2011;23(3):1124–37. 10.1105/tpc.110.082792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krauss A, Marschner H. Influence of nitrogen nutrition, daylength and temperature on contents of gibberellic and abscisic acid and on tuberization in potato plants. Potato research. 1982;25(1):13–21. [Google Scholar]
- 61.Matsuo T, Yoneda T, Itoo S. Identification of free cytokinins and the changes in endogenous levels during tuber development of sweet potato (Ipomoea batatas Lam.). Plant and cell physiology. 1983;24(7):1305–12. [Google Scholar]
- 62.Nakatani M, Komeichi M. Changes in the endogenous level of zeatin riboside, abscisic acid and indole acetic acid during formation and thickening of tuberous roots in sweet potato. Jpn J Crop Sci. 1991;60(1):91–100. [Google Scholar]
- 63.Nakatani M, Komeichi M, Watanabe Y. Tuber sink potential in sweet potato (Ipomoea batatas Lam.) II. Estimation of tuber sink potential of cultivars using single leaf grafts. Jpn J Crop Sci. 1988;57(3):544–52. [Google Scholar]
- 64.Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. [DOI] [PubMed] [Google Scholar]
- 65.Moore B, Zhou L, Rolland F, Hall Q, Cheng W-H, Liu Y-X, et al. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300(5617):332–6. [DOI] [PubMed] [Google Scholar]
- 66.Balasubramanian R, Karve A, Moore BD. Actin-based cellular framework for glucose signaling by Arabidopsis hexokinase1. Plant signaling & behavior. 2008;3(5):322–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang C, Ding S, Zhang H, Du H, An L. CIPK7 is involved in cold response by interacting with CBL1 in Arabidopsis thaliana. Plant science. 2011;181(1):57–64. 10.1016/j.plantsci.2011.03.011 [DOI] [PubMed] [Google Scholar]
- 68.Schmidt R, Caldana C, Mueller-Roeber B, Schippers JH. The contribution of SERF1 to root-to-shoot signaling during salinity stress in rice. Plant signaling & behavior. 2014;9(1):e27540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chikano H, Ogawa M, Ikeda Y, Koizumi N, Kusano T, Sano H. Two novel genes encoding SNF1-related protein kinases from Arabidopsis thaliana: differential accumulation of AtSR1 and AtSR2 transcripts in response to cytokinins and sugars, and phosphorylation of sucrose synthase by AtSR2. Molecular and General Genetics MGG. 2001;264(5):674–81. [DOI] [PubMed] [Google Scholar]
- 70.Cosgrove DJ. Growth of the plant cell wall. Nature reviews molecular cell biology. 2005;6(11):850–61. [DOI] [PubMed] [Google Scholar]
- 71.Cosgrove DJ. Plant expansins: diversity and interactions with plant cell walls. Current opinion in plant biology. 2015;25:162–72. 10.1016/j.pbi.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hur YS, Um JH, Kim S, Kim K, Park HJ, Lim JS, et al. Arabidopsis thaliana homeobox 12 (ATHB12), a homeodomain-leucine zipper protein, regulates leaf growth by promoting cell expansion and endoreduplication. New Phytologist. 2015;205(1):316–28. 10.1111/nph.12998 [DOI] [PubMed] [Google Scholar]
- 73.Villordon AQ, Ginzberg I, Firon N. Root architecture and root and tuber crop productivity. Trends in plant science. 2014;19(7):419–25. 10.1016/j.tplants.2014.02.002 [DOI] [PubMed] [Google Scholar]
- 74.Orman-Ligeza B, Parizot B, Gantet PP, Beeckman T, Bennett MJ, Draye X. Post-embryonic root organogenesis in cereals: branching out from model plants. Trends in plant science. 2013;18(8):459–67. 10.1016/j.tplants.2013.04.010 [DOI] [PubMed] [Google Scholar]
- 75.Xie Q, Frugis G, Colgan D, Chua N-H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes & Development. 2000;14(23):3024–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal. 2006;46(4):601–12. [DOI] [PubMed] [Google Scholar]
- 77.Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, et al. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. The Plant Cell. 2007;19(1):270–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. The Plant Cell. 2005;17(11):2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhong R, Richardson EA, Ye Z-H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta. 2007;225(6):1603–11. [DOI] [PubMed] [Google Scholar]
- 80.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29(7):644–52. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. [DOI] [PubMed] [Google Scholar]
- 82.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic acids research. 2008;36(suppl 1):D480–D4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iseli C, Jongeneel CV, Bucher P, editors. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. ISMB; 1999. [PubMed] [Google Scholar]
- 84.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3(7):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(JPG)
(TIF)
ACT2: Actin-2; ACT7: Actin-7; REFA1: Elongation factor 1-alpha; TUBA1: Tubulin alpha-1 chain; UBQ10: Polyubiquitin 10.
(TIF)
(XLS)
(DOC)
(DOC)
Data Availability Statement
All RNA-seq files are available from the NCBI BioProject database (accession number PRJNA309919).