SUMMARY
Chromatin-mediated processes influence the development and progression of breast cancer. Using murine mammary carcinoma-derived tumorspheres as a functional readout for an aggressive breast cancer phenotype, we performed a loss-of-function screen targeting sixty epigenetic regulators. We identified the Polycomb protein Cbx8 as a key regulator of mammary carcinoma both in vitro and in vivo. Accordingly, Cbx8 is overexpressed in human breast cancer and correlates with poor survival. Our genomic analyses revealed that Cbx8 positively regulates Notch signaling by maintaining H3K4me3 levels on Notch-network gene promoters. Ectopic expression of Notch receptors partially rescues tumorsphere formation in Cbx8-depleted cells. We find that Cbx8 associates with non-PRC1 complexes containing the H3K4 methyltransferase complex component WDR5, which together regulate Notch gene expression. Thus, our study implicates a key non-canonical role for Cbx8 in promoting breast tumorigenesis.
INTRODUCTION
Chromatin modifiers have been increasingly reported as regulators of tumorigenicity (Shen and Laird, Vardabasso et al., 2014). Recent studies also suggest that tumor heterogeneity and tumor cell plasticity may be under epigenetic control (Easwaran et al., 2014). For example, in the context of breast cancer, EZH2 promotes the expansion of breast tumor initiating cells and KDM5B/JARID1B acts as an oncogene in luminal breast tumor cells (Chang et al., 2011, Yamamoto et al., 2014). Such epigenetic regulators, which mediate reversible changes at the chromatin level, may be targetable therapeutically (Campbell and Tummino, 2014). However, the epigenetic mechanisms at play in the development of breast cancer remain poorly understood.
Polycomb Repressive Complexes (PRC1 and PRC2) regulate chromatin states during development and have been linked to cancer (Aloia et al., 2013, Mills, 2010, Whitcomb et al., 2007). PRC2 performs its repressive function by catalyzing H3K27 methylation and PRC1 mediates gene silencing through mono-ubiquitylation of H2AK119 in an H3K27me3-dependent or -independent manner (Tavares et al., 2012). Canonical PRC1 is comprised of four subunits, including a Ring E3 ubiquitin ligase, Polyhomeotic, Posterior sex combs, and Polycomb (which binds H3K27me3 through its chromodomain) (Whitcomb et al., 2007). However, variant PRC1 complexes have been identified in mammals, which contain distinct Polycomb Group family members and/or alternate proteins, such as chromatin modifying enzymes and transcription factors (Gao et al., 2012).
Mammalian genomes encode five Polycomb orthologues known as the Chromobox (Cbx) members Cbx2, 4, 6, 7 and 8 (Bernstein et al., 2006, Whitcomb et al., 2007). We and others demonstrated that Cbx7 is the predominant Polycomb protein expressed in mouse embryonic stem cells (ESC), where it maintains self-renewal and pluripotency; while Cbx8 promotes ESC differentiation by repressing pluripotency genes (Morey et al., 2012, O'Loghlen et al., 2012). Both CBX7 and CBX8 have been linked to cancer through their ability to bypass senescence by repressing the Ink4a/Arf locus and cooperate with oncogenes to initiate hematopoietic malignancies (Dietrich et al., 2007, Gil et al., 2004, Scott et al., 2007, Tan et al., 2011). CBX8 is altered in a number of cancers including glioblastoma and esophageal squamous cell carcinoma (Li et al., 2013, Zhang et al., 2015), however a role for CBX8 in breast cancer remains unclear.
The Notch signaling pathway regulates normal mammary gland development and plays a key role in mammary progenitor cell maintenance (Reedijk, 2012). In breast cancer, upregulation of Notch receptors and ligands has been shown to correlate with high-grade tumors and poor patient prognosis, and confer drug resistance, particularly in the triple-negative subtype that lacks targeted therapies (Magnani et al., 2013, Reedijk et al., 2005). In addition, Notch signaling is required for tumor initiating properties (D'Angelo et al., 2015, Farnie et al., 2007). Genomic alternations of Notch gene loci appear rarely in solid tumors, and Notch hyperactivation may be driven by epigenetic events (Reedijk, 2012). Thus, targeting the Notch pathway through chromatin regulation may be an important avenue to treat breast cancer.
Using tumorspheres (TS) to enrich for highly tumorigenic cell populations, we have conducted an RNA interference (RNAi) screen to uncover epigenetic factors essential for breast tumorigenicity and identified the Polycomb ortholog Cbx8. Cbx8 promotes TS formation, mammary tumorigenesis in vivo and is upregulated in human breast tumors. We further demonstrate that Cbx8 sustains Notch gene expression by maintaining the transcriptionally active histone modification H3K4me3 on Notch gene promoters, as well as other Notch-network genes. We find that Cbx8 interacts with Wdr5, a core component of H3K4 methyltransferase complexes, and that loss of Wdr5 phenocopies Cbx8 loss. Collectively, the functional and biochemical studies presented here demonstrate a non-canonical role for Cbx8 in breast cancer through activation of genes involved in Notch signaling.
RESULTS
Mammary tumorspheres enrich for tumorigenic cells and provide a robust screening system
In order to identify chromatin regulators required for breast tumorigenicity we used TS culture, which enriches for cells with tumor initiating properties (Dontu et al., 2003, Kurpios et al., 2013). We utilized the mammary carcinoma mouse model MMTV-Myc, which generates heterogeneous and highly aggressive tumors (Andrechek et al., 2009, Bosch et al., 2012) and reproducibly generates TS (Figure 1A). By culturing cells from MMTV-myc tumors in bulk (adherent) or TS conditions, we detected an increase of CD49f+/CD24− population, suggesting enrichment of cells associated with basal subtype characteristics (Figure S1A). Further, RNA-seq analysis of bulk versus TS cultures revealed a distinct high-grade tumor and basal subtype gene expression program in TS (Figure S1B, S1C and Table S1). Importantly, we demonstrated that TS cells are more tumorigenic than bulk cells through in vivo mammary fat pad injections at limiting dilutions (Figure 1B). This suggests that by culturing mammary tumor cells as TS, we enrich for a cell population with higher tumorigenic potential. Because we observed that propagating MMTV-Myc TS was quite robust in comparison to TS from other tumor models (e.g. MMTV-neu model; data not shown), we used this model for pooled RNAi screens, which requires selection over time to allow effective competition of shRNAs.
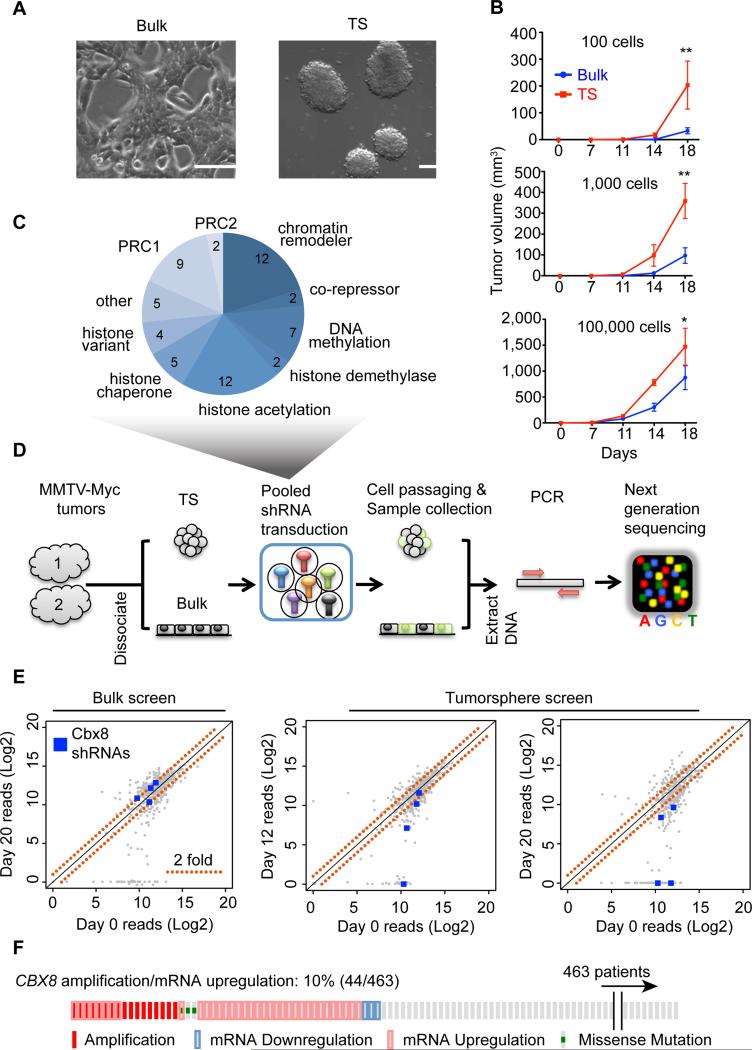
Figure 1. Functional RNAi screen targeting epigenetic factors in TS.
(A) Bright field images of MMTV-Myc bulk and TS cells. Scale bar = 100 μm. (B) In vivo tumor growth (mammary fat pad) of bulk and TS MMTV-Myc cells at 1 × 102, 1 × 103 and 1 × 105 cells per injection. Mean ± s.e.m (n=4 injections per group), *p<0.05; **p<0.001. (C) Pie chart depicting the composition of the chromatin-focused shRNA library used in the TS screen. Numbers indicate genes per category (total = 60). (D) Mammary TS RNAi screening strategy. (E) Scatter plots of normalized shRNA reads of bulk and TS screens at the indicated time points. Significantly depleted Cbx8 shRNAs (p<0.1) are highlighted in blue; red dotted lines represent 2-fold change. (F) CBX8 alterations in breast cancer patients (n=463) from TCGA datasets (Cancer Genome Atlas, 2012).
TS loss-of-function screen identifies a dependency on Cbx8
We developed a functional screen in TS culture using lentiviral transduction of a pool of shRNAs, followed by high-throughput sequencing. We created and utilized an shRNA library targeting 60 epigenetic factors (Figure 1C and Table S2), averaging 7 shRNAs per gene (total of 452 shRNAs). Cells were dissociated from two transplanted MMTV-Myc tumors and cultured as TS, which were maintained in suspension during the entire screening process to maintain tumorigenic properties. Two independent TS cultures from each tumor were cultured to serve as technical replicates. In addition, we performed the screen in bulk cells as a control for shRNAs that affect proliferation or survival. Bulk and TS cells were collected at three time points (baseline, day 12 and day 20), genomic DNA extracted, the shRNA pool amplified by PCR and subjected to high-throughput sequencing analysis (Figure 1D).
Over 90% of shRNAs were present (>500 reads) at baseline, which were used as a reference for comparison with later time points. In addition, the average reads between the two tumors showed high correlation as they clustered together at each time point using unsupervised hierarchical clustering (Figure S1D). The screen produced 18% of shRNAs with significant TS-specific depletions (Figure S1E and Table S3). The candidates were then further filtered by the following criteria: (1) genes with >2 shRNAs present in the library at baseline and (2) >33% shRNAs significantly changed. The resulting hits were ranked by their percent of genomic alterations from The Cancer Genome Atlas (TCGA) datasets for breast cancer (Figure S1F). The Polycomb family member Cbx8 was amongst the top compelling candidates, which showed significant TS-specific shRNA depletion at both early and late time points (Figure 1E) and is amplified and/or upregulated transcriptionally in 10% of breast tumors (Figure 1F).
Cbx8 promotes a tumorigenic phenotype in breast cancer cells
We validated Cbx8 as a candidate using two individual shRNAs that were contained within the shRNA pool (Figure 2A,B). In addition, we knocked down human CBX8 in four distinct human breast cancer cell lines including MCF7 (luminal), T47D (luminal), MDA-MB-157 (basal) and MDA-MB-231-Luc (basal) (Figure 2C). We observed that knock down of Cbx8 in both mouse and human cells significantly decreased TS formation (Figure 2B, 2C). These results not only validate our TS screening approach, but also extend the mouse mammary carcinoma findings to human breast cancer cells.
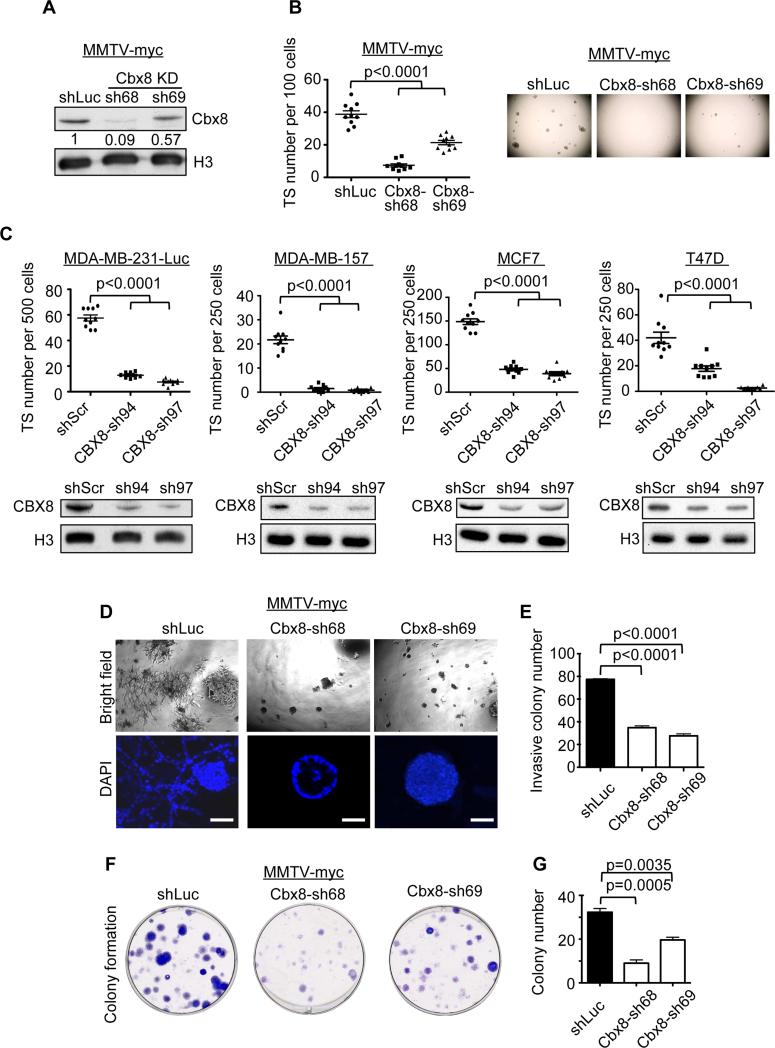
Figure 2. Cbx8 sustains tumorigenic phenotypes of mammary carcinoma cells.
(A) Immunoblot of Cbx8 knockdown using two individual shRNAs in MMTV-Myc cells. Histone H3 used as loading control; numbers indicate relative intensity of bands normalized to H3. (B) RNAi screen validation of TS formation upon Cbx8 knockdown (left). Mean ± s.e.m (n=10). Representative images of individual 96 wells are shown (right). (C) TS formation in human breast cancer cell lines upon CBX8 knockdown (top). Mean ± s.e.m (n=10). Immunoblot of control and CBX8 knockdown cells (bottom). Histone H3 used as loading control. (D) Bright field and confocal images of control and Cbx8 knockdown MMTV-Myc cells plated on Matrigel. Scale bars = 50 μm. (E) Quantification of invasive colonies in (D). Mean ± s.e.m (n=3). (F) Clonogenic ability of control and Cbx8 knockdown MMTV-Myc cells. (G) Quantification of clonogenicity in (F). Mean ± s.e.m (n=3).
Next, we performed functional assays to assess the effect of Cbx8 depletion in both mouse and human cell lines. We cultured cells in extracellular matrix (Matrigel) to examine their invasiveness versus differentiation capacity in 3D culture (Debnath et al., 2003). For example, non-tumorigenic MCF10A cells form highly organized acinar structures, whereas MDA-MB-231-Luc cells formed disorganized structures that invade the extracellular matrix (Figure S2A). By knocking down Cbx8 in MMTV-Myc and MDA-MB-231-Luc cells, we observed a significant reduction of invasive colonies, with some displaying cavitation, which is typical of acini morphogenesis (Figure 2D, 2E, S2B and S2C). We also performed clonogenicity assays to examine the ability of cells to initiate colonies at low density. Using control (shLuc or shScr) and Cbx8 knockdown in MMTV-Myc and MDA-MB-231-Luc cells, we observed a requirement of Cbx8 for colony initiation in vitro (Figure 2F, 2G, S2D and S2E). While Cbx8 has been reported to repress Ink4a/Arf, we did not observe senescence-like growth arrest (Figure S2F and S2G) or the induction of senescence markers upon Cbx8 knockdown (Figure S2H and S2I). In addition, we did not observe clear perturbation of Epithelial-To-Mesenchymal (EMT) markers and luminal/basal markers upon Cbx8 loss (Figure S2H and S2I). This suggests that Cbx8 functions to regulate alternative gene expression programs in breast cancer.
Cbx8 is required for tumor growth in vivo and correlates with poor survival
We next investigated the role of Cbx8 in mediating tumorigenesis in vivo. To do so, only 1,000 control and Cbx8 knockdown MMTV-Myc cells were injected into mouse mammary fat pads and monitored over a period of 24 days. Cbx8 knockdown significantly reduced tumor growth as measured by both tumor size and weight (Figure 3A), suggesting that Cbx8 is indeed required for tumor growth in vivo.
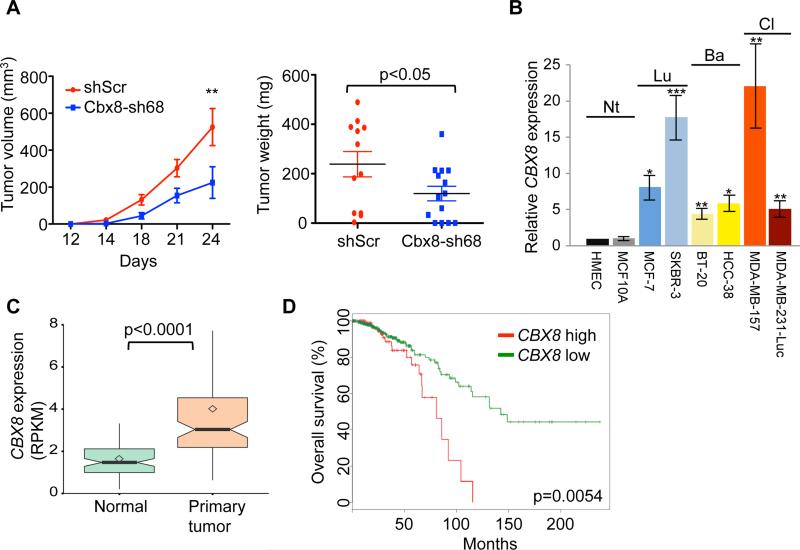
Figure 3. Cbx8 promotes mammary tumorigenesis in vivo.
(A) Tumor growth upon mammary fat pad injection of 1 × 103 control and Cbx8 knockdown MMTV-Myc cells; tumor size and tumor weight (day 24, end point) shown. Mean ± s.e.m (n=13 for shScr, n=15 for shCbx8). **p<0.001. (B) RT-qPCR analysis of CBX8 mRNA expression in 2 normal and 6 breast cancer cell lines, including non-tumorigenic (Nt), luminal (Lu) and basal (Ba) molecular subtypes. GAPDH was used for normalization. Mean ± s.e.m (n=3). *p<0.05, **p<0.01, ***p<0.001 (all tested to HMEC). (C) Boxplot of CBX8 mRNA expression in paired normal tissue and primary tumor (n=94); RNA-seq from TCGA. Square dots = sample mean; bold lines = median; boxes = 25-75 percentiles;. whiskers = 1.5x IQR; notches = 95% CI. Outliers are not shown. (D) Kaplan-Meier graph representing the probability of cumulative overall survival in breast cancer patients (n=597). Tumors were grouped by top 25% (CBX8 high) and lower 75% CBX8 (CBX8 low) expression. Logrank test p value is shown. Patient data from (C) and (D) extracted from TCGA (Cancer Genome Atlas, 2012).
We next extended our studies to human breast cancer. Interestingly, breast cancer is one of the most common cancer types with CBX8 genomic alterations (Figure S3A) (Gao et al., 2013). First, we examined the expression levels of CBX8 by qPCR in a panel of breast cancer cell lines that includes non-tumorigenic cells (human mammary epithelial cells and MCF10A) as well as luminal and basal subtypes. All of the tumorigenic cells displayed a significant increase of CBX8 levels regardless of molecular subtype (Figure 3B). Next, we examined the TCGA datasets to identify correlations between CBX8 expression and the occurrence of breast cancer. CBX8 copy number amplification correlates with high mRNA levels, suggesting that the increased expression can be partially explained by copy number gain (Figure S3B). Moreover, CBX8 mRNA levels are higher in primary tumors versus normal tissues (Figure 3C). Similar to breast cancer cell lines (Figure 3B), increased levels of CBX8 in patient samples does not correlate with a specific subtype (Figure S3C), suggesting that CBX8 is a general regulator of breast tumorigenesis. Finally, from TCGA datasets and two other studies (Cancer Genome Atlas, 2012, Hu et al., 2006b, Loi et al., 2008), high CBX8 expression predicts poor prognosis (Figure 3D, S3D and S3E).
Cbx8 maintains a stem cell-like gene expression program and Notch signaling
To further understand the role of Cbx8 in promoting tumorigenesis, we conducted transcriptional profiling by RNA sequencing (RNA-seq). By comparing mRNA expression between control and Cbx8 knockdown MMTV-Myc cells, we found 188 genes upregulated (‘Cbx8 KD up’) and 259 genes down-regulated (‘Cbx8 KD down’) with greater than two-fold change (Figure 4A and Table S1). Gene Ontology (GO) analysis revealed that Cbx8 KD up genes are involved in stimulus response and nucleosome assembly, whereas Cbx8 KD down genes are involved in developmental processes (Figure S4A and Table S1). Using GSEA, we found a significant anti-correlation of Cbx8 knockdown with fetal mammary stem cell (fMaSC) and embryonic stem cell (ESC) gene signatures (Figure 4B) (Ben-Porath et al., 2008, Spike et al., 2012, Wong et al., 2008). Collectively, these data indicate that Cbx8 maintains a stem cell-like gene expression program.
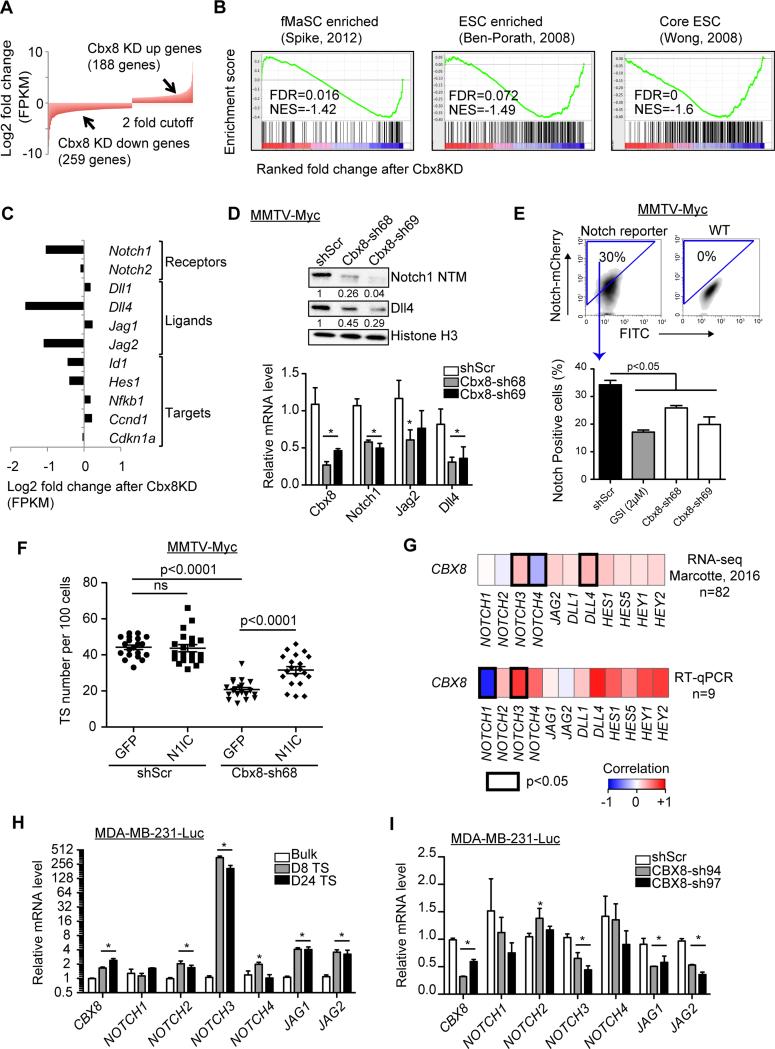
Figure 4. Cbx8 regulates stem cell-like gene expression including Notch signaling.
(A) Representation of genes up- and down-regulated (>2 fold) after Cbx8 knockdown in MMTV-Myc cells, as analyzed by RNA-seq. (B) Gene set enrichment analysis (GSEA) of gene expression changes upon Cbx8 knockdown in MMTV-Myc cells using published stem cell gene signatures. (C) Representation of Notch-network gene expression changes (RNA-seq) upon Cbx8 knockdown in MMTV-Myc cells. (D) Top: Immunoblotting of Notch1 NTM (cleaved intracellular/transmembrane domain) and Dll4 in MMTV-Myc whole cell lysate. Histone H3 used as loading control; numbers indicate relative intensity of bands normalized to H3. Bottom: RT-qPCR of control and Cbx8 knockdown MMTV-Myc cells. Rpl7 was used for normalization. Mean ± s.e.m (n=3). *p<0.05 compared to shScr. (E) Notch-mCherry reporter activity in control and Cbx8 knockdown MMTV-Myc TS cells (bottom). Notch-mCherry positive population was determined by FACS (top); 2μM of GSI; PF-384014 was used as positive control for Notch inhibition. Mean ± s.e.m (n=3). (F) TS formation upon ectopic Notch expression in control and Cbx8 knockdown MMTV-Myc cells (n=20). ns = not significant. (G) Heatmap showing the Spearman correlation of mRNA expression between CBX8 and NOTCH genes from RNA-seq in 82 human mammary cell lines (top) (Marcotte et al., 2016) and RT-qPCR from 9 human mammary cell lines (bottom; eight from Figure 3B, and MDA-MB-231 cells). Significant correlations (p<0.05) are marked with black boxes. (H) RT-qPCR of Notch gene expression in bulk, day 8 and day 24 TS of MDA-MB-231-Luc cells. GAPDH was used for normalization. Mean ± s.e.m (n=3). (I) RT-qPCR of Notch gene expression in control and CBX8 knockdown MDA-MB-231-Luc cells. GAPDH was used for normalization. Mean ± s.e.m (n=3). *p<0.05 compared to shScr.
We next performed network analysis using MetaCore software. In addition to several known oncogenic factors that are altered, such as Wnt signaling components (Figure S4B), we found the Notch signaling network to be most enriched upon Cbx8 KD down (Table S4). Specifically, we observed that the Notch1 receptor and its ligands Dll4 and Jag2 were downregulated (>2 fold) in Cbx8 KD MMTV-Myc cells (Figure 4C). This was confirmed by immunoblotting and RT-qPCR with two individual shRNAs (Figure 4D). To determine whether this translates into a diminution of Notch signaling, we utilized a Notch reporter in MMTV-Myc TS. Indeed, Cbx8 loss resulted in significant decrease of Notch activity and one shRNA (Cbx8-sh69) produced a similar level of reduction as treatment with a gamma secretase inhibitor (GSI) (Figure 4E).
We reasoned that if Cbx8 regulates Notch signaling then ectopic Notch1 expression should rescue the TS defect caused by Cbx8 loss. To test this, we knocked down Cbx8 in MMTV-Myc cells followed by ectopic expression of N1IC (Notch1 intracellular domain) and tested the ability of the cells to form TS. Knockdown efficiency of Cbx8 and Notch expression was validated by RT-qPCR and immunoblotting (Figure S4C, S4D). As expected, we observed decreased TS formation upon Cbx8 depletion. However, when N1IC was expressed in Cbx8 knockdown cells, TS formation was partially restored (Figure 4F). Together, these data indicate that Notch signaling is an important downstream effector of Cbx8-mediated tumorigenesis.
To determine if the relation between Cbx8 and Notch holds true in human breast cancer, we associated the expression of NOTCH-network genes to the levels of CBX8 using RNA-seq data from a panel of 82 human breast cell lines (Marcotte et al., 2016) and validated by RT-qPCR using 9 of these cell lines. We found that CBX8 positively correlates with several NOTCH receptors, ligands and target genes (Figure 4G). Here, CBX8 most significantly correlated with NOTCH3 of the four NOTCH receptors. Further, we observed a significant upregulation of NOTCH3 receptor, as well as JAG1 and JAG2 ligand, upon TS formation of MDA-MB-231-Luc cells compared to bulk cells (Figure 4H). In accordance, knockdown of CBX8 in MDA-MB-231-Luc cells significantly decreased NOTCH3, JAG1 and JAG2 expression (Figure 4I). Furthermore, CBX8 knockdown in three additional human breast cancer cell lines showed that JAG2 is consistently downregulated upon CBX8 loss, while NOTCH3 decrease occurred more modestly (Figure S4E). Collectively, our data suggest that Cbx8 positively regulates Notch-network genes in both human and mouse breast cancer cells, with differential Notch receptor-ligand regulation.
Cbx8 is required for Notch expression in vivo
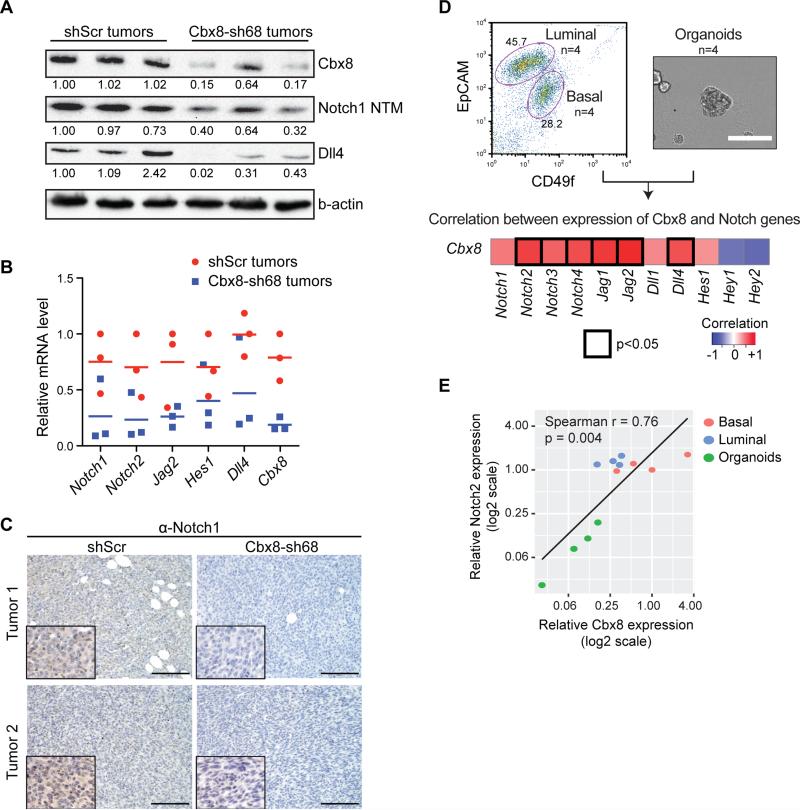
We next examined the levels of Notch-network genes in transplanted tumors derived from control and Cbx8 knockdown MMTV-myc cells. Cbx8 knockdown tumors showed reduction of Notch1 and Dll4 protein levels (Figure 5A). Moreover, we found a decrease in expression of Notch1, Dll4 and Jag2, genes that were identified by RNA-seq analysis (Figure 4C), amongst others (Figure 5B). Immunostaining of these tumors (see Figure 3A), showed a significant reduction of Notch1 signal in Cbx8 knockdown tumors as compared to controls (Figure 5C). These data suggest that Cbx8 regulates Notch gene expression in vivo.
Figure 5. Cbx8 promotes Notch expression in mammary tumors and mammary epithelial cells.
(A) Immunoblotting of Cbx8, Notch1 NTM and Dll4 in whole cell lysate of MMTV-Myc tumors (n=3 individual tumors). B-actin was used as loading control; numbers indicate relative intensity of bands normalized to b-actin. (B) RT-qPCR of Notch gene expression in control and Cbx8 knockdown MMTV-Myc tumors (see Figure 3). Individual data points and means are shown (n=3 individual tumors). Rpl7 was used for normalization. (C) Notch1 immunostaining of control and Cbx8 knockdown MMTV-Myc tumors. Representative images were taken (20x) with nuclei insets (40x) from two individual tumors. Scale bar = 100 μm. (D) Top: FACS analysis of mouse mammary epithelial cells for luminal and basal populations (left). Bright field image showing mammary organoids (right). Scale bar = 200 μm. Bottom: heatmap representing the Spearman correlation of mRNA expression between Cbx8 and Notch genes in mouse mammary gland and organoid culture (n=4 for each luminal, basal and organoids; total n=12). Significant correlations (p<0.05) are marked with black boxes. (E) Scatter plot representing the expression correlation between Cbx8 and Notch2 from (D).
We sought to extend our observations to the epithelial cells of the normal mammary gland. Notch has been reported to repress the mammary stem cell state, while promoting differentiation of the luminal lineage in mice (Bouras et al., 2008). Interestingly, Cbx8 and Notch receptor-ligand expression positively correlates in mammary epithelial cells. In fact, by correlating the levels of Cbx8 and the Notch-network genes from isolated luminal and basal populations, together with organoid cultures that enrich for mammary epithelial stem cells (MaSC), we identified a strong correlation between Notch receptors/ligands and Cbx8 (Figure 5D, S5A). For example, organoid cultures of MaSC, which are low for Notch2 expression, are also low for Cbx8 (Figure 5E). Conversely, basal and luminal populations have higher levels of both (Figure 5E). Moreover, knockdown and overexpression of Cbx8 in mammary organoid cultures leads to decreased and increased levels of Notch2, respectively (Figure S5B, C). Thus, we find that Notch signaling is also regulated by Cbx8 in normal mammary epithelial cells,.
Cbx8 maintains H3K4me3 at Notch-network gene loci
To gain insight into the mechanism by which Cbx8 regulates Notch-network gene expression, we performed chromatin immunoprecipitation coupled to high-throughput sequencing (ChIP-seq) for histone modifications associated with gene expression in control and Cbx8 knockdown MMTV-Myc cells. We examined H3K27me3, the repressive modification directly bound by PRC1 complexes, as well as H3K27ac and H3K4me3, which mark actively transcribed genes.
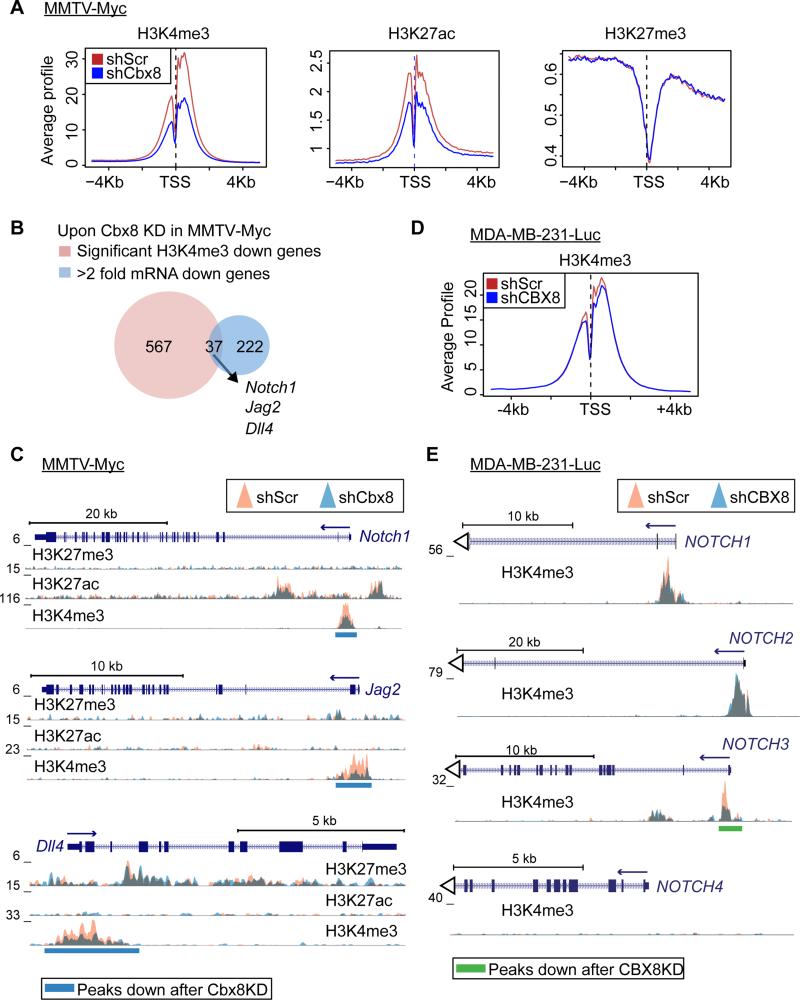
Strikingly, Cbx8 loss resulted in a global decrease of H3K4me3 and H3K27ac, whereas H3K27me3 did not show global changes (Figure 6A and S6A). Using SICER-df, which identifies regions with significantly changed ChIP signals between samples, we found regions with significant changes (FDR<1x10−8 and fold change >1.8) upon Cbx8 knockdown. By mapping these significantly altered regions to genes, we found H3K4me3 changes to be the most evident with over 600 genes showing reduced H3K4me3 level near their transcription start site (TSS) (Figure S6B and Table S5). By overlapping these genes with Cbx8 KD down genes, we found that Notch1, Jag2 and Dll4 displayed both decreased mRNA expression and significant loss of H3K4me3 at their TSS (Figure 6B). By examining the profile of these loci, we observed significant H3K4me3 loss at their TSS without significant changes in H3K27ac or H3K27me3 (Figure 6C). Of note, genes with H3K4me3 increase were also identified in our ChIP-seq analysis (Figure S6C).
Figure 6. Cbx8 maintains H3K4me3 at Notch-network gene loci.
(A) Average enrichment signal of H3K4me3, H3K27ac and H3K27me3 at all annotated TSS (−5kb to +5kb) in control and Cbx8 knockdown MMTV-Myc cells. (B) Venn-diagram representing genes with reduced mRNA expression (>2 fold decrease) and significant H3K4me3 reduction (FDR<1×10−8 and >1.8 fold decrease) upon Cbx8 knockdown in MMTV-Myc cells. Overlapping genes indicated. (C) Overlay of H3K27me3, H3K27ac and H3K4me3 (fold enrichment over input) at Notch1, Jag2 and Dll4 loci in control (orange) and Cbx8 knockdown (blue) MMTV-Myc cells. Significantly decreased regions (FDR=1×10−8) underscored (blue bars). (D) Average enrichment signal of H3K4me3 at all annotated TSS (−5kb to +5kb) in control and CBX8 knockdown MDA-MB-231-Luc cells. (E) Overlay of H3K4me3 at NOTCH1, NOTCH2, NOTCH3 and NOTCH4 in control and CBX8 knockdown MDA-MB-231-Luc cells. Significantly decreased regions (FDR=2×10−3) underscored (green bars).
To extend these observations to human breast cancer cells, we performed H3K4me3 ChIP-seq in control and CBX8 knockdown MDA-MB-231-Luc cells (Figure 6D and S6D). Consistent with our findings of CBX8-regulated genes in human cells, NOTCH3 showed a significant decrease of H3K4me3 at its TSS (Figure 6E). Such a decrease was not observed at the NOTCH1, 2 and 4 loci (Figure 6E), confirming NOTCH3 as an effector of CBX8 in MDA-MB-231-Luc cells. Together, our data suggest that CBX8 regulates specific NOTCH receptor expression through regulation of H3K4me3, and indicates a non-canonical role for CBX8 by promoting gene activation.
Cbx8 acts in a non-canonical fashion in breast cancer
To dissect the molecular mechanism of Cbx8 function, we generated epitope tagged Cbx8 stable MMTV-Myc and MDA-MB-231-Luc cell lines. Cbx8 localizes primarily to the chromatin fraction (Figure S7A and S7B) and interacts with PRC1 members, including Ring1b and Bmi1, suggesting that Cbx8 can be incorporated into canonical PRC1 complexes (Figure 7A and S7C). By eliminating H3K27me3 via EZH1/2 inhibitor treatment (Konze et al., 2013), we observed a modest, but not complete, reduction of chromatin-bound Cbx8 in both cell lines (Figure S7D and S7E). These data suggest that Cbx8 has the potential to function through canonical PRC1 in these cells.
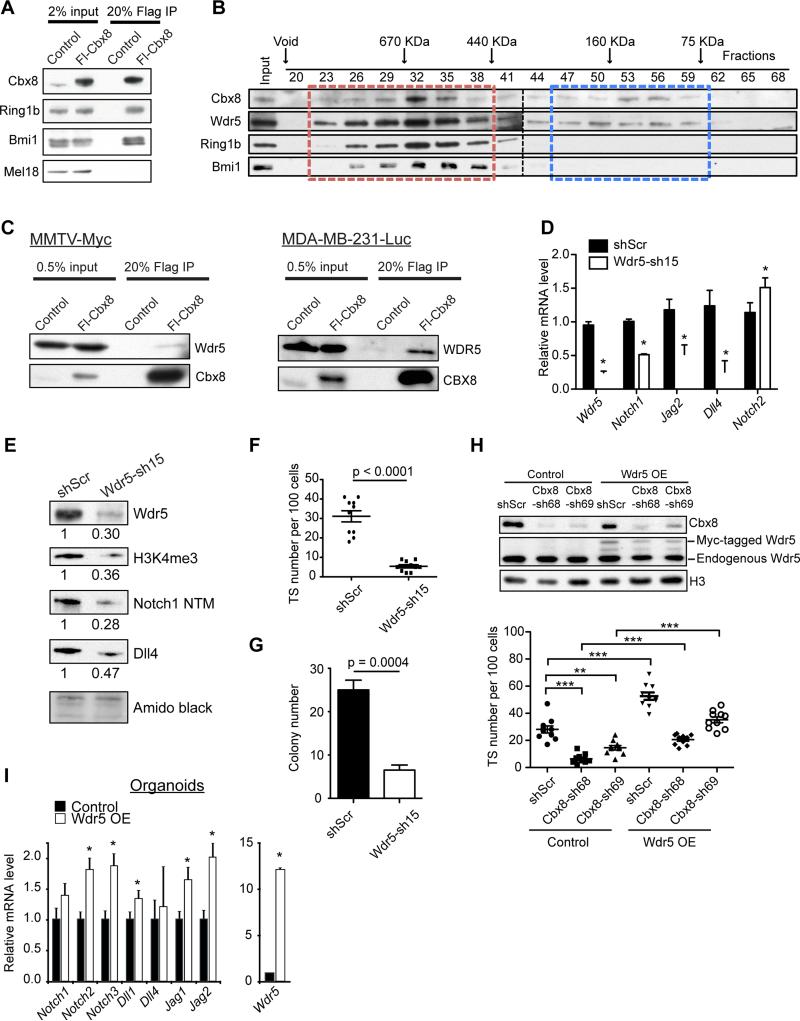
Figure 7. Cbx8 interacts with Wdr5 in breast cancer cells.
(A) Co-immunoprecipitation of Flag-Cbx8 with endogenous PRC1 members in MMTV-Myc cells. (B) Size exclusion chromatography followed by immonublotting of Cbx8-associated complexes in MMTV-Myc cells. Cbx8-containing PRC1 and non-PRC1 complexes are indicated by red and blue boxes, respectively. (C) Co-immunoprecipitation of Flag-Cbx8 with Wdr5 in MMTV-Myc and MDA-MB-231-Luc cells. (D) RT-qPCR of control and Wdr5 knockdown MMTV-Myc cells. Rpl7 was used for normalization. Mean ± s.e.m (n=3). *p<0.05 compared to shScr. (E) Immunoblotting of control and Wdr5 knockdown MMTV-Myc whole cell lysate. Amido black staining of histones used as loading control; numbers indicate relative intensity of bands normalized to amido black. (F) TS formation of control and Wdr5 knockdown MMTV-Myc cells. Mean ± s.e.m (n=10). (G) Clonogenic ability of control and Wdr5 knockdown MMTV-Myc cells. Mean ± s.e.m (n=4). (H) Immunoblots (top) and TS formation (bottom) upon ectopic Myc-tagged Wdr5 expression in control and Cbx8 knockdown MMTV-Myc cells (n=10). Histone H3 used as loading control. **p<0.01, ***p<0.0001. (I) RT-qPCR of Notch-network genes in control and Wdr5 overexpressed mouse mammary organoids. Rpl7 was used for normalization. Mean ± s.e.m (n=3). *p<0.05 compared to control.
To test whether Cbx8 acts to promote tumorigenesis in the context of PRC1, we knocked down Ring1b in MMTV-Myc cells. Loss of Ring1b did not compromise TS formation, clonogenicity, or proliferation in MMTV-Myc cells (Figure S7F-H). This is consistent with the fact that while Ring1b shRNAs were present in our library, they were not depleted in our TS screen. Furthermore, Ring1b knockdown did not reduce Notch-network gene expression (Figure S7I). This suggests that while Cbx8 can participate in PRC1 complexes, its role in promoting breast cancer may be independent of canonical PRC1.
To further investigate Cbx8-containing complexes, we performed size exclusion chromatography. By screening for endogenous Cbx8-containing complexes, we identified multiple Cbx8 sub-complexes in MMTV-Myc cells (Figure 7B). We observed a canonical PRC1 complex, consisting of Cbx8, Bmi1 and Ring1b, as well as an uncharacterized Cbx8 complex lacking PRC1 members. Here, Cbx8 co-fractionated with Wdr5, a core member of the MLL methyltransferase complexes, which methylates H3K4. In addition, we observed an interaction between Cbx8 and Wdr5 in both MMTV-Myc and MDA-MB-231-Luc cells (Figure 7C). While the Cbx8-Wdr5 interaction is not as robust as the canonical Cbx8-PRC1 interactions (Figure 7A), these results suggest that Cbx8 has gene-activating roles, as we have observed for Notch-network genes.
Functionally, Wdr5 knockdown significantly impaired TS formation, clonogenicity, invasiveness, as well as the expression of specific Notch-network genes in both MMTV-Myc and MDA-MB-231-Luc cells (Figure 7D-G and S7J-N), suggesting that loss of Wdr5 recapitulates the effects of Cbx8 depletion. Moreover, ectopic expression of Wdr5 enhances TS formation and rescues TS deficiency in Cbx8 knockdown cells (Figure 7H). Finally, by using organoid cultures, which have low expression of Notch receptors/ligands (Figure S5A), Wdr5 overexpression resulted in increased levels of Notch-network genes (Figure 7I). Together, these data suggest that Cbx8 and Wdr5 act cooperatively to regulate normal mammary stem cells and breast tumorigenesis through regulation of Notch-network genes. Therefore, our studies implicate a non-PRC1 dependent role for Cbx8 in regulating H3K4me3 in the context of breast cancer.
DISCUSSION
A role for Cbx8 in breast cancer
Through a chromatin-focused RNAi screen using TS, we have unveiled a role for Cbx8 in promoting breast cancer; Cbx8 is required for tumorigenesis and malignant phenotypes of breast cancer cells. CBX8 is overexpressed in primary breast tumors and high CBX8 expression in patients correlates with poor outcome. We found that Cbx8 exerts its function in breast cancer through maintaining gene expression of Notch signaling components. Together, our data point towards a pivotal role for Cbx8 in promoting breast tumorigenesis.
Data from TCGA show that CBX8 is altered in a broad range of cancer types.. Among these, breast cancer has one of the highest rates of CBX8 genomic alteration, predominately amplification (Figure S3A). This suggests that mammary epithelial cells are sensitive to CBX8 dysregulation. Given that breast tissue contains various populations of stem and progenitor cells (Visvader and Stingl, 2014), and that our data suggest Cbx8 maintains tumorigenesis via a stem-like gene expression program, investigating the role of Cbx8 in normal mammary development and stem/progenitor cells might shed light on the role of Cbx8 in breast tumor initiation.
Cbx8 acts in a non-canonical manner in breast cancer
Here, we find that Cbx8 function is not entirely repressive and can activate transcription in a PRC1-independent fashion. First, our ChIP-seq data demonstrate (i) that Cbx8 is required for maintaining H3K4me3, a histone modification found at the TSS of active genes (Bernstein et al., 2005, Gaspar-Maia et al., 2011) and (ii) a positive correlation between H3K4me3 levels and mRNA expression of Notch-network genes. Secondly, knock down of an integral PRC1 member, Ring1b, failed to phenocopy Cbx8 loss-of-function in breast cancer cells. Finally, we identified a non-PRC1 complex containing Cbx8 that co-fractionates with Wdr5, and further validated Wdr5 as a Cbx8 interacting protein. These interactions may account for the non-canonical functions of Cbx8 in breast cancer. However, we cannot exclude a role for canonical PRC1 complex in tumorigenesis.
In agreement with our findings, recent evidence suggests that Cbx8 is associated with gene activation in certain cellular contexts. In human fibroblasts, 38% of CBX8 binding regions contain H3K4me3, and 28% of CBX8 targets are actively transcribed (Pemberton et al., 2014). In ESC, Cbx8 is transiently bound to lineage genes for activation during early ESC differentiation (Creppe et al., 2014). In addition, CBX8 can induce leukemogenesis by cooperating with MLL-AF9 fusion protein and Tip60 to activate HOX gene transcription. This function of CBX8 is likely PRC1-independent, as RING1B and BMI1 knockdown did not recapitulate the effect of CBX8 loss (Tan et al., 2011). Together, these data suggest that CBX8 can activate gene expression in either a PRC1-dependent or -independent fashion. Collectively, we propose that CBX8's role in activating gene expression could be transient in normal cells, but become deregulated and stable in tumor cells.
A role for Cbx8 in tumor initiating cells?
Breast cancer contains a population of stem-like cells with higher tumorigenic potential called breast tumor initiating cells (BTIC) (Al-Hajj et al., 2003). The presence of such cells is associated with drug resistance and poor patient survival (Creighton et al., 2009, Pece et al., 2010). Consistent with previous studies (Dontu et al., 2003, Kurpios et al., 2013), we show that TS culture functionally enriches for cells with BTIC properties. Here, we uncover that Cbx8 is required for TS formation, as well as other BTIC properties such as the ability to initiate colonies at low cell confluency and to form tumors in vivo at low cell numbers. Furthermore, we show that Cbx8 regulates Notch signaling, a pathway required to maintain BTIC populations (Yamamoto et al., 2013). Our data also suggests that Cbx8 regulates other known BTIC pathways, such as Wnt signaling (Jang et al., 2015). Together, these data implicate a potential role for Cbx8 in mediating BTIC populations through Notch signaling and other oncogenic pathways for future exploration.
Epigenetic regulation of Notch in breast cancer
It is established that aberrant Notch signaling confers breast carcinogenesis, therapy resistance and the maintenance of BTIC populations (Reedijk, 2012). In fact, transgenic overexpression of all four Notch receptors in the mouse mammary gland leads to tumor formation (Gallahan and Callahan, 1997, Hu et al., 2006a, Politi et al., 2004). Therefore, Notch inhibition may serve as a novel therapeutic option for breast tumors. However, inhibition of Notch signaling in the clinical setting has been largely unsuccessful due to high toxicity and low specificity of GSIs, which inhibit Notch receptor maturation. In addition, despite the wealth of knowledge regarding Notch signaling and Notch target genes, little is known about the upstream regulation of Notch receptors and ligands. Our study has revealed a novel mechanism of Notch epigenetic regulation in breast cancer. In light of the recent development of compounds that inhibit Cbx function (Ren et al., 2015, Simhadri et al., 2014, Stuckey et al., 2016), our studies provide support for targeting Cbx8 as an alternative strategy to attenuate Notch signaling in breast cancer. Specificity of inhibitory compounds for Cbx orthologues will be essential for such therapeutic targeting.
EXPERIMENTAL PROCEDURES
Cell culture
MMTV-Myc cells were derived from spontaneous mammary tumors of MMTV-Myc transgenic mice, and cultured in DMEM/F12 (Cellgro) with 2% FBS, 10μg/ml insulin, 1X glutamine, 10mM HEPES, 1X nonessential amino acids and 1X Penicillin/Streptomycin. MCF7, T47D, MDA-MB-157 and MDA-MB-231-Luc cells were cultured in DMEM with 10% FBS and 1X Penicillin/Streptomycin. MCF10A cells were cultured as described (Debnath et al., 2003). TS were cultured in DMEM/F12 (Cellgro) or MEBM (Lonza) with 20ng/ml EGF, 20ng/ml bFGF, 1X Gem21 supplement (Gemini Bio-products) and 1X Penicillin/Streptomycin on Ultra-low attachment plates (Corning) at a density <1×106 cells per 10ml media. TS were dissociated with trypsin into single cells and passaged every 4 days.
TS RNAi screen
The chromatin shRNA library targeting 60 genes (total of 452 shRNAs) was cloned into pLKO.1 lentiviral vector with its puromycin resistance gene replaced with GFP in order to monitor transduction efficiencies. shRNA sequences were designed using the TRC algorithm. The library was produced and sequenced to ensure proper coverage of shRNAs. For screening, 100K MMTV-Myc cells were injected into the mammary fat pad of two female FvB/N mice (10-16 weeks old). Tumors were harvested two weeks later, dissociated into single cells and cultured as TS (1×106 cells per 10ml media) or as bulk (adherent) cells for three days. TS were then dissociated into single cells and 5×106 cells were transduced with the shRNA library at a low multiplicity of infection (<1) to ensure a single shRNA vector per cell. After transduction, the cells were kept in suspension with rotation for 4 hours at 37°C and subsequently replated as TS. Two days later a fraction of cells from both conditions were analyzed by flow cytometry, and <30% of cells were GFP-positive, corresponding to single vector copy. TS cells were passaged as above every four days to prevent aggregation and ensure selection. Bulk cells were passaged every two days. For both conditions, two cell cultures from each tumor were passaged independently to serve as technical replicates (total = 4 replicates). Cells from both conditions were collected at day 0, 12 and 20 and genomic DNA was extracted. The integrated shRNAs were amplified by PCR with primers containing multiplexing barcodes and adaptors and sequenced on the Illumina HiSeq 2000 (ISMMS Genomic Core Facility). Analyses and plots of the sequencing data were conducted in Microsoft Excel and R statistical environment (R Foundation for Statistical Computing, http://www.R-project.org). The hits were selected based on the change of shRNA reads at day 12 and/or 20 vs. day 0. P-values were calculated by comparing shRNA read numbers of the 4 replicates (t test). Hit selection criteria are depicted in Figure S1E, and top 10 candidates are listed in Figure S1F.
TS assays
Cells were trypsinized into single cells and cultured in Ultra-low attachment 96-well plates in DMEM/F12 with 1.2% methylcellulose (to prevent cell aggregation), with supplements as described above. Cells were seeded at low density (1000-2500 cell/ml) to ensure TS are derived from a single cell. After 8-12 days, TS larger than 200 μm were counted manually.
Native ChIP-sequencing
Native ChIP-seq for H3K27me3 (Millipore, 07-449), H3K27ac (Abcam, ab4729) and H3K4me3 (Abcam, ab1012) were performed in control (shScramble) and Cbx8 knockdown cells (Cbx8-sh68 for MMTV-Myc cells; CBX8-sh94 for MDA-MB-231-Luc cells) as described (Gaspar-Maia et al., 2013). Input DNA was used to control for background. High throughput sequencing on all samples was performed using Illumina HiSeq 2500 with 100nt single-end sequencing.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M. Mahajan, O. Jabado and H. Shah from the ISMMS Genomics Core Facility, members of the Bernstein lab, S. Aaronson lab, I. Aifantis lab, G. Peters lab, F. Hof lab, G. de Haan lab, D. Placantonakis lab, J. Wang lab, S. Frye lab, I. Lemischka lab, M. Walsh, V. Gouon-Evans, J. Jin, W. Guo, N. Bansal and M.S. Sosa.for advice, reagents and technical support.. This work was supported by the Office of Research Infrastructure of the NIH (award number S10OD018522), the Department of Defense Breast Cancer Research Program BC100975 and New York Stem Cell Foundation – Druckenmiller fellowship to A.G.M., NCI T32 T32CA078207-11 to Z.Q., Helmsley Trust Award to G.M., NIH Pathfinder Award (DP2DK083052-01) and Juvenile Diabetes Research Foundation (JDRF-17-2010-770) to B.D.B., R01 CA82783 and R01CA184016 to R.P., Samuel Waxman Cancer Research Foundation Tumor Dormancy Program to J.A.A.-G., E.F.F. and E.B., The JJR and Mary Kay Foundations to E.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
C-Y.C., A.G.M, and E.B. conceived of this study. C-Y.C. performed the RNAi screen with assistance from A.G.M.. G.M. and B.D.B. generated the pooled shRNA library and assisted in sequencing and screen analysis. C-Y.C., E.F.F. and A.B. performed Matrigel and mouse xenograft studies. C-Y.C. performed functional assays of all knockdown experiments, TCGA, GSEA and Metacore analyses. A.G.M. performed organoid studies and RT-qPCR. C-Y.C., Z.S. and E.C. performed immunoblots. C-Y.C. performed RNA-seq and ChIP-seq with help of A.G.M.. Z.R., Z.S. and C-Y.C. performed size exclusion chromatography with the support of R.P.. Z.S. performed IPs and EZH2 inhibition experiments. Z.Q. performed immunohistochemistry. C-Y.C., A.G.M, E.F.F., B.D.B. and E.B. designed experiments and interpreted results. C-Y.C., A.G.M. and E.B. wrote the manuscript with input from all coauthors.
ACCESSION NUMBERS
The GEO accession number for the RNA-seq and ChIP-seq data reported in this paper is GSE71077.
Competing financial interests: The authors declare no competing financial interests.
REFERENCES
- AL-HAJJ M, WICHA MS, BENITO-HERNANDEZ A, MORRISON SJ, CLARKE MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALOIA L, DI STEFANO B, DI CROCE L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–34. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- ANDRECHEK ER, CARDIFF RD, CHANG JT, GATZA ML, ACHARYA CR, POTTI A, NEVINS JR. Genetic heterogeneity of Myc-induced mammary tumors reflecting diverse phenotypes including metastatic potential. Proc Natl Acad Sci U S A. 2009;106:16387–92. doi: 10.1073/pnas.0901250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEN-PORATH I, THOMSON MW, CAREY VJ, GE R, BELL GW, REGEV A, WEINBERG RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERNSTEIN BE, KAMAL M, LINDBLAD-TOH K, BEKIRANOV S, BAILEY DK, HUEBERT DJ, MCMAHON S, KARLSSON EK, KULBOKAS EJ, 3RD, GINGERAS TR, SCHREIBER SL, LANDER ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- BERNSTEIN E, DUNCAN EM, MASUI O, GIL J, HEARD E, ALLIS CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–9. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOSCH A, BERTRAN SP, LU Y, GARCIA A, JONES AM, DAWSON MI, FARIAS EF. Reversal by RARalpha agonist Am580 of c-Myc-induced imbalance in RARalpha/RARgamma expression during MMTV-Myc tumorigenesis. Breast Cancer Res. 2012;14:R121. doi: 10.1186/bcr3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOURAS T, PAL B, VAILLANT F, HARBURG G, ASSELIN-LABAT M-LL, OAKES SR, LINDEMAN GJ, VISVADER JE. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell stem cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- CAMPBELL RM, TUMMINO PJ. Cancer epigenetics drug discovery and development: the challenge of hitting the mark. J Clin Invest. 2014;124:64–9. doi: 10.1172/JCI71605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CANCER GENOME ATLAS N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHANG CJ, YANG JY, XIA W, CHEN CT, XIE X, CHAO CH, WOODWARD WA, HSU JM, HORTOBAGYI GN, HUNG MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CREIGHTON CJ, LI X, LANDIS M, DIXON JM, NEUMEISTER VM, SJOLUND A, RIMM DL, WONG H, RODRIGUEZ A, HERSCHKOWITZ JI, FAN C, ZHANG X, HE X, PAVLICK A, GUTIERREZ MC, RENSHAW L, LARIONOV AA, FARATIAN D, HILSENBECK SG, PEROU CM, LEWIS MT, ROSEN JM, CHANG JC. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–5. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CREPPE C, PALAU A, MALINVERNI R, VALERO V, BUSCHBECK M. A Cbx8-containing polycomb complex facilitates the transition to gene activation during ES cell differentiation. PLoS Genet. 2014;10:e1004851. doi: 10.1371/journal.pgen.1004851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'ANGELO RC, OUZOUNOVA M, DAVIS A, CHOI D, TCHUENKAM SM, KIM G, LUTHER T, QURAISHI AA, SENBABAOGLU Y, CONLEY SJ, CLOUTHIER SG, HASSAN KA, WICHA MS, KORKAYA H. Notch Reporter Activity in Breast Cancer Cell Lines Identifies a Subset of Cells with Stem Cell Activity. Mol Cancer Ther. 2015 doi: 10.1158/1535-7163.MCT-14-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEBNATH J, MUTHUSWAMY SK, BRUGGE JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- DIETRICH N, BRACKEN AP, TRINH E, SCHJERLING CK, KOSEKI H, RAPPSILBER J, HELIN K, HANSEN KH. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J. 2007;26:1637–48. doi: 10.1038/sj.emboj.7601632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONTU G, ABDALLAH WM, FOLEY JM, JACKSON KW, CLARKE MF, KAWAMURA MJ, WICHA MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EASWARAN H, TSAI HC, BAYLIN SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 2014;54:716–27. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FARNIE G, CLARKE RB, SPENCE K, PINNOCK N, BRENNAN K, ANDERSON NG, BUNDRED NJ. Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst. 2007;99:616–27. doi: 10.1093/jnci/djk133. [DOI] [PubMed] [Google Scholar]
- GALLAHAN D, CALLAHAN R. The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4). Oncogene. 1997;14:1883–90. doi: 10.1038/sj.onc.1201035. [DOI] [PubMed] [Google Scholar]
- GAO J, AKSOY BA, DOGRUSOZ U, DRESDNER G, GROSS B, SUMER SO, SUN Y, JACOBSEN A, SINHA R, LARSSON E, CERAMI E, SANDER C, SCHULTZ N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAO Z, ZHANG J, BONASIO R, STRINO F, SAWAI A, PARISI F, KLUGER Y, REINBERG D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–56. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GASPAR-MAIA A, ALAJEM A, MESHORER E, RAMALHO-SANTOS M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GASPAR-MAIA A, QADEER ZA, HASSON D, RATNAKUMAR K, LEU NA, LEROY G, LIU S, COSTANZI C, VALLE-GARCIA D, SCHANIEL C, LEMISCHKA I, GARCIA B, PEHRSON JR, BERNSTEIN E. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat Commun. 2013;4:1565. doi: 10.1038/ncomms2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIL J, BERNARD D, MARTINEZ D, BEACH D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- HU C, DIEVART A, LUPIEN M, CALVO E, TREMBLAY G, JOLICOEUR P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006a;168:973–90. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HU Z, FAN C, OH DS, MARRON JS, HE X, QAQISH BF, LIVASY C, CAREY LA, REYNOLDS E, DRESSLER L, NOBEL A, PARKER J, EWEND MG, SAWYER LR, WU J, LIU Y, NANDA R, TRETIAKOVA M, RUIZ ORRICO A, DREHER D, PALAZZO JP, PERREARD L, NELSON E, MONE M, HANSEN H, MULLINS M, QUACKENBUSH JF, ELLIS MJ, OLOPADE OI, BERNARD PS, PEROU CM. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006b;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JANG GB, KIM JY, CHO SD, PARK KS, JUNG JY, LEE HY, HONG IS, NAM JS. Blockade of Wnt/beta-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci Rep. 2015;5:12465. doi: 10.1038/srep12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KONZE KD, MA A, LI F, BARSYTE-LOVEJOY D, PARTON T, MACNEVIN CJ, LIU F, GAO C, HUANG XP, KUZNETSOVA E, ROUGIE M, JIANG A, PATTENDEN SG, NORRIS JL, JAMES LI, ROTH BL, BROWN PJ, FRYE SV, ARROWSMITH CH, HAHN KM, WANG GG, VEDADI M, JIN J. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8:1324–34. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KURPIOS NA, GIRGIS-GABARDO A, HALLETT RM, ROGERS S, GLUDISH DW, KOCKERITZ L, WOODGETT J, CARDIFF R, HASSELL JA. Single unpurified breast tumor-initiating cells from multiple mouse models efficiently elicit tumors in immune-competent hosts. PLoS One. 2013;8:e58151. doi: 10.1371/journal.pone.0058151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI G, WARDEN C, ZOU Z, NEMAN J, KRUEGER JS, JAIN A, JANDIAL R, CHEN M. Altered expression of polycomb group genes in glioblastoma multiforme. PLoS One. 2013;8:e80970. doi: 10.1371/journal.pone.0080970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOI S, HAIBE-KAINS B, DESMEDT C, WIRAPATI P, LALLEMAND F, TUTT AM, GILLET C, ELLIS P, RYDER K, REID JF, DAIDONE MG, PIEROTTI MA, BERNS EM, JANSEN MP, FOEKENS JA, DELORENZI M, BONTEMPI G, PICCART MJ, SOTIRIOU C. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAGNANI L, STOECK A, ZHANG X, LANCZKY A, MIRABELLA AC, WANG TL, GYORFFY B, LUPIEN M. Genome-wide reprogramming of the chromatin landscape underlies endocrine therapy resistance in breast cancer. Proc Natl Acad Sci U S A. 2013;110:E1490–9. doi: 10.1073/pnas.1219992110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARCOTTE R, SAYAD A, BROWN KR, SANCHEZ-GARCIA F, REIMAND J, HAIDER M, VIRTANEN C, BRADNER JE, BADER GD, MILLS GB, PE'ER D, MOFFAT J, NEEL BG. Functional Genomic Landscape of Human Breast Cancer Drivers, Vulnerabilities, and Resistance. Cell. 2016;164:293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLS AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10:669–82. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOREY L, PASCUAL G, COZZUTO L, ROMA G, WUTZ A, BENITAH SA, DI CROCE L. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- O'LOGHLEN A, MUNOZ-CABELLO AM, GASPAR-MAIA A, WU HA, BANITO A, KUNOWSKA N, RACEK T, PEMBERTON HN, BEOLCHI P, LAVIAL F, MASUI O, VERMEULEN M, CARROLL T, GRAUMANN J, HEARD E, DILLON N, AZUARA V, SNIJDERS AP, PETERS G, BERNSTEIN E, GIL J. MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell. 2012;10:33–46. doi: 10.1016/j.stem.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PECE S, TOSONI D, CONFALONIERI S, MAZZAROL G, VECCHI M, RONZONI S, BERNARD L, VIALE G, PELICCI PG, DI FIORE PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- PEMBERTON H, ANDERTON E, PATEL H, BROOKES S, CHANDLER H, PALERMO R, STOCK J, RODRIGUEZ-NIEDENFUHR M, RACEK T, DE BREED L, STEWART A, MATTHEWS N, PETERS G. Genome-wide co-localization of Polycomb orthologs and their effects on gene expression in human fibroblasts. Genome Biol. 2014;15:R23. doi: 10.1186/gb-2014-15-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POLITI K, FEIRT N, KITAJEWSKI J. Notch in mammary gland development and breast cancer. Semin Cancer Biol. 2004;14:341–7. doi: 10.1016/j.semcancer.2004.04.013. [DOI] [PubMed] [Google Scholar]
- REEDIJK M. Notch signaling and breast cancer. Adv Exp Med Biol. 2012;727:241–57. doi: 10.1007/978-1-4614-0899-4_18. [DOI] [PubMed] [Google Scholar]
- REEDIJK M, ODORCIC S, CHANG L, ZHANG H, MILLER N, MCCREADY DR, LOCKWOOD G, EGAN SE. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–7. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- REN C, MOROHASHI K, PLOTNIKOV AN, JAKONCIC J, SMITH SG, LI J, ZENG L, RODRIGUEZ Y, STOJANOFF V, WALSH M, ZHOU MM. Small-molecule modulators of methyl-lysine binding for the CBX7 chromodomain. Chem Biol. 2015;22:161–8. doi: 10.1016/j.chembiol.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCOTT CL, GIL J, HERNANDO E, TERUYA-FELDSTEIN J, NARITA M, MARTINEZ D, VISAKORPI T, MU D, CORDON-CARDO C, PETERS G, BEACH D, LOWE SW. Role of the chromobox protein CBX7 in lymphomagenesis. Proc Natl Acad Sci U S A. 2007;104:5389–94. doi: 10.1073/pnas.0608721104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEN H, LAIRD PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMHADRI C, DAZE KD, DOUGLAS SF, QUON TT, DEV A, GIGNAC MC, PENG F, HELLER M, BOULANGER MJ, WULFF JE, HOF F. Chromodomain antagonists that target the polycomb-group methyllysine reader protein chromobox homolog 7 (CBX7). J Med Chem. 2014;57:2874–83. doi: 10.1021/jm401487x. [DOI] [PubMed] [Google Scholar]
- SPIKE BT, ENGLE DD, LIN JC, CHEUNG SK, LA J, WAHL GM. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell. 2012;10:183–97. doi: 10.1016/j.stem.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STUCKEY JI, DICKSON BM, CHENG N, LIU Y, NORRIS JL, CHOLENSKY SH, TEMPEL W, QIN S, HUBER KG, SAGUM C, BLACK K, LI F, HUANG XP, ROTH BL, BAUGHMAN BM, SENISTERRA G, PATTENDEN SG, VEDADI M, BROWN PJ, BEDFORD MT, MIN J, ARROWSMITH CH, JAMES LI, FRYE SV. A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1. Nat Chem Biol. 2016;12:180–187. doi: 10.1038/nchembio.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAN J, JONES M, KOSEKI H, NAKAYAMA M, MUNTEAN AG, MAILLARD I, HESS JL. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20:563–75. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAVARES L, DIMITROVA E, OXLEY D, WEBSTER J, POOT R, DEMMERS J, BEZSTAROSTI K, TAYLOR S, URA H, KOIDE H, WUTZ A, VIDAL M, ELDERKIN S, BROCKDORFF N. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–78. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VARDABASSO C, HASSON D, RATNAKUMAR K, CHUNG CY, DUARTE LF, BERNSTEIN E. Histone variants: emerging players in cancer biology. Cell Mol Life Sci. 2014;71:379–404. doi: 10.1007/s00018-013-1343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VISVADER JE, STINGL J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–58. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITCOMB SJ, BASU A, ALLIS CD, BERNSTEIN E. Polycomb Group proteins: an evolutionary perspective. Trends Genet. 2007;23:494–502. doi: 10.1016/j.tig.2007.08.006. [DOI] [PubMed] [Google Scholar]
- WONG DJ, LIU H, RIDKY TW, CASSARINO D, SEGAL E, CHANG HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–44. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMAMOTO M, TAGUCHI Y, ITO-KUREHA T, SEMBA K, YAMAGUCHI N, INOUE J. NF-kappaB non-cell-autonomously regulates cancer stem cell populations in the basal-like breast cancer subtype. Nat Commun. 2013;4:2299. doi: 10.1038/ncomms3299. [DOI] [PubMed] [Google Scholar]
- YAMAMOTO S, WU Z, RUSSNES HG, TAKAGI S, PELUFFO G, VASKE C, ZHAO X, MOEN VOLLAN HK, MARUYAMA R, EKRAM MB, SUN H, KIM JH, CARVER K, ZUCCA M, FENG J, ALMENDRO V, BESSARABOVA M, RUEDA OM, NIKOLSKY Y, CALDAS C, LIU XS, POLYAK K. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell. 2014;25:762–77. doi: 10.1016/j.ccr.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG L, ZHOU Y, CHENG C, CUI H, CHENG L, KONG P, WANG J, LI Y, CHEN W, SONG B, WANG F, JIA Z, LI L, LI Y, YANG B, LIU J, SHI R, BI Y, ZHANG Y, WANG J, ZHAO Z, HU X, YANG J, LI H, GAO Z, CHEN G, HUANG X, YANG X, WAN S, CHEN C, LI B, TAN Y, CHEN L, HE M, XIE S, LI X, ZHUANG X, WANG M, XIA Z, LUO L, MA J, DONG B, ZHAO J, SONG Y, OU Y, LI E, XU L, WANG J, XI Y, LI G, XU E, LIANG J, YANG X, GUO J, CHEN X, ZHANG Y, LI Q, LIU L, LI Y, ZHANG X, YANG H, LIN D, CHENG X, GUO Y, WANG J, ZHAN Q, CUI Y. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet. 2015;96:597–611. doi: 10.1016/j.ajhg.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.