Abstract
Photoreceptor cell degeneration leads to visual impairment and blindness in several types of retinal disease. However, the discovery of safe and effective therapeutic strategies conferring photoreceptor cell protection remains challenging. Targeting distinct cellular pathways with low doses of different drugs that produce a functionally synergistic effect could provide a strategy for preventing or treating retinal dystrophies. We took a systems pharmacology approach to identify potential combination therapies using a mouse model of light-induced retinal degeneration. We showed that a combination of U.S. Food and Drug Administration–approved drugs that act on different G protein (guanine nucleotide–binding protein)–coupled receptors (GPCRs) exhibited synergistic activity that protected retinas from light-induced degeneration even when each drug was administered at a low dose. In functional assays, the combined effects of these drugs were stimulation of Gi/o signaling by activating the dopamine receptors D2R and D4R, as well as inhibition of Gs and Gq signaling by antagonizing D1R and the α1A-adrenergic receptor ADRA1A, respectively. Moreover, transcriptome analyses demonstrated that such combined GPCR-targeted treatments preserved patterns of retinal gene expression that were more similar to those of the normal retina than did higher-dose monotherapy. Our study thus supports a systems pharmacology approach to identify treatments for retinopathies, an approach that could extend to other complex disorders.
INTRODUCTION
The pathophysiology of disparate diseases often reflects complex mechanisms resulting from intricate subcellular and cellular interactions, necessitating the development of therapeutic strategies that address multiple targets. Systems pharmacology integrates aspects of systems biology with next-generation experimental approaches to develop novel drug treatments for complex disorders. It enables the discovery of treatments through a mechanism-based combination of multiple therapies directed at different cellular targets to produce the desired effect. Thus, each individual drug in a combination can be administered at a reduced dose that synergistically maximizes therapeutic benefits while minimizing the side effects characteristic of monotherapy at a much higher dose (1–5).
The retina is a complex tissue composed of multiple types of sensory neurons and supporting cells that collectively contribute to normal vision. Photoreceptor cell death is a central pathological manifestation of several different vision-threatening retinal degenerative disorders, including retinitis pigmentosa (RP), Stargardt disease, and age-related macular degeneration (AMD) (6, 7). Exposure to bright light is also associated with retinal degeneration (8, 9) and is widely used as a model to investigate the protective potential of therapeutics against photoreceptor cell loss. Changes in second-order neurons, such as bipolar and horizontal cells, also occur as a consequence of photoreceptor degeneration in RP patients (10). After the loss of photoreceptor cells in animal models of RP, deterioration of bipolar cell dendrites and retraction of horizontal cell processes in the outer plexiform layer (OPL) have been documented (11–13). In addition, the presence of ectopic Müller cell bodies in the outer retina produces reactive gliosis. Uncontrolled gliosis leads to the formation of a fibrotic seal that can diminish the efficacy of cellular and bionic interventional strategies, such as stem cell transplantation and retinal prosthesis implants, to rescue a degenerating retina (12, 14).
G protein (guanine nucleotide–binding protein)–coupled receptors (GPCRs) constitute a large family of transmembrane proteins that regulate intracellular signaling essential for cellular homeostasis, playing critical roles in the pathophysiology of multiple biological processes and serving as targets for 30 to 50% of clinically used drugs (15–17). Multiple GPCRs and their respective intracellular signaling pathways are implicated in the pathogenesis of light-induced retinal degeneration in animal models (18, 19). Here, we tested the hypothesis that combination therapy derived from a systems pharmacology approach could achieve a protective effect against retinal degeneration. Our results showed that exposure to bright light caused complex cellular impairment in the mouse retina, disrupting neighboring bipolar and horizontal cells, as well as inducing photoreceptor cell degeneration. Combination therapeutic regimens consisting of GPCR-targeted, U.S. Food and Drug Administration (FDA)–approved drugs (approved for various primary indications) protected retinal photoreceptor cells against light-induced loss by stimulating GPCR signaling through the Gi/o family of G proteins and antagonizing GPCR signaling through the Gq and Gs families of G proteins. Furthermore, bipolar and horizontal cells were also protected even when the individual drugs were given at doses that are subeffective doses required for monotherapeutic effectiveness. Finally, transcriptome analysis provided molecular evidence supporting the protective effects of the combination therapies.
RESULTS
Bright light exposure impairs OPL and photoreceptor terminal morphology in Abca4−/−Rdh8−/− mice
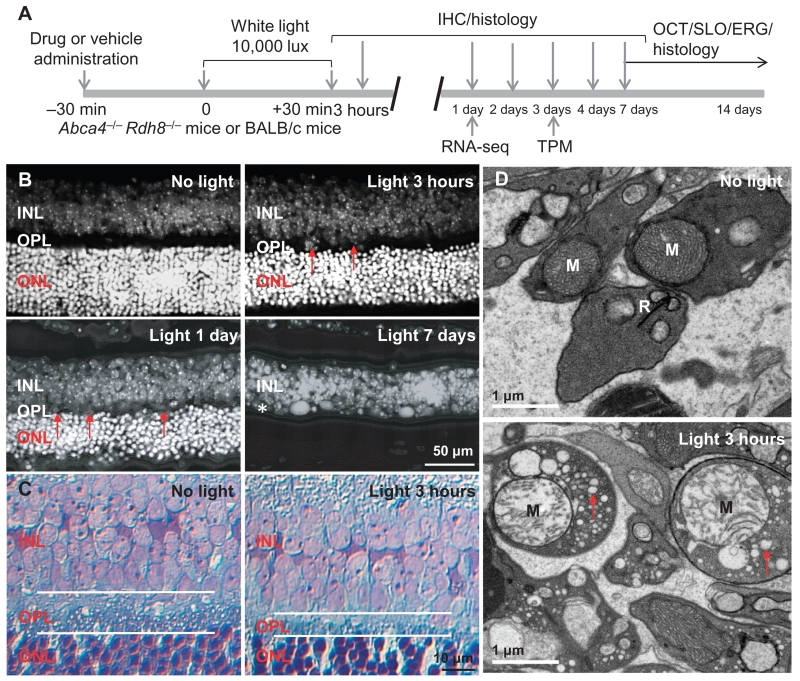
Abca4−/−Rdh8−/− mice lack adenosine triphosphate–binding cassette, subfamily A (ABC1), member 4 (Abca4) and retinol dehydrogenase 8 (Rdh8), and exhibit increased susceptibility to light-induced photoreceptor degeneration (18). In addition to the loss of photoreceptors induced by exposure to bright light at 10,000 lux for 30 min (Fig. 1, A and B), histological examination of the retinas revealed thinning of the OPL that accompanied the onset of photoreceptor cell loss. The OPL is a network of synapses of photoreceptors, bipolar cells, and horizontal cells. We observed sporadic thinning of the OPL in retinas 3 hours after light exposure and detected uniform thinning 1 day later (Fig. 1B). Most photoreceptor cells had disappeared 7 days after light exposure, and the distinct demarcation between the inner nuclear layer (INL), which represents packed bipolar, horizontal, and amacrine cells, and the outer nuclear layer (ONL), which represents photoreceptors, was no longer apparent. Histological examination at higher resolution consistently demonstrated narrowing of the OPL 3 hours after light exposure (Fig. 1C). Transmission electron microscopy revealed damaged mitochondria, cytoplasmic vacuolization, and the loss of synaptic ribbons in photoreceptor synaptic terminals, which are major cellular constituents of the OPL, 3 hours after light exposure when only sporadic photoreceptor cell death had begun (Fig. 1D and fig. S1A). These results indicated that light exposure caused rapid impairment of photoreceptor synaptic terminals and disruption of OPL morphology.
Fig. 1. Disruption of the OPL and damage of photoreceptor synaptic terminals in bright light–exposed retinas of Abca4−/−Rdh8−/− mice.
(A) Schematic of the experimental protocol. Four- to 6-week-old male and female pigmented Abca4−/−Rdh8−/− mice or BALB/c mice were exposed to bright light at 10,000 lux for 30 min, followed by the procedures as indicated. IHC, immunohistochemistry; OCT, optical coherence tomography; SLO, scanning laser ophthalmoscopy; ERG, electroretinogram; TPM, two-photon microscopy; RNA-seq, RNA sequencing. (B) After 4′,6-diamidino-2-phenylindole (DAPI) staining, retinal cryosections from Abca4−/−Rdh8−/− mice were observed under a fluorescence microscope. Retinas were collected from Abca4−/−Rdh8−/− mice at the time points indicated including 3 hours, 1 day, and 7 days after light exposure. Red arrows mark areas showing the thinning of the OPL. (C) Toluidine blue–stained retinal sections from Abca4−/−Rdh8−/− mice unexposed to bright light or at 3 hours after light exposure. White lines outline the OPL. (D) Electron microscopy of retinal sections from Abca4−/−Rdh8−/− mice unexposed to bright light or at 3 hours after light exposure. M, mitochondrion; R, synaptic ribbon in photoreceptor synaptic terminal. Red arrows, cytoplasmic vacuolation.
To evaluate other changes in the OPL after exposure to bright light, we performed immunohistochemistry. We used antibodies recognizing the synaptic vesicle protein synaptophysin to label rod and cone terminals (19). We observed a broad distribution of synaptophysin in the OPL of mice that had not been exposed to bright light, and this staining was disrupted 1 day after exposure to bright light (Fig. 2A).
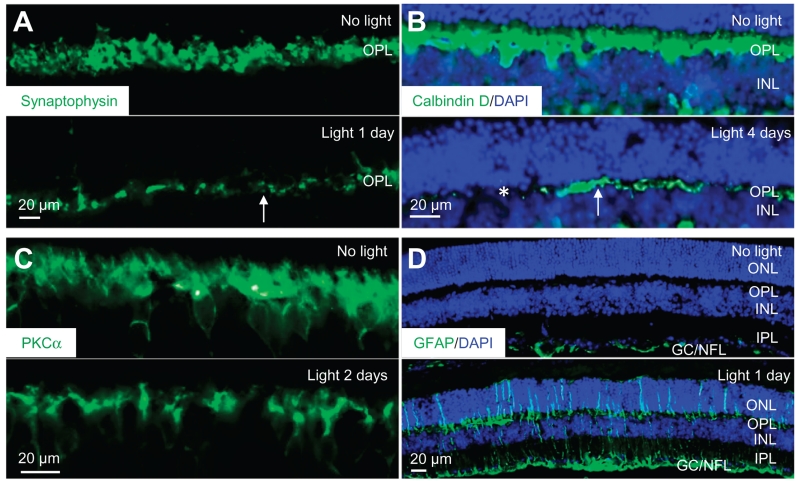
Fig. 2. Impaired photoreceptor synaptic terminals, horizontal cell morphology, bipolar cell dendrites, and retinal gliosis in bright light–exposed Abca4−/−Rdh8−/− mice.
Retinal cryosections were obtained from Abca4−/−Rdh8−/− mice either unexposed to bright light or after exposure to light at the indicated days. (A to D) Abundance of synaptophysin, calbindin D, PKCα (protein kinase Cα), and GFAP (glial fibrillary acidic protein) (all of which are green) was then revealed by immunohistochemistry and observed by fluorescence microscopy. DAPI counterstaining (blue) was performed to visualize the retinal structure in (B) and (D). IPL, inner plexiform layer; GC, ganglion cell; NFL, nerve fiber layer. White arrows in (A) and (B) indicate representative areas with diminished synaptophysin or calbindin D. Asterisks in (B) identify areas where calbindin D immunoreactivity was barely detected.
Abca4−/−Rdh8−/− mice bred into an albino background that lack pigmentation in the retina and choroid also exhibited susceptibility to bright light–induced retinal damage (fig. S1). Similar to the pigmented AAbca4−/−Rdh8−/− mice, light induced a reduction in synaptophysin labeling in the retinas of the albino mice (fig. S1B). These results confirmed that bright light exposure damaged photoreceptor synaptic terminals in mouse retinas.
Secondary neurons are impaired in light-exposed Abca4−/−Rdh8−/− mice
In addition to photoreceptor cell synaptic terminals, the OPL consists of horizontal cell projections and bipolar cell dendrites, which represent secondary neurons. We used calbindin D as a marker to assess any changes in horizontal cells in response to bright light (20) and PKCα to assess light-induced changes in bipolar cells (21). Calbindin D staining was reduced in the retinas of light-exposed Abca4−/−Rdh8−/− mice such that, by 4 days after exposure to bright light, calbindin D immunoreactivity was no longer present in the central retina and only residual staining was detected in the peripheral retina (Fig. 2B). These results indicated a light-induced loss of horizontal cell projections. We observed similar changes in calbindin D staining in albino Abca4−/−Rdh8−/− mice after exposure to bright light; however, these mice had diminished retinal calbindin D abundance by 2 days after bright light exposure (fig. S1C). The reduction in PKCα staining (Fig. 2C) indicated that bipolar cell dendrites were compromised 2 days after exposure to bright light in the retinas of Abca4−/−Rdh8−/− mice.
In response to retinal injury, Müller cells undergo reactive gliosis characterized by enhanced transcription and ensuing protein abundance of GFAP (22), an intermediate filament protein that is present in Müller cells but normally only in those Müller cells in the retinal layers containing GCs and the NFL. We detected GFAP throughout the IPL (which is the retinal layer composed of bipolar cell axons, amacrine cell dendrites, and GC branches), the INL, and the ONL starting 1 day after exposure to bright light (Fig. 2D).
Diverse GPCR-modulating compounds preserve retinal morphology and function in Abca4−/−Rdh8−/− mice exposed to bright light
The pathogenesis of light-induced retinopathy in Abca4−/−Rdh8−/− mice involves changes in the activities of GPCRs that are coupled to Gq, Gs, or Gi (23, 24). In particular, those activating Gq or Gs contribute to retinopathy (23), and those activating Gi protect photoreceptor cells from light-induced death (23). Therefore, we evaluated the effect of pharmacological compounds that either antagonize Gq/11- or Gs-coupled receptors or stimulate activated Gi/o-coupled receptors on bright light–induced retinopathy in Abca4−/−Rdh8−/− mice. To identify appropriate pharmacological compounds, we predicted the abundance of specific GPCRs from their transcript abundance in mouse and human retinas (Table 1). Sixteen compounds with activity on 11 of the 20 GPCRs with abundant transcripts in the retinas produced complete protection against bright light–induced photoreceptor degeneration (Table 2). When administered by intraperitoneal injection to Abca4−/−Rdh8−/− mice, these 16 drugs preserved retinal morphology, as revealed by OCT imaging (fig. S2) and H&E (hematoxylin and eosin) staining (fig. S3), after the exposure of the mice to bright light. Furthermore, administration of any of these 16 drugs individually preserved retinal function, as assessed by ERG recordings (fig. S4). These results provided evidence that pharmacologically targeting GPCRs protects photoreceptor cells under light conditions that induced retinal degeneration.
Table 1. Expression of GPCR genes encoding potential therapeutic targets for retinal degeneration in the Abca4−/− Rdh8−/− mouse model of bright light–induced retinopathy.
FPKM, normalized fragments per kilobase of exon per million mapped reads.
| Gene* | Mouse retina (FPKM) | Human retina (FPKM) | Receptor | G protein signaling | Agonist | Antagonist |
|---|---|---|---|---|---|---|
| Drd4 | 241.78 | 139.49 | Dopamine receptor D4 | Gi | + | |
| Gabbr1 | 40.24 | 35.38 | γ-Aminobutyric acid B receptor 1 | Gi | + | |
| Drd2 | 23.1 | 26.33 | Dopamine receptor D2 | Gi | + | |
| Adora1 | 18.26 | 13.55 | Adenosine A1 receptor | Gi | + | |
| Crhr1 | 6.49 | 12.76 | Corticotropin-releasing hormone receptor 1 | Gs | + | |
| S1pr1 | 11.21 | 11.78 | Sphingosine-1-phosphate receptor 1 | Gi | + | |
| Drd1a | 9.49 | 8.45 | Dopamine receptor D1a | Gs | + | |
| Smo | 6.35 | 5.91 | Smoothened | Gi | + | |
| Ednrb | 1.94 | 5.77 | Endothelin receptor type B | Gq | + | |
| Grm1 | 3.52 | 4.97 | Metabotropic glutamate receptor 1 | Gq | + | |
| Adrb1 | 20.18 | 3.84 | β1-Adrenergic receptor | Gs | + | |
| Adora2b | 2.53 | 3.83 | Adenosine A2B receptor | Gs and Gq | + | |
| Hrh3 | 19.12 | 3.75 | Histamine receptor H3 | Gi | + | |
| Grm4 | 4.93 | 3.1 | Metabotropic glutamate receptor 4 | Gi | + | |
| Grm8 | 6.43 | 2.04 | Metabotropic glutamate receptor 8 | Gi | + | |
| Grm7 | 3.95 | 1.16 | Metabotropic glutamate receptor 7 | Gi | + | |
| Adra1a † | 0.45 | 0.20 | α1A-Adrenergic receptor | Gq | ||
| Adra1b † | 1.60 | 1.12 | α1B-Adrenergic receptor | Gq | ||
| Adra1d † | 1.72 | 1.08 | α1D-Adrenergic receptor | Gq | ||
| Adrb2 | 1.03 | 0.98 | β2-Adrenergic receptor | Gs | + | |
| Tacr1 | 2.06 | 0.95 | Tachykinin receptor 1 | Gq | + | |
| Ptger1 | 14.88 | 0.94 | Prostaglandin E receptor 1 | Gq | + | |
| S1pr2 | 1.13 | 0.63 | Sphingosine-1-phosphate receptor 2 | Gq, Gs, and Gi |
Genes in bold, italicized font encode receptors targeted by the drugs displaying efficacy in this study.
Data from previous study (23).
Table 2. Summary of GPCR-modulating compounds that conferred retinal morphological and functional protection in bright light–exposed Abca4−/−/Rdh8−/− mice.
Morphological protection was assessed as morphological preservation of the retina by OCT imaging 7 days after exposure to bright light. Complete protection represented the retinas with intact ONL morphology similar to those of mice unexposed to bright light, with ONL thicknesses of ≥50 μm at 0.45 mm away from the optic nerve head (ONH) in the temporal retina. Functional protection was assessed by ERG analyses of retinal function in Abca4−/−Rdh8−/− mice 10 days after bright light exposure. Protection data were collected from at least five mice from each experimental group unless otherwise specifically indicated. ERG data are means ± SD, and statistical analyses were performed with either Student’s t test or analysis of variance (ANOVA). P < 0.05 was considered statistically significant.
| Agent | Major action(s) | G protein category |
|---|---|---|
| SCH 23390 hydrochloride | Dopamine receptor D1 and D5 antagonist | Gs |
| Rotigotine hydrochloride | Dopamine receptor D2 and D3 agonist | Gi |
| 2-Bromo-α-ergocryptine methanesulfonate salt | Dopamine receptor D2 and D3 agonist | Gi |
| Sumanirole maleate | Dopamine receptor D2 agonist | Gi |
| B-HT 920 | Dopamine receptor D2, α2-adrenergic receptor agonist | Gi |
| Ro 10-5824 dihydrochloride | Dopamine receptor D4 agonist | Gi |
| YM 202074 | mGluR1 receptor antagonist | Gq |
| Cinnabarinic acid | mGluR4 receptor agonist | Gi |
| Doxazosin* | Adrenergic receptor α1 antagonist | Gq |
| Tamsulosin* | Adrenergic receptor α1 antagonist | Gq |
| MTP tartrate | Adrenergic receptor β1 antagonist | Gs |
| ICI 118,551 hydrochloride | Adrenergic receptor β1 antagonist | Gs |
| GS 6201 | Adenosine A2B receptor antagonist | Gs |
| PSB 1115 | Adenosine A2B receptor antagonist | Gs |
| SC 19220 | EP1 prostanoid receptor antagonist | Gq |
| CP 154526 | Corticotropin-releasing factor 1 receptor antagonist | Gs |
| L-733060 | Tachykinin NK1 receptor antagonist | Gq |
| CP 96345 | Tachykinin NK1 receptor antagonist | Gq |
Compounds identified from previous study (23).
Bromocriptine or metoprolol protects the retinas of Abca4−/−Rdh8−/− mice against bright light–induced degeneration
Among the 16 compounds that protected retinas of Abca4−/−Rdh8−/− mice against exposure to bright light, we selected the 2 with FDA-approved counterparts—the synthetic ergot derivative bromocriptine (BRM) and the β1 receptor antagonist metoprolol (MTP)—for further evaluation in the light-exposed Abca4−/−Rdh8−/− mouse model. We administered either BRM or MTP by intraperitoneal injection before light exposure at doses of 2.5 or 10 mg/kg body weight (bw), respectively. As the controls for these experiments, we used light-exposed mice injected with dimethyl sulfoxide (DMSO), the solvent used for the drugs. DMSO administered intraperitoneally does not cause retinal degeneration (25). We assessed retinal morphology, including photoreceptor protection, by labeling the cone cell sheath with peanut agglutinin (PNA) 7 days after light exposure. PNA labeling revealed that the ONL was severely disrupted in retinas of light-exposed, DMSO-treated Abca4−/−Rdh8−/− mice such that PNA labeling was barely detected in photoreceptor cells (Fig. 3A). However, pretreatment with either BRM or MTP resulted in the preservation of the ONL and photoreceptor cells detected by PNA staining. SLO imaging also revealed many autofluorescent spots in the retinas of light-exposed Abca4−/−Rdh8−/− mice (26, 27), an indication of light damage, whereas the retinas of BRM- or MTP-treated mice lacked these autofluorescent spots (Fig. 3B) and were similar to the retinas of mice that had not been exposed to bright light.
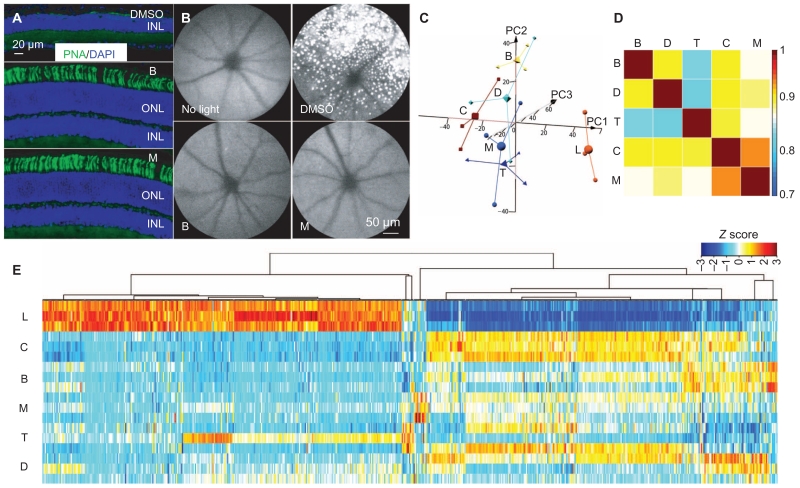
Fig. 3. Retina protection conferred by monotherapy.
(A) Retinal expression of PNA (green) along with DAPI counterstaining (blue) was examined in cryosections collected from light-exposed Abca4−/−Rdh8−/− mice pre-treated with DMSO, BRM, or MTP. (B) Autofluorescent spots in the retinas of Abca4−/−Rdh8−/− mice imaged after exposure to bright light. Abca4−/−Rdh8−/− mice were pretreated as indicated and exposed to bright light, and then SLO was performed to visualize autofluorescence in retinas of Abca4−/−Rdh8−/− mice. (C to E) Total RNA from the indicated experimental groups (n = 3 per group) was isolated from Abca4−/−Rdh8−/− mice 1 day after light exposure, along with mice unexposed to bright light. Total RNA was then subjected to RNA-seq analysis. (C) Three-dimensional (3D) PCAs of all expressed transcripts are shown for the control mice unexposed to bright light (C), light-exposed, DMSO-pretreated mice (L), and mice pre-treated with either BRM (B), MTP (M), DOX (D), or TAM (T). Small shapes represent individual samples, and large shapes are the centroid representations for each group. (D) Pearson’s correlation plots of differentially expressed transcripts from retinas of mice unexposed to bright light and the indicated pretreatment groups. Scale bar indicates correlation co-efficient with identity orange (correlation coefficient, 1). (E) Gene expression clustering of the differentially expressed transcripts is shown for mice with the indicated pretreatments along with clustering from mice unexposed to light and DMSO-pretreated mice exposed to light. Data are shown for the three independent samples in each treatment. Scale bar represents the Z score indicating up-regulation (orange) and down-regulation (blue).
Light-exposed BALB/c mice served as an additional model. We subjected these mice to the same pretreatment with either BRM or MTP and performed OCT imaging 7 days after light exposure. The ONL was severely disrupted in the retinas of light-exposed, DMSO-treated BALB/c mice (fig. S5, A and B), and pretreatment with either BRM or MTP preserved the ONL (fig. S5, A and B). Moreover, we detected only residual PNA staining in the outer retina of light-exposed, DMSO-treated BALB/c mice (fig. S5C), in contrast to the nearly normal pattern of PNA staining observed in both BRM- and MTP-treated mice. Thus, BRM or MTP protected cone photoreceptor cells from light-induced damage based on morphological assessment. These results validated the retinal protection conferred by either BRM or MTP in two different models of light-induced retinal degeneration.
To examine the impact of these drug regimens at the molecular level, we used global transcriptome analysis by RNA-seq. Retinas were collected from mice that had not been exposed to bright light and 1 day after exposure to light coupled with pretreatment with DMSO or individual drugs, including BRM, MTP, TAM (tamsulosin), and DOX (doxazosin). TAM and DOX are ADRA1 antagonists identified in a previous study as photoreceptor-protective agents against light-induced degeneration (23). Retinal RNA samples from three mice (six eyes) for each condition were sequenced to a mean depth of 46.9 ± 5.4 million paired-end reads with a genomic alignment percentage of 91.4 ± 1.0 and transcriptomic alignment percentage of 79.2 ± 1.4 (fig. S6A). After normalization, read depths and count distributions were examined for outliers (fig. S6B). The data set included 18,483 expressed transcripts after eliminating transcripts with very low expression (<1 FPKM).
Principal components analysis (PCA) performed on the normalized FPKM values from the entire expressed data set revealed good biological replication (Fig. 3C). Each condition resulted in a different pattern within the principal component space, indicating differential gene expression profiles associated with normal retinas, light-exposed retinas, and light-exposed retinas from mice receiving the different treatments. Data from the retinas from the DMSO-pretreated and light-exposed animals, the control animals that were not exposed to light, as well as the BRM-pretreated and light-exposed animals were the most consistent across the individual mice. Data from the DOX-, MTP-, or TAM-pretreated and light-exposed animals were more variable. However, retinas of control mice that had not been exposed to bright light and DMSO-pretreated and light-exposed mice had the largest separation in gene expression along principal component 1 (PC1) (Fig. 3C), consistent with retinal damage producing large changes in global gene expression. Pearson’s correlation was performed using the expression values from the differentially expressed genes and showed that pretreatment with individual drug regimens produced shifts in global gene expression toward that exhibited by mice that had not been exposed to bright light (correlation coefficient >0.8) (Fig. 3D). These findings suggested that, although the different pharmacological agents produced variable differential gene expression profiles in light-exposed retinas, overall each tended to shift the gene expression pattern to one similar to the pattern in control undamaged retina, indicating protective effects against light-induced retinal damage (Fig. 3D). MTP produced a gene expression pattern most similar to the undamaged control (not light exposed) (Fig. 3D), indicating that MTP conferred the most protection. Cluster analysis of the 622 differentially expressed transcripts that were significantly stimulated or repressed by bright light exposure showed that the profiles from the retinas from the drug-treated and light-exposed mice showed some individual variation within their respective groups but were overall more similar to the retinas of the control mice than to the damaged retinas from the DMSO-treated and light-exposed mice (Fig. 3E and fig. S6C).
G protein activation assays reveal a mechanistic basis for retinal-protective adrenergic and dopaminergic ligands
To assess how BRM, MTP, DOX, and TAM influence GPCR signaling to exert a protective effect against retinal degeneration, we analyzed their ability to activate G proteins. We used a real-time kinetic assay (28) in which G protein activation by GPCRs is quantified by monitoring changes in the bioluminescence resonance energy transfer (BRET) signal between Venus-tagged Gβγ and its luciferase-tagged reporter in living cells (Fig. 4A). The ability of ligands to increase in the BRET signal indicated agonistic activity [Fig. 4, B (left) and C (top)]. Conversely, the antagonistic activities were ascertained by the ability of ligand pretreatment to inhibit BRET signal induction by the physiological agonist dopamine or noradrenaline [Fig. 4, B (right) and C (bottom)]. Because the four synthetic drugs are adrenergic and dopaminergic ligands (Table 2), we focused on dopamine D2, D4, D1, and ADRA1 as representative Gi/o-, Gs-, and Gq-coupled receptors, which are encoded by genes that were prominently expressed in Abca4−/−Rdh8−/− mouse retina (Table 1) (23). The β1-adrenergic receptor ADRB1 is the target of MTP. We excluded ADRB1 from the group of receptors tested because, with the exception of BRM, which is a weak antagonist (29), it lacks high-affinity interactions with the test compounds. All possible ligand-GPCR pairs were examined.
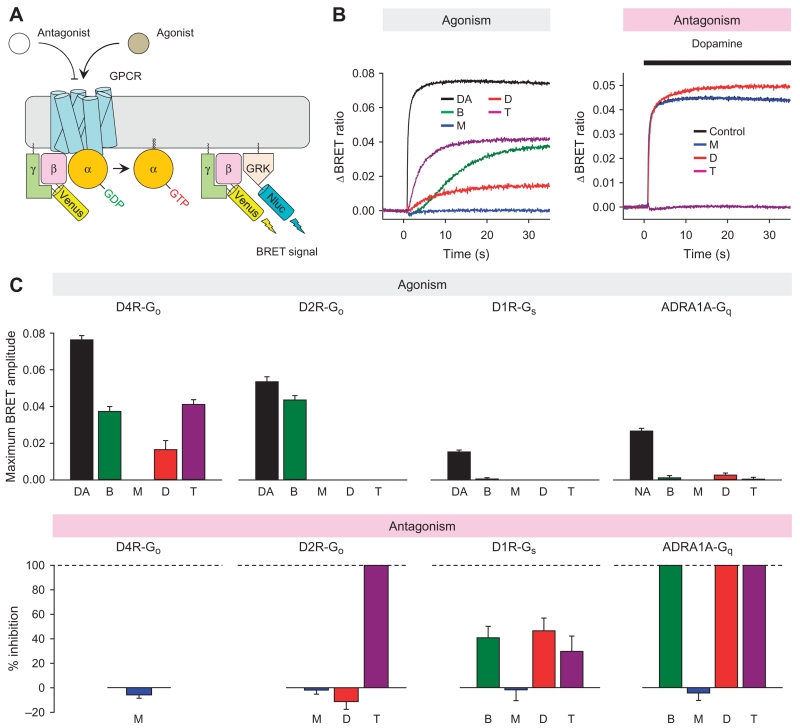
Fig. 4. Characterizing the activity of BRM, MTP, DOX, and TAM on dopamine and adrenaline receptors.
(A) Schematic representation of the BRET assay used to evaluate actions of drugs on selected GPCRs. Activation of a GPCR by agonist leads to the dissociation of inactive heterotrimeric G proteins into active GTP-bound Gα and Venus-Gβγ subunits. The free Venus-Gβγ then interacts with the Gβγ-effector mimetic masGRK3ct-Nluc to produce the BRET signal. GDP, guanosine diphosphate; GRK, G protein–coupled receptor kinase; Nluc, nanoluciferase. (B) Left: Representative BRET signal traces recorded upon activation of D4R with indicated drugs in cells expressing Gαo and Venus-Gβγ, illustrating the agonism assay. Right: Representative traces of D2R responses to 1 μM dopamine in the absence (control) or presence of indicated drugs, illustrating antagonism assay. Note that, in the antagonism assay shown, the black trace overlaps with the red trace and therefore is not visible. (C) Top: Quantification of agonistic activity of ligands on D2R, D4R, D1R, and ADRA1A. D2R-Go, D4R-Go, D1R-Gs, and ADRA1A-Gq signaling were reconstituted in HEK293T/17 cells by transient transfection separately. Maximum amplitudes induced by dopamine (DA; 100 μM), noradrenaline (NA; 100 μM), BRM (25 μM), MTP (100 μM), DOX (50 μM), and TAM (100 μM) were plotted as bar graphs. Bottom: Quantification of antagonistic activity of ligands. To examine the antagonistic activity, cells were preincubated with each of the ligands, and then 1 μM dopamine for dopamine receptors or 1 μM noradrenaline for the adrenaline receptor was applied. In these experiments, only ligands that did not show any agonistic effect were examined for each receptor–G protein combination. The activity is plotted as percent inhibition. Data are means ± SD. Results are representative of two independent experiments each performed with 12 replicates.
Representative BRET data are shown in Fig. 4B, and quantification of the activity analysis is shown in Fig. 4C. As predicted from reported studies (29–32), BRM activated D4R and D2R, as well as inhibited ADRA1A. However, BRM had no significant agonistic effect on D1R and, instead, exhibited partial antagonism at this receptor. As expected, DOX was a full antagonist for ADRA1A (33), but DOX also exhibited a partial agonistic effect on D4R and had a partial antagonistic effect on D1R. The ADRA1 antagonist TAM demonstrated complex activity on the four receptors. In addition to ADRA1A, TAM exhibited antagonistic activity on D2R and acted as a partial antagonist toward D1R. TAM also had a partial agonistic effect on D4R. We observed no agonistic or antagonistic effects of MTP on any of the four GPCRs tested, likely consistent with the lack of unexpected pharmacology beyond its selective antagonism at Gs-coupled β-adrenergic receptors (34).
Although BRM and TAM had opposite effects on D2R, our data showed that both of these ligands activated D4R and inhibited D1R and ADRA1A. Because we detected almost 10-fold higher expression of transcripts for D4R than D2R and the transcript analysis suggested that D4R is the most abundant GPCR in the retina among those examined (Table 1), we predict that D4R is a predominant source of protective Gi/o signaling in the mouse retina. Thus, we hypothesized that, in the retina, these drugs would activate protective Gi/o signaling through D4R and inhibit adverse effects of Gs and Gq signaling by antagonizing D1R and ADRA1A receptors.
Combined pretreatments improve protection of Abca4−/−Rdh8−/− mouse retina against bright light–induced degeneration
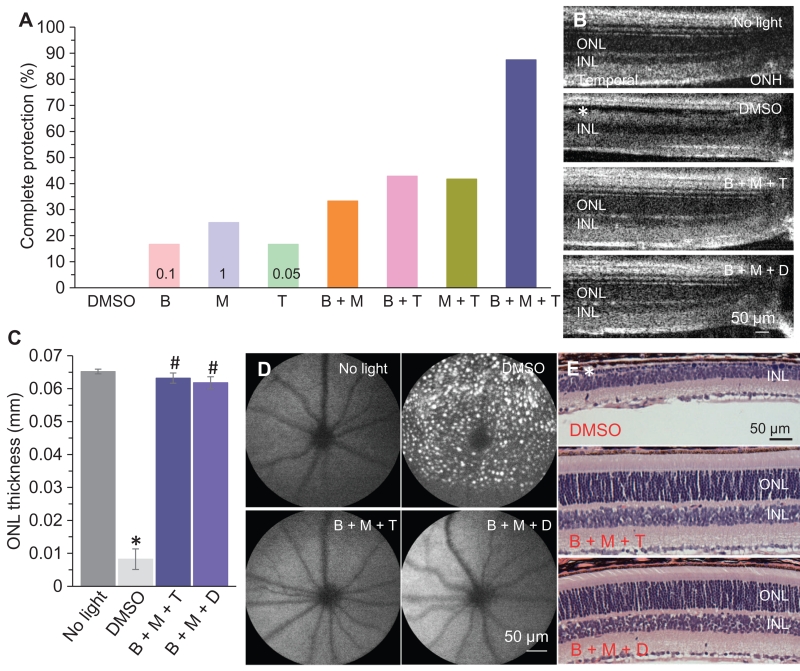
The complex effects of BRM, DOX, and TAM on GPCR signaling observed in the BRET-based signaling assays led us to hypothesize that combinations of retinal-protective drugs at individual subeffective doses could achieve synergistic effects by concurrent modulation of distinct pathways and that these synergistic effects might provide better preservation of retinal integrity. Although MTP was not a ligand for any of the four GPCRs tested in the functional assay, we included MTP, because it exhibited remarkable protection against light-induced retinal degeneration. To test this concept, we compared the protective effect of each of the FDA-approved drugs BRM, MTP, and TAM individually with their protective effect at lower doses in combination. We performed OCT imaging to determine the percentage of mice showing complete protection of retinal structure, as defined by indistinguishable ONL morphology from that of mice unexposed to light. Each drug individually conferred dose-dependent retinal protection in light-exposed Abca4−/−Rdh8−/− mice (fig. S7). Complete retinal protection occurred when BRM, MTP, and TAM were administered individually at doses of 1, 10, and 2.5 mg/kg bw, respectively, but this was reduced to only 17, 25, and 17% of light-exposed Abca4−/−Rdh8−/− mice when each drug was given individually at the lower doses of 0.1, 1, and 0.05 mg/kg bw, respectively (Table 3). We then used these subeffective doses to evaluate retinal protection conferred by combined regimens with these drugs in light-exposed Abca4−/−Rdh8−/− mice. Subeffective doses of the combination of all three drugs (BRM, MTP, and TAM) produced complete retinal protection in 88% of light-exposed mice, whereas complete protection was observed in fewer light-exposed Abca4−/−Rdh8−/− mice pretreated with subeffective combinations of either BRM + MTP, BRM + TAM, or MTP + TAM (Table 3). Conversely, the percentage of light-exposed Abca4−/−Rdh8−/− mice showing no retinal protection was reduced with the combination treatments compared with the individual treatments at subeffective doses (Table 3). When the combination of BRM, MTP, and TAM was administered at subeffective doses, only 4% of light-exposed Abca4−/−Rdh8−/− mice exhibited no retinal protection (fig. S8 and Table 3). Similar results were observed when DOX, an ADRA1 antagonist that was previously shown to protect retina against light-induced degeneration (23), was used in combination with BRM and MTP (fig. S9 and Table 3). Therefore, improved retinal protection could be attained by combined pretreatment with suboptimal doses of individual drugs with different mechanisms of action involving separate GPCRs.
Table 3. Retinal protection conferred by individual and combination therapies targeting GPCRs.
Either BRM, MTP, or TAM was administered individually at a subeffective dose to Abca4−/−Rdh8−/− mice or in a combined pretreatment consisting of two or three of these compounds, each at its subeffective dose. Doses for either single or combined pretreatments were 0.1, 1, and 0.05 mg/kg bw for BRM, MTP, and TAM, respectively. Protection was assessed as morphological preservation of the retina by OCT imaging 7 days after exposure to bright light. Complete protection represented the retinas with intact ONL morphology similar to those of mice unexposed to bright light, with ONL thicknesses of ≥50 μm at 0.45 mm away from the ONH in the temporal retina. No protection represented retinas exhibiting morphology similar to that of light-exposed and vehicle-treated mice, with ONL thickness of ≤20 μm at 0.45 mm away from the ONH in the temporal retina. Partial protection defined retinas manifesting a reduction in the thickness of the ONL between 20 and 50 μm measured 0.45 mm away from the ONH in the temporal retina. This was followed by exposure to bright light and OCT imaging performed 7 days later. Percentages of mice manifesting no protection of retinal structures were calculated for each condition.
| Drug | Percent of mice with complete protection | Percent of mice with no protection |
|---|---|---|
| BRM | 16.7 | 66.7 |
| MTP | 25 | 41.7 |
| TAM | 16.7 | 58.3 |
| DOX | 0 | 100 |
| BRM + MTP | 33.3 | 25 |
| BRM + TAM | 42.86 | 42.86 |
| MTP + TAM | 41.7 | 16.7 |
| BRM + MTP + TAM | 87.5 | 4.17 |
| BRM + DOX | 50 | 16.7 |
| MTP + DOX | 33.3 | 41.7 |
| BRM + MTP + DOX | 80 | 5 |
OCT imaging revealed preservation of retinal structures in mice pretreated with combinations of BRM, MTP, and TAM, or BRM, MTP, and DOX with each drug at an individual subeffective dose (Fig. 5, A to C). This was in distinct contrast to the damaged photoreceptor structures noted in light-exposed, DMSO-treated Abca4−/−Rdh8−/− mice. SLO imaging revealed that either of these combination treatments prevented retinas from developing retinal autofluorescence associated with damage induced by exposure to bright light (Fig. 5D). Examination of retinal gross histology further confirmed the structural protection conferred by these combined pretreatments (Fig. 5E).
Fig. 5. Combination pretreatments improve retinal morphological protection against bright light exposure in Abca4−/−Rdh8−/− mice.
(A) BRM, MTP, or TAM was administered either individually at the indicated subeffective dose (in mg/kg bw) to Abca4−/−Rdh8−/− mice or as a combined pretreatment as indicated, each at its subeffective dose. The mice were then exposed to bright light, and OCT imaging was performed 7 days later. Percentages of mice manifesting complete protection of retinal structures were calculated for each condition. For individual drug treatment, combination treatment with BRM and MTP, and combination treatment with MTP and TAM, n = 12 mice per group. For combination of BRM and TAM, n = 21 per group. For combination of BRM, MTP, and TAM, n = 24 per group. (B) Retinal OCT images of Abca4−/−Rdh8−/− mice either unexposed to bright light or exposed to bright light after pretreatment with DMSO or the indicated drug combinations at the subeffective doses for each. Images were obtained 7 days after light exposure. Asterisk indicates impaired ONL structure. (C) The thickness of the ONL was determined from OCT images at 0.45 mm away from the ONH in the region of the retinas in the direction of the temples from five mice 7 days after light exposure. *P < 0.05, compared to no light; #P < 0.05, compared to DMSO (independent samples t test). (D) SLO was performed to image autofluorescent spots in the retinas of Abca4−/−Rdh8−/− mice unexposed to bright light or exposed to light after the indicated pretreatment. SLO imaging was performed 9 days after light exposure. (E) Gross morphology of retinas was examined after H&E staining of paraffin sections collected from light-exposed Abca4−/−Rdh8−/− mice pretreated as indicated. Retinal tissue was obtained 10 days after light exposure. Asterisk indicates severely diminished ONL. All morphological analyses in (B) to (E) were performed with at least five mice.
Combined pretreatments protect against bright light–induced damage to retinal morphology and function in Abca4−/−Rdh8−/− and BALB/c mice
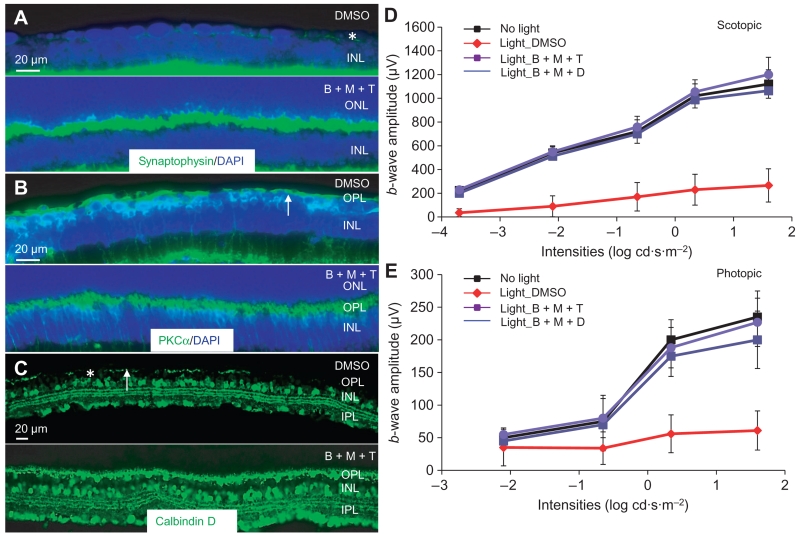
Protective effects of combined pretreatments against light-induced photoreceptor degeneration in Abca4−/−Rdh8−/− mice were further examined by staining for the synaptic vesicle protein synaptophysin. Abundant and well-organized staining of synaptophysin in photoreceptor synaptic terminals was evident in light-exposed Abca4−/−Rdh8−/− mice pretreated with BRM, MTP, and TAM (Fig. 6A). This was in contrast with the diminished residual amount of synaptophysin detected in the outer retina of light-exposed, DMSO-treated mice (Fig. 6A). Similarly, PKCα staining indicated that bipolar and horizontal cell morphologies were maintained in retinas of light-exposed mice pretreated with the combination regimen. Uniform arbor-shaped bipolar cell dendrites were preserved in light-exposed Abca4−/−Rdh8−/− mice pretreated with a combination of BRM, MTP, and TAM, each at a suboptimal individual dose (Fig. 6B). In addition, the combination treatment prevented the disintegrated horizontal cell morphology detected with calbindin D staining in light-exposed, DMSO-treated Abca4−/− Rdh8−/− mice (Fig. 6C). Our results therefore provide evidence that combined pretreatment protected mouse retinas against light-induced damage to photoreceptor, bipolar, and horizontal cells.
Fig. 6. Combination pretreatment preserves OPL morphology and retinal function in light-exposed Abca4−/−Rdh8−/− mice.
Seven days after bright light exposure, cryosections were prepared from the eye cups of light-exposed Abca4−/−Rdh8−/− mice pretreated with DMSO or a combination of BRM (0.1 mg/kg bw), MTP (1 mg/kg bw), and TAM (0.05 mg/kg bw) (B + M + T). (A to C) The abundance of synaptophysin, PKCα, and calbindin D (each shown in green) was examined by immunohistochemistry. DAPI counterstaining (blue) was performed in (A) and (B) to visualize the retinal structure. White arrows in (B) and (C) denote regions of diminished staining of PKCα or calbindin D. Asterisk indicates a representative area showing diminished ONL and residual staining for synaptophysin (A) or the absence of calbindin D immunoreactivity (C). (D and E) Scotopic and photopic b-wave amplitudes were analyzed after ERG recordings were performed in Abca4−/−Rdh8−/− mice either unexposed to bright light or exposed to bright light and pretreated with either DMSO, a combination of BRM (0.1 mg/kg bw), MTP (1 mg/kg bw), and TAM (0.05 mg/kg) (B + M + T), or a combination of BRM (0.1 mg/kg bw), MTP (1 mg/kg bw), and DOX (1 mg/kg) (B + M + D). Data are means ± SD from five mice.
Functional assessments of the retina by ERG revealed that both scotopic and photopic b-wave amplitudes were decreased in mice exposed to bright light and pretreated with DMSO, indicating impaired retinal function (Fig. 6, D and E). Both scotopic and photopic b-wave amplitudes in the Abca4−/−Rdh8−/− mice not exposed to bright light were similar to those in the Abca4−/−Rdh8−/− mice exposed to bright light but pretreated with subeffective dose combinations of BRM, MTP, and TAM, or BRM, MTP, and DOX. These results indicated that combined pretreatments preserved retinal function in Abca4−/−Rdh8−/− mice exposed to bright light.
To confirm the retinal protection of combined treatments, we also examined the effects of the combined GPCR modulators in light-exposed BALB/c mice, a wild-type mouse strain that is susceptible to bright light–induced retinal degeneration and accessible to TPM because of its albino features (fig. S10). OCT imaging of these mice demonstrated full protection of the thickness of the ONL (fig. S10A). Staining anti-GFAP antibody indicated that activation of the Müller cells was diminished (fig. S10B). Enlargement of photoreceptor cells is an early manifestation of light-induced retinal degeneration (27). Thus, we performed TPM imaging, which revealed that the combined pretreatments significantly diminished the number of enlarged photoreceptor outer segments compared to the number observed in DMSO-pretreated, light-exposed BALB/c mice (fig. S10C). In concordance with the morphological findings, ERG analyses revealed the preservation of retinal function after either combined pretreatment regimen in light-exposed BALB/c mice. Whereas light exposure resulted in decreased scotopic b-wave amplitudes in DMSO-treated BALB/c mice, retinal function was preserved with combinations of BRM, MTP, and TAM, or BRM, MTP, and DOX, each at suboptimal doses (fig. S10D).
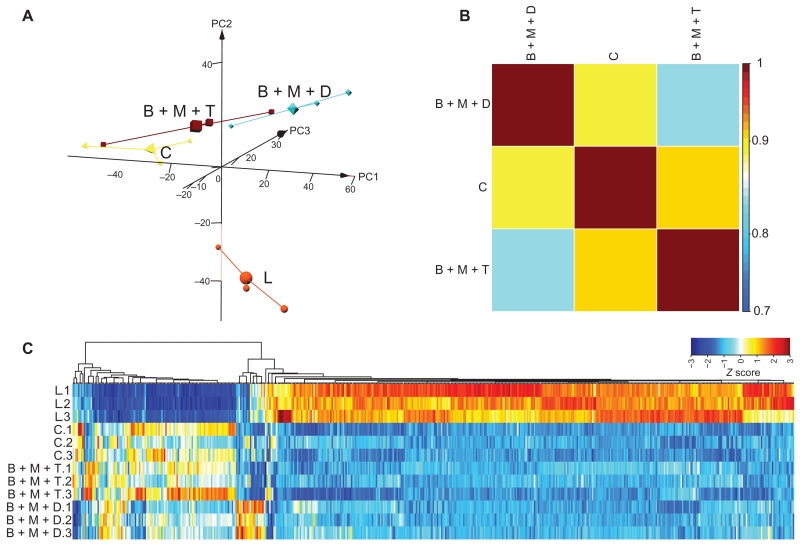
Transcriptomic analysis shows that combination pretreatments preserve an undamaged retina gene expression profile
As a molecular indication of protection, we evaluated the effect of the combined pretreatments with BRM, MTP, and TAM, or BRM, MTP, and DOX on global retinal gene expression (fig. S6A). Read depths and count distributions were examined for outliers after normalization (fig. S6C). The data set included 17,982 expressed transcripts after eliminating transcripts with very low expression (<1 FPKM). Good biological replication was revealed by PCA of the transcriptome data (Fig. 7A). The largest separation in gene expression along PC1 was observed between retinas from mice that had not been exposed to bright light and light-exposed mice pretreated with DMSO. Both combined treatments clustered closer to mice that had not been exposed to bright light (Fig. 7A). Pearson’s coefficient analysis showed that pretreatment with either combination shifted global gene expression toward that exhibited by mice that had not been exposed to bright light (correlation coefficient >0.8) (Fig. 7B). Cluster analysis of the 521 differentially expressed transcripts demonstrated a gene signature consistent with protection from light damage by either regimen, although the BRM, MTP, and TAM regimen produced a pattern that was more similar to the undamaged control than did the BRM, MTP, and DOX regimen (Fig. 7C and fig. S6C).
Fig. 7. RNA-seq analysis of retinal gene expression in Abca4−/−Rdh8−/− mice pretreated with drug combinations.
Total RNA from the indicated experimental groups (n = 3 per group) was isolated from Abca4−/−Rdh8−/− mice 1 day after light exposure, along with mice unexposed to bright light. Total RNA was then subjected to RNA-seq analysis. (A) 3D PCAs of all expressed transcripts are shown for the control mice unexposed to bright light (C), light-exposed, DMSO-pretreated mice (L), and mice pretreated with combinations of either BRM, MTP, and TAM (B + M + T) or BRM, MTP, and DOX (B + M + D). (B) Pearson’s correlation plots of the differentially expressed transcripts from retinas of mice unexposed to bright light or subjected to combination therapy before light exposure. Scale bar represents correlation coefficient. (C) Gene expression clustering of differentially expressed transcripts is shown for mice with the indicated combined pre-treatments along with clustering from mice unexposed to light and DMSO-pretreated mice exposed to light. Scale bar represents the Z score indicating up-regulation (orange) and down-regulation (blue).
Although single drugs acting on different GPCRs provided morphological protection of the retina when administered at high doses, the resulting transcriptome profiles differed among them (Fig. 3, D and E). Gene set enrichment analysis (GSEA) of pathways associated with each therapeutic demonstrated that 44 gene sets were significantly up-regulated in light-exposed, DMSO-treated mice as compared to mice unexposed to bright light (table S1). BRM (table S2), MTP (table S3), TAM (table S4), and DOX (table S5) pretreatment resulted in significant down-regulation of 29, 36, 32, and 28, respectively, of the gene sets associated with bright light exposure. Among pathways up-regulated by bright light exposure, those implicated in the pathogenesis of light-induced retinal degeneration genes in these pathways were down-regulated by each individual therapeutic agent (Table 4). Six pathways were down-regulated in retinas from DMSO-treated, light-exposed mice compared to retinas from mice that had not been exposed to bright light, and pretreatment with either BRM, MTP, TAM, or DOX prevented several of these changes in gene expression (Table 5). These results provided further evidence supporting the retinal protection conferred by each of these compounds.
Table 4. Effect of single and combination therapies on gene sets associated with retinal degeneration pathways.
GSEA identified pathways associated with retinal degeneration in retinas from light-exposed mice. NES, normalized enrichment score.
| Pathways | Experiment 1: Change in light-exposed retinas NES (P)* |
BRM NES (P)† |
MTP NES (P)† |
TAM NES (P)† |
DOX NES (P)† |
Experiment 2: Change in light-exposed retinas NES (P)* |
BRM + MTP + TAM NES (P)† |
BRM + MTP + DOX NES (P)† |
|---|---|---|---|---|---|---|---|---|
| APOPTOSIS | 1.83 (0) | 1.75 | 1.61 | 1.60 | 1.75 | 1.88 (0) | 1.81 (0) | 1.89 (0) |
| (0.0018) | (0.0055) | (0.011) | (0.0018) | |||||
| P53 SIGNALING | 2.29 (0) | 2.06 (0) | 2.04 (0) | 1.98 (0) | 2.18 (0) | 1.71 (0.0039) | 1.98 (0) | 1.69 (0.0064) |
| CYTOKINE-CYTOKINE | 2.50 (0) | 2.31 (0) | 2.30 (0) | 2.29 (0) | 2.42 (0) | 2.18 (0) | 2.13 (0) | 2.04 (0) |
| RECEPTOR | ||||||||
| INTERACTIONS | ||||||||
| CHEMOKINE | 1.62 | 1.67 | 1.44 | 1.39 | 1.41 | 1.71 (0) | 1.57 | 1.67 |
| SIGNALING | (0.0042) | (0.0018) | (0.02) | (0.0199) | (0.018) | (0.0102) | (0.00144) | |
| TOLL-LIKE RECEPTOR | 2.14 (0) | 2.20 (0) | 2.05 (0) | 1.83 (0) | 1.99 (0) | 2.10 (0) | 2.10 (0) | 2.21 (0) |
| SIGNALING |
Comparison of the transcriptomes from vehicle-treated and light-exposed retinas against those from the mice unexposed to bright light.
Comparison of the transcriptomes from vehicle-treated and light-exposed retinas against those from the mice treated by indicated drug(s).
Table 5. Effect of single and combination therapies on gene sets in pathways down-regulated in retinas from DMSO-treated, light-exposed mice.
Pathways were identified by GSEA. Only those pathways down-regulated in at least 50% of treatments are included.
| Pathways | Experiment 1: Change in light-exposed retinas NES (P)* |
BRM NES (P)† |
MTP NES (P)† |
TAM NES (P)† |
DOX NES (P)† |
Experiment 2: Change in light-exposed retinas NES (P)* |
BRM + MTP + TAM NES (P)† |
BRM + MTP + DOX NES (P)† |
|---|---|---|---|---|---|---|---|---|
| PHOTOTRANSDUCTION | −2.71 (0) | −2.42 (0) | −2.74 (0) | −2.79 (0) | −2.74 (0) | −2.70 (0) | −2.78 (0) | −2.31 (0) |
| OXIDATIVE | −1.78 (0) | −1.52 | −1.40 | −1.63 | −1.33 | −1.38 | ||
| PHOSPHORYLATION | (0.010) | (0.019) | (0.0022) | (0.0362) | (0.0229) |
Comparison of the transcriptomes from vehicle-treated and light-exposed retinas against those from the mice unexposed to bright light.
Comparison of the transcriptomes from vehicle-treated and light-exposed retinas against those from the mice treated by indicated drug(s).
Although individually the therapeutic GPCR-targeting agents partially prevented the abnormal transcriptome profile in these models of photoreceptor degeneration, treatment with a combination of drugs, each administered at an individual subeffective dose, engendered a more normal landscape of mRNAs in the retina (Fig. 7, B and C). As revealed by GSEA of an independent experiment, exposure to bright light resulted in significant up-regulation of 45 gene sets (table S6). Most (37 and 31) of these gene sets were significantly down-regulated by combined pretreatment with BRM, MTP, and TAM (table S7), or BRM, MTP, and DOX (table S8), respectively. These sets included genes expected to be involved in pathways that would contribute to retinal degeneration (Table 4). In addition, the bright light–induced down-regulation of gene set related to phototransduction involved in physiological signaling of light stimuli at normal light levels (table S6) was prevented by pretreatment with the combination of BRM, MTP, and TAM (table S7), or BRM, MTP, and DOX (table S8). Moreover, when genes for each relevant signaling pathway were examined, for example, those affecting apoptosis and phototransduction, each treatment exhibited a similar impact on gene expression within the same set of genes (Tables 6 and 7).
Table 6. Genes of core enrichment in the apoptosis pathway of different experimental groups.
Both of the DMSO vs. no light conditions produced an increase in expression of all of the listed genes in the first condition compared to the second. All other conditions produced a decrease in expression of all of the listed genes. In particular, the “DMSO vs. no light” columns represented controls for each set of experiments and DMSO induced an increase in the expression of the listed genes compared with the no light control.
| DMSO vs. no light |
BRM vs. DMSO |
MTP vs. DMSO |
TAM vs. DMSO |
DOX vs. DMSO |
DMSO vs. no light* |
BRM + MTP + TAM vs. DMSO |
BRM + MTP + DOX vs. DMSO |
|---|---|---|---|---|---|---|---|
| Fas | Myd88 | Fas | Bid | Fas | Myd88 | Myd88 | Tnfrsf1a |
| Ripk1 | Ripk1 | Myd88 | Il1r1 | Myd88 | Tnfrsf1a | Fas | Myd88 |
| Myd88 | Il1r1 | Bid | Tnfrsf1a | Il1r1 | Il1r1 | Tnfrsf1a | Ripk1 |
| Il1r1 | Tnfrsf1a | Ripk1 | Casp6 | Ripk1 | Ripk1 | Ripk1 | Fas |
| Tnfrsf1a | Bid | Il1r1 | Tradd | Trp53 | Irak1 | Il1r1 | Il1r1 |
| Bid | Trp53 | Trp53 | Myd88 | Tnfrsf1a | Rela | Ntrk1 | Prkx |
| Trp53 | Prkx | Tnfrsf1a | Trp53 | Tradd | Trp53 | Trp53 | Rela |
| Capn2 | Fas | Prkx | Akt2 | Tnfrsf10b | Bid | Bid | Map3k14 |
| Prkx | Tradd | Capn2 | Fas | Irak1 | Akt2 | Rela | Trp53 |
| Apaf1 | Casp3 | Irak2 | Ripk1 | Bid | Prkx | Map3k14 | Ntrk1 |
| Tradd | Apaf1 | Casp6 | Il1rap | Chuk | Fas | Traf2 | Bid |
| Casp6 | Irak1 | Nfkbia | Casp3 | Casp3 | Map3k14 | Akt2 | Akt2 |
| Ntrk1 | Birc2 | Tradd | Rela | Casp6 | Ntrk1 | Bax | Irak1 |
| Tnfrsf10b | Ikbkb | Il1rap | Ntrk1 | Il1rap | Tradd | Nfkbia | Ikbkb |
| Il1rap | Nfkbia | Rela | Irak2 | Apaf1 | Apaf1 | Prkx | Bax |
| Prkar2b | Ntrk1 | Irak1 | Nfkb1 | Capn2 | Nfkbia | Bcl2l1 | Apaf1 |
| Irak1 | Il1rap | Casp3 | Nfkbia | Prkar2b | Tnfrsf10b | Tnfrsf10b | Casp3 |
| Rela | Capn2 | Akt2 | Capn2 | Prkx | Capn2 | Irak1 | Nfkbia |
| Casp3 | Rela | Ntrk1 | Prkar2b | Nfkbia | Akt1 | Bad | Traf2 |
| Nfkbia | Pik3cb | Irak2 | Bax | Akt1 | Bcl2l1 | ||
| Chuk | Pik3cb | Traf2 | Tradd | Aifm1 | |||
| Ppp3ca | Il1rap | Irak2 | Pik3r3 | ||||
| Prkacb | Capn1 | ||||||
| Capn2 |
The controls are from two separate experiments (single and combined treatment) that were performed. Each experiment has its own control and light-exposed mice. The differences represent the variation of light damage from one experiment to the other.
Table 7. Genes of core enrichment in the phototransduction pathway of different experimental groups.
Both of the DMSO vs. no light conditions produced an increase in expression of all of the listed genes in the first condition compared to the second. All other conditions produced a decrease in expression of all of the listed genes. In particular, the “DMSO vs. no light” columns represented controls for each set of experiments and DMSO induced an increase in the expression of the listed genes compared with the no light control.
| DMSO vs. no light |
BRM vs. DMSO |
MTP vs. DMSO |
TAM vs. DMSO |
DOX vs. DMSO |
DMSO vs. no light |
BRM + MTP + TAM vs. DMSO |
BRM + MTP + DOX vs. DMSO |
|---|---|---|---|---|---|---|---|
| Grk1 | Grk1 | Gucy2e | Guca1b | Gucy2e | Gucy2e | Guca1b | Guca1b |
| Gucy2e | Cnga1 | Grk1 | Pde6g | Grk1 | Grk1 | Gucy2e | Grk1 |
| Cnga1 | Rgs9 | Guca1b | Gucy2e | Cnga1 | Guca1b | Rcvrn | Rgs9 |
| Guca1b | Pde6a | Cnga1 | Guca1a | Rgs9 | Guca1a | Grk1 | Rcvrn |
| Pde6a | Slc24a1 | Pde6a | Grk1 | Gngt1 | Rgs9 | Rho | Pde6a |
| Rgs9 | Gngt1 | Rgs9 | Rcvrn | Pde6a | Pde6a | Gnat1 | Slc24a1 |
| Gngt1 | Guca1b | Rcvrn | Gngt1 | Slc24a1 | Slc24a1 | Guca1a | Gnat1 |
| Guca1a | Rcvrn | Guca1a | Pde6b | Pde6b | Rcvrn | Rgs9 | Guca1a |
| Rcvrn | Pde6b | Slc24a1 | Pde6a | Guca1b | Rho | Pde6g | Rho |
| Slc24a1 | Gnb1 | Pde6b | Rgs9 | Guca1a | Gnat1 | Slc24a1 | Cnga1 |
| Pde6b | Gucy2f | Gngt1 | Cnga1 | Gucy2f | Gucy2f | Pde6a | Pde6b |
| Rho | Gnat1 | Gnat1 | Rho | Gnb1 | Cnga1 | Gngt1 | Gucy2f |
| Pde6g | Guca1a | Gnat2 | Gnat1 | Rcvrn | Pde6g | Cnga1 | Pde6g |
| Gnat1 | Gnat2 | Rho | Slc24a1 | Pde6g | Gnat2 | Pde6b | Gnat2 |
| Gucy2f | Rho | Pde6g | Gnb1 | Gnat1 | Pde6b | Gucy2f | Gngt1 |
| Gnb1 | Pde6g | Gucy2f | Gnat2 | Rho | Gngt1 | Gnat2 | Gnb1 |
| Gnat2 | Gnb1 | Gucy2f | Gnat2 | Gnb1 | Gnb1 |
Notably, when the retinal transcriptomes from different single and combined pretreatments were compared to those of mice that had not been exposed to bright light, a down-regulated phototransduction gene set was detected for most of the treatments except for the combination of BRM, MTP, and TAM (Table 8). Moreover, when considering retinal expression of apoptosis and p53 signaling pathway genes, only mice pretreated with combined BRM, MTP, and TAM were statistically indistinguishable from mice that had not been exposed to bright light (Table 8). These results suggest that combined pretreatment with BRM, MTP, and TAM was most effective at maintaining phototransduction, apoptosis, and p53 signaling pathways at their normal physiological levels.
Table 8. Effect of different therapies on signaling pathways associated with the altered retinal transcriptome in response to light-induced retinal degeneration.
The transcriptome-associated pathways in light-exposed Abca4−/−Rdh8−/− mice were compared to those in Abca4−/−Rdh8−/− mice that had not been exposed to bright light. NOM P, nominal P value.
| Treatment | Gene set name | NES | NOM P |
|---|---|---|---|
| BRM | P53 SIGNALING PATHWAY | 1.75 | 0.00277 |
| PHOTOTRANSDUCTION | −2.74 | 0 | |
| MTP | P53 SIGNALING PATHWAY | 1.50 | 0.01504 |
| PHOTOTRANSDUCTION | −2.32 | 0 | |
| TAM | P53 SIGNALING PATHWAY | 1.51 | 0.01966 |
| PHOTOTRANSDUCTION | −1.94 | 0.00166 | |
| DOX | APOPTOSIS | 1.57 | 0.01075 |
| P53 SIGNALING PATHWAY | 1.95 | 0 | |
| PHOTOTRANSDUCTION | −2.49 | 0 | |
| BRM, MTP, and DOX | APOPTOSIS | 1.40 | 0.04657 |
| PHOTOTRANSDUCTION | −1.83 | 0.00301 |
DISCUSSION
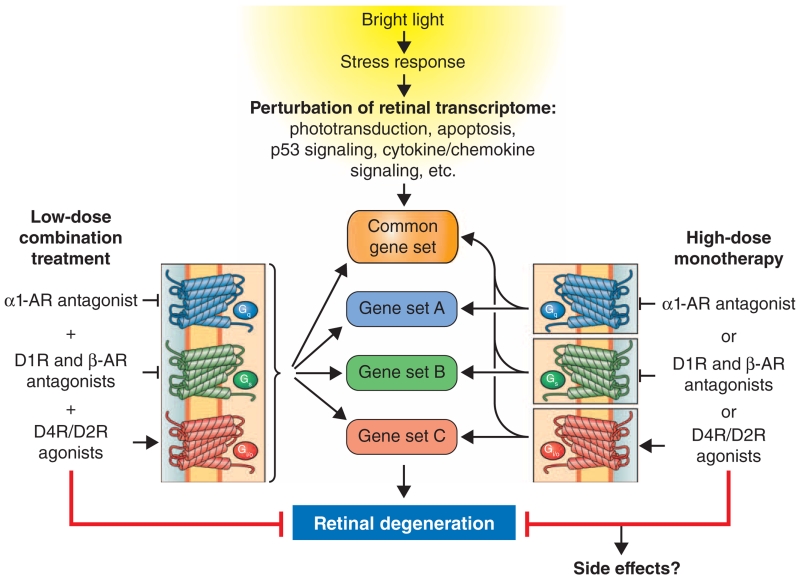
We used a systems pharmacology approach to evaluate combinations of FDA-approved drugs that achieve protection in mouse models of bright light–induced retinal degeneration. The drugs were administered by intraperitoneal injection, but other delivery systems should be considered in the future. We identified BRM and MTP as GPCR-targeting drugs that protected the retina against phototoxicity, similar to DOX and TAM, as we have shown in a previous study (5). BRET-based GPCR signaling assays demonstrated that combination treatment with these drugs is generally expected to promote signaling through Gi/o-coupled dopamine D4R and D2R, as well as attenuate D1R-mediated Gs and ADRA1-mediated Gq signaling, in addition to the known stimulatory effects on β1-adrenergic receptors, which are Gs-coupled (35). Consistent with a coordinated effect of these compounds on GPCR signaling, we observed that administration of combined treatments consisting of BRM, MTP, and TAM, or BRM, MTP, and DOX with each component dosed at an individual subeffective level resulted in morphological and functional protection of photoreceptor cells. In addition, bipolar and horizontal cells were also preserved by these combined treatments. Transcriptome analyses demonstrated that combination treatments also exhibited an enhanced capability to preserve a genome-wide gene expression pattern in the retina similar to that of un-damaged retinas compared to the patterns resulting from individual component therapeutics (Fig. 8).
Fig. 8. Pharmacological treatments protect against bright light–induced alterations in the retinal transcriptome and retinal degeneration.
Bright light exposure causes retinal degeneration that is associated with perturbation of retinal transcriptome homeostasis manifested as dysregulation of multiple gene sets, including but not limited to down-regulation of the phototransduction pathway and up-regulation of the apoptosis pathway, p53 signaling, cytokine-cytokine receptor interactions, and chemokine signaling. Pretreatment with individual drugs that act as antagonists at Gs-coupled GPCRs, agonists at Gi-coupled GPCRs, or antagonists at Gq-coupled GPCRs results in retinal protection. We found that, when each drug is delivered at lower subeffective doses, combinations of these drugs acting synergistically upon different GPCRs protected retinas from bright light–induced degeneration. The retinal protection by different treatment modalities, namely, monotherapy at high doses or combined treatments consisting of subeffective doses of drugs, not only prevents light-induced changes in common gene sets but also affects gene sets specific to each treatment. Combined treatment results in improved preservation of the retinal transcriptome compared to that conferred by monotherapy and may also offer the benefit of reduced side effects because of lower doses required for effectiveness.
Complex diseases are not easily managed and are typically confounded by aging. For example, multiple distinct cellular pathways have been associated with AMD and should be targeted for successful treatment (36). Monotherapies typically have partial effects even at high doses. Polypharmacology that takes advantage of diverse and unrelated targets may effectively deal with the initial problem but at the risk of multiple unwanted side effects. This risk could be acceptable for treating terminal diseases or chronic infections (for example, cancer or HIV), but for diseases such as blindness, polypharmacology is probably not the best solution. Systems pharmacology differs from polypharmacology in that it modifies several pathways that culminate in a common response, whether mediated by enzymes, second messengers, or channels. An advantageous solution is the modulation of pharmacologically accessible GPCRs, because their overlapping downstream pathways can elicit synergistic effects. Therefore, suboptimal doses of different GPCR modulators can be used in concert to achieve an enhanced desired effect. Low doses of combined GPCR-targeted drugs can also prevent massive internalization of their receptors and enable prolonged therapy. For chronic diseases, where prophylaxis would be most beneficial, systems pharmacology could play a major role. As examples of neuronal diseases, rod and cone retinopathies such as Stargardt disease, AMD, or RP could benefit greatly from systems pharmacology approaches focused on long-term preservation of cone function.
Biological insights derived from transcriptome analyses and gene regulatory networks (37) associated with the pharmacological actions of drugs can provide an important reference for future clinical evaluation of different treatment regimens. Moreover, in addition to gaining insights into retinal protective changes at the transcriptome level, we can also infer possible off-target effects of drugs and drug interactions. These advantages should make transcriptome analyses another standard approach in the evaluation of drug therapy.
In summary, we present a systems pharmacology approach to treat mouse models of human retinopathies that cause the death of rod and cone photoreceptor cells. We demonstrated that the therapies identified by systems pharmacology preserve photoreceptor cells and other neuronal cells in the retina, as well as the overall structure, function, and transcriptional integrity of the retina. Moreover, we showed that, at the molecular level, the combined effect of our systems pharmacology–based treatments is to activate Gi/o signaling through D2R and D4R, as well as to inhibit adverse effects of Gs and Gq signaling by antagonistic effects on D1R and ADRA1A receptors, respectively.
The GSEA analyses of the transcriptomic data suggested a link between the GPCR signaling and the JAK (Janus kinase)/STAT (signal transducer and activator of transcription) pathway, a pathway implicated in retinal degeneration (38). The GSEA data are also consistent with the evidence linking adrenergic receptor signaling and the JAK/STAT pathway. In cultured human vascular smooth muscle cells, the α1 agonist phenylephrine induces tyrosine phosphorylation of JAK2 and STAT1, indicating activation of this pathway (39). This stimulation occurs through an interaction of the α1B-adrenergic receptor with JAK2 and STAT1.
Because GPCRs are pharmacologically accessible targets, the application of systems pharmacology could benefit the treatment of other complex diseases. For example, in another visual impairment disorder, RPE65 (retinal pigment epithelium 65)–related (type 2) Leber congenital amaurosis, gene transfer can rescue the direct enzymatic defect that initiates the disease, but it does not prevent the ensuing pathology (40–42), indicating that (i) multiple therapeutic approaches could be needed to inhibit the progression of this chronic disease, and (ii) methods are needed to determine whether a chosen therapy normalizes signaling within the targeted cells. Our study illustrates the power of transcriptome analysis to optimize pharmacologic interventions intended to retain normal tissue signaling under pathological conditions. The sensitivity and scope of the transcriptome analysis as a pharmacological assay to assess global tissue health are unprecedented. Adapting this methodology could pave the way for successfully treating other complex and intractable diseases.
MATERIALS AND METHODS
Animals
Male and female Abca4−/−Rdh8−/− (18) and BALB/cJ mice (The Jackson Laboratory) at 4 to 6 weeks of age were used for the current study. Mice were genotyped by well-established methods with the following primers: Abca4 wild type, 5′-GCCCAGTGGTCGATCTGTCTAGC-3′ and 5′-CGGACACAAAGGCCGCTAGGACCACG-3′; Abca4 mutant, 5′-CCACAGCACACATCAGCATTTCTCC-3′ and 5′-TGCGAGGCCAGAGGCCACTTGTGTAGC-3′; Rdh8 wild type, 5′-CTTCAAAGTCAGTGGTGACTGGG-3′ and 5′-GCTATCCAGCTGCGACAATTC-3′; Rdh8 mutant, 5′-TCCGCCTTGGAAACCTGAGCCAGAAG-3′ and 5′-TGCGAGGCCAGAGGCCACTTGTGTAGC-3′. Only Rd8 mutation–free mice with the Leu variation at amino acid 450 of RPE65 were used. Abca4−/−Rdh8−/− mice were maintained with either pigmented 129Sv/Ev or C57BL/6 mixed backgrounds, and their siblings were used for most experiments. Either pigmented C57BL/6J or albino C57BL/6J (C57BL/6J−Tyrc−2J/J) mice from The Jackson Laboratory and their littermates were used as wild-type controls. BALB/c mice were obtained from The Jackson Laboratory. All mice were housed and maintained in a 12-hour light (≤10 lux)/12-hour dark cyclic environment in the Animal Resource Center at the School of Medicine, Case Western Reserve University (CWRU). Bright light–induced retinal damage was generated by exposing Abca4−/−Rdh8−/− mice or dark-adapted BALB/c mice to white light delivered at 10,000 lux (150-W spiral lamp, Commercial Electric) for 30 min. Abca4−/−Rdh8−/− mouse pupils were dilated with 1% tropicamide before light exposure, whereas tropicamide was not needed for BALB/c mice. All animal handling procedures and experimental protocols were approved by the Institutional Animal Care and Use Committee at CWRU and conformed to recommendations of both the American Veterinary Medical Association (AVMA) Panel on Euthanasia and the Association for Research in Vision and Ophthalmology.
Chemicals
2-Bromo-α-ergocryptine methanesulfonate salt (BRM) was purchased from Sigma-Aldrich. Aprepitant was obtained from Selleck. All the other compounds tested were purchased from Tocris Biosciences, including SCH 23390 hydrochloride, rotigotine hydrochloride, sumanirole maleate, B-HT 920, Ro 10-5824 dihydrochloride, YM 202074, cinnabarinic acid, MTP, ICI 118,551 hydrochloride, GS 6201, PSB 1115, SC 19220, CP 154526, L-733060, CP 96345, TAM, DOX, A 412997 dihydrochloride, AMN 082 dihydrochloride, SKF 97541, Rac BHFF, SEW 2871, purmorphamine, (R)-(−)-α-methylhistamine dihydrobromide, methimepip dihydrobromide, VU 0155041 sodium salt, antalarmin hydrochloride, NBI 35965 hydrochloride, BQ 788 sodium salt, BAY36-7620, 3-MATIDA, MPEP hydrochloride, MRS 1754, SC 51322, SC 19220, and JTE 013.
Complementary DNA constructs for GPCR signaling assays
Dopamine D1, dopamine D4, and α1A-adrenergic receptors were purchased from cDNA Resource Center (Bloomsburg University, Bloomsburg, PA). Plasmid encoding the Flag-tagged, long isoform of the D2 dopamine receptor was a gift from A. Kovoor (University of Rhode Island). pCMV5 plasmids encoding GαoA were gifts from H. Itoh (Nara Institute of Science and Technology, Japan). Plasmids encoding Venus 156-239-Gβ1 and Venus 1-155-Gγ2 were gifts from N. Lambert (Georgia Regents University) (43). Plasmids encoding RGS9-2, Gβ5S, R7BP, and masGRK3ct-Nluc were previously described (28, 44).
BRET assay for G protein activation in live cells
BRET experiments were performed as previously reported with slight modifications (28, 45). Briefly, 293T/17 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, minimum Eagle’s medium nonessential amino acids, 1 mM sodium pyruvate, and antibiotics [penicillin (100 U/ml) and streptomycin (100 μg/ml)] at 37°C in a humidified incubator containing 5% CO2. Cells were transfected with Lipofectamine LTX transfection reagents. BRET measurements were performed using a microplate reader (POLARstar Omega, BMG Labtech) equipped with two emission photomultiplier tubes. All measurements were performed at room temperature. The BRET signal is determined by calculating the ratio of the light emitted by Gβ1γ2-Venus (535 nm) over the light emitted by masGRK3ct-Nluc (475 nm). The average baseline value recorded before agonist stimulation was subtracted from BRET signal values, and the resulting difference (ΔBRET ratio) was obtained.
Mouse treatments
All indicated treatments were administered by intraperitoneal injection 30 min before bright light exposure. Compounds and their tested doses were as follows: SCH 23390 hydrochloride, 5 and 10 mg/kg bw; rotigotine hydrochloride, 5 mg/kg bw; BRM, 0.1, 0.25, 1, 2.5, and 10 mg/kg bw; sumanirole maleate, 10 mg/kg bw; B-HT 920, 10 mg/kg bw; Ro 10-5824 dihydrochloride, 50 mg/kg bw; YM 202074, 20 mg/kg bw; cinnabarinic acid, 10 mg/kg bw; MTP, 1, 2.5, and 10 mg/kg bw; ICI 118,551 hydrochloride, 10 mg/kg bw; GS 6201, 50 mg/kg bw; PSB 1115, 50 mg/kg bw; SC 19220, 10 mg/kg bw; CP 154526, 10 mg/kg bw; L-733060, 10 mg/kg bw; CP 96345, 25 mg/kg bw; TAM, 0.05, 0.5, and 2.5 mg/kg bw; DOX, 1, 2.5, and 10 mg/kg bw; A 412997 dihydrochloride, 10 mg/kg bw; AMN 082 dihydrochloride, 12.5 mg/kg bw; SKF 97541, 5 and 10 mg/kg bw; SEW 2871, 10 and 50 mg/kg bw; purmorphamine, 20 mg/kg bw; (R)-(−)-α-methylhistamine dihydrobromide, 10 mg/kg bw; methimepip dihydrobromide, 50 mg/kg bw; VU 0155041 sodium salt, 10 mg/kg bw; antalarmin hydrochloride, 10 mg/kg bw; NBI 35965 hydrochloride, 20 mg/kg bw; BQ 788 sodium salt, 1 mg/kg bw; BAY36-7620, 10 and 25 mg/kg bw; 3-MATIDA, 10 mg/kg bw; MPEP hydrochloride, 20 mg/kg bw; MRS 1754, 50 mg/kg bw; SC 51089, 20 mg/kg bw; and JTE 013, 5 mg/kg bw. All compounds were dissolved in DMSO and delivered in a total volume of 50 μl per injection.
Spectral domain OCT
Ultrahigh-resolution spectral domain OCT (Bioptigen) was performed for in vivo imaging of mouse retinas as previously described (23). Briefly, mice were injected with an anesthetic cocktail consisting of ketamine (6 mg/ml) and xylazine (0.44 mg/ml) at a dose of 10 μl/g bw, and pupils were dilated with 1% tropicamide before spectral domain OCT imaging. Five frames of OCT images were acquired in the B-mode and then averaged. For quantitative measurements addressing early changes in the illuminated retinas and assessing the retinal protection of tested compounds, thicknesses of the ONL or those of the outer segment and inner segment layers were measured 0.45 mm away from the ONH in the temporal retina, where the most severe damage was found in bright light–exposed Abca4−/−Rdh8−/− mice. Complete protection was defined by those retinas with intact structures similar to those of mice without exposure to bright light and with ONL thicknesses of ≥50 μm at the measured sites. No protection was defined by retinas exhibiting morphology similar to that of light-exposed and vehicle-treated mice, but with an ONL thickness of ≤20 μm at the measured sites. Partial protection defined retinas manifesting a reduction in the thickness of the ONL between 20 and 50 μm measured at these sites. For OCT measurements performed in BALB/cJ mice, the thickness of the ONL was measured at 0.15, 0.25, 0.35, and 0.45 mm away from the ONH in both the superior and the inferior retina.
SLO imaging
SLO (Heidelberg Engineering) was carried out for whole fundus imaging of mouse retinas invivo. Mice were anesthetized, and their pupils were dilated as described above. SLO images were then acquired in the autofluorescence mode.
Histology
For examination of gross retinal histology, mouse eyes were enucleated and then fixed in 4% paraformaldehyde and 0.5% glutaraldehyde before paraffin sectioning. Paraffin sections (5 μm thick) were stained by H&E. Eye cups were fixed in 2% glutaraldehyde and 4% paraformaldehyde and then processed for Epon embedding. Sections (1 μm thick) were stained with toluidine blue for histological examination. Light microscopy (Leica) was performed to observe the H&E- or toluidine blue–stained sections. Electron microscopic examination was used for some Epon-embedded sections as previously described (46).
Immunohistochemistry
Eye cups were fixed in 4% paraformaldehyde and processed for cryosectioning. Sections (12 μm thick) were then used for immunohistochemical processing. Primary antibodies used for immunohistochemistry included rabbit anti-synaptophysin antibody (Abcam), rabbit anti–calbindin D antibody (Abcam), mouse anti-PKCα antibody (Abcam), rabbit anti-GFAP antibody (Dako), and fluorescein isothiocyanate–conjugated anti-PNA antibody (Sigma). Secondary antibodies included Alexa Fluor 488–conjugated goat anti-rabbit immunoglobulin G (IgG) or Alexa Fluor 488–conjugated goat anti-mouse IgG.
Electroretinograms
ERGs were obtained as previously described (47). Briefly, dark-adapted mice were anesthetized, and their pupils were dilated as described in previous sections. Contact lens electrodes, a reference electrode, and a ground electrode were positioned on both corneas, ear, and tail, respectively. ERGs were then recorded with a UTAS-E 3000 (LKC Technologies Inc.).
TPM imaging
TPM imaging was performed as previously published (27, 48). Briefly, a Leica TCS SP5 confocal MP system equipped with a tunable Vision-S laser (Coherent) delivering 75-fs pulses at an 80-MHz pulse repetition frequency was used for TPM imaging. Before eye enucleation, mice were anesthetized by intraperitoneal injection of an anesthetic mixture containing ketamine (6 mg/ml) and xylazine (0.44 mg/ml) at a dose of 10 μl/g bw and then euthanized in compliance with the AVMA Guidelines for the Euthanasia of Animals and approval by the CWRU Institutional Animal Care and Use Committee. Images of the retina and RPE in the enucleated, intact mouse eyes were obtained with a 20× magnification, 1.0 numerical aperture water immersion objective. Laser light power at the sample was maintained at 5 to 10 mW. TPM 3D reconstructions of raw retinal images were analyzed offline with a Leica LAS AF 3.0.0.
Strand-specific RNA-seq
Transcriptome analyses of the human retina were carried out as previously described (49). Total RNA from mouse retinas was isolated with TRIzol (Invitrogen) following the manufacturer’s protocol. The quality of isolated RNA was assessed with a Bioanalyzer RNA 6000 Nano assay (Agilent Technologies). Strand-specific mRNA sequencing libraries were constructed from 100 ng of total RNA with the TruSeq Stranded mRNA Library Prep Kit (Illumina), and 125-base paired-end sequence reads were generated on the HiSeq 2500 platform (Illumina). Base calls were generated with RTAv1.18.64 software, and sequencing reads passing Illumina’s chastity filter were used for further analysis. Illumina adapter trimming was performed with Trimmomatic v0.33 (50). Quality control of trimmed Fastq files was achieved with FastQC v0.11.2 software (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Transcript quantitation was accomplished with eXpress v1.5.1 (51, 52) streaming pass filter reads aligned to Ensembl v78 transcriptome annotation using Bowtie2 v2.2.6 (53) as previously described (54). BAM files were generated by aligning to the GRCm38.p3/Ensembl v78 assembly and annotation with TopHat2 v2.1.0 (55) as previously noted (54). This study also used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (http://hpc.nih.gov).
RNA-seq differential gene expression analysis
Single and combined pretreatment differential expression studies were performed separately. Initially, analyses involved filtering out low- and/or non-expressing transcripts and retaining transcripts expressing ≥1.0 FPKM (from eXpress output) in all replicates of any group. Effective counts from the eXpress output of transcripts that passed FPKM filtering were TMM (trimmed mean of M values)–normalized with edgeR v3.10.2 (56, 57). Differential expression analyses comparing all drug treatments for light damage were performed with limma v3.24.15 (58). Briefly, a generalized linear model was set up with untreated light damage as a reference, dispersion estimation was performed with the voom function, and appropriate contrast statistics were used with the eBayes function. Transcripts featuring greater than a twofold change and a false discovery rate (FDR) of less than 0.05 in any comparison were considered to be differentially expressed and thus were used for further analysis.
Secondary analyses
All secondary analyses were performed in the R statistical environment (www.r-project.org). PCAs were accomplished with normalized log2 FPKM values and visualized with the pca3d v0.8 package. Pearson’s correlation analyses were performed with normalized log2 FPKM values of differentially expressed transcripts. Expression clustering of differentially expressed transcripts was done with affinity propagation that used the “corSimMat” function in apcluster v1.4.1 (59) after data Z score standardization. Gene Ontology (GO) analysis involved the functional annotation clustering method of DAVID v6.7 (60, 61) with GOTERM_BP_5 as implemented in RDAVIDWebService (62). To reduce redundancy normally associated with GO analyses, we selected only clusters having an enriched term <0.01 FDR, and a representative of each cluster was then selected by choosing the most significant end-of-branch term. Biological pathways associated with experimental groups under comparison, for example, retinal expression profiles from DMSO-treated, light-exposed mice compared to mice unexposed to bright light, were obtained by GSEA (63, 64). Gene sets with a normalized enrichment score of >0.5 and a nominal P value of <0.05 were considered as significantly enriched.
Statistical analyses
Results were collected from at least five mice from each experimental group unless otherwise specifically indicated. Data are means ± SEM, and statistical analyses were performed with either Student’s t test or ANOVA. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank L. T. Webster Jr. for helpful comments on this article and N. K. Skamangas for technical support. We thank A. Kovoor, H. Itoh, and N. Lambert for providing reagents. This research used the computational resources of the HPC Biowulf cluster at the NIH (http://hpc.nih.gov).
Funding: This work was supported by the NIH (grant EY018139 to K.A.M.; grants EY021126 and U01 EY025451 to K.P.), the National Natural Science Foundation of China (grant 81473732 to Y.C.), the Department of Veterans Affairs (grant IK2BX002683 to P.D.K.), and the Intramural Research Program of the National Eye Institute (grants EY000474 and EY000546 to A.S.). This work was also supported in part by the Arnold and Mabel Beckman Foundation and the Canadian Institute for Advanced Research. K.P. receives funding as the John H. Hord Professor of Pharmacology.
Footnotes
www.sciencesignaling.org/cgi/content/full/9/438/ra74/DC1
Fig. S1. Abca4−/−Rdh8−/− mice exposed to bright light exhibit pyknosis of photoreceptor cells, diminished synaptophysin in the OPL, and altered horizontal cell morphology.
Fig. S2. Pharmacological pretreatments targeting different GPCRs preserve retinal structure in bright light–exposed Abca4−/−Rdh8−/− mice.
Fig. S3. Variable preservation of retinal morphology by pharmacological pretreatments targeting different GPCRs in bright light–exposed Abca4−/−Rdh8−/− mice.
Fig. S4. Pharmacological pretreatment affecting various GPCRs preserves retinal function in bright light–exposed Abca4−/−Rdh8−/− mice.
Fig. S5. BRM or MTP pretreatment protects retinas of BALB/c mice from bright light–induced degeneration.
Fig. S6. Transcriptome analysis of the retina.
Fig. S7. Dose-dependent protection of retinal morphology in Abca4−/−Rdh8−/− mice by BRM, MTP, and TAM pretreatment.
Fig. S8. Combined pretreatments improve retinal morphological protection against bright light–exposed Abca4−/−Rdh8−/− mice.
Fig. S9. Combined treatments with subeffective doses of individual drugs exhibit improved retinal morphological protection in bright light–exposed Abca4−/−Rdh8−/− mice.
Fig. S10. Combined pretreatments protect retinas of BALB/c mice from bright light–induced photoreceptor degeneration.
Table S1. Retinal gene sets significantly altered in response to bright light exposure in Abca4−/−Rdh8−/− mice.
Table S2. Retinal gene sets beneficially regulated by BRM pretreatment.
Table S3. Retinal gene sets beneficially affected by MTP pretreatment.
Table S4. Retinal gene sets beneficially affected by TAM pretreatment.
Table S5. Retinal gene sets beneficially affected by DOX pretreatment.
Table S6. Significantly altered retinal gene sets as a result of bright light exposure in Abca4−/−Rdh8−/− mice.
Table S7. Retinal gene sets beneficially affected as a result of combined pretreatment with BRM, MTP, and TAM.
Table S8. Retinal gene sets beneficially affected as a result of combined pretreatment with BRM, MTP, and DOX.
Author contributions: Y.C. A.S., K.A.M., and K.P. conceived and designed the experiments. Y.C. conducted experiments for morphological assessments, screened GPCR antagonists/agonists, optimized single and combination treatments, and performed gene expression analyses. G.P. quantified the changes in photoreceptors by TPM. I.M. and K.A.M. carried out the BRET-based G protein activation experiments. S.G. assessed retinal function by ERG. H.J. validated the expression of genes with real-time polymerase chain reaction. Z.D. maintained mouse colonies and helped with mouse treatments. P.D.K. helped with clinical pharmacology. L.G. performed RNA-seq experiments. M.J.B. conducted RNA-seq data analyses. Y.C., T.S.K., A.S., and K.P. wrote the paper. K.P. and T.S.K. coordinated and oversaw the research project. All authors discussed the results and commented on the article.
Competing interests: K.P. is an inventor of U.S. patent nos. 8722669 (“Compounds and methods of treating ocular disorders”) and 20080275134 (“Methods for treatment of retinal degenerative disease”) issued to CWRU, whose values may be affected by this publication. CWRU may license this technology for commercial development. K.P. is the chief scientific officer of Polgenix Inc.
Data and materials availability: Transcriptomic data are available from the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) with accession no. GSE79061.
REFERENCES AND NOTES
- 1.Westerhoff HV, Nakayama S, Mondeel TDGA, Barberis M. Systems pharmacology: An opinion on how to turn the impossible into grand challenges. Drug Discov. Today Technol. 2015;15:23–31. doi: 10.1016/j.ddtec.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Hansen J, Zhao S, Iyengar R. Systems pharmacology of complex diseases. Ann. N. Y. Acad. Sci. 2011;1245:E1–E5. doi: 10.1111/j.1749-6632.2011.06382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyengar R. Complex diseases require complex therapies. EMBO Rep. 2013;14:1039–1042. doi: 10.1038/embor.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silverman EK, Loscalzo J. Developing new drug treatments in the era of network medicine. Clin. Pharmacol. Ther. 2013;93:26–28. doi: 10.1038/clpt.2012.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao S, Nishimura T, Chen Y, Azeloglu EU, Gottesman O, Giannarelli C, Zafar MU, Benard L, Badimon JJ, Hajjar RJ, Goldfarb J, Iyengar R. Systems pharmacology of adverse event mitigation by drug combinations. Sci. Transl. Med. 2013;5:206ra140. doi: 10.1126/scitranslmed.3006548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wenzel A, Grimm C, Samardzija M, Remé CE. Molecular mechanisms of light-induced photoreceptor apoptosis and neuroprotection for retinal degeneration. Prog. Retin. Eye Res. 2005;24:275–306. doi: 10.1016/j.preteyeres.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Curcio CA, Medeiros NE, Millican CL. Photoreceptor loss in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 1996;37:1236–1249. [PubMed] [Google Scholar]
- 8.Taylor HR, Muñoz B, West S, Bressler NM, Bressler SB, Rosenthal FS. Visible light and risk of age-related macular degeneration. Trans. Am. Ophthalmol. Soc. 1990;88:163–178. [PMC free article] [PubMed] [Google Scholar]
- 9.Organisciak DT, Darrow RM, Barsalou L, Darrow RA, Kutty RK, Kutty G, Wiggert B. Light history and age-related changes in retinal light damage. Invest. Ophthalmol. Vis. Sci. 1998;39:1107–1116. [PubMed] [Google Scholar]
- 10.Milam AH, Li Z-Y, Fariss RN. Histopathology of the human retina in retinitis pigmentosa. Prog. Retin. Eye Res. 1998;17:175–205. doi: 10.1016/s1350-9462(97)00012-8. [DOI] [PubMed] [Google Scholar]
- 11.Strettoi E, Pignatelli V. Modifications of retinal neurons in a mouse model of retinitis pigmentosa. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11020–11025. doi: 10.1073/pnas.190291097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips MJ, Otteson DC, Sherry DM. Progression of neuronal and synaptic re-modeling in the rd10 mouse model of retinitis pigmentosa. J. Comp. Neurol. 2010;518:2071–2089. doi: 10.1002/cne.22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barhoum R, Martínez-Navarrete G, Corrochano S, Germain F, Fernandez-Sanchez L, de la Rosa EJ, de la Villa P, Cuenca N. Functional and structural modifications during retinal degeneration in the rd10 mouse. Neuroscience. 2008;155:698–713. doi: 10.1016/j.neuroscience.2008.06.042. [DOI] [PubMed] [Google Scholar]
- 14.Marc RE, Jones BW, Watt CB, Strettoi E. Neural remodeling in retinal degeneration. Prog. Retin. Eye Res. 2003;22:607–655. doi: 10.1016/s1350-9462(03)00039-9. [DOI] [PubMed] [Google Scholar]
- 15.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein–coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 17.Tyndall JDA, Sandilya R. GPCR agonists and antagonists in the clinic. Med. Chem. 2005;1:405–421. doi: 10.2174/1573406054368675. [DOI] [PubMed] [Google Scholar]
- 18.Maeda A, Maeda T, Golczak M, Chou S, Desai A, Hoppel CL, Matsuyama S, Palczewski K. Involvement of all-trans-retinal in acute light-induced retinopathy of mice. J. Biol. Chem. 2009;284:15173–15183. doi: 10.1074/jbc.M900322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhingra NK, Ramamohan Y, Raju TR. Developmental expression of synaptophysin, synapsin I and syntaxin in the rat retina. Brain Res. Dev. Brain Res. 1997;102:267–273. doi: 10.1016/s0165-3806(97)00085-0. [DOI] [PubMed] [Google Scholar]
- 20.Hamano K, Kiyama H, Emson PC, Manabe R, Nakauchi M, Tohyama M. Localization of two calcium binding proteins, calbindin (28 kD) and parvalbumin (12 kD), in the vertebrate retina. J. Comp. Neurol. 1990;302:417–424. doi: 10.1002/cne.903020217. [DOI] [PubMed] [Google Scholar]
- 21.Greferath U, Grünert U, Wässle H. Rod bipolar cells in the mammalian retina show protein kinase C-like immunoreactivity. J. Comp. Neurol. 1990;301:433–442. doi: 10.1002/cne.903010308. [DOI] [PubMed] [Google Scholar]
- 22.Dyer MA, Cepko CL. Control of Müller glial cell proliferation and activation following retinal injury. Nat. Neurosci. 2000;3:873–880. doi: 10.1038/78774. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Palczewska G, Mustafi D, Golczak M, Dong Z, Sawada O, Maeda T, Maeda A, Palczewski K. Systems pharmacology identifies drug targets for Stargardt disease– associated retinal degeneration. J. Clin. Invest. 2013;123:5119–5134. doi: 10.1172/JCI69076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Okano K, Maeda T, Chauhan V, Golczak M, Maeda A, Palczewski K. Mechanism of all-trans-retinal toxicity with implications for Stargardt disease and age-related macular degeneration. J. Biol. Chem. 2012;287:5059–5069. doi: 10.1074/jbc.M111.315432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bian M, Du X, Cui J, Wang P, Wang W, Zhu W, Zhang T, Chen Y. Celastrol protects mouse retinas from bright light-induced degeneration through inhibition of oxidative stress and inflammation. J. Neuroinflammation. 2016;13:50. doi: 10.1186/s12974-016-0516-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohno H, Chen Y, Kevany BM, Pearlman E, Miyagi M, Maeda T, Palczewski K, Maeda A. Photoreceptor proteins initiate microglial activation via Toll-like receptor 4 in retinal degeneration mediated by all-trans-retinal. J. Biol. Chem. 2013;288:15326–15341. doi: 10.1074/jbc.M112.448712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeda A, Palczewska G, Golczak M, Kohno H, Dong Z, Maeda T, Palczewski K. Two-photon microscopy reveals early rod photoreceptor cell damage in light-exposed mutant mice. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E1428–E1437. doi: 10.1073/pnas.1317986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masuho I, Ostrovskaya O, Kramer GM, Jones CD, Xie K, Martemyanov KA. Distinct profiles of functional discrimination among G proteins determine the actions of G protein–coupled receptors. Sci. Signal. 2015;8:ra123. doi: 10.1126/scisignal.aab4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millan MJ, Maiofiss L, Cussac D, Audinot V, Boutin J-A, Newman-Tancredi A. Differential actions of antiparkinson agents at multiple classes of monoaminergic receptor. I. A multivariate analysis of the binding profiles of 14 drugs at 21 native and cloned human receptor subtypes. J. Pharmacol. Exp. Ther. 2002;303:791–804. doi: 10.1124/jpet.102.039867. [DOI] [PubMed] [Google Scholar]
- 30.Vila E, Badia A, Jane F. Effects of bromocriptine on catecholamine receptors mediating cardiovascular responses in the pithed rat. J. Auton. Pharmacol. 1985;5:125–130. doi: 10.1111/j.1474-8673.1985.tb00113.x. [DOI] [PubMed] [Google Scholar]
- 31.McPherson GA. In vitro selectivity of lisuride and other ergot derivatives for α1- and α2-adrenoceptors. Eur. J. Pharmacol. 1984;97:151–155. doi: 10.1016/0014-2999(84)90525-9. [DOI] [PubMed] [Google Scholar]
- 32.Gibson A, Samini M. The effects of bromocriptine on pre-synaptic and post-synaptic α-adrenoceptors in the mouse vas deferens. J. Pharm. Pharmacol. 1979;31:826–830. doi: 10.1111/j.2042-7158.1979.tb13674.x. [DOI] [PubMed] [Google Scholar]
- 33.Hamilton CA, Reid JL, Vincent J. Pharmacokinetic and pharmacodynamic studies with two α-adrenoceptor antagonists, doxazosin and prazosin in the rabbit. Br. J. Pharmacol. 1985;86:79–87. doi: 10.1111/j.1476-5381.1985.tb09437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werkö L, Aurell M, Johnsson G, Sannerstedt R, Svedmyr N. Studies with a new cardioselective beta-blocker, metoprolol. Adv. Clin. Pharmacol. 1976;11:19–25. [PubMed] [Google Scholar]
- 35.Frielle T, Collins S, Daniel KW, Caron MG, Lefkowitz RJ, Kobilka BK. Cloning of the cDNA for the human β1-adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 1987;84:7920–7924. doi: 10.1073/pnas.84.22.7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A. Age-related macular degeneration: Genetics and biology coming together. Annu. Rev. Genomics Hum. Genet. 2014;15:151–171. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H-J, Ratnapriya R, Cogliati T, Kim J-W, Swaroop A. Vision from next generation sequencing: Multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease. Prog. Retin. Eye Res. 2015;46:1–30. doi: 10.1016/j.preteyeres.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samardzija M, Wenzel A, Aufenberg S, Thiersch M, Remé C, Grimm C. Differential role of Jak-STAT signaling in retinal degenerations. FASEB J. 2006;20:2411–2413. doi: 10.1096/fj.06-5895fje. [DOI] [PubMed] [Google Scholar]
- 39.Sasaguri T, Teruya H, Ishida A, Abumiya T, Ogata J. Linkage between α1 adrenergic receptor and the Jak/STAT signaling pathway in vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2000;268:25–30. doi: 10.1006/bbrc.1999.2066. [DOI] [PubMed] [Google Scholar]
- 40.Cideciyan AV, Jacobson SG, Beltran WA, Sumaroka A, Swider M, Iwabe S, Roman AJ, Olivares MB, Schwartz SB, Komáromy AM, Hauswirth WW, Aguirre GD. Human retinal gene therapy for Leber congenital amaurosis shows advancing retinal degeneration despite enduring visual improvement. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E517–E525. doi: 10.1073/pnas.1218933110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobson SG, Aleman TS, Cideciyan AV, Sumaroka A, Schwartz SB, Windsor AM, Traboulsi EI, Heon E, Pittler SJ, Milam AH, Maguire AM, Palczewski K, Stone EM, Bennett J. Identifying photoreceptors in blind eyes caused by RPE65 mutations: Prerequisite for human gene therapy success. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6177–6182. doi: 10.1073/pnas.0500646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cideciyan AV, Aleman TS, Boye SL, Schwartz SB, Kaushal S, Roman AJ, Pang J, Sumaroka A, Windsor EAM, Wilson JM, Flotte TR, Fishman GA, Heon E, Stone EM, Byrne BJ, Jacobson SG, Hauswirth WW. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15112–15117. doi: 10.1073/pnas.0807027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hollins B, Kuravi S, Digby GJ, Lambert NA. The c-terminus of GRK3 indicates rapid dissociation of G protein heterotrimers. Cell. Signal. 2009;21:1015–1021. doi: 10.1016/j.cellsig.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martemyanov KA, Yoo PJ, Skiba NP, Arshavsky VY. R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J. Biol. Chem. 2005;280:5133–5136. doi: 10.1074/jbc.C400596200. [DOI] [PubMed] [Google Scholar]
- 45.Masuho I, Martemyanov KA, Lambert NA. Monitoring G protein activation in cells with BRET. Methods Mol. Biol. 2015;1335:107–113. doi: 10.1007/978-1-4939-2914-6_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maeda A, Maeda T, Golczak M, Palczewski K. Retinopathy in mice induced by disrupted all-trans-retinal clearance. J. Biol. Chem. 2008;283:26684–26693. doi: 10.1074/jbc.M804505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maeda A, Maeda T, Imanishi Y, Kuksa V, Alekseev A, Bronson JD, Zhang H, Zhu L, Sun W, Saperstein DA, Rieke F, Baehr W, Palczewski K. Role of photoreceptor-specific retinol dehydrogenase in the retinoid cycle in vivo. J. Biol. Chem. 2005;280:18822–18832. doi: 10.1074/jbc.M501757200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palczewska G, Dong Z, Golczak M, Hunter JJ, Williams DR, Alexander NS, Palczewski K. Noninvasive two-photon microscopy imaging of mouse retina and retinal pigment epithelium through the pupil of the eye. Nat. Med. 2014;20:785–789. doi: 10.1038/nm.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mustafi D, Kevany BM, Genoud C, Bai X, Palczewski K. Photoreceptor phagocytosis is mediated by phosphoinositide signaling. FASEB J. 2013;27:4585–4595. doi: 10.1096/fj.13-237537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts A, Schaeffer L, Pachter L. Updating RNA-Seq analyses after re-annotation. Bioinformatics. 2013;29:1631–1637. doi: 10.1093/bioinformatics/btt197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaewkhaw R, Kaya KD, Brooks M, Homma K, Zou J, Chaitankar V, Rao M, Swaroop A. Transcriptome dynamics of developing photoreceptors in three-dimensional retina cultures recapitulates temporal sequence of human cone and rod differentiation revealing cell surface markers and gene networks. Stem Cells. 2015;33:3504–3518. doi: 10.1002/stem.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bodenhofer U, Kothmeier A, Hochreiter S. APCluster: An R package for affinity propagation clustering. Bioinformatics. 2011;27:2463–2464. doi: 10.1093/bioinformatics/btr406. [DOI] [PubMed] [Google Scholar]
- 60.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 61.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fresno C, Fernández EA. RDAVIDWebService: A versatile R interface to DAVID. Bioinformatics. 2013;29:2810–2811. doi: 10.1093/bioinformatics/btt487. [DOI] [PubMed] [Google Scholar]
- 63.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 64.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.