Abstract
The proteins encoded by the two major breast cancer susceptibility genes, BRCA1 and BRCA2, work in a common pathway of genome protection. However, the two proteins work at different stages in the DNA damage response (DDR) and in DNA repair. BRCA1 is a pleiotropic DDR protein that functions in both checkpoint activation and DNA repair, whereas BRCA2 is a mediator of the core mechanism of homologous recombination. The links between the two proteins are not well understood, but they must exist to explain the marked similarity of human cancer susceptibility that arises with germline mutations in these genes. As discussed here, the proteins work in concert to protect the genome from double-strand DNA damage during DNA replication.
The greatest risk factor for breast and ovarian cancer is inheritance of a mutation in one of the breast cancer susceptibility genes, BRCA1 or BRCA2. BRCA1 and BRCA2 are tumour suppressor genes, the coding regions of which show no homology to previously described proteins or to each other. If one copy of either gene is mutated in the germ line, the result is hereditary breast and ovarian cancer (HBOC) syndrome, which is inherited in an autosomal-dominant manner. This syndrome is associated with not only early-onset breast cancer but also an increased risk of ovarian, pancreatic, stomach, laryngeal, fallopian tube and prostate cancer. HBOC syndrome accounts for 5–7% of all cases of breast cancer, and individuals with HBOC syndrome have a lifetime risk of developing breast cancer of 50–80%, and of 30–50% for ovarian cancer (TABLE 1). The estimated frequency of developing other common malignancies associated with a mutation in BRCA2 is only 0.1% for prostate cancer and 0.5% for pancreatic cancer, although the relative risk is significantly increased (up to 20-fold for prostate cancer and 10-fold for pancreatic cancer)1.
Table 1.
BRCA1 and BRCA2 functions: their domains and binding partners
| Function | Domain | Direct binding | Indirect binding | Refs |
|---|---|---|---|---|
| BRCA1 | ||||
| Recruitment to DNA damage sites | BRCT | Abraxas | RAP80 | 19,20, 108,109 |
| DNA end resection | BRCT and RING? | CtIP | MRN complex | 13,14,22 |
| G2/M checkpoint | BRCT | Abraxas | RAP80 | 20,21 |
| BRCT | CtIP | MRN complex | 13 | |
| SCD (S1423 and S1524 phosphorylation) | ATM | MRN complex | 111 | |
| S-phase checkpoint | SCD (S1387 phosphorylation) | ATM | MRN complex | 112 |
| BRCT | BRIP1 | TOPBP1 | 40 | |
| Repair during DNA replication | BRCT | BRIP1 | TOPBP1 | 110 |
| HR | Coiled-coil and S988 phosphorylation | PALB2 | BRCA2 | 16–18,25 |
| BRCA2 | ||||
| HR | BRC | RAD51 | 46,50,52 | |
| DBD | DSS1 | 43–46 | ||
| N terminus | PALB2 | BRCA1 | 16–18,25 | |
| C terminus | RAD51 | CDK2 | 53 | |
ATM, ataxia-telangiectasia mutated; BRIP1, BRCA1-interacting protein C-terminal helicase 1; CDK2, cyclin-dependent kinase 2; CtIP, CtBP-interacting protein; DBD, DNA-binding domain; DSS1, deleted in split-hand/split-foot syndrome; HR, homologous recombination; MRN, MRE11, RAD50 and Nijmegen breakage syndrome protein 1 (NBS1); PALB2, partner and localizer of BRCA2; SCD, SQ/TQ cluster domain; TOPBP1, DNA topoisomerase 2-binding protein 1.
In addition to having similar disease phenotypes, both proteins are known to function in homologous recombination (HR), a vital DNA repair process that uses the undamaged sister chromatid to carry out high-fidelity repair of predominantly replication-associated DNA double-strand breaks (DSBs). HR appears to be the major mechanism for protecting the integrity of the genome in proliferating cells, because other DSB repair pathways are error-prone and generate chromosome deletions and translocations2. A curious feature of HBOC syndrome is that BRCA1-associated breast cancer is more often oestrogen-receptor (ER) negative, whereas BRCA2-associated breast cancers have the same distribution of cancer subtypes as found sporadically. In hereditary breast cancer for which there is no evidence of a BRCA1 or BRCA2 mutation, mutations in the DNA damage response (DDR) kinases CHK2 or ataxia-telangiectasia mutated (ATM) might account for the breast cancer predisposition3,4. Additional inherited mutations may also occur in other members of the BRCA1–BRCA2–HR pathway, such as partner and localizer of BRCA2 (PALB2) and BRCA1-interacting protein C-terminal helicase 1 (BRIP1; also known as BACH1 or FANCJ), but currently the frequency of mutations in these genes in hereditary breast cancer is low. In the absence of known germline predisposition for breast cancer, mutations in BRCA1 and BRCA2 are uncommon in sporadic breast cancer.
Linking the biochemistry of BRCA1 and BRCA2 function to a common pathway of genome protection often creates more questions than answers. This Perspective discusses how the BRCA1 and BRCA2 proteins function biochemically and how this is related to their observed roles. We ask from what type of DNA damage are these proteins protecting the cell? How do the two proteins communicate in a common pathway of genome protection? And why are breast and ovarian epithelial cells preferentially susceptible to tumorigenesis? We also discuss the association of breast cancer subtypes with BRCA1 and BRCA2 deficiency.
Maintaining genome integrity
One aspect of maintaining genomic integrity is mediated by a cellular network of signalling events (the DDR) that is triggered in response to genotoxic stress. The DDR to DSBs involves sensors that can detect broken ends, effectors that execute repair and mediators that facilitate interactions between sensors and effectors (FIG. 1). The DDR also includes the activation of checkpoints that delay the cell cycle before or during replication (G1/S or intra-S-phase checkpoints) or before cell division (G2/M checkpoint) to ensure that genetic errors are not transmitted to subsequent generations by allowing time for DNA repair. DSBs are considered to be the most threatening form of DNA damage, as the integrity of both strands of the DNA duplex is compromised simultaneously. DSBs can occur as by-products of DNA replication or during exposure to ionizing radiation and other genotoxic compounds. In mammalian cells, DSBs are repaired by HR (which is mostly error-free), or by non-homologous end-joining (NHEJ; which is error-prone). The genome is particularly susceptible to DNA damage during replication because damage on a single strand can be converted to double-strand damage and lead to replication fork collapse. In the absence of an intact HR pathway, these replication-associated DSBs can result in chromosome rearrangements and hence genomic instability.
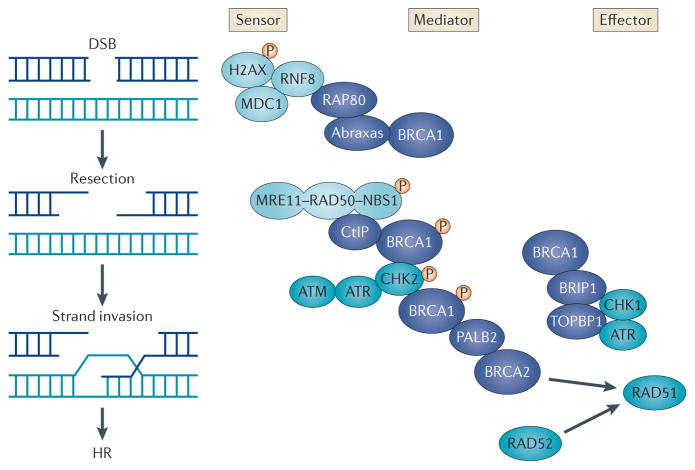
Figure 1. Molecular mechanisms of the DNA damage response.
In response to DNA double-strand breaks (DSBs) or replication fork collapse (not shown), sensors (light blue) detect the damage, and signalling mediators recruit or activate effectors that repair the damage and activate cell cycle checkpoints. BRCA1-containing macro-complexes (dark blue) are crucial mediators of the DNA damage response. The BRCA1–abraxas–RAP80 complex associates with ubiquitylated histones near the sites of DNA damage; this is dependent on phosphorylation of histone H2AX (γH2AX), mediator of DNA damage checkpoint protein 1 (MDC1) and RING finger protein 8 (RNF8). The BRCA1–CtBP-interacting protein (CtIP) complex associates with the MRN complex (which is comprised of MRE11, RAD50 and Nijmegen breakage syndrome protein 1 (NBS1)), which senses DSBs and is responsible for DSB resection. The BRCA1–partner and localizer of BRCA2 (PALB2)–BRCA2 complex is important in mediating RAD51-dependent homologous recombination (HR). CHK2-dependent phosphorylation of S988 in BRCA1 appears to be required for the BRCA1–PALB2–BRCA2 effector complex, which is important in RAD51-mediated HR. The BRCA1–BRCA1-interacting protein C-terminal helicase 1 (BRIP1)–DNA topoisomerase 2-binding protein 1 (TOPBP1) complex is associated with DNA repair during replication and may help mediate ataxia telangiectasia and Rad3-related (ATR)–CHK1 signalling, but its precise function is unknown. DNA damage is also recognized by ataxia-telangiectasia mutated (ATM) and ATR kinases, which phosphorylate BRCA1, BRCA1-associated proteins and p53 and mediate signalling to form macro-complexes and activate cell cycle checkpoints.
HR repairs DSBs during the S and G2 phases of the cell cycle, when an intact sister chromatid can serve as a template for repair; it is also pivotal in maintaining replication fidelity. The protection of the genome by HR involves damage recognition by the kinases ATM and ataxia telangiectasia and Rad3-related (ATR), signal mediation by CHK2 and BRCA1, and initiation of repair by the effectors BRCA2 and RAD51. There are also several facilitators of the HR pathway, such as PALB2 and BRIP1, and each of these facilitators is a predisposing factor for HBOC syndrome when mutated, which suggests that it is the BRCA1–BRCA2–HR pathway that suppresses tumorigenesis.
BRCA1 functional domains and binding partners
BRCA1 is a versatile protein that links DNA damage sensing and DDR effectors. BRCA1 interacts with tumour suppressors, DNA repair proteins and cell cycle regulators through its various functional domains and thereby has diverse roles in multiple DNA repair pathways (particularly HR, NHEJ and single-strand annealing (SSA)) and in checkpoint regulation5,6. BRCA1 contains an amino-terminal RING domain that has E3 ubiquitin ligase activity (which catalyses protein ubiquitylation) and a BRCT domain that facilitates phospho-protein binding (FIG. 2a). Many inherited cancer-associated BRCA1 mutations have been found within the RING and BRCT domains, indicating that both domains are involved in suppressing breast and ovarian cancer7–9. BRCA1 E3 ubiquitin ligase activity is enhanced when associated with the RING domain of its partner protein, BRCA1-associated RING domain protein 1 (BARD1)10. The BRCA1–BARD1 heterodimer generates polyubiquitin chains at unconventional K6 linkages that do not appear to signal for protein degradation, but may instead mediate downstream signalling events through mechanisms that are still unclear10–13. The tumour suppressor function of the E3 ubiquitin ligase activity has been questioned recently by the observation that in a knock-in mouse model expressing an E3-ligase defective mutant of BRCA1, the development of tumours was suppressed to the same extent as when wild-type BRCA1 was expressed. BRCA1 ubiquitylation of CtBP-interacting protein (CtIP; also known as RBBP8), a protein involved in DNA DSB resection through its association with the MRN complex (which is comprised of MRE11, RAD50 and Nijmegen breakage syndrome protein 1 (NBS1; also known as nibrin)), may have a role in DSB repair pathway choice, as CtIP-dependent resection promotes HR and inhibits NHEJ14. Why the E3-ligase function appears to be dispensable for tumour suppression has not yet been satisfactorily answered, as many tumour-producing mutations are located in the RING domain, suggesting that there may be another function of the protein associated with the RING domain that has yet to be defined.
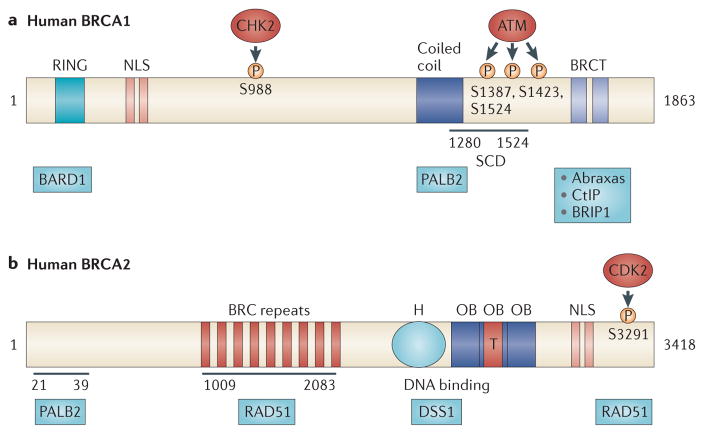
Figure 2. BRCA1 and BRCA2 functional domains.
a | The BRCA1 amino terminus contains a RING domain that associates with BRCA1-associated RING domain protein 1 (BARD1) and a nuclear localization sequence (NLS). The central region of BRCA1 contains a CHK2 phosphorylation site on S988 (REF. 25). The carboxyl terminus of BRCA1 contains: a coiled-coil domain that associates with partner and localizer of BRCA2 (PALB2); a SQ/TQ cluster domain (SCD) that contains approximately ten potential ataxia-telangiectasia mutated (ATM) phosphorylation sites and spans amino acid residues 1280–1524; and a BRCT domain that binds ATM-phosphorylated abraxas, CtBP-interacting protein (CtIP) and BRCA1-interacting protein C-terminal helicase 1 (BRIP1). The BRCA1–abraxas complex is associated with BRCA1 recruitment to sites of DNA damage19,20,108,109. The BRCA1–BRIP1 complex, which also contains DNA topoisomerase 2-binding protein 1 (TOPBP1), is associated with DNA repair during replication110. The BRCA1–CtIP complex promotes ataxia-telangiectasia and Rad3-related (ATR) activation and homologous recombination (HR) by associating with the MRN complex (which is comprised of MRE11, RAD50 and Nijmegen breakage syndrome protein 1 (NBS1)) and facilitating DNA double-strand break resection22. The central region of BRCA1, which contains the SCD, is phosphorylated by ATM. This phosphorylation is important for BRCA1-mediated G2/M and S-phase checkpoint activation, as expression of a BRCA1 mutant that lacks three of the phosphorylation sites (S1387, S1423 and S1524) fails to rescue defective checkpoint activation and ionizing radiation hypersensitivity in a BRCA1-deficient cell line111,112. b | The N terminus of BRCA2 binds PALB2 at amino acids 21–39 (REF. 68). BRCA2 contains eight BRC repeats between amino acid residues 1009 and 2083 that bind RAD51. The BRCA2 DNA-binding domain contains a helical domain (H), three oligonucleotide binding (OB) folds and a tower domain (T), which may facilitate BRCA2 binding to both single-stranded DNA and double-stranded DNA46. This region also associates with deleted in split-hand/split-foot syndrome (DSS1)42,44,45. The C terminus of BRCA2 contains an NLS and a cyclin-dependent kinase (CDK) phosphorylation site at S3291 that also binds RAD51 (REF. 53).
The BRCT phosphopeptide-binding motif, which is conserved in multiple DDR proteins, is responsible for the association of BRCA1 with proteins phosphorylated on serine in SXXF motifs by ATM. The BRCA1- interacting proteins include abraxas, BRIP1 and CtIP. The binding of these proteins make up separate BRCA1 macro-protein complexes that have distinct and overlapping functions in the DDR15 (FIG. 1; TABLE 1). A fourth BRCA1-containing complex mediated through the BRCA1 coiled-coil domain is composed of PALB2 and BRCA2 and is specifically involved in DSB repair by HR16–18. How these multiple BRCA1 complexes work in a coordinated manner is still unclear. It will be interesting to uncover whether one BRCA1-containing complex is replaced with a different BRCA1-containing complex during the DDR, or whether BRCA1 functions as a platform on which initial DNA damage sensing proteins assemble and disassemble and subsequent repair proteins associate. As many of the current investigations have been performed using global DNA damaging agents, disentangling which BRCA1 complex functions at different steps in the pathway will be difficult. The study of a single, site-specific damage locus may help to shed light on the crosstalk between the various BRCA1 macro-complexes and their assembly and disassembly.
BRCA1 and HR
BRCA1 is directly involved in HR-mediated repair of DSBs19–21. BRCA1 binds to DSBs through its association with the abraxas–RAP80 macro-complex, which associates with ubiquitylated histones at DNA DSBs19 (FIG. 1). Next, BRCA1 is involved in processing DSBs through its interaction with CtIP and the MRN complex (FIG. 1). The BRCA1–CtIP complex promotes CtIP-mediated 5′-end resection of DSBs14, which is abrogated by three independent tumour-associated mutations in the BRCT domain of BRCA1 (REFS 22,23). BRCA1 is also required for RAD51 recruitment to the sites of DNA damage through its interactions with PALB2 and BRCA2 (FIG. 1). This interaction appears to be dependent on CHK2-mediated phosphorylation of S988 on BRCA1 (S.N.P., unpublished observations). Importantly, knock-in mice expressing an S971A mutant (in mice, S971 corresponds to human S988) develop mammary and endometrial tumours after treatment with DNA damaging agents24. BRCA1-deficient human cells expressing BRCA1-S988A have defects in HR but retain normal checkpoint function and resistance to ionizing radiation, implying that the HR function of BRCA1 is distinct from its other functions in the DDR and that this mutation causes a dissociation of function25. Brca1-null embryonic stem (ES) cells from mice expressing the human BRCA1-S988A mutant also displayed no difference in cell cycle profiles or sensitivity to DNA damaging agents compared to those expressing wild-type human BRCA1 (REF. 26). In this study, HR function was not analysed, and sensitivity to DNA damaging agents was measured by growth assay rather than clonogenic survival. The extent to which deficiency in HR contributes to cell survival after ionizing radiation, even for cells in S phase, remains a point of debate — HR is likely to be more important for surviving replication errors than for surviving exogenous DNA damage resulting in DSBs, in which case other repair pathways can be used. Similarly, tumour suppression is likely to be related to fixing replication errors rather than repairing DSBs, which is why HR and tumour suppression are closely linked.
BRCA1 and other DNA repair pathways
BRCA1 may also function in other DNA repair pathways, including NHEJ and SSA. The role of BRCA1 in NHEJ is somewhat controversial, as BRCA1 has been observed to facilitate27,28, suppress29,30 or have no effect on NHEJ31. These wide-ranging observations may be attributed to the variety of assays used to measure NHEJ and the possibility that BRCA1 has different roles in the various subtypes of NHEJ. BRCA1 may be involved in HR, NHEJ and SSA through its interaction with the MRN complex, which is required for DNA end resection before all three repair processes32,33. BRCA1 recruitment to DSBs is facilitated by a DNA damage-induced interaction of the BRCA1 N terminus with the NHEJ protein KU80, thereby providing another mechanism for BRCA1 accumulation at DSBs34. A recent study has proposed that a critical function of BRCA1 is to remove NHEJ proteins such as p53-binding protein 1 (53BP1) from DSBs35 to prevent aberrant end-joining and to regulate the choice between HR and NHEJ. A BRCA1 exon 11 deletion mutant exhibited decreased SSA in addition to decreased HR, providing further evidence that BRCA1 functions in both SSA and HR36. The relevance of SSA in genome stability and tumour suppression is not known, but is likely to be limited. Like NHEJ, SSA is also mutagenic, producing deletions and insertions at sites of long repeat sequences.
BRCA1 and checkpoint activation
The BRCA1–BARD1 complex is involved in the activation of G1/S, S-phase and G2/M checkpoints (TABLE 1). The G1/S-checkpoint requires phosphorylation of BRCA1 by ATM or ATR, which facilitates phosphorylation of p53 on S15. p53-S15 phosphorylation is necessary for transcriptional induction of the cyclin dependent kinase (CDK) inhibitor p21 and ionizing radiation-induced G1/S checkpoint activation37. BRCA1–BARD1 depletion compromises the induction of p21 and activation of the G1/S checkpoint in response to ionizing radiation38. The exact mechanism of BRCA1–BARD1 control of the S-phase and G2/M checkpoints is not well characterized39. The BRCA1–BRIP1–DNA topoisomerase 2-binding protein 1 (TOPBP1) macro-complex appears to be necessary for the S-phase checkpoint in response to stalled or collapsed replication forks40, whereas the BRCA1–abraxas–RAP80 macro-complex appears to be involved in the G2/M checkpoint in response to ionizing radiation-induced DNA damage (TABLE 1).
The G2/M checkpoint is defective in cells lacking functional BRCA1, BARD1, RAP80 or abraxas20,21 (TABLE 1). Interestingly, partial defects in G2/M checkpoint activation and p53 stabilization were observed in mouse embryonic fibroblasts (MEFs) from knock-in mice expressing the cancer-associated BRCA1-S971A mutation (which is the equivalent to human BRCA1-S988A)26. The checkpoint defect was mild and disappeared 4 hours after treatment with ionizing radiation. Furthermore, after treatment with an alkylating agent, but not ionizing radiation, the levels of p53 were reduced in MEFs expressing BRCA1-S971A24, whereas in human HCC1937 cells expressing BRCA1-S988A, no detectable defect in the G2/M checkpoint was observed (S.N.P., unpublished observations). The stability of p53 has not been tested in BRCA1-deficient human cells and may need to be measured to verify whether, in human cells, BRCA1-S988A is a true separation-of-function mutation. These additional functions of BRCA1 in checkpoints and in stabilizing p53 may contribute to genomic stability, which is primarily determined by the role of BRCA1 in HR. Likewise, this broader range of BRCA1 functions may account for the severity and pattern of genomic instability found in BRCA1-deficient tumours relative to BRCA2-deficient tumours.
BRCA2 functional domains and binding partners
In contrast to the multifunctional activities of BRCA1, the primary function of BRCA2 is in HR (TABLE 1). BRCA2 mediates the recruitment of the recombinase RAD51 to DSBs; RAD51 recruitment is not only essential for HR but is also responsible for the tumour-suppressive function of this repair process41 (FIG. 1). BRCA2 contains a DNA-binding domain (DBD) that binds single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) and eight BRC repeats that bind RAD51 (FIG. 2b). The DBD contains five components: a 190-amino-acid α-helical domain, three oligonucleotide binding (OB) folds that are ssDNA-binding modules, and a tower domain (TD) that protrudes from OB2 and binds dsDNA42. The helical domain, OB1 and OB2 also associate with deleted in split-hand/split-foot syndrome (DSS1), which has been linked to BRCA2 protein stabilization42–45. Ustilago maydis Brh2, the BRCA2 homologue in corn smut, binds to DNA at the resected ends of a DSB, where both dsDNA and ssDNA exist46, presumably facilitating RAD51 filament formation at that site47. This implies that BRCA2 mediates RAD51 filament formation at the appropriate sites of ssDNA and prevents it from binding to dsDNA, as supported by recent biochemical data using purified BRCA2 (REF. 48).
Point mutations within BRC repeats that compromise interactions with RAD51 are found in individuals with HBOC syndrome49. The BRC repeats have subtle differences in sequence and bind RAD51 with varying affinity by mimicking the structure of RAD51 monomers50. In addition to facilitating the recruitment of RAD51 to ssDNA, the BRC repeats accelerate replication protein A (RPA)-displacement from ssDNA by RAD51 (REF. 51), block RAD51 nucleation at dsDNA and facilitate RAD51 filament formation on ssDNA by maintaining the active ATP-bound form of RAD51 on ssDNA52. The binding of RAD51 by the C terminus of BRCA2 has been shown to be dependent on CDK activity53,54. This association, however, appears to be dispensable for HR in vivo and may be important for the disassembly of RAD51 complexes to facilitate mitotic entry2. Whether this association is important for suppression of breast and ovarian tumorigenesis is unknown.
Much of our knowledge of BRCA2 comes from studying portions of the BRCA2 protein or investigating BRCA2 orthologues, such as Brh2 in U. maydis and BRC-2 in Caenorhabditis elegans46,52,55–57. Insight into the details of mammalian BRCA2 function had been hampered by the inability to isolate the full-length protein, which is 3,418 amino acids in humans. Recently, however, three independent groups have successfully purified and functionally validated full-length human BRCA2 (REFS 48,58,59). These studies showed that the stoichiometry of RAD51 binding was 6–7 to 1, and that the protein could indeed catalyse many steps of the RPA to RAD51 transition, as had been predicted by the genetic studies. Future studies using the full-length BRCA2 protein will help to further our understanding of the structure and function of BRCA2.
BRCA2, HR and replication fidelity
Overwhelming evidence suggests that the primary function of BRCA2 is to facilitate HR (TABLE 1). BRCA2-deficient cells are defective in recruiting RAD51 to sites of DSBs and in repairing DSBs by HR60. Although human and mouse cells expressing a BRCA2 loss-of-function truncation mutant display some defects in replication and checkpoint control, BRCA2 is not essential for these processes61,62. Recently, BRCA2-deficient hamster cells treated with hydroxyurea (which causes replication fork stalling and collapse) were shown to have defects in maintaining the length of the nascent strand of DNA, perhaps because BRCA2 protects the nascent strand from degradation at stalled replication forks2. Thus, in addition to its role in repair by HR, these results imply a second role for BRCA2 in protecting the replication fork. The critical evidence supporting these two functions of BRCA2 was a dissociation-of-function mutant, S3291A, the expression of which allows normal DSB-induced HR but results in defective protection of replication forks. Without the dissociation-of-function mutant, the reduced length of the nascent strand after replication fork stalling could be secondary to deletions in the sister chromatid arising either from defective HR repair of the collapsed replication fork or from defective daughter-strand gap (DSG) repair by HR. We favour the view that the activity of BRCA1 and BRCA2 can occur at DSBs created behind the replication fork (in addition to replication fork collapse) as a consequence of gaps on the nascent strand that are created by single-strand lesions on the parental strand (FIG. 3).
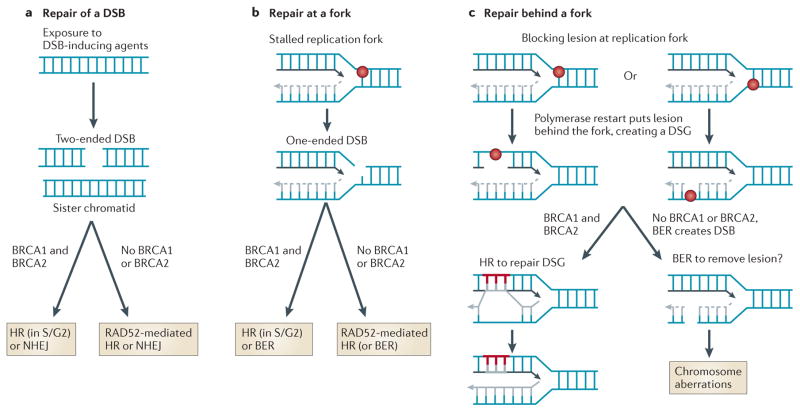
Figure 3. Homologous recombination at different types of DNA damage.
Exogenous agents produce DNA double-strand breaks (DSBs) with two ends (a), whereas during replication, blocking lesions on the template strand can produce either one-ended DSBs (b) or daughter-strand gaps (DSGs) (c), both of which are preferentially repaired in the S and G2 phases of the cell cycle by the BRCA1–BRCA2-mediated homologous recombination (HR) pathway. During replication, template strand lesions may be repaired either behind the fork or at the fork. Gaps associated with lesions on the parental strand behind the fork cannot be removed by base excision repair (BER) because an undamaged template is needed for repair; therefore, HR is the only available pathway for the repair of DSGs. Cells lacking functional BRCA1 or BRCA2 exhibit an abundance of chromatid breaks, which indicates an attempt to repair DSGs in the absence of a functional BRCA-mediated HR pathway. DSBs that remain unrepaired behind a replication fork can also produce chromatid breaks or aberrant junctions or exchanges. Hence, BRCA1 and BRCA2 have crucial roles in the repair of replication-associated lesions at or behind the replication fork. NHEJ, non-homologous end-joining.
Links between BRCA1 and BRCA2
Although germline Brca1 or Brca2 heterozygous mutations in mice do not have a strong phenotype, most homozygous mutations are embryonically lethal63,64. Both Brca1 and Brca2 homozygous mouse embryos are hypersensitive to ionizing radiation and have widespread chromosome and chromatid aberrations, which indicates error-prone repair of chromatid breaks. Interestingly, the phenotype of BRCA-deficient mouse embryos mimics the phenotype of mice with inactivating mutations of Rad51 (REF. 65). Therefore, these mouse models were the initial evidence to indicate that BRCA1 and BRCA2 function in a common pathway of RAD51-mediated HR63,64.
In humans, the tumours that develop in patients with germline heterozygous mutations in BRCA1 or BRCA2 are defective in HR-mediated repair. The germline heterozygous cells must be haplo-insufficient, but this functional defect has so far been difficult to identify or investigate at the cellular level. Defective HR can also be found in sporadic breast cancers despite the absence of a germline mutation in one of the crucial members of the HR pathway, as shown by array-comparative genomic hybridization (aCGH) studies66 and functional evaluations of human breast cancer cells67. However, the nature of how these functional defects in HR occur is not yet clear. Nevertheless, the common defect in HR indicates that the function of the BRCA1–BRCA2 pathway in mediating HR is important for tumour suppression in inherited and sporadic breast cancer.
PALB2 connects BRCA1 and BRCA2
PALB2 binds directly to both BRCA1 and BRCA2 and thereby provides a physical link between the two proteins16–18 (FIG. 1). The N-terminal coiled-coil domain of PALB2 interacts with the coiled-coil domain of BRCA1, and the C terminus of PALB2 interacts with the N terminus of BRCA2 (REF. 68) (FIG. 2b). The interaction of PALB2 with BRCA2 was shown to be essential for loading RAD51 onto RPA-bound ssDNA69. Furthermore, the BRCA1–PALB2 interaction is a prerequisite for the recruitment of BRCA2 and RAD51 to the site of DNA damage and for HR, but had no impact on BRCA1-mediated S-phase checkpoint activation17,18. Depletion of PALB2 expression in cells phenocopied BRCA2 deficiency and abrogated the interaction between BRCA1 and BRCA2. Additionally, BRCA1 phosphorylation on S988 by CHK2 promotes formation of the BRCA1–PALB2–BRCA2 complex (FIG. 1), which may explain why mutating this site abrogates HR25. It is unknown whether there are other regulators of the BRCA1–PALB2–BRCA2 complex.
BRCA1 and BRCA2 function in a common pathway
Repair by HR can be triggered at ionizing radiation-induced two-ended DSBs or at one-ended DSBs that are generated by the cleavage of replication forks that have been stalled secondary to a blocking lesion. In some instances, single-strand lesions that do not produce a strand break are bypassed by the replication machinery and replication is restarted downstream of the lesion, leaving behind a region of ssDNA or a DSG in which no DSB end is present70 (FIG. 3). Replication-associated one-ended DSBs or DSGs recruit BRCA1, PALB2, BRCA2 and RAD51. Brca1-mutant mice exhibit telomere dysfunction, chromosome translocations and chromatid aberrations71, and Brca2-mutant mice accumulate chromatid breaks and aberrant chromatid exchanges61, providing further evidence in support of a BRCA1–BRCA2-mediated HR response to replication-associated DNA damage. Similarly, Brca1- and Brca2-mutant mice develop thymic lymphomas61,71, which is a common tumour that arises in mice owing to defective DSB repair. Moreover, conditional expression of homozygous Brca1 or Brca2 mutants in mammary epithelium was sufficient to generate mammary cancers72.
BRCA1 is involved in DDR signalling, checkpoint activation and HR and may also play a role in other DNA repair processes, such as NHEJ and SSA. Conversely, BRCA2 is primarily involved in HR. The human syndromes associated with BRCA1 or BRCA2 germline mutations are almost identical, and the only common functional link between the two BRCA proteins is the HR pathway. Therefore, it seems reasonable to conclude that the HR pathway is crucial for protecting the genome and that this pathway is disrupted in tumours arising in these mutation carriers. The Fanconi anaemia pathway — which includes BRCA2 — has some functional overlap with the BRCA1–BRCA2 pathway, but the human syndrome Fanconi anaemia (which is caused by defects in members of the Fanconi anaemia pathway) is markedly different to HBOC syndrome and is characterized by anaemia, skeletal abnormalities and predisposition to squamous cell carcinomas. In addition, the inheritance of Fanconi anaemia is autosomal recessive (owing to the inheritance of two hypomorphic alleles), whereas HBOC syndrome shows autosomal-dominant inheritance, with loss of the second allele (loss of heterozygosity (LOH)) occurring in the cancers that arise in mutation carriers. The effect of a defective BRCA1 or BRCA2 allele in the germ line must cause haploinsufficiency of HR to trigger the subsequent genetic alterations that result in cancer. Presumably, haplo-insufficiency from a single defective allele has different biological impacts compared with biallelic inheritance of hypomorphic alleles. However, the total number of families in the world that have been diagnosed with Fanconi anaemia caused by defects in BRCA1–BRCA2 pathway genes (BRCA2 (also known as FANCD1), BRIP1 (also known as FANCJ and BACH1), PALB2 (also known as FANCN), RAD51C (also known as FANCO) or SLX4 (also known as FANCP)) is small, and whether these patients show all the characteristic features of Fanconi anaemia has been debated73.
In the BRCA1–BRCA2-mediated HR pathway, BRCA1 functions upstream of BRCA2, the function of which is dependent on BRCA1. In mammalian cells, HR can also occur through an alternative, BRCA1–BRCA2-independent, RAD52-dependent pathway. When BRCA2 function is disrupted in a tumour cell, RAD52 helps the cell to stay viable. Indeed, cells that are simultaneously depleted of RAD52 and BRCA1, RAD52 and PALB2 (S.N.P., unpublished observations) or RAD52 and BRCA2 (REF. 74) exhibit synthetic lethality. Taken together, these results support the hypothesis that BRCA1 and BRCA2 are connected in a common HR pathway that functions to repair DSBs, collapsed DNA replication forks or DSGs. Germline mutations in genes involving this common HR pathway are all associated with HBOC syndrome, suggesting that this pathway is the crucial tumour suppressor activity. Mutations in BRCA1 and BRCA2 are the predominant cause of HBOC syndrome, with ATM and CHK2 mutations being less common. Mutations in PALB2 that lead to HBOC syndrome are very rare compared to mutations in BRCA1 and BRCA2 (REF. 75).
BRCA1 and BRCA2 in tumorigenesis
Common genetic alterations are associated with heterozygous BRCA1 or BRCA2 mutations, and these include loss of the wild-type BRCA1 or BRCA2 allele (LOH), loss of TP53 (which encodes p53), and loss of ATM or CHK2 function. These additional alterations may allow cells to bypass checkpoint controls and evade apoptosis, and thereby initiate tumorigenesis. The fact that both BRCA1 and BRCA2 mutation carriers display these similar somatic alterations further confirms that their role in HR-mediated repair is important for tumour suppression.
Loss of the wild-type BRCA allele
When a tumour develops in patients with HBOC syndrome caused by BRCA mutation, loss of the wild-type BRCA allele (LOH) was initially always reported, as would be expected of a tumour suppressor. However, more recently this dogma has been questioned. Initial studies of Brca1+/− and Brca2+/− mice showed no increase in tumour formation compared with wild-type mice76, but subsequent studies using Brca1+/−Trp53+/− mice showed a slight increase in mammary carcinoma incidence compared with Trp53+/− mice77. All tumours in this study retained BRCA1 protein expression, which rules out epigenetic silencing. Similar results were also observed in Brca2+/− mice: mammary tumours developed in a p53-deficient background78; however, in this study, epigenetic silencing cannot be ruled out because levels of BRCA2 protein were not measured. A study of breast cancer tissue samples from patients with HBOC syndrome caused by BRCA mutation showed that out of 18 cases in which LOH was observed, 11 patients showed loss of the mutant rather than the wild-type allele, suggesting that loss of the wild-type BRCA allele is not required for tumorigenesis79. By contrast, all ovarian cancer tissue samples tested showed LOH of the wild-type BRCA allele79. LOH was also not observed in a small study of pancreatic cancer tissue samples from carriers of the Icelandic founder mutation BRCA2-999del5, which produces a truncated form of BRCA2 (REFS 80,81). However, it is important to keep in mind the methodological issues about measuring LOH in microdissected tumour samples by PCR, because a small amount of contaminating normal tissue could produce a wild-type allele. In addition, epigenetic silencing of the wild-type allele, which cannot be detected by PCR, must also be considered. In the absence of LOH, haploinsufficiency of BRCA activity may cause enough genomic instability to promote tumorigenesis. Although some data suggest that loss of the wild-type allele may not be required for all BRCA-associated tumorigenesis, in most cases LOH does occur. However, whether BRCA heterozygosity promotes loss of the wild-type allele or whether loss occurs randomly is still unclear.
BRCA LOH is thought to occur by either deletion or gene conversion. When LOH does occur, a germline mutation in BRCA1 results in loss of the wild-type BRCA1 allele but not the wild-type BRCA2 allele, and vice versa. The similar pattern of LOH observed in patients carrying a BRCA1 or a BRCA2 mutation and the fact that only one of the BRCA genes is affected in an individual (and not both genes in one individual) suggests that tumorigenesis in BRCA1 and BRCA2 mutation carriers is primarily caused by functional inactivation of either BRCA protein, and we suggest that it is the deficiency in HR that leads to tumorigenesis (as discussed above).
Loss of p53 expression
p53 plays a vital part in maintaining genomic integrity by regulating the transcription of target genes that are involved in cell cycle arrest, apoptosis and DNA repair82. Multiple studies suggest that the loss of p53 cooperates with the loss of BRCA1 or BRCA2 in tumorigenesis78,83–86. TP53 mutations are present in 30–50% of human cancers, and they occur in tumours with BRCA1 or BRCA2 mutations with greater frequency than in sporadic tumours with wild-type BRCA1 and BRCA2 (REFS 83,87). A susceptibility to develop breast carcinomas is also a clinical feature of Li–Fraumeni syndrome, which is caused by germline mutation of TP53 and is associated with predisposition to develop several types of cancer. Loss of Trp53 delays embryonic lethality by 2 to 3 days in Brca1−/− or Brca2−/− mice88. Loss of Trp53 also partially rescues the embryonic lethality of Palb2−/− mice16,17,69,89. Presumably, chromosome breaks caused by loss of BRCA function activate p53-dependent checkpoint controls and/or apoptosis to prevent tumour formation. Selective pressures then favour the proliferation of cells with loss of p53 function.
In support of this idea, T cells from T cell lineage-specific BRCA2-deficient mice have an accumulation of chromosome aberrations that result in increased p53-mediated apoptosis90. However, p53 mutants (such as T150I, G199R and R202S) that were identified specifically in tumours from BRCA1 or BRCA2 mutation carriers retain the transactivation, checkpoint and apoptotic activities of wild-type p53, but they still fail to suppress transformation and exhibit gain of function transforming activity in rat embryo fibroblasts91,92. Future studies may reveal p53 functions that are uniquely impaired in BRCA-deficient cells. The rarity of these mutants in human cancer and their multiple occurrences in BRCA-associated breast tumours suggests that these novel p53 mutants are selected for during malignant progression in the unique genetic background of BRCA1- or BRCA2-mutation-associated tumours. Therefore, the common HR defect in both BRCA1- and BRCA2-deficient cells may be responsible for the selection of these specific p53 mutants.
Loss of ATM or CHK2 function
Consistent with being in the same signalling pathway as p53, loss of Atm or Chek2 also rescues the embryonic lethality of Brca1 mutant mice and leads to the development of multiple tumours, although at a lower frequency compared to mice with Brca1 and Trp53 mutations93. In addition, ATM expression can be aberrantly reduced or lost in tumours expressing BRCA1 or BRCA2 mutants compared with sporadic tumours without BRCA1 or BRCA2 mutations94. These data suggest that the genomic instability caused by heterozygous BRCA mutations may lead to the selection of ATM-deficient cells. Although ATM, BRCA1 and BRCA2 are in a common signalling pathway, additional loss of ATM activity may contribute to a selective growth advantage95. This apparently paradoxical finding makes sense when one considers that ATM functions in multiple DDR signalling pathways. Alternatively, in genetically unstable BRCA-deficient tumours, random gains and losses occur across the genome — secondary to genetic instability — and ATM loss could occur secondary to global instability.
Breast and ovarian tissue tropism
Why breast and ovarian cancer?
Given that BRCA1 and BRCA2 protect the genome from errors that arise during DNA replication, it is logical that cells driven to replicate would develop potentially oncogenic genetic alterations in the absence of BRCA1 or BRCA2 function. However, most cancers are driven to grow and divide, so this feature alone does not determine why there is a major predisposition to breast and ovarian cancer in individuals who lack functional BRCA1 or BRCA2. One common feature is that breast and ovarian epithelial cells are subject to strong growth signals by hormonal stimulation during the normal menstrual cycle. The question then becomes: what features of hormonally triggered growth make it vulnerable to genetic instability in the context of BRCA1 or BRCA2 deficiency? A number of theories have surfaced to explain the tissue specificity of HBOC syndrome, but as yet none is definitive.
One hypothesis relates to the connection between BRCA1 function and the regulation of ER signalling96, whereby BRCA1 represses the transcription of hormone-mediated signalling factors and therefore functions in growth control. Early in BRCA1 research, many transcriptional effects of BRCA1 were described, including co-activation and co-repression of target genes, but the importance of these effects has lessened over time, suggesting that some of the initial observations were a product of overexpression studies97. A recent report has suggested that the effect of BRCA1 is to maintain heterochromatin, and the loss of Brca1 in mice could be reversed by expressing histone 2A fused to ubiquitin98. Some of the reported effects of BRCA1 on transcription may actually be due to effects on chromatin structure. However, these findings do not explain why germline BRCA2 mutations have the same tissue predisposition.
Our preferred theory, which is still speculative but reflects ongoing work in our laboratory, is that hormonally driven growth during each menstrual cycle produces reactive oxygen species, which cause measurable oxidative DNA damage99–102. The consequence of oxidative DNA damage is the production of a subset of lesions that cause DNA replication stress and result in one-ended DSBs or DSGs. In other words, oxidative DNA damage can produce replication stress that demands the use of the BRCA1–BRCA2–HR pathway. This explanation would account for the common features of BRCA1 and BRCA2 predisposing to breast and ovarian cancer.
Subtypes of breast cancer
An additional unsolved mystery is why BRCA1 mutation carriers develop predominantly (but not exclusively) ER-negative tumours, whereas BRCA2 mutation does not favour the development of any particular subtype of breast cancer103 (TABLE 2). However, the breast cancers that arose in Ashkenazi Jewish women with founder mutations in BRCA1 or BRCA2 were mostly triple-negative (ER-negative, progesterone receptor (PR)-negative and ERBB2-non-amplified)104, so the association with ER expression and BRCA genes is not rigid. These observations pose the question of why there should be a difference in the biological subtypes of breast cancer when the two proteins appear to be working in a common pathway of DNA repair (TABLE 3).
Table 2.
Human cancers arising in BRCA1 or BRCA2 mutation carriers
| Cancer type | BRCA1 mutations | BRCA2 mutations | Notes |
|---|---|---|---|
| Breast | 70–80% lifetime risk | 50–60% lifetime risk | Breast and ovarian cancer is the dominant cancer predisposition in BRCA1 and BRCA2 mutation carriers. BRCA1 mutation carriers develop breast and ovarian cancer at a younger age than BRCA2 mutation carriers113 |
| Ovarian | 50% lifetime risk | 30% lifetime risk | Breast and ovarian cancer is the dominant cancer predisposition in BRCA1 and BRCA2 mutation carriers. LOH of the wild-type BRCA allele is always found |
| Prostate | Ashkenazi Jewish founder mutations are associated with increased risk | 20-fold increased risk | <1% of BRCA2 mutation carriers have prostate cancer. Prostate cancer is even rarer in BRCA1 mutation carriers, except in members of the Ashkenazi Jewish population with BRCA1 mutations |
| Pancreatic | Anecdotal evidence and case reports only | Tenfold increased risk | <1% of BRCA2 mutation carriers have pancreatic cancer. No incidence has been clearly documented in BRCA1 mutation carriers |
| Gastric | None reported | Limited reports | It is unclear whether stomach cancer is associated with BRCA2 mutations |
| Others | None reported | Brain, medulloblastoma, pharyngeal, CLL and AML | Fanconi anaemia subtype D1 (caused by BRCA2 mutations) is associated with cancer of the central nervous system |
| Fallopian tube | Observed, but rare | Rare | This cancer type is like ovarian cancer, but it is a rare cancer overall and is still uncommon in BRCA mutation carriers |
AML, acute myeloid leukaemia; CLL, chronic lymphocytic leukaemia; LOH, loss of heterozygosity.
Table 3.
Characteristics of BRCA1- and BRCA2-mutation-associated breast cancers
| Phenotype | BRCA1 | BRCA2 | Notes |
|---|---|---|---|
| ER expression | Negative in 80–90% | Positive in 60–65% | One of the major mysteries to be solved |
| PR expression | Predominantly negative | Positive in the majority of cases | Less complete data relative to ER expression |
| ERBB2 amplification | Usually absent | ~15% have amplification | ERBB2 amplification can occur in BRCA mutation carriers |
| Early onset | Highly prevalent between 30 and 50 years of age | Less prevalent between 40 and 70 years of age | |
| Lobular cancers | Less likely | As frequent as in sporadic breast cancer (~15%) | |
| High grade | Likely | Common | More common than sporadic cancers |
| Basal markers | Frequent | Less common | Tumours have cytokeratin profile of basal or myoepithelial markers |
| HR function | Defective | Defective | Some debate over the frequency of LOH for the wild-type allele |
| Prognosis relative to sporadic cancer at the same stage | No difference overall. Local recurrence in the breast is increased with conservative surgery and radiation therapy | No difference |
ER, oestrogen receptor; HR, homologous recombination; LOH, loss of heterozygosity; PR, progesterone receptor.
One explanation is that there is no difference in the predisposition to genetic alterations between carriers of BRCA1 or BRCA2 mutations, but that the true connection to the ER-negative subtype is the occurrence of co-inherited mutations (or even polymorphisms) with the BRCA1 mutant haplotype. This idea has been studied in some detail, and at present there is no candidate co-inherited mutation or polymorphism to support this hypothesis. A second explanation is that the role of BRCA1 in transcriptional co-activation (or co-repression), which is a function not shared by BRCA2, produces changes in gene expression that are sufficient to change the expression of the ER bio-marker. ER-negative tumours have a characteristic gene expression profile105, and so it would be intriguing to determine whether the profile of BRCA1-associated ER-negative and sporadic ER-negative tumours is similar. However, for this hypothesis to be correct, a mechanistic connection between BRCA1-dependent transcription and the transcriptional profile of ER-negative tumours should be established. A third explanation is that a different mutational spectrum is induced by BRCA1 heterozygosity compared with BRCA2 heterozygosity, which could potentially be due to the differences in DNA repair found with BRCA1 deficiency (which could include defects in SSA and NHEJ). Analyses using aCGH show some similarities between BRCA1- and BRCA2-associated cancers, including large deletions and amplifications66. However, some differences are also detectable66, such as the locus specificity of the alterations. The importance of these alterations in specific loci in terms of how they arise or the resulting cellular consequences is not understood. A final explanation is that the cell-of-origin of ER-negative tumours is more susceptible to alterations in BRCA1, whereas BRCA2 haplo-insufficiency predisposes to loss of the second allele in all cell lineages, which would therefore result in the same biomarker profile as sporadic cancers. All of these explanations need more supportive evidence and should be the focus of future studies into the molecular genetics of breast cancer.
One intriguing clue regarding BRCA mutations and tissue tropism is that male breast cancer, which is almost always ER-positive, is more strongly associated with BRCA2 mutations106. A second clue is that the range of cancer types observed in BRCA2 mutation carriers is broader (BRCA2 mutation carriers can develop prostate and pancreatic cancer, among others) than the range observed in BRCA1 mutation carriers. The mechanistic explanation here could be linked to the cellular stresses to which the epithelial cells are exposed, although why this selects for differences in the repair pathway response to stress is not clear. BRCA2-deficient cells have been reported to be capable of carrying out SSA, whereas BRCA1-deficient cells cannot36. However, the role of SSA in the maintenance of genome integrity is not clear. In addition, recent work has suggested that 53BP1 mediates the genetic and chromosome rearrangements specifically in BRCA1-deficient cells, as loss of 53BP1 lessens the severity of the repair defect caused by loss of BRCA1 (but not loss of BRCA2)35. In this study, 53BP1 protected DSB ends from 5′-end resection, which initiates HR. If this was the entire explanation for the effect of 53BP1, this protein should also protect against the defective HR seen with BRCA2 defects, but this does not seem to be the case. Instead, the specificity towards BRCA1 would suggest that 53BP1 is protecting the genome from defective SSA, which could be more important in genome stability than currently considered. In conclusion, although we have stressed the role of BRCA1 and BRCA2 functioning in a common pathway of DNA repair, there may be subtle differences between BRCA1- and BRCA2-deficient cells that account for this curious difference in the range of cancers that develop.
Sporadic breast and ovarian cancers
We and others have reported that HR defects occur in sporadic breast cancers as well as the cancers arising in carriers of BRCA1 or BRCA2 mutations66,67,107. The extent of this type of DNA repair defect in other types of cancer is not known, as specific assays are needed to detect this phenotype. Our approach has been to test the functional integrity of the HR pathway directly in ex vivo human tumour samples by examining the formation of nuclear foci of RAD51 and BRCA1 induced by ionizing radiation67 (such nuclear foci indicate the recruitment of DDR proteins to a site of DNA damage). Others have suggested that a ‘BRCA-like’ phenotype exists if tumour cells show the characteristic large-region gains and losses that are also seen in BRCA-deficient tumours (FIG. 4). We have initiated a more comprehensive assessment of the frequency and pathological associations of HR pathway defects both in primary breast tumours and in established breast cancer cell lines. We have found that there is a significant prevalence of HR defects in sporadic breast cancer (S.N.P., unpublished observations). In addition, we have observed the same phenotype in breast cancer cell lines, none of which has known BRCA1 or BRCA2 mutations or changes in BRCA1 and BRCA2 expression. The sporadic nature of these tumours with BRCA-like features would make the explanation less likely to be an unknown genetic alteration and more likely to be an epigenetic acquired event in tumorigenesis. However, the implications are profound — the number of patients who might be suitable for therapeutic strategies to target defects in HR would be substantially expanded.
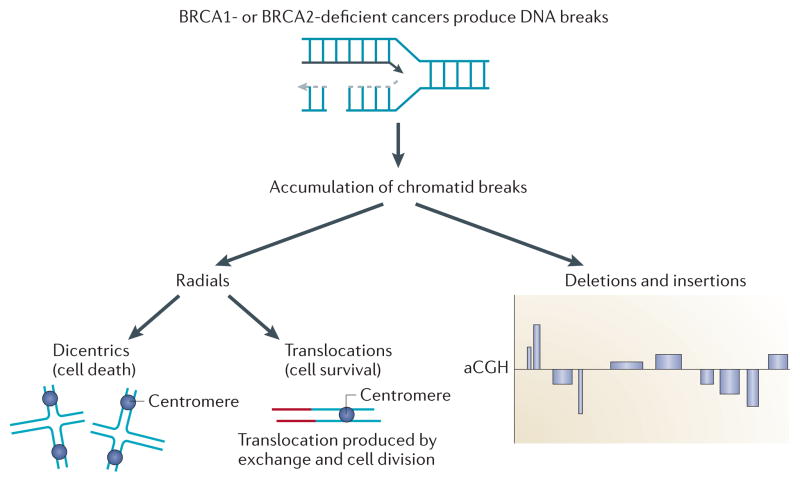
Figure 4. BRCA-deficient cells accumulate chromatid breaks and chromatid exchanges.
In the absence of BRCA1 or BRCA2 function, chromatid breaks accumulate, resulting in aberrant chromatid exchanges or other processes involving illegitimate end-joining. If two chromatid breaks are joined to produce a chromosome structure containing two centromeres, a dicentric quadri-radial chromosome is formed, which leads to cell death at mitosis. If an exchange is made with a chromatid fragment without a centromere, processing and cell division can produce a viable cell with a translocation. All of the hallmarks of BRCA-deficient cancers can be explained by the production of chromatid breaks and illegitimate end-joining. Without exchange events between different chromosomes, interstitial deletions, terminal deletions and insertions of chromosome fragments can originate from the chromatid break. In the absence of homologous recombination, the resulting phenotypes can be seen either by spectral karyotyping or by array-comparative genomic hybridization (aCGH), which detects large losses and gains across the genome.
Conclusions and perspectives
BRCA1 and BRCA2 function in the DDR during S and G2 phase by mediating HR to maintain replication fidelity. The loss of BRCA1 or BRCA2 function in normal cells results in growth defects, which are required, in combination with the subsequent loss of other DDR mediators, for tumour development. The genetic instability resulting from these growth defects and the loss of DDR mediators leads to multiple genetic gains and losses, but understanding which are the crucial secondary targets and which are nonspecific changes is one of the key research challenges in understanding the aetiology of cancers associated with BRCA loss. The observations that defects in HR can be acquired rather than inherited support the view that genetic instability provides a selective advantage to breast cancer cells. Both mechanisms result in genetic instability, which is perhaps triggered by oxidative metabolism producing replication stress. Therefore, the number of BRCA1–BRCA2–HR pathway-defective breast cancers may be 4 to 5 times greater than originally thought, making the number of patients who are amenable to DNA repair targeting strategies much higher than previously estimated.
Acknowledgments
The authors are supported by grants from the US National Cancer Institute and the Susan G. Komen For The Cure foundation.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Foulkes WD. Molecular origins of cancer: inherited susceptibility to common cancers. N Engl J Med. 2008;359:2143–2153. doi: 10.1056/NEJMra0802968. [DOI] [PubMed] [Google Scholar]
- 2.Schlacher K, et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Futreal PA, et al. A census of human cancer genes. Nature Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rahman N, Stratton MR. The genetics of breast cancer susceptibility. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- 5.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nature Rev Mol Cell Biol. 2010;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng CX, Brodie SG. Roles of BRCA1 and its interacting proteins. Bioessays. 2000;22:728–737. doi: 10.1002/1521-1878(200008)22:8<728::AID-BIES6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 7.Friedman LS, et al. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nature Genet. 1994;8:399–404. doi: 10.1038/ng1294-399. [DOI] [PubMed] [Google Scholar]
- 8.Couch FJ, Weber BL. Mutations and polymorphisms in the familial early-onset breast cancer (BRCA1) gene. Breast Cancer Information Core. Hum Mutat. 1996;8:8–18. doi: 10.1002/humu.1380080102. [DOI] [PubMed] [Google Scholar]
- 9.Shattuck-Eidens D, et al. A collaborative survey of 80 mutations in the BRCA1 breast and ovarian cancer susceptibility gene. Implications for presymptomatic testing and screening. JAMA. 1995;273:535–541. [PubMed] [Google Scholar]
- 10.Wu LC, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nature Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 11.Sato K, et al. Nucleophosmin/B23 is a candidate substrate for the BRCA1-BARD1 ubiquitin ligase. J Biol Chem. 2004;279:30919–30922. doi: 10.1074/jbc.C400169200. [DOI] [PubMed] [Google Scholar]
- 12.Nishikawa H, et al. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J Biol Chem. 2004;279:3916–3924. doi: 10.1074/jbc.M308540200. [DOI] [PubMed] [Google Scholar]
- 13.Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair. 2009;8:1009–1017. doi: 10.1016/j.dnarep.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci USA. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009;7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 21.Kim H, Huang J, Chen J. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nature Struct Mol Biol. 2007;14:710–715. doi: 10.1038/nsmb1277. [DOI] [PubMed] [Google Scholar]
- 22.Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 23.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 24.Kim SS, et al. Uterus hyperplasia and increased carcinogen-induced tumorigenesis in mice carrying a targeted mutation of the Chk2 phosphorylation site in Brca1. Mol Cell Biol. 2004;24:9498–9507. doi: 10.1128/MCB.24.21.9498-9507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang S, Biswas K, Martin BK, Stauffer S, Sharan SK. Expression of human BRCA1 variants in mouse ES cells allows functional analysis of BRCA1 mutations. J Clin Invest. 2009;119:3160–3171. doi: 10.1172/JCI39836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong Q, Boyer TG, Chen PL, Lee WH. Deficient nonhomologous end-joining activity in cell-free extracts from Brca1-null fibroblasts. Cancer Res. 2002;62:3966–3970. [PubMed] [Google Scholar]
- 28.Zhong Q, Chen CF, Chen PL, Lee WH. BRCA1 facilitates microhomology-mediated end joining of DNA double strand breaks. J Biol Chem. 2002;277:28641–28647. doi: 10.1074/jbc.M200748200. [DOI] [PubMed] [Google Scholar]
- 29.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 30.Snouwaert JN, et al. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a brca1 transgene. Oncogene. 1999;18:7900–7907. doi: 10.1038/sj.onc.1203334. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, et al. Nonhomologous end-joining of ionizing radiation-induced DNA double-stranded breaks in human tumor cells deficient in BRCA1 or BRCA2. Cancer Res. 2001;61:270–277. [PubMed] [Google Scholar]
- 32.Fu YP, et al. Breast cancer risk associated with genotypic polymorphism of the nonhomologous end-joining genes: a multigenic study on cancer susceptibility. Cancer Res. 2003;63:2440–2446. [PubMed] [Google Scholar]
- 33.Paull TT, Cortez D, Bowers B, Elledge SJ, Gellert M. Direct DNA binding by Brca1. Proc Natl Acad Sci USA. 2001;98:6086–6091. doi: 10.1073/pnas.111125998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei L, et al. Rapid recruitment of BRCA1 to DNA double-strand breaks is dependent on its association with Ku80. Mol Cell Biol. 2008;28:7380–7393. doi: 10.1128/MCB.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siliciano JD, et al. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabbro M, et al. BRCA1-BARD1 complexes are required for p53Ser-15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage. J Biol Chem. 2004;279:31251–31258. doi: 10.1074/jbc.M405372200. [DOI] [PubMed] [Google Scholar]
- 39.Xu B, Kim S, Kastan MB. Involvement of Brca1 in S-phase and G2-phase checkpoints after ionizing irradiation. Mol Cell Biol. 2001;21:3445–3450. doi: 10.1128/MCB.21.10.3445-3450.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenberg RA, et al. Multifactorial contributions to an acute DNA damage response by BRCA1/ BARD1-containing complexes. Genes Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 42.Yang H, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 43.Li J, et al. DSS1 is required for the stability of BRCA2. Oncogene. 2006;25:1186–1194. doi: 10.1038/sj.onc.1209153. [DOI] [PubMed] [Google Scholar]
- 44.Kojic M, Yang H, Kostrub CF, Pavletich NP, Holloman WK. The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2003;12:1043–1049. doi: 10.1016/s1097-2765(03)00367-8. [DOI] [PubMed] [Google Scholar]
- 45.Kristensen CN, Bystol KM, Li B, Serrano L, Brenneman MA. Depletion of DSS1 protein disables homologous recombinational repair in human cells. Mutat Res. 2010;694:60–64. doi: 10.1016/j.mrfmmm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA–ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 47.Powell SN, Willers H, Xia F. BRCA2 keeps Rad51 in line. High-fidelity homologous recombination prevents breast and ovarian cancer? Mol Cell. 2002;10:1262–1263. doi: 10.1016/s1097-2765(02)00789-x. [DOI] [PubMed] [Google Scholar]
- 48.Thorslund T, et al. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nature Struct Mol Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venkitaraman AR. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu Rev Pathol. 2009;4:461–487. doi: 10.1146/annurev.pathol.3.121806.151422. [DOI] [PubMed] [Google Scholar]
- 50.Pellegrini L, et al. Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 51.Wooster R, et al. Identification of the breast cancer susceptibility gene. BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 52.Carreira A, et al. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Esashi F, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 54.Ayoub N, et al. The carboxyl terminus of Brca2 links the disassembly of Rad51 complexes to mitotic entry. Curr Biol. 2009;19:1075–1085. doi: 10.1016/j.cub.2009.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saeki H, et al. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc Natl Acad Sci USA. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nature Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 57.Petalcorin MI, Sandall J, Wigley DB, Boulton SJ. CeBRC-2 stimulates D-loop formation by RAD-51 and promotes DNA single-strand annealing. J Mol Biol. 2006;361:231–242. doi: 10.1016/j.jmb.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 58.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nature Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan SS, et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complexin vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 61.Patel KJ, et al. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 62.Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–879. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- 63.Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 2006;25:5885–5897. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 64.Deng CX, Scott F. Role of the tumor suppressor gene Brca1 in genetic stability and mammary gland tumor formation. Oncogene. 2000;19:1059–1064. doi: 10.1038/sj.onc.1203269. [DOI] [PubMed] [Google Scholar]
- 65.Tsuzuki T, et al. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stefansson OA, et al. Genomic profiling of breast tumours in relation to BRCA abnormalities and phenotypes. Breast Cancer Res. 2009;11:R47. doi: 10.1186/bcr2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Willers H, et al. Utility of DNA repair protein foci for the detection of putative BRCA1 pathway defects in breast cancer biopsies. Mol Cancer Res. 2009;7:1304–1309. doi: 10.1158/1541-7786.MCR-09-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10:990–996. doi: 10.1038/embor.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 70.Nagaraju G, Scully R. Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair. 2007;6:1018–1031. doi: 10.1016/j.dnarep.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McPherson JP, et al. A role for Brca1 in chromosome end maintenance. Hum Mol Genet. 2006;15:831–838. doi: 10.1093/hmg/ddl002. [DOI] [PubMed] [Google Scholar]
- 72.Deng CX, Brodie SG. Knockout mouse models and mammary tumorigenesis. Semin Cancer Biol. 2001;11:387–394. doi: 10.1006/scbi.2001.0394. [DOI] [PubMed] [Google Scholar]
- 73.Alter BP, Kupfer G. Fanconi anemia. GeneReviews. 2011 [online], http://www.ncbi.nlm.nih.gov/books/NBK1401/
- 74.Feng Z, et al. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci USA. 2011;108:686–691. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hellebrand H, et al. Germline mutations in the PALB2 gene are population specific and occur with low frequencies in familial breast cancer. Hum Mutat. 2011;32:e2176–e2188. doi: 10.1002/humu.21478. [DOI] [PubMed] [Google Scholar]
- 76.Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nature Genet. 1996;12:191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 77.Cressman VL, et al. Mammary tumor formation in p53- and BRCA1-deficient mice. Cell Growth Differ. 1999;10:1–10. [PubMed] [Google Scholar]
- 78.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nature Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 79.King TA, et al. Heterogenic loss of the wild-type BRCA allele in human breast tumorigenesis. Ann Surg Oncol. 2007;14:2510–2518. doi: 10.1245/s10434-007-9372-1. [DOI] [PubMed] [Google Scholar]
- 80.Skoulidis F, et al. Germline Brca2 heterozygosity promotes KrasG12D-driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell. 2010;18:499–509. doi: 10.1016/j.ccr.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 81.Thorlacius S, et al. A single BRCA2 mutation in male and female breast cancer families from Iceland with varied cancer phenotypes. Nature Genet. 1996;13:117–119. doi: 10.1038/ng0596-117. [DOI] [PubMed] [Google Scholar]
- 82.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nature Rev Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 83.Ramus SJ, et al. Increased frequency of TP53 mutations in BRCA1 and BRCA2 ovarian tumours. Genes Chromosomes Cancer. 1999;25:91–96. doi: 10.1002/(sici)1098-2264(199906)25:2<91::aid-gcc3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 84.Rowley M, et al. Inactivation of Brca2 promotes Trp53-associated but inhibits KrasG12D-dependent pancreatic cancer development in mice. Gastroenterology. 2011;140:1303–1313.e3. doi: 10.1053/j.gastro.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McAllister KA, et al. Spontaneous and irradiation-induced tumor susceptibility in BRCA2 germline mutant mice and cooperative effects with a p53 germline mutation. Toxicol Pathol. 2006;34:187–198. doi: 10.1080/01926230600611794. [DOI] [PubMed] [Google Scholar]
- 86.Liu X, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA. 2007;104:12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schuyer M, Berns EM. Is TP53 dysfunction required for BRCA1-associated carcinogenesis? Mol Cell Endocrinol. 1999;155:143–152. doi: 10.1016/s0303-7207(99)00117-3. [DOI] [PubMed] [Google Scholar]
- 88.Hakem R, de la Pompa JL, Elia A, Potter J, Mak TW. Partial rescue of Brca15–6 early embryonic lethality by p53 or p21 null mutation. Nature Genet. 1997;16:298–302. doi: 10.1038/ng0797-298. [DOI] [PubMed] [Google Scholar]
- 89.Bouwman P, et al. Loss of p53 partially rescues embryonic development of Palb2 knockout mice but does not foster haploinsufficiency of Palb2 in tumour suppression. J Pathol. 2011;224:10–21. doi: 10.1002/path.2861. [DOI] [PubMed] [Google Scholar]
- 90.Cheung AM, et al. Loss of Brca2 and p53 synergistically promotes genomic instability and deregulation of T-cell apoptosis. Cancer Res. 2002;62:6194–6204. [PubMed] [Google Scholar]
- 91.Crook T, et al. p53 mutation with frequent novel codons but not a mutator phenotype in BRCA1- and BRCA2-associated breast tumours. Oncogene. 1998;17:1681–1689. doi: 10.1038/sj.onc.1202106. [DOI] [PubMed] [Google Scholar]
- 92.Smith PD, et al. Novel p53 mutants selected in BRCA-associated tumours which dissociate transformation suppression from other wild- type p53 functions. Oncogene. 1999;18:2451–2459. doi: 10.1038/sj.onc.1202565. [DOI] [PubMed] [Google Scholar]
- 93.Cao L, et al. ATM-Chk2-p53 activation prevents tumorigenesis at an expense of organ homeostasis upon Brca1 deficiency. EMBO J. 2006;25:2167–2177. doi: 10.1038/sj.emboj.7601115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tommiska J, et al. The DNA damage signalling kinase ATM is aberrantly reduced or lost in BRCA1/BRCA2-deficient and ER/PR/ERBB2-triple-negative breast cancer. Oncogene. 2008;27:2501–2506. doi: 10.1038/sj.onc.1210885. [DOI] [PubMed] [Google Scholar]
- 95.Sullivan A, et al. Concomitant inactivation of p53 and Chk2 in breast cancer. Oncogene. 2002;21:1316–1324. doi: 10.1038/sj.onc.1205207. [DOI] [PubMed] [Google Scholar]
- 96.Fan S, et al. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science. 1999;284:1354–1356. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- 97.Somasundaram K, et al. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CiP1. Nature. 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 98.Zhu Q, et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sipe HJ, Jr, Jordan SJ, Hanna PM, Mason RP. The metabolism of 17β-estradiol by lactoperoxidase: a possible source of oxidative stress in breast cancer. Carcinogenesis. 1994;15:2637–2643. doi: 10.1093/carcin/15.11.2637. [DOI] [PubMed] [Google Scholar]
- 100.Malins DC, Holmes EH, Polissar NL, Gunselman SJ. The etiology of breast cancer. Characteristic alteration in hydroxyl radical-induced DNA base lesions during oncogenesis with potential for evaluating incidence risk. Cancer. 1993;71:3036–3043. doi: 10.1002/1097-0142(19930515)71:10<3036::aid-cncr2820711025>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 101.Lavigne JA, et al. The effects of catechol-O-methyltransferase inhibition on estrogen metabolite and oxidative DNA damage levels in estradiol-treated MCF-7 cells. Cancer Res. 2001;61:7488–7494. [PubMed] [Google Scholar]
- 102.Hamada J, et al. Increased oxidative DNA damage in mammary tumor cells by continuous epidermal growth factor stimulation. J Natl Cancer Inst. 2001;93:214–219. doi: 10.1093/jnci/93.3.214. [DOI] [PubMed] [Google Scholar]
- 103.Armes JE, et al. Distinct molecular pathogeneses of early-onset breast cancers in BRCA1 and BRCA2 mutation carriers: a population-based study. Cancer Res. 1999;59:2011–2017. [PubMed] [Google Scholar]
- 104.Comen E, et al. Relative contributions of BRCA1 and BRCA2 mutations to “triple-negative” breast cancer in Ashkenazi Women. Breast Cancer Res Treat. 2011;129:185–190. doi: 10.1007/s10549-011-1433-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bane AL, et al. BRCA2 mutation-associated breast cancers exhibit a distinguishing phenotype based on morphology and molecular profiles from tissue microarrays. Am J Surg Pathol. 2007;31:121–128. doi: 10.1097/01.pas.0000213351.49767.0f. [DOI] [PubMed] [Google Scholar]
- 107.Holstege H, et al. BRCA1-mutated and basal-like breast cancers have similar aCGH profiles and a high incidence of protein truncating TP53 mutations. BMC Cancer. 2010;10:654. doi: 10.1186/1471-2407-10-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu Z, Wu J, Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nature Struct Mol Biol. 2007;14:716–720. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- 109.Sobhian B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cantor SB, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 111.Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of Brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 112.Xu B, O’Donnell AH, Kim ST, Kastan MB. Phosphorylation of serine 1387 in Brca1 is specifically required for the Atm-mediated S-phase checkpoint after ionizing irradiation. Cancer Res. 2002;62:4588–4591. [PubMed] [Google Scholar]
- 113.Krainer M, et al. Differential contributions of BRCA1 and BRCA2 to early-onset breast cancer. N Engl J Med. 1997;336:1416–1421. doi: 10.1056/NEJM199705153362003. [DOI] [PubMed] [Google Scholar]