Abstract
In general, a long-lasting immune response to viruses is achieved when they are infectious and replication-competent. In the mouse, the neutralizing antibody response to Friend murine leukemia virus is contributed by an allelic form of the enzyme Apobec3 (abbreviated A3). This is counterintuitive, because A3 directly controls viremia before the onset of adaptive anti-viral immune responses. It suggests that A3 also affects the antibody response directly. Here we studied the relative size of cell populations of the adaptive immune system as a function of A3 activity. We created a transgenic mouse that expresses all seven human A3 enzymes (hA3) and compared it to wild-type and mouse A3 (mA3)-deficient mice. A3 enzymes decreased the number of marginal zone (MZ) B cells, but not the number of follicular B or T cells. When mA3 was knocked out, the retroelement hitchhiker-1 and sialyl transferases encoded by genes close to it were overexpressed three and two orders of magnitude, respectively. We suggest that A3 shifts the balance, from the fast antibody response mediated by MZ B cells with little affinity maturation, to a more sustained germinal center B-cell response, which drives affinity maturation and, thereby, a better neutralizing response.
Keywords: Recovery from Friend virus 3 (Rfv3), sialyl transferases, hitchhiker-1, Apobec3, B cell
Introduction
Apobec3 (A3) enzymes restrict exogenous and endogenous retroelements by a variety of mechanisms [1], including hypermutating the first-strand cDNA of endogenous mouse leukemia virus (MLV) [2], aggregating retroelement mRNA [3–7], and decreasing the activity and fidelity of reverse transcriptase [8]. As a result, these enzymes prevent the spread of infection by exogenous retroviruses, and help maintain the integrity of the cellular genome and transcriptome by suppressing insertional mutagenesis of endogenous retroelements. In the mouse, an allele of A3 encodes the dominant resistance gene Recovery from Friend virus 3 (Rfv3) [9, 10], which also contributes to the neutralizing antibody response to Friend MLV (F-MLV). This function is at odds with the classical role of A3, wherein A3 directly controls viremia before the onset of an anti-viral antibody immune response. In general, long-lasting neutralizing antibody responses are achieved by infectious, replication-competent particles. To explain this paradox, the neutralizing antibody response of Rfv3 has been attributed to an indirect effect of A3. A3 may, for example, contribute to the hypermutation of antibody genes [11], cause retrovirus particles to be noninfectious and, thereby, help them evade a form of “high-zone tolerance,” or inhibit early F-MLV-induced injury of critical cell types, such as B and T cells [12].
More generally, A3 may also directly change the relative size of cell populations involved in the antibody response. For example, A3 might shift the balance from the fast immune response mediated by marginal zone (MZ) B cells with little affinity maturation [13], to a more sustained germinal center B cell response, characterized by hypermutation, which drives affinity maturation and, thereby, a better neutralizing response. In mice, MZ B cells reside mainly in the outer white pulp of the spleen, where they are ideally located to respond to blood-borne pathogens. The cells have an “activated” phenotype, are long-lived, and are considered to belong to a distinct naïve B-cell lineage, separate from that of B-1 cells and mature follicular (FO) B cells [14]. Together with B-1 B cells, MZ B cells are thought to be the bridge between innate and adaptive immune responses and a source of “natural antibodies,” i.e., the antibodies that make up some of the serum Ig and contain part of the immunological record of previous antigen exposures [15]. MZ B cells respond vigorously to blood-borne pathogens [16].
To study the effect of A3 on the adaptive immune system, we determined the relative size of cell populations as a function of graded A3 activity. The mouse has only one A3 family member (mA3), while humans have seven (hA3) and are therefore more effective in restricting retroelements. Human A3 proteins are effective inhibitors of MLV [17, 18]. One of these, human A3G, also sequesters autonomous and non-autonomous retroelement RNAs in cytoplasmic high-molecular-mass complexes away from the nuclear enzymes necessary for replication [3–7]. We created a transgenic mouse that expresses all seven hA3 enzymes, and compared it to mA3-sufficient and -deficient mice. We found a striking inverse correlation between the number of functional A3 genes and the number of MZ B cells. In mice with both mA3 and hA3 enzymes, few MZ B cells were left. Conversely, the lack of any (mouse or human) A3 protein expanded the MZ B cell population. A3 also controlled the steady-state mRNA level of an endogenous retroelement that, in the absence of A3, was overexpressed by 3 orders of magnitude. This retroelement, in turn, drives sialic transferases, which are important in MZ development and antibody response.
Results
Mice with all seven hA3 genes
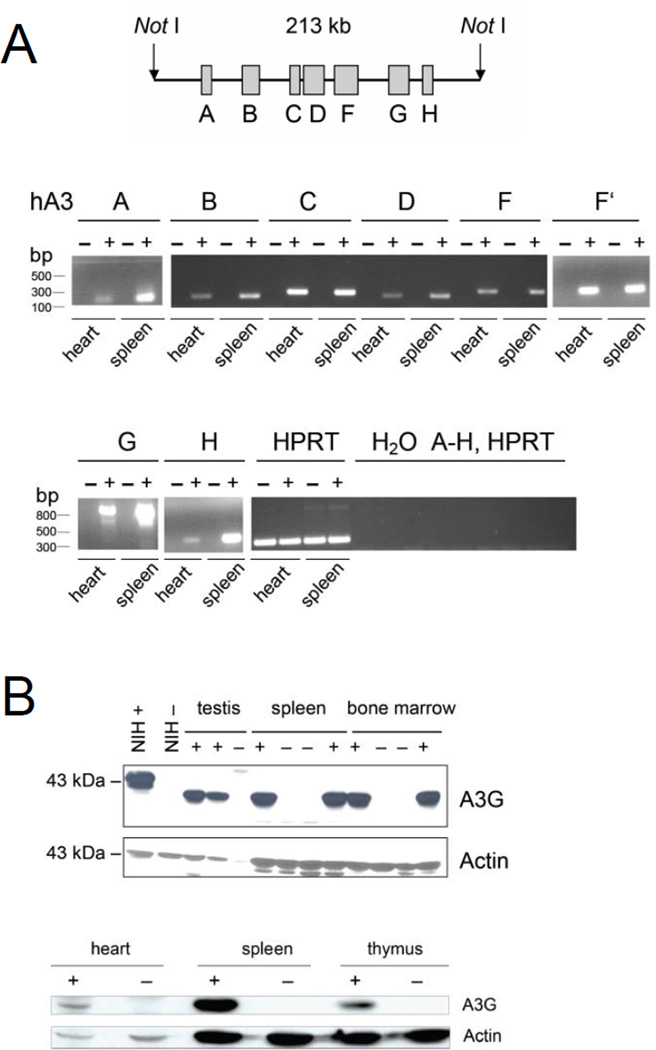
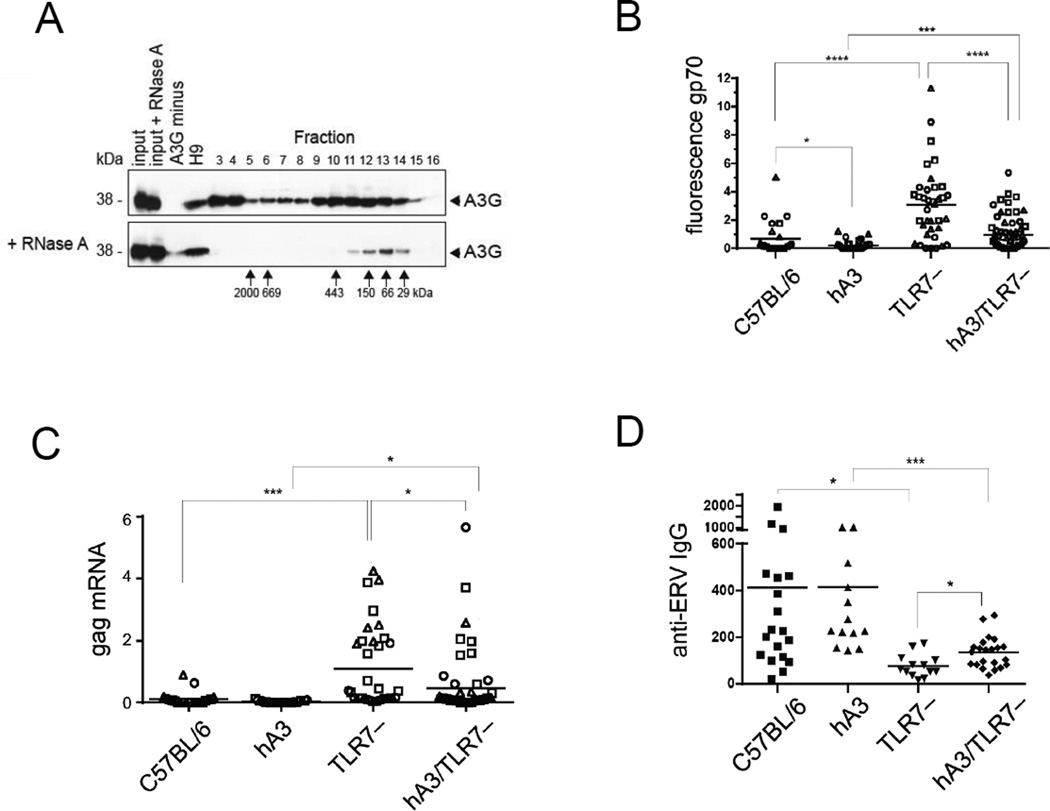
We generated mice that are transgenic for all seven human A3 (hA3) genes contained in a 213-kb Not1 fragment of a bacmid (Fig. 1A). In these mice, mRNAs encoding the seven A3 enzymes are expressed in heart and spleen, which are the (only) tissues we tested (Fig. 1A). In western blot analysis, we found that APOBEC3G protein (A3G) is synthesized in testis, bone marrow, thymus, spleen and heart (Fig. 1B). Because A3G in human cells forms high-molecular-mass complexes that restrict Alu and LINE-1 (non-autonomous and autonomous retrotransposons, respectively) by aggregating their mRNA, we tested this aggregation activity in cells of our hA3 mouse. Because in most normal tissues, retroelements are repressed, we used LPS to induce them in cultures of spleen cells. We then separated the cell lysates on a size-exclusion fast protein liquid chromatography (FPLC) column (Fig. 2A). As in human cells, hA3G in mouse cells formed large complexes that fell apart after RNase A treatment, indicating that complex formation is mediated by RNA (Fig. 2A). This showed that the transgenic A3G functioned physiologically in mouse cells.
Figure 1.
Expression of human APOBEC3 enzymes in the hA3 transgenic mouse. (A) A schematic of the 213-kb Not1 fragment containing the seven-member human A3 family is shown (top). Genes A3A through A3H are shown; A3E is a pseudogene. Real-time RT-PCR of cDNA from hearts and spleens of hA3 transgenic (+) and nontransgenic (−) littermates was performed (bottom). The agarose gel displays the amplicons from the genes indicated in the schematic. For A3F, two different primer pairs were used. Numbers to the left of the gels are size standards (bp). (B) Western blot analysis of A3G protein. Cell lysates of tissues from hA3 transgenic (+) and nontransgenic (−) tissues were electrophoresed, and the gel blots were developed with antibody to A3G. NIH+, NIH/3T3 cell line transfected with a tagged hA3 construct; NIH−, without construct. Actin was used as loading control. Numbers to the left of the gels are size standards (kDA). (A and B) Two founder mice were selected and analyzed in A and B.
Figure 2.
Inhibition of murine endogenous retroelements by hA3 transgenes. (A) The formation of A3G complexes in hA3 transgenic mice was evaluated by fractionation according to size by gel filtration of A3G, followed by western blot. Untreated cell lysate, devoid of nuclei (top) and lysates treated with RNase A before fractionation on FPLC (bottom) are shown. (Upon fractionation, the protease inhibitors are separated away from proteins, which are thus partially degraded and yield less intense bands; apparently RNA is protective.) Numbers above panels indicate fractions of separation by FPLC; numbers to the left and below panels indicate molecular mass standards of the SDS gel run and fractions of the FPLC run, respectively. Western blots of fractions of FPLC eluates were developed with polyclonal antibody to A3G. Input: untreated lysate of cells from hA3 transgenic mice; input + RNase A: RNase A-treated lysate of hA3 transgenic mice; A3G minus: lysed A3G-negative cells; H9: lysed cells of A3G-positive cell line H9; fractions 3–16: fractionated lysates of LPS- and IL-4-activated B lymphocytes from hA3 transgenic mice. One of three experiments shown. (B) Thymus, bone marrow, and spleen cells from C57BL/6 mice (average age 4.6 months), hA3 mice (6.1 months), TLR7−/− mice (denoted TLR7–; 4.6 months) and hA3TLR7−/− (5.0 months) were harvested to evaluate suppression of endogenous MLV gp70 expression by flow cytometry with anti-MLV-gp70 (83A25) antibody. Each symbol represents an individual cell population: Bone marrow, square; spleen, triangle; thymus, circle. Number of mice: C57BL/6: spleen and bone marrow, 9; thymus, 6; hA3: spleen, 24; bone marrow, 21; thymus, 24; TLR7−/−: spleen, 13; bone marrow, 11; thymus, 13; hA3TLR7−/−: spleen, 13; bone marrow, 8; thymus, 11) Student’s t test was applied after the specific predictions were made by preceding experiments: C57BL/6 wt vs. hA3, * p = 0.03; C57BL/6 vs. TLR7−/−, and TLR7−/− vs. hA3/TLR7−/−, **** p < 0.0001; hA3BL/6 vs. hA3TLR7−/−, *** p = 0.0007. (C) The suppression of MLV gag mRNA expression in tissues from C57BL/6, hA3, TLR7−/− and hA3TLR7−/− mice was measured by real-time qPCR. Values are normalized to tubulin. Numbers of C57BL/6 mice: spleen, 8; bone marrow, 8; thymus, 6. hA3BL/6 mice: spleen, 8; bone marrow, 8; thymus, 9. TLR7−/− mice: spleen, 10; bone marrow, 13; thymus, 11. hA3/TLR7−/− mice: spleen, 19; bone marrow, 20; thymus, 19. Student’s t test: hA3/TLR7−/− vs. TLR7−/−, * p = 0.01; hA3 vs. hA3/TLR7−/−, * p = 0.04; C57BL/6 vs. TLR7−/−, *** p = 0.001. (D) The levels of IgG serum antibody specific for endogenous retrovirus (anti-ERV) of MLV type in the presence or absence of TLR7 and/or hA3, was tested by ELISA specific for MLV (test antigen was a collection of isolated virus of different genotypes). Number of mice and average age, respectively: C57BL/6, n = 19, 4.5 months; hA3BL/6, n = 13, 4.9 months; TLR7−/−, n = 13, 4.6 months; hA3TLR7−/−, n = 22, 5.0 months). Student’s t test: TLR7−/− vs. hA3/TLR7−/−, * p = 0.01; C57BL/6 vs. TLR7−/−, * p = 0,02; hA3 vs. hA3/TLR7−/−, *** p = 0.0003. Data in (B–D) are from single experiments, which were repeated once (but not displayed).
Inhibition of endogenous retrovirus by hA3
A3G also efficiently inhibits MLV [17, 18]. Although not all mouse strains produce replication-competent ecotropic virus, all of them contain proviral genomes that can express viral proteins when transcribed. In the C57BL/6 strain, for example, the single infectious ecotropic provirus cannot replicate owing to a premature termination codon in its DNA polymerase [19]. However, it expresses envelope protein and can replicate upon recombination [20]. Using flow cytometry and real-time qRT-PCR, we measured the surface expression of retroviral envelope protein gp70 and MLV gag mRNA, respectively (Fig. 2 B and C). Cells from various tissues from C57BL/6 mice in the presence or absence of Toll-like receptor 7 (TLR7) [21] and/or hA3 were tested. In wild-type mice, gp70 is expressed at very low levels (Fig. 2B). This expression reflects proviral transcriptional and translational activities. It is low because retroviral replication is stifled by an immune response [20, 22]. However, hA3 further reduces the gp70 expression to essentially zero (Fig. 2B). The effect of hA3 is seen more clearly in the absence of TLR7. TLR7 is necessary for an effective immune response. Without this receptor, a weakened response allows recombinant virus to replicate and increase its proviral copy number in the mouse genome [20, 22]. As a result, both gp70 protein and MLV gag RNA expression in the cells are increased (Fig. 2 B and C). This increase is eliminated, however, when the TLR7 deficiency is complemented by the hA3 transgenes (Fig. 2 B and C). Therefore, we conclude that hA3 inhibits the replication of endogenous MLV. Curiously, rather than further decreasing titers of free anti-MLV antibody in mice lacking TLR7, hA3 slightly increased anti-MLV titers (Fig. 2D). This is consistent with the positive influence of mA3 on the F-MLV-specific neutralizing antibody response [9], which we discuss below.
Percentage of MZ B cells depends on A3 activity
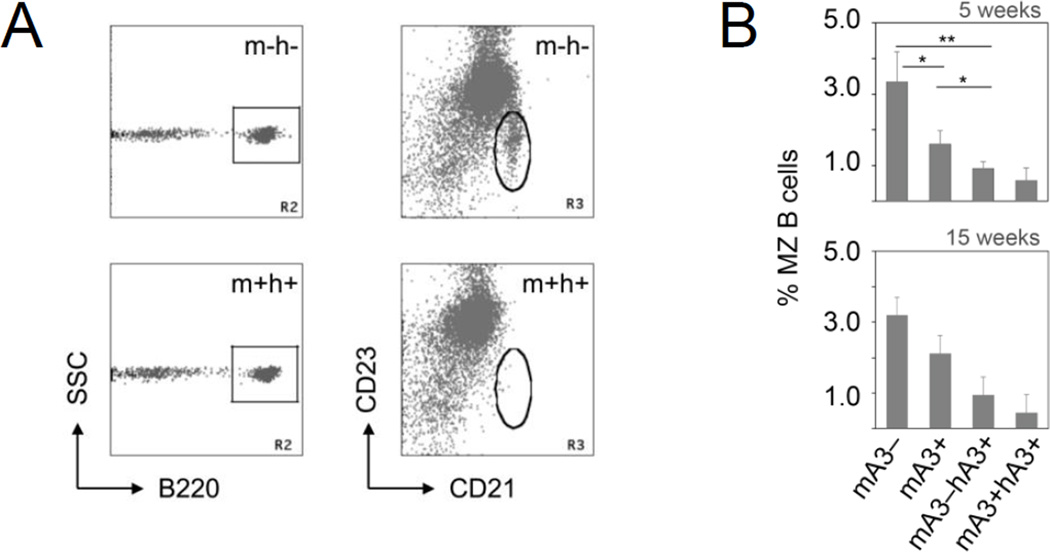
In the A3 mice of the various genotypes, there was no difference in the relative size of the T-cell compartment of the transgenic mice and their non-transgenic littermates. Nor was there any obvious difference between pro-, pre- and recirculating mature B cells in the bone marrow and recirculating B cells in the blood and spleen of the mice we compared. For total splenic B cells, the percentages were 26.7 ± 0.6 (± standard deviation) for mA3-deficient mice; 26.6 ± 1.3 for wild type; 22.6 ± 1.7 for mA3−/−hA3+ mice; and 24.9 ± 4.3 for mA3+/+hA3+ mice. However, we found a clear difference in the size of the MZ B-cell compartment (Fig. 3). We identified the MZ B cells by the surface markers B220, CD23lo [low] and CD21hi [high], and counted them in 5- and 15-week-old C57BL/6 mice that were mA3+/+ (wild-type) or had the hA3 transgenes (Fig. 3). In the transgenic hA3+ mice, the percentage of MZ B cells in the total B220-positive B-cell population was reduced to one-third that in the mA3+/+ mice. We found a similar reduction in hA3+ mice on a BALB/c background, and also with mice of mixed genetic backgrounds, as they were taken at various stages of backcrossing from the original hA3+ 129 strain to the C57BL/6 or BALB/c strain (not shown).
Figure 3.
Effect of APOBEC3 enzyme activity on the frequency of MZ B cells. (A) Flow cytometry plots of B cells from the spleen of a mA3-deficient C57BL/6 mouse (mA3−hA3−) and from a human APOBEC3 cluster (hA3) transgenic (mA3+hA3+) C57BL/6 mouse. Triple-stained spleen cells from littermates were gated for B220-positive cells (R2 gate, square; SSC, side scatter) and analyzed for CD21 (x-axis) and CD23 (y-axis) abundance. R3 gate (oval) enumerates the MZ B cells. (B) The percentage (y-axis) of B220+, CD21hi, and CD23lo MZ B cells in the spleens of mA3-deficient (mA3−), wild-type (mA3+), mA3-deficient/hA3 transgenic (mA3−hA3+) and hA3 transgenic wild-type mice (mA3+hA3+) are shown. Data for 5-week-old littermates (n = 3, 4, 4, and 3, respectively) were analyzed for the respective genotypes, resulting in the following p values (Student’s t test): mA3− vs. mA3+, * p = 0.012; mA3− vs. mA3−hA3+, ** p = 0.002; mA3− vs. mA3+hA3+, ** p = 0.006; mA3+ vs. mA3−hA3+, p = 0.015; mA3+ vs. mA3+hA3+, p = 0.013 (top). Data for 15-week-old littermates (n = 2, 7, 5, and 6, respectively) are also shown; because they are almost identical to the data with 5-week-old mice, the same statistics apply. Error bars, standard deviation. One of 10 experiments shown.
If the reduction in cell numbers in the hA3+ mice was due to a direct or indirect retroelement-inhibitory activity, and not due to an effect of the integration site of the transgene, then MZ cells ought to be increased in the mA3-deficient mice. At 5 weeks, A3-deficient mice had twice as many MZ B cells as did wild-type mice (Fig. 3 A and B), and at 15 weeks they had 50% more than wild-type mice (Fig. 3B). In mA3-deficient mice, as expected, the more efficient hA3 enzymes compensated for the deficiency and reduced the numbers of MZ B cells relative to those in the wild-type mice (Fig. 3B). In conclusion, there is a striking inverse correlation between the number of functional A3 genes and the number of MZ B cells.
Cell intrinsic vs. environmental effects of A3 on percentage of MZ B cells
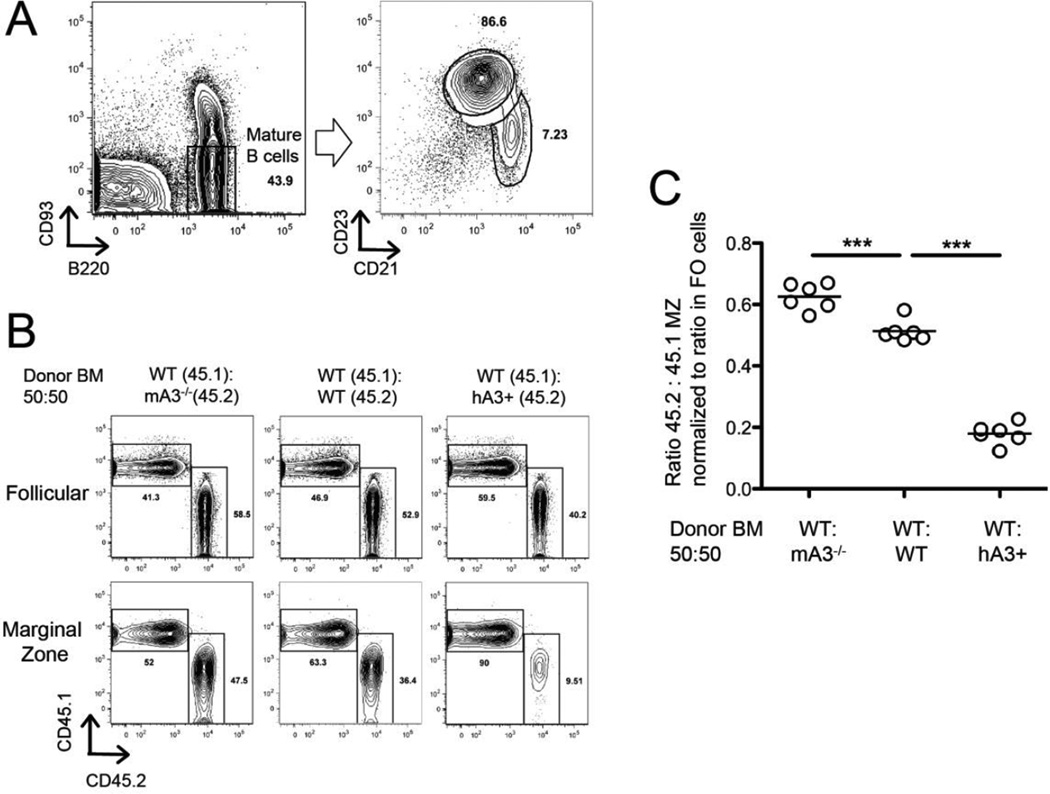
The pronounced effect of A3 expression on MZ B-cell numbers in the spleen could be intrinsic to MZ B cells or their precursors. Alternatively, the effect could be extrinsic; for example, increased numbers of (auto)antigens might interact with the antigen receptors on immature B cells to influence the choice of cell fate between MZ and FO B-cell types, or might act on MZ cells to promote their expansion or survival. To distinguish between these two broad models, we rescued lethally irradiated wild-type C57BL/6 mice with injection of equal mixtures of allotype-marked congenic bone marrow cells from mA3+hA3+CD45.1 and mA3−hA3−CD45.2 mice (Fig. 4). Because the FO B- cell population was normal in the mouse strains with different numbers of A3 genes, we used their relative numbers as the controls for reconstitution efficiency; as a second control, we used the numbers of allotype-marked T cells. If the effect of A3 expression on MZ numbers was cell-intrinsic, then CD45.2 cells should dominate in the MZ B-cell compartment, whereas FO B cells and T cells would be expressed at a value close to 50%. If, on the other hand, the effect was cell-extrinsic, then the two genotypes of MZ B cells should be equally represented but the total number of MZ B cells could be affected.
Figure 4.
Frequency of MZ B cells in bone marrow chimeras. Bone marrow cells of wt C57BL/6, mA3−/−, or mA3+hA3+ were mixed at a 50:50 ratio with allelically marked wt bone marrow cells. (A) Flow cytometry and gating strategy for MZ B cells, defined as B220+, CD93−, CD21hi, CD23lo; and for FO cells, B220+, CD93−, CD21lo, CD23hi. (B) Flow cytometry of MZ B cells, differentiated by the allelic markers CD45.1 (y-axis) and CD45.2 (x-axis). (C) Ratio of the allelically marked MZ cells (normalized to the ratio in the FO B cells) in three different kinds of chimeras (x-axis). *** p < 0.001 (Student’s t test). One set of experiments shown; experiments repeated once (not included here).
In these experiments, we identified MZ B cells as CD93−, CD23lo, or CD21hi (Fig. 4A) and differentiated them as CD45.1+ or CD45.2+ (Fig. 4B). We found that the cells lacking A3 enzymes dominated in the MZ B-cell compartment, and cells expressing both mA3 and hA3 were in the minority (Fig. 4C). Because their environment is presumed to be the same in each individual mouse, but the cells with the different A3 genotypes respond differently, we conclude that the effect of A3 activity is intrinsic to the MZ cells.
Inhibition of hitchhiker-1 mRNA by A3
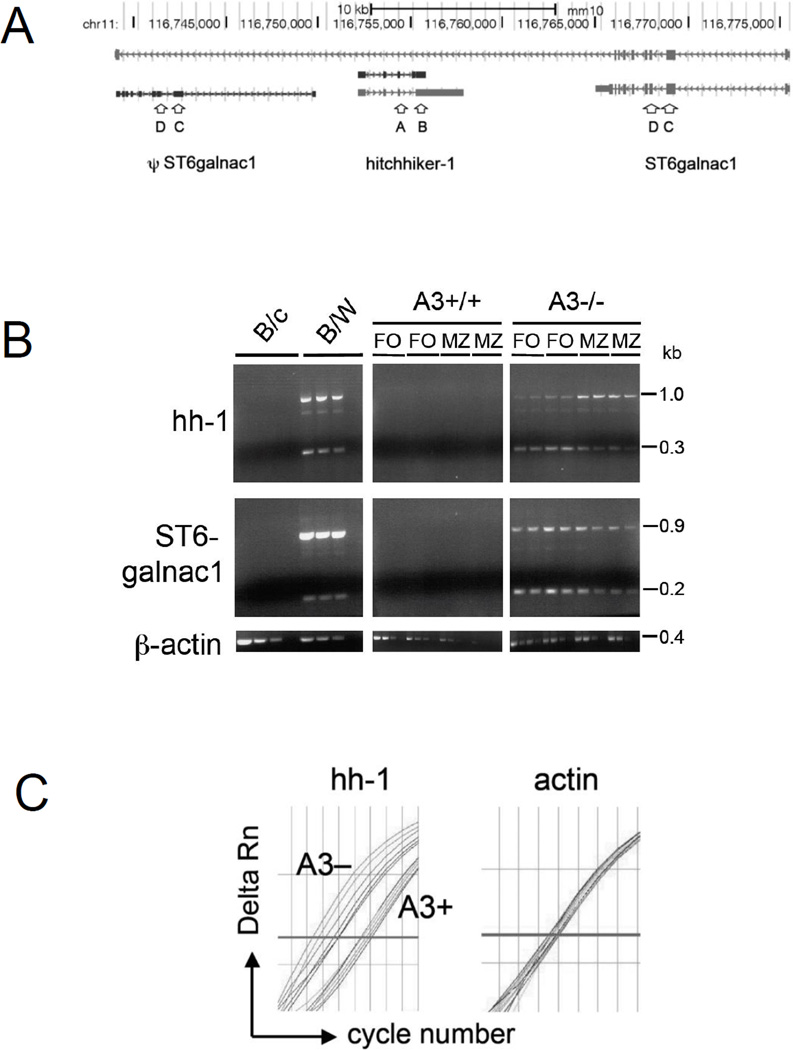
It was surprising that, in mice that contain both mA3 and hA3 enzymes, there were few MZ B cells left, as if the development, or expansion, of MZ B cells depended on retroelements. To gain detailed insight into the changes that resulted from varying the number of A3 enzymes, we utilized a deep sequencing approach to compare the transcriptomes of FACS-sorted MZ B cells from mA3-deficient, mA3-sufficient, and hA3 transgenic mice. In pairwise comparisons, we mainly considered normalized count values of at least 2 counts/million (cpm). For example, although BAFF-R was well expressed, with 234.2, 252.2, and 269.1 cpm in the MZ B cells from k.o., wt, and hA3+ mice, respectively, there was little difference between the genotypes. However, the MLV-like retroelement hitchhiker-1 (hh-1; Fig. 5A) was among the 10 most affected loci, and in the mA3- mice, this locus was the most highly expressed (highest cpm) of the 10 loci. In the absence of A3, hh-1 was overexpressed 3,000-fold relative to wild-type MZ B cells, and cells with both mA3 and hA3 expressed no hh-1 mRNA (Table 1).
Figure 5.
Expression of hitchhiker-1 and ST6galnac1. (A) Depiction of the locus containing ST6galnac1 (transcribed from right to left) and hitchhiker-1 (from left to right), adapted from UCSC Genome Browser, mm10 assembly of the C57BL/6 mouse genome. ψST6galnac1 is a pseudogene that is not spliced. (B) Semi-quantitative PCR of hitchhiker and ST6galnac1; cDNA from splenic B cells of BALB/c and B/W mice, or from FO B cells and MZ B cells of A3+/+ (wild-type) and A3−/− C57BL/6 littermates (two mice each). cDNAs encoding hh-1 and ST6galnac1 from A3+/+ and A3−/− mice were used undiluted or diluted 1:2; cDNAs from BALB/c and B/W mice were undiluted or diluted 1:2 and 1:4. β-actin cDNA from BALB/c and B/W mice was diluted 1:100, 1:1,000, 1:10,000, and 1:100,000, and β-actin cDNA from A3−/− and wild-type mice was undiluted or diluted 1:10, 1:100, and 1:1,000. One experiment representative of two experiments is shown (C) qPCR amplification plot of hh-1 (left) and β-actin (right) DNA from FO cells of three A3-deficient (A3−) and three age-matched wt C57BL/6 (A3+) mice; duplicate measurements for each mouse, i.e., 6 curves per genotype (A3− left, A3+ right). y-axis, Delta Rn, normalized reporter fluorescence signal minus baseline; x-axis, cycle number; bold line, threshold. One experiment representative of six experiments is shown.
Table 1.
The 10 transcripts from MZ B cells that are most affected by A3
| Gene | Transcript | A3−/− cpm |
A3+/+ cpm |
hA3+ cpm |
hA3+ vs A3−/− |
hA3+ vs A3+/+ |
A3−/− vs A3+/+ |
||
|---|---|---|---|---|---|---|---|---|---|
| CCNB1IP1 | 10.10562447 | 0 | 0 | 0 | NA | Inf | |||
| COL6A4 | 11.45304106 | 0 | 0.018839996 | 0.001644978 | Inf | Inf | |||
| CPNE4 | AK016705,AK014396 | 2.566507801 | 0 | 0 | 0 | NA | Inf | ||
| CYGB + GM11744 | 12.9608644 | 0 | 0 | 0 | NA | Inf | |||
| hitchhiker-1 | AF118847,NR_003364 | 131.3570774 | 0.044353687 | 0 | 0 | 0 | 2961.581922 | ||
| 4931429I11RIK | AK169524 | 130.8918979 | 0.06653053 | 0.018839996 | 0.000143936 | 0.2831782 | 1967.395977 | ||
| ST6GALNAC2 | 60.02420121 | 0.06653053 | 0.169559963 | 0.00282486 | 2.548603798 | 902.2053609 | |||
| ST6GALNAC1 | BC145374,BC145373,AK155119 | 22.34465855 | 0.110884217 | 0 | 0 | 0 | 201.5134261 | ||
| A630020A06 | NR_045740 | 4.539510674 | 0.044353687 | 0.320279929 | 0.070553844 | 7.221044094 | 102.3479892 | ||
| NFIB | AK148573,AK087480,AK034261 | 4.475347979 | 0.243945278 | 0.320279929 | 0.071565369 | 1.312917108 | 18.34570446 | ||
Genes and other transcripts with their sequence cpm from A3−/−, A3+/+ (wild type), and hA3+ mice. The last three columns compare mice with different genotypes. Genes of the hh-1–ST6galnac locus are highlighted in blue.
Using semi-quantitative RT-PCR to amplify transcripts from splenic B cells, we confirmed the results from deep sequencing for hh-1. With primers A and B (Fig. 5B, hybridizing with exons 3 and 4, respectively), an unspliced transcript (a 1-kb band in Fig. 3B) [23] and a spliced transcript (0.3 kb) of hh-1 were amplified in A3−/− spleen cells (on C57BL/6 background) (Fig. 5B), but not in A3+/+ spleen cells from C57BL/6 littermates and BALB/c mice. For the C57BL/6 mice, we differentiated MZ and FO cells by sorting them before isolating RNA and found that, in the A3-deficient mice, MZ cells had higher steady-state levels of unspliced hh-1 RNA than did FO cells. Unlike in the FO cells, the ratio of unspliced to spliced RNA was greater than one (Fig. 5B). We confirmed by qPCR that indeed there was 2.2 times more unspliced hh-1 message in MZ than in FO cells (triplicate measurements of pools of FO and MZ cells from 4 mice each, with and without A3). In A3-deficient mice, the average ddCt for hh-1 (β-actin as a control) was 1.13 ± 0.28 (standard deviation, calculated with error propagation) for FO and MZ cells. There was also 250 times more unspliced hh-1 mRNA in the A3-deficient than in the –sufficient MZ cells. Because almost no hh-1 mRNA was produced in A3-sufficient littermates, 35 amplification cycles were needed to reach the threshold, which was at the limit of reliable measurements. But with a ddCt = 0.01 ± 0.78, there was apparently no difference between MZ and FO cells.
The hh-1 locus is amplified in NZB and NZW mice and contains a mouse mammary tumor virus-like retroelement, MTV-3. As a consequence, these mice (over)express the locus even in the presence of A3 [23]. In our semi-quantitative RT-PCR experiments, we thus included RNA from spleen cells of (NZBxNZW) F1 mice (abbreviated B/W mice), which also resulted in the 1-kb band from the unspliced hh-1 transcript and the 0.3-kb band from the spliced transcript (Fig. 5B). The faint band at 0.7 kb may result from the alternative exon 3a of hh-1, with a weak splice site. The 0.7-kb band is also present, albeit still weaker, in A3-deficient cells from C57BL/6 mice (Fig. 5B).
High mRNA abundance of genes linked to hitchhiker-1 in A3-deficient B cells
The 3’ end of hh-1 is located 7 kb from the 3’ end of the short form of the gene ST6galnac1 (in the opposite orientation) and embedded in its long form (Fig. 5A). Upstream of hh-1, and also embedded in the long form of ST6galnac1, there is a pseudogene (transcript ENSMUST00000106377) that is similar to ST6galnac1 but does not encode a protein. In A3- mice, the ST6galnac1 transcripts were increased 200-fold, and the nearby (approximately 50 kb, promoter to promoter) ST6galnac2 was induced 900-fold (Table 1). Mxra7, which is located approximately 76 kb downstream of hh-1 (promoter to promoter), was induced from 0 cpm in MZ cells from both hA3 and mA3 mice to 0.3 cpm in A3-deficient mice. We propose that A3 inhibits hh-1, and that in the absence of A3 the high-level transcription of hh-1 in turn increases the transcription of ST6galnac1, ST6galnac2 and Mxra7.
In semi-quantitative RT-PCR, using primers C and D (binding to exons 2 and 3 of ST6galnac1, respectively; Fig. 5A), we confirmed our RNAseq findings (Table 1) for ST6galnac1. These primers bind to both ST6galnac1 and the similar pseudogene. As a result, the strong 0.9-kb band from B/W mice in Figure 5B presumably originated from the unspliced transcript of the pseudogene, and the weaker 0.2-kb band originated from the protein-encoding gene [23]. The same-size bands were present in cDNA from FO and MZ cells of A3−/− mice (Fig. 5B) but not in those of A3+/+ C57BL/6 or BALB/c mice (Fig 5B). However, in the A3−/− mice, the bands were of nearly equal intensity, which, considering the size difference, indicates that there was more spliced than unspliced transcript.
Increase in copy number of hh-1 in A3-deficient mice
The increased steady-state level of hh-1 mRNA may be explained, in part, by an increased DNA copy number. Using primers that span part of an intron of hh-1 in qPCR, we measured the copy number in DNA of A3-deficient and –sufficient mice. The A3-deficient mice had over fivefold more hh-1 copies than A3-sufficient mice (Fig. 5C; duplicate qPCR measurements of three mice of each genotype). While the number of cell cycles needed to reach the preset threshold varied only by 0.7 cycles between A3-sufficient mice, it varied by 1.5 cycles between A3-deficient mice (Fig. 5C). In comparison, there was minimal variation for the β-actin gene in mice of both genotypes (Fig. 5C). Apparently, A3 inhibits the retrotransposition of hh-1, either in the germ line, or in somatic tissue, or both.
Discussion
We have shown that, compared to wild-type mice, the number of MZ B cells is greatly increased in the absence of A3 and reduced by the presence of hA3 genes. We also found that in the absence of A3, a region at the distal end of chromosome 11 is overexpressed by up to three orders of magnitude. In the C57BL/6 mouse, this locus consists of ST6galnac2 and the retroelement hitchhiker-1 (transcript BC018473), embedded in the long nuclear transcript of ST6galnac1, in the opposite transcriptional orientation (Fig. 5A) [23]. hh-1 got its name from the fact that, in the genome of some mouse strains other than C57BL/6, hh-1 “hitches a ride” with the mouse mammary tumor provirus MTV-3 by extending its RNA to proceed at least 10 kb into the juxtaposed genomic territory [24]. Downstream of the Mtv-3-hh-1 locus, there is a second copy of hh-1, but without Mtv-3 attached. C57BL/6 mice lack Mtv-3-hh-1 but have hh-1. If we assume that A3 acts on hh-1 rather than on ST6galnac1, ST6galnac2, and Mxra7, then it is the transcription of those three genes that is helped by the hh-1 transcript rather than the converse. But how does A3 decrease the steady-state level of hh-1 mRNA 3,000-fold? Does A3 prevent replication and, thereby, an increase in copy number, or does it prevent transcription of hh-1? Over stretches of 56–187 amino acids, the hh-1 sequence is 31–45% similar to the gag and pol genes of MLV. However, owing to premature termination codons and frame shifts, no functional protein is synthesized [23]; thus, replication competence has to be acquired via enzymes borrowed from other retroelements.
The approximately fivefold increase in DNA copy number of the hh-1 retroelement in A3-deficient mice likely contributes to its overexpression in A3-deficient B cells. This would not, however, explain the increase in steady-state levels of the three genes proximate to the germ line hh-1 locus. Unless the whole ST6galnac2– ψST6galnac1–hh-1–ST6galnac1 locus is amplified in the absence of A3, the 3,000-fold increase in the steady-state level of (spliced plus unspliced) hh-1 mRNA is likely to be also due to increased transcription, RNA stability, or both. We conclude this from the converse effect of hA3, which decreases the wild-type level of hh-1 mRNA to zero. We propose that A3 inhibits transcription of hh-1, perhaps by binding to the nascent hh-1 transcript and thus inactivating the genomic locus in a manner similar to the mechanism of restriction of Alu and LINE-1 elements [3–7]. As intriguing as the mechanism of hh-1 restriction is the question of how hh-1 influences its linked genes ST6galnac1, ST6galnac2, and Mxra7. There is a large variety of mechanisms by which de novo inserted retroviruses affect transcription of nearby genes or genes in which they are inserted [25]; however, there is no simple mechanism for how increased mRNA of a germ line retroelement would increase the steady-state levels of different proximate genes.
With the hh-1–ST6galnac locus so highly overexpressed in the absence of A3, we wonder how the enzymes ST6galnac1 and ST6galnac2 can influence the generation and/or expansion of MZ B cells, but not that of FO cells. We cannot exclude the possibility that this overexpression is just a bystander effect. We presume that, of the thousands of retroelements that live within or near genes, there will be others similar to hh-1. For example, in the absence of A3, the abundance of transcript AK169524 is increased 2,000-fold (Table 1). Retroelement transcripts that are undefined or filtered out by the current databases may also have effects similar to that of hh-1. However, of the seven transcripts that increased from 0 to >10 cpm, three were ST6galnac2, ST6galnac1 and hh-1. Therefore, we believe that the enzyme activities of ST6galnac1 and ST6galnac2 help increase the percentage of MZ B cells. Although the function of these enzymes in the immune system is not known, the enzyme ST6Gal1, with similar activity, affects B-cell antigen receptor signaling [26] and, thereby, likely affects MZ B-cell development. ST6Gal1 produces the ligand Siaα2-6Galβ1-4GlcNAc (Sia6LacNAc) for the CD22 lectin molecule expressed on B lymphocytes, which itself also carries Sia6LacNAc. Both CD22-deficient mice and mice with a mutation in sialate:O-acetyl esterase, an enzyme that specifically removes acetyl moieties from the sialic acid, have a marked reduction in splenic MZ B cells [27, 28]. In the presence of A3, an underexpressed ST6GalNAc1 may play no role in the synthesis of ligands for CD22. However, in the absence of A3, or in mice with an allele encoding A3 with weaker activity, ST6GalNAc1 or ST6GalNAc2, or both, may produce more ligand and thereby increase the number of MZ B cells. Similarly, when the locus is overexpressed, as in B/W mice, there are higher than normal percentages of MZ B cells [29].
In our mice transgenic for the human A3 enzymes, endogenous MLV is restricted and APOBEC3G is aggregated, presumably by endogenous retrotransposon mRNA. In the absence of TLR7, the IgG response to (endogenous) MLV is (slightly) increased in the hA3 mice, which seems counterintuitive, considering the mechanism by which A3 restricts retrovirus. If A3 reduces the number of replicating viruses, one might expect a concomitant decrease in the immune response. The A3-mediated neutralizing antibody response to F-MLV was attributed to the release of noninfectious virus particles [9, 12] and to the mutational activity of A3 on antibody genes [11]. It is not immediately clear how noninfectious virus particles would improve an immune response. It was suggested that the lower early viral antigenic load would prevent a form of “high-zone tolerance,” or that the Friend virus would damage cell types critical for the humoral immune response, and that less virus would cause less damage and thus a better response [9]. In another proposal, A3 would have a role in shaping the antibody repertoire [9], for example, by hypermutating antibody genes, as recently demonstrated [11].
We propose two additional, not mutually exclusive, possibilities of how the restriction by A3 shapes the antibody repertoire to Friend virus. First, it suppresses MZ B cells, which mediate a fast immune response, but with low-affinity antibodies. Because an increase in MZ B cells would also decrease the antigen concentration available for FO cells, the low-affinity antibody response would come at the expense of a response in the germinal center with better neutralizing antibodies. Second, concomitant with an increase in the number of MZ B cells, the classical T-cell independent antibody response, mediated by the multivalent antigenic structure of virus, is inhibited due to the sialic acid transfer onto viral glycoproteins. Sialylated antigens are poorly immunogenic and can induce tolerance to subsequent challenge with immunogenic antigen [30]. By preventing hh-1 and its effect on the sialic transferases from being expressed, A3 would thus prevent tolerogenic viral antigens from being produced.
Materials and methods
Transgenic Mice
For the generation of the hA3 transgenic mice, the 213-kb Not1 fragment of the bacmid RP11-63H6 (E2026) containing all seven functional A3 enzymes was used. We verified the genomic structure, gel-purified the Not1 fragment, and injected it into zygotes of the FVB strain. We obtained 11 transgenic founders, which were backcrossed onto BALB/c and C57BL/6 mice. Experiments in this paper were with mice backcrossed to C57BL/6 for 19 generations. The A3-deficient mice [31] on C57BL/6 background were provided by Dr. M. Peterlin, UCSF.
Fast Protein Liquid Chromatography (FPLC)
Cells were lysed in FPLC lysis buffer (50 mM HEPES, pH 7.4, 125 mM NaCl, 0.2% NP-40, 5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, EDTA-free Complete Mini protease inhibitors); nuclei were removed and the remainder was passed over a Superose 6 HR 10/30 column (GE Healthcare) that separates molecules according to size using an AKTA purifier (GE Healthcare). FPLC running buffer: 50 mM HEPES, pH 7.4, 125 mM NaCl, 1 mM dithiothreitol, 10% glycerol. Protein fractions were collected and subjected to SDS-PAGE. In some cases, cell lysates were treated with 100 µg/ml RNase A (Roche) and RNase inhibitors (Promega) before fractionation.
Sorting MZ and FO B Cell Populations
B cells were isolated from the spleens and stained with B220-PE (BD Pharmingen), CD23-allophycocyanin (Southern Biotech), and CD21/CD35-FITC (BD Pharmingen). MZ B cells were sorted by gating on B220+, CD21hi, and CD23lo fluorescence intensity; and FO B cells were sorted by gating on B220+, CD21lo, and CD23hi.
RNAseq
MZ B cells were sorted from the spleens of mA3−/−, mA3+/+ and hA3+ mice, and three mice each were pooled per genotype. RNA was isolated and treated with DNase I on the mini prep column as described by the manufacturer (Qiagen). At least 300 ng of RNA was used for the RNAseq, performed by the UCSF Genomics Core Facility.
Mixed Bone Marrow Chimeras
Wild-type (wt) C57BL/6 recipient mice (8–10 weeks old) were lethally irradiated with two 3.3-Gy doses of total body radiation (separated by 3 h intervals). Bone marrow cells were flushed from the femur and tibia of wt C57BL/6, mA3−/−, and hA3-transgenic mice (all CD45.2) and each genotype was mixed at a 50:50 ratio with wt Boy/J (CD45.1) bone marrow cells. Cells were suspended in RPMI containing 2% FBS and penicillin-streptomycin at 2.5 × 107 cells/ml, and injected (5 × 106 total cells in 200 µl/mouse) retro-orbitally into recipient mice immediately following the second dose of radiation. Recipients were analyzed 8–12 weeks after bone marrow transplantation using CD45.1 and CD45.2 allelic markers to distinguish donor cells of each genotype by flow cytometry.
RT-PCR
For cDNA synthesis, B cells were isolated from the splenocytes of BALB/c, B/W and C57BL/6 mice using negative B cell isolation kits (StemCell Technologies). For A3−/− mice and A3+/+ littermates on a C57BL/6 background, the B cells were further stained and sorted for FO B cell and MZ B cell populations. Total RNA was isolated with the Qiagen mini prep kit (Qiagen) and treated with DNaseI (Invitrogen). The cDNA was synthesized using oligo(dT)20 and SuperScript III (Invitrogen).
Primers and conditions for Semi-Quantitative PCR. hh-1: forward 5’-CAA AGT GAC TCC TAG CCA CG-3’ (primer #12 in ref. [23]); reverse 5’-CTC CCT TCA TGG AAT CCC AAG-3’ (primer #13 in ref. [23]). ST6galnac1: forward 5’-GAA GTG CTC TAG ACG GTT CCA-3’ (primer #7 in [23]); reverse 5’-GAG TTC AAG TCT GAG CCT CG-3’ (primer #8 in [23]). β-actin: forward 5’-AGA CTT CGA GCA GGA GAT GG-3’; reverse 5’-CAC AGA GTA CTT GCG CTC AG-3’. PCR conditions: 3 min at 94°C; then 30 cycles: 1 min 94°C, 1 min at 55°C, 1 min at 72°C; after the cycles 10 min at 72°C, and thereafter at 4°C.
Primers and conditions for RT-qPCR of hh-1 mRNA: forward 5’-CTC CAA TAT GCA GAT GAC CTC-3’; reverse 5’-CTC CCT TCA TGG AAT CCC AAG-3’; FAM-labeled MGB probe: 5’-TGT GGC ATT CCG GTT GCA GAG T-3’. PCR conditions: 2 min at 50°C; 10 min 95°C; 40 cycles of 0.15 min at 95°C and 1 min at 60°C.
Primers and conditions for qPCR of hh-1 DNA: forward 5’-CAA AGT GAC TCC TAG CCA CG-3’; reverse 5’-GGA AAG AGA CTC GTT ACA CA-3’; FAM-labeled MGB probe: 5’-CAG CCT GGT GCT AAA CAA TAC C-3’. PCR conditions: 2 min at 50°C; 10 min 95°C; 40 cycles of 0.15 min at 95°C and 1 min at 60°C.
Acknowledgments
We thank Pieter de Jong for the BAC clone RP11-63H6 (E2026) containing the hA3 linkage group; Michael Bösl for injecting the transgene into zygotes and producing the transgenic mice; Matija Peterlin for the A3-deficient mice; Leonard Evans for hybridoma and antibody 83A25; Stefan Weinberger and Edith Roth for expert technical assistance; Philip Coffino, Martin Hoyt and Allen Henderson for assistance with the FPLC fractionation; Joshua Pollack and Andrea Barczak for carrying out the RNAseq; Cliff Wang for suggestions; and Rafael Wabl and Mary McKenney for editing the manuscript. Supported by grants from the NIH (R01AI041570, R21AI107101) and the Lupus Research Institute to MW; from the U.S.–Israel Binational Science Foundation to DE and MW; from the Fritz Thyssen foundation to PY; and from the IZKF Erlangen and the DFG (FOR832) to HMJ.
Footnotes
Conflict of Interest: The authors declare no financial or commercial conflict of interest.
References
- 1.Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langlois MA, Kemmerich K, Rada C, Neuberger MS. The AKV murine leukemia virus is restricted and hypermutated by mouse APOBEC3. J Virol. 2009;83:11550–11559. doi: 10.1128/JVI.01430-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, et al. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 5.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 7.Metzner M, Jäck H-M, Wabl M. LINE-1 retroelements complexed and inhibited by AID. PLoS ONE. 2012 doi: 10.1371/journal.pone.0049358. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boi S, Kolokithas A, Shepard J, Linwood R, Rosenke K, Van Dis E, Malik F, et al. Incorporation of mouse APOBEC3 into murine leukemia virus virions decreases the activity and fidelity of reverse transcriptase. J Virol. 2014;88:7659–7662. doi: 10.1128/JVI.00967-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santiago ML, Montano M, Benitez R, Messer RJ, Yonemoto W, Chesebro B, Hasenkrug KJ, et al. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science. 2008;321:1343–1346. doi: 10.1126/science.1161121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeda E, Tsuji-Kawahara S, Sakamoto M, Langlois MA, Neuberger MS, Rada C, Miyazawa M. Mouse APOBEC3 restricts friend leukemia virus infection and pathogenesis in vivo. J Virol. 2008;82:10998–11008. doi: 10.1128/JVI.01311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halemano K, Guo K, Heilman KJ, Barrett BS, Smith DS, Hasenkrug KJ, Santiago ML. Immunoglobulin somatic hypermutation by APOBEC3/Rfv3 during retroviral infection. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1403361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith DS, Guo K, Barrett BS, Heilman KJ, Evans LH, Hasenkrug KJ, Greene WC, et al. Noninfectious retrovirus particles drive the APOBEC3/Rfv3 dependent neutralizing antibody response. PLoS Pathog. 2011;7:e1002284. doi: 10.1371/journal.ppat.1002284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerutti A, Cols M, Puga I. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat Rev Immunol. 2013;13:118–132. doi: 10.1038/nri3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliver AM, Martin F, Gartland GL, Carter RH, Kearney JF. Marginal zone B cells exhibit unique activation, proliferative and immunoglobulin secretory responses. Eur J Immunol. 1997;27:2366–2374. doi: 10.1002/eji.1830270935. [DOI] [PubMed] [Google Scholar]
- 15.Pillai S, Cariappa A, Moran ST. Marginal zone B cells. Annu Rev Immunol. 2005;23:161–196. doi: 10.1146/annurev.immunol.23.021704.115728. [DOI] [PubMed] [Google Scholar]
- 16.Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat Rev Immunol. 2009;9:767–777. doi: 10.1038/nri2656. [DOI] [PubMed] [Google Scholar]
- 17.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 18.Doehle BP, Schafer A, Wiegand HL, Bogerd HP, Cullen BR. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J Virol. 2005;79:8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak CA. Viewpoint on Emv2, the onlhy endogenous ecotropic murine leukemia virus of C57BL/6 mice. Retrovirology. 2012;9:25. doi: 10.1186/1742-4690-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young GR, Eksmond U, Salcedo R, Alexopoulou L, Stoye JP, Kassiotis G. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491:774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 22.Yu P, Lubben W, Slomka H, Gebler J, Konert M, Cai C, Neubrandt L, et al. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity. 2012;37:867–879. doi: 10.1016/j.immuni.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 23.Fu G, Haywood ME, Morley BJ. Representational difference analysis in a lupus-prone mouse strain results in the identification of an unstable region of the genome on chromosome 11. Nucleic Acids Res. 2002;30:1394–1400. doi: 10.1093/nar/30.6.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stegalkina SS, Guerrero A, Walton KD, Liu X, Robinson GW, Hennighausen L. Transcription originating in the long terminal repeats of the endogenous mouse mammary tumor virus MTV-3 is activated in Stat5a-null mice and picks Up hitchhiking exons. J Virol. 1999;73:8669–8676. doi: 10.1128/jvi.73.10.8669-8676.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sokol M, Wabl M, Ruiz IR, Pedersen FS. Novel principles of gamma-retroviral insertional transcription activation in murine leukemia virus-induced end-stage tumors. Retrovirology. 2014;11:36. doi: 10.1186/1742-4690-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hennet T, Chui D, Paulson JC, Marth JD. Immune regulation by the ST6Gal sialyltransferase. Proc Natl Acad Sci U S A. 1998;95:4504–4509. doi: 10.1073/pnas.95.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samardzic T, Marinkovic D, Danzer CP, Gerlach J, Nitschke L, Wirth T. Reduction of marginal zone B cells in CD22-deficient mice. Eur J Immunol. 2002;32:561–567. doi: 10.1002/1521-4141(200202)32:2<561::AID-IMMU561>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 28.Cariappa A, Takematsu H, Liu H, Diaz S, Haider K, Boboila C, Kalloo G, et al. B cell antigen receptor signal strength and peripheral B cell development are regulated by a 9-O-acetyl sialic acid esterase. J Exp Med. 2009;206:125–138. doi: 10.1084/jem.20081399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuster H, Martin T, Marcellin L, Garaud JC, Pasquali JL, Korganow AS. Expansion of marginal zone B cells is not sufficient for the development of renal disease in NZBxNZW F1 mice. Lupus. 2002;11:277–286. doi: 10.1191/0961203302lu191oa. [DOI] [PubMed] [Google Scholar]
- 30.Duong BH, Tian H, Ota T, Completo G, Han S, Vela JL, Ota M, et al. Decoration of T-independent antigen with ligands for CD22 and Siglec-G can suppress immunity and induce B cell tolerance in vivo. J Exp Med. 2010;207:173–187. doi: 10.1084/jem.20091873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okeoma CM, Lovsin N, Peterlin BM, Ross SR. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature. 2007;445:927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]