Abstract
Whether the human tumor virus, Epstein–Barr Virus (EBV), promotes breast cancer remains controversial and a potential mechanism has remained elusive. Here we show that EBV can infect primary mammary epithelial cells (MECs) that express the receptor CD21. EBV infection leads to the expansion of early MEC progenitor cells with a stem cell phenotype, activates MET signaling and enforces a differentiation block. When MECs were implanted as xenografts, EBV infection cooperated with activated Ras and accelerated the formation of breast cancer. Infection in EBV-related tumors was of a latency type II pattern, similar to nasopharyngeal carcinoma (NPC). A human gene expression signature for MECs infected with EBV, termed EBVness, was associated with high grade, estrogen-receptor-negative status, p53 mutation and poor survival. In 11/33 EBVness-positive tumors, EBV-DNA was detected by fluorescent in situ hybridization for the viral LMP1 and BXLF2 genes. In an analysis of the TCGA breast cancer data EBVness correlated with the presence of the APOBEC mutational signature. We conclude that a contribution of EBV to breast cancer etiology is plausible, through a mechanism in which EBV infection predisposes mammary epithelial cells to malignant transformation, but is no longer required once malignant transformation has occurred.
Highlights
-
•
Mammary epithelial cells, but not breast cancer cells, express CD21 and can be infected by EBV.
-
•
EBV infection of MECs rewires mitogenic signaling with activation of the MET pathway and gene expression changes (EBVness).
-
•
EBV infection of MECs lowers the threshold for malignant transformation in xenografts.
-
•
Human tumors with ‘EBVness’ are associated with poor prognosis and presence of remnant EBV DNA (but not RNA).
Research in context:
This study shows that Epstein–Barr Virus (EBV) infection occurs in breast epithelial cells (MECs), but not in breast cancer cells. EBV infection of MECs leads to changes in gene expression, activation of oncogenic signaling via c-MET and lowers the threshold for cancerous transformation. In human breast cancer, these gene expression changes, termed ‘EBVness’, are associated with high grade and the finding of remnant EBV DNA. EBV infection of breast epithelial cells may increase the odds of later breast cancer formation in predisposed individuals, which could potentially be avoided by a childhood vaccine against EBV.
1. Introduction
The human tumor virus, Epstein–Barr Virus (EBV), is causally associated with approximately 200,000 malignancies worldwide annually. The risk of cancer linked to EBV infection recently prompted a joint call by the NCI and the NIAID for development of an EBV vaccine (Cohen et al., 2011). EBV is typically transmitted early in life as a subclinical illness. When delayed until early adulthood symptoms of infectious mononucleosis often occur (Evans, 1971). Globally, > 90% of the adult population has been infected (Evans, 1971, Cohen, 2000). In general, most humans tolerate latent EBV infection without adverse effects. However, in certain individuals, EBV has been linked to the etiology of cancers, including African Burkitt lymphoma (Epstein et al., 1964, Epstein and Barr, 1964); Hodgkin's disease, nasopharyngeal carcinoma (NPC); gastric adenocarcinoma; and leiomyosarcoma (Niedobitek et al., 2001). Epithelial cancers associated with EBV vary markedly in viral prevalence, from nearly 100% of NPCs to about 10% of gastric carcinomas (Gulley, 2001) and also differ in the patterns of viral genes expressed. As exposure to virus frequently precedes the manifestation of cancer by years, and only a minority of individuals exposed to EBV will develop a related cancer, it is difficult to establish a causal role.
An association of EBV infection with breast cancer has been reported from India (Joshi et al., 2009), China (Peng et al., 2014, He et al., 2012), Northern Africa (Fina et al., 2001, Hachana et al., 2011b) and southern Europe (Marrao et al., 2014, Mazouni et al., 2011, Labrecque et al., 1995). However, no mechanism of infection has been identified. Two recent analyses of the RNAseq data in the TCGA dataset did not show evidence for actively transcribed virus (Khoury et al., 2013a, Tang et al., 2013). These results argued against a role of active EBV infection for the growth of established breast cancers, but did not rule out a mechanism of oncogenesis where viral infection contributes to transformation but is no longer required once a tumor has established itself. In this report we show that EBV can infect primary human mammary epithelial cells (MECs) through CD21 leading to phenotypic changes consistent with transformation. These immortalized MECs infected with EBV cooperatively (with activated Ras) increase tumor formation in vivo, recapitulating a multistep tumorigenesis in an established animal model. Significantly, when a transcriptional profile based on cellular gene expression in EBV-positive xenograft tumors was used to interrogate different human breast cancer databases, a subset of high grade breast tumors was identified in which EBV DNA, but not viral RNA, was detected by FISH.
2. Methods
2.1. Cell lines
Immortalized human mammary epithelial cell lines (HMLE and HMEC) were provided by Robert Weinberg, MIT. HMLE was generated from normal human primary mammary epithelial that overexpress hTERT (Elenbaas et al., 2001). HMEChtert, in short H:MEC, had been generated from primary mammary epithelial cells (MECs) that were immortalized with human telomerase (hTERT) and SV40 Large T antigen (Zhao et al., 2003). MCF10A cells were from the ATCC. The EBV producing AKATA cells were a gift of Dr. Lindsey Hutt-Fletcher, Louisiana State University Health Sciences Center, and cultured as described (Kuhn-Hallek et al., 1995).
2.2. Cell culture, EBV generation, infection and generation of cell lines
MCF-10A mammary epithelial cells and HMEC were cultured as described (Debnath et al., 2003). The isolation of primary human mammary epithelial cells (PMECs) was done according to a protocol approved by the Institutional Review Board (IRB) at Beth Israel Deaconess Medical Center. PMECs were isolated from reduction mammoplasty specimens, and cultured in MEGM supplemented with 20 ng/ml EGF, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, 1% bovine serum albumin, and 2% calf serum, as described (Burga et al., 2009).
Recombinant EBV was isolated from AKATA cells that generate GFP-labeled EBV (GFP-EBV) defective of viral thymidine kinase (TK). Reactivation of virus from AKATA cells was induced by adding Goat anti-human immunoglobulin G at a final concentration of 50 μg/ml to the culture medium, as described (Huang et al., 2003). GFP-control cells were generated using retroviral transduction (pBABE-GFP), followed by Neomycin selection at 100 μg/ml initially and 50 μg/ml for maintenance. To generate EBV-infected MECs, MECs were incubated with GFP-EBV for 2 h, after which the cells were washed. At 72 h post infection, selection with Neomycin was started. Care was taken to culture the GFP-control MECs exactly like the GFP-EBV cells.
For overexpression, vHRasV12D was subcloned into the pBabe retroviral vector (Addgene #17,756). Production of retroviruses and transduction of MECs was performed as described previously (Stewart et al., 2003). Following infection, the cells were selected using zeocin (vHRasV12D). Cells were used up to three weeks after selection. Fresh cell lines were made for each group of experiments and experiments were performed following at least two separate infections.
2.3. Flow cytometry
Anti-CD21-PE antibody (HB5, eBioscience) was used for CD21 detection. Anti-CD24-PE and anti CD44-APC antibodies (eBioscience) were used to detect progenitor cells. Anti-CD49f–APC (eBioscience) and anti-EpCAM-FITC (Biolegend) antibody were used for detection of mammary epithelial lineage. Isotype matched antibodies were used as negative controls. Flow cytometry was performed on a LSRII cytometer (BD Biosciences) and data were analyzed with FlowJo.
2.4. Competitive inhibition of EBV infection
HMECs were cultured in 6 well dishes to 40% confluency for the blocking experiments as described in (Fingeroth et al., 1984). Briefly, cell were pretreated with HB5 (mouse anti-CD21 antibody) or normal mIgG2a (40 μg/ml) for 30 min, followed by 3 PBS washes, and then goat F(ab’)2 fragments (50 μg/ml) to mouse IgG was added and incubated for another 30 min followed by another 3 washes with PBS, before EBV were added into the culture medium and incubated for another 4 h to infect the cells.
2.5. Mammosphere culture
Mammosphere cultures were performed as described (Dontu et al., 2003). Single-cell suspensions were plated in ultra-low attachment plates (Corning, Costar) in DMEM/F-12 HAM medium containing bFGF (20 ng/ml, EGF (20 ng/ml), heparin (4 μg/ml) and B-27 supplement (1:50 dilution, Invitrogen). The Mammospheres were cultured for two weeks. Mammospheres with diameter > 75 μm were counted.
2.6. 3D differentiation culture
10 day mammospheres were resuspended in Matrigel (Gibco) and transferred to culture slides, allowed to solidify and then covered with differentiation medium (MEGM medium (Clonetics) supplemented with 10 ng/ml FGF, 4 ng/ml heparin and 2 μg/ml prolactin) for additional 10–14 days, as described (Mani et al., 2008).
2.7. Tumorigenesis assays
Aliquots of indicated numbers of cells were injected subcutaneously into the mammary fat pad of 5-week-old NOD/SCID mice (Jackson Laboratories). Mice were examined at least twice weekly for evidence of tumor growth. After tumors were detected, tumor size was measured every two days. All studies involving mice were approved by the Institutional Animal Care and Use Committee at Beth Israel Deaconess Medical Center and performed in accordance with the protocol.
2.8. Immunoblotting
Primary monoclonal LMP1 antibody (1:500) (Abcam), Pan-H-Ras antibody (1:1000) (BD), Alpha-Tubulin antibody (1:10,000) (Sigma), and polyclonal CD21 antibody (1:500) (Abcam), MET-antibody (1:500) (Santa Cruz), p-METantibody (1:500) (Cell signaling), STAT3 (1:500) (Cell signaling), p-STAT3 (1:500) (Cell signaling) antibody were used.
2.9. Immunohistochemistry
Immunohistochemical staining for Ki67, p-Met, pSTAT3, K5/6, K18, ALDH1, p63, Vimentin w done as described previously (Burga et al., 2009, Burga et al., 2011). Immunolabeling was visualized with DAB solution (Vector Laboratories), followed by counterstaining with hematoxylin.
2.10. phospho-RTK scan
The p-RTK arrays were performed according to the manufacturer's recommendations (cell signaling Human Phospho-RTKArrayKit, Fluorescent Readout, #7949). The signal was developed using DyLight 680®-linked Streptavidin and then scanned with the Odyssey Infrared Imager (LI-COR). The intensity was read out by Genepix Pro 6.0 (Molecular Devices) and generated the heatmap by MeV4.7 (Saeed et al., 2006).
2.11. In situ hybridization
EBER in situ hybridization (Fig. 4C) was performed at the Dana Farber Cancer Center Specialized Histopathology Core Facility. Fish of LMP1 (type 1 probeset) and glycoprotein gH (BXFL2) (type 6 probeset) in patients' tumor tissue sections (Fig. 7) were performed following the instructions for the Affymetrix QuantiGene View RNA ISH Tissue Assay kit. The LMP1 probe covers bases 70–1115 of the LMP1 gene. The BXFL2 probe covers bases 450–1358 of the BXFL2 gene. As there are no introns in these viral genes, probes can target both EBV DNA and RNA. ISH stained tissue sections were scanned at 400 × resolution using a Zeiss LSM501 laser scanning microscope at the Ragon Institute and Tissue FAX software. They were scored for LMP1 and BFXL2 by three different physicians (XY, GP and GMW).
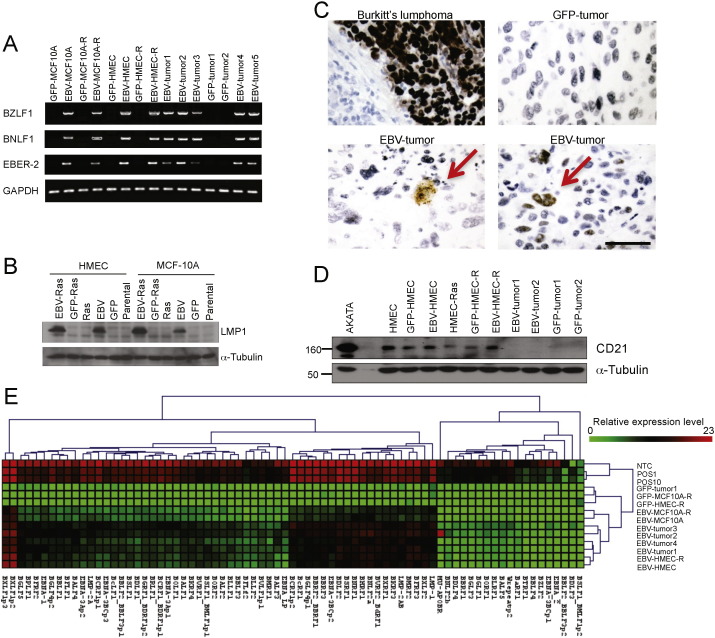
Fig. 4.
Evidence for EBV-infection of MECs and MEC-derived breast cancers. (A) PCR amplification of EBV genes BZLF1, BNLF1 and EBER-2 in DNA extracted from EBV-HMEC-R or GFP-HMEC-R and xenograft tumors as indicated. (B) Cell lysates from EBV-infected and control HMEC or MCF10A cells were subjected to immunoblotting with anti-LMP1 antibodies. (C) In situ hybridization of EBV-expressed RNAs (EBERs) in xenograft tumors derived from control (upper left) and EBV-infected HMECs (lower part of panel). A Burkitt's lymphoma (upper left panel) served as positive control. EBV-associated breast cancers derived from EBV-HMECs stained only infrequently positive for EBERS, with some EBERS positivity in tumor areas with cell death (left bottom). The red arrows indicate the cells with EBERS staining in the xenografts. (D) Immunoblotting of tumor lysates for CD21. (E) Heat map of viral gene expression. polyA + RNA was extracted from cell lines and xenograft tumors as indicated, and viral gene expression was measured by Q-PCR, normalized to 5 reference genes and displayied relative to the average level of 3 EBV negative samples (GFP-Tumor1, GFP-MCF-10A-R, GFP-HMEC-R). NTC, POS1 and POS10 were positive controls. Scale bars represent 100 μm.
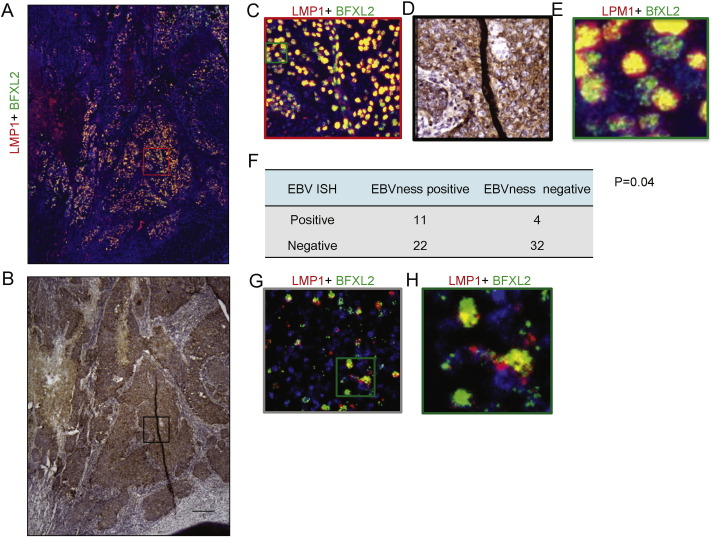
Fig. 7.
EBVness correlates with the presence of EBV DNA in breast cancer. (A–E) Fluorescent in situ hybridization of EBV DNA detected by probes against LMP1 or BXLF2 in breast cancer tissues or Burkitt's lymphoma. After in situ hybridization, the entire slide was scanned at high resolution suitable for virtual microscopy (A) Low power view of EBV DNA in situ hybridization of the entire tumor tissue section. (B) Anti-cytokeratin 5/6 stain of the same tissue section. (C) Medium power detail view (red box in A) and corresponding anti-cytokeratin 5/6 stain (D). (E) High power view of the green box in C. (F) Correlation of EBV DNA in situ hybridization with EBVness in 69 cases of breast cancer tissues. (G) EBV DNA in situ hybridization in Burkitt's lymphoma as a positive control. (H) High power view of the green image in G.
RNAse A and RNAse 1 were used to treat controls of Burkitt's lymphoma before hybridization to confirm the probes are RNAse resistant and target to viral DNA.
2.12. LMP1 RNA interference and MET inhibition
The siRNA target sequence to LMP1 mRNA is AAGAGCCUUCUCUGUCCACU (Guasparri et al., 2008). A control scrambled siRNA duplex was obtained from Dharmacon (Scramble II Duplex). Transfection of siRNA was performed using HiPerFect Transfection Reagent (Qiagen). For MET inhibition, the inhibitor XL184 (Chemietek) was dissolved in DMSO and introduced into the culture medium at the concentrations indicated in Fig. 5.
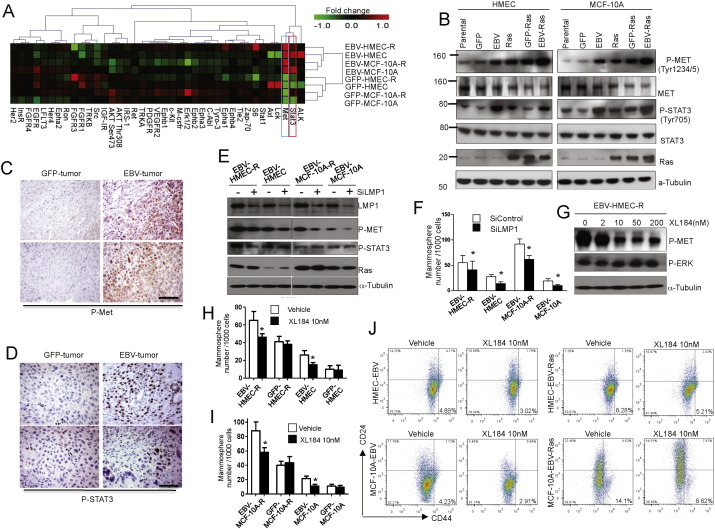
Fig. 5.
Viral LMP1 expression induces expansion of the progenitor cell population and METphosphorylation. (A) Receptor tyrosine kinase signaling in EBV-infected and control MECs. Lysates from cells as indicated were analyzed using a RTK-array. A heat map was generated from the log2-transformed fluorescence intensities of phospho-proteins. (B) Immunoblotting of MET and STAT3 phosphorylation in cell lines. (C, D) Immunoblotting of MET(C) or STAT3 (D) phosphorylation in EBV- and GFP-derived tumors. (E) Extinction of LMP1 using siRNA decreases MET but not STAT3 phosphorylation. (F) Extinction of LMP1 using siRNA decreases mammosphere formation of EBV-infected MECs. (G) Inhibition of METphosphorylation with Cabozatinib (XL 184). (H, I) Treatment with Cabozatinib decreases mammosphere formation in EBV-infected, but not in control HMECs (H) or MCF10A (G). (J) Decrease of the CD24low/CD44high population in response to met-inhibitor Cabozatinib (XL 184). Bar graphs display results of experimental triplicates ± SD. Scale bars represent 100 μm. * indicates p ≤ 0.05. See also Fig. S3.
2.13. Microarray analysis
RNA was extracted using a total RNA isolation mini kit (Agilent). Microarray expression profiles were collected using Affymetrix GeneChip human HT_HG-U133 +_PM. Affymetrix CEL files were analyzed with BRB-ArrayTools (Biometrics Research Branch, National Cancer Institute). Array data from our experimentally derived tumors can be accessed in the GEO database; (http://www.ncbi.nlm.nih.gov/geo/GSE81068). Four datasets obtained from NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) with GEO Series accession numbers GSE1456, GSE2990 GSE4922 and GSE2304 were reanalyzed together with our raw data. Raw data were processed using Robust Multi-Array (RMA) analysis (Irizarry et al., 2003). Signal intensities were normalized, background corrected, and bottom-trimmed at signal intensity of 50. Genes were filtered out if their log intensity variation percentile was < 25% and/or, if they were absent in > 85% of the experiments.
2.14. Hierarchical clustering
Gene expression data from EBV-infected and GFP-infected cells or tumors were used for unsupervised hierarchical clustering with the average linkage method as implemented in the BRB Array Tools Version 3.6. All genes were median-centered across the experiments. Differential expression between the EBV-infected and GFP-labeled cells or tumors was analyzed by BRB-ArrayTools. Statistical significance was set at p ≤ 0.01 at a false discovery rate of 0.15; and at p ≤ 0.001 at a false discovery rate of 0.05. Pathway analysis was performed using the gene set comparison tool of BRB-ArrayTools. We analyzed all pre-defined Biocarta pathways (obtained through the NCI public database) for differential expression between the EBV infected and the GFP labeled cells. The statistical significance for differential expression of each pathway was estimated using the functional class scoring method (Pavlidis et al., 2004). In brief, a p-value was computed for each gene in each pathway and then the set of p-values for each pathway was expressed as LS score (mean negative natural logarithm of the p-values of the respective single gene univariate test) and the Kolmogorov–Smirnov (KS) score (Pavlidis et al., 2004). For each pathway, we tested the null hypothesis that the list of differentially expressed genes from each pathway was random. N genes (equal to the number of genes of the pathway) were randomly selected from the project gene list, and the LS and KS statistics and their random distribution were computed (100,000 random selections). The LS (KS) permutation p-value was defined as the proportion of random simulations for which the LS (KS) statistic was larger than the LS (KS) statistic computed for the pathway with the original gene list. Statistical significance was set at 0.005.
2.15. Generation of the EBVness signature
To generate the EBVness signature, our data set of EBV infected and GFP labeled cells or tumors was used as the training set. The training was performed with BRB-ArrayTools. The most differentially expressed genes between EBV-infected and GFP labeled samples (p < 0.001) were used as input in the diagonal linear discriminant model with in a leave-one-out cross validation process. This generated a function with weight for each input gene and gave a cut off value to distinguish EBV-like tumor from none EBV-like tumor. The signature value for each sample was calculated using the function F(x) = weight(n) × gene(n) for all n genes in of the signature based on their expression in the training set.
2.16. Correlation of EBVness with the APOBEC mutational signatures
This analysis was done using TCGA Breast cancer data. Tumors for which mutational signatures have been analyzed were grouped into APOBEC-positive (positive for signature 2 and/or 13) and APOBEC-negative cases (absence of signature 2 and 13) according to (Nik-Zainal et al., 2014, Alexandrov et al., 2015). ComBat software was used for batch effect cancellation of the expression array data (Agilent platform). The expression values of probes corresponding to each of the 37 genes of the EBVness signature were extracted for each of the cases for which mutational signatures were available. To obtain the expression level for each of the 37 genes, the expression values for the probes corresponding to each gene were averaged.
2.17. Survival analysis
In order to evaluate whether the gene expression signatures associated with EBV infection provided clinically relevant information, we used four independent, clinically annotated, publicly available microarray datasets from NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) with GEO Series accession numbers GSE1456, GSE2990 GSE4922 and GSE2034 (HG-U133A platform). Gene signatures were mapped from HT_HG-U133 + _PM to HG-U133A platforms using the Affymetrix ‘best match’ tool. We tested whether these signatures would predict disease-free survival (DFS) and overall survival (OS) using the Kaplan–Meier survival model.
2.18. EBV transcription profile
Herpes virus transcription was profiled as described (Papin et al., 2005). mRNA was enriched using a poly A tract mRNA isolation system (Promega) and subjected to reverse transcription with transcriptor first strand cDNA synthesis kit (Roche). Real time qPCR was conducted using 2xSYBR (Roche) on an LC480. Data were normalized to cellular reference genes and analyzed by unsupervised clustering. Raw threshold cycle values CT were normalized to the median of reference genes (ACTIN, GAPDH, HPRT) and EBV positive samples compared to negative control samples. The relative expression level for each gene were clustered by gene with MeV4.7 (Saeed et al., 2006).
2.19. Quantitative RT-PCR
RNA from cells was extracted with a Total RNA isolation mini kit (Agilent). cDNA was prepared with transcriptor first strand cDNA synthesis kit (Roche) and PCR was carried out with iQ SYBR Green Supermix (Bio-Rad). Samples were run on the QIAGEN Rotor-Gene Q real-time cycler. GAPDH was used as an internal control. The following primers were used:
GAPDH:
Forward CATGAGAAGTATGACAACAGCCT;
Reverse AGTCCTTCCACGATACCAAAGT.
E-cadherin:
Forward TGCCCAGAAAATGAAAAAGG;
Reverse GTGTATGTGGCAATGCGTTC.
N-caderin:
Forward ACAGTGGCCACCTACAAAGG;
Reverse CCGAGATGGGGTTGATAATG.
Fibronectin 1:
Forward CAGTGGGAGACCTCGAGAAG;
Reverse TCCCTCGGAACATCAGAAAC.
Vimentin:
Forward GAGAACTTTGCCGTTGAAGC;
Reverse GCTTCCTGTAGGTGGCAATC.
2.20. Amplification of EBV DNA
PCR to detect EBV DNA was done as described (Bonnet et al., 1999a). Briefly, DNA samples were subjected to PCR using Taq DNA polymerase, specific oligonucleotides listed below. The standard cycle procedure was performed for 35 cycles with the annealing temperature at 58 °C for BZLF1, 55 °C for BNLF1, 60 °C for EBER-2 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Two microliters of the BZLF1–PCR product was taken for a second round of PCR using internal primers.
BZLF1:
ZES2 103,226–103,207 AGGGGAGATGTTAGACAGGT.
ZAS2 102,041–102,060 AGTATGCCAGGAGTAGAACA.
ZES 103180–103,161 GCCACCTTTGCTATCTTTGC.
ZEAS 102187–102,207 AGGCGTGGTTTCAATAACGG.
BNLF1:
LMP2CS 168,373–168,392 CTAGCGACTCTGCTGGAAAT.
LMP2CAS 168,075–168,056 GAGTGTGTGCCAGTTAAGGT.
EBER-2:
EBER-2S 6969–6988 CCCTAGTGGTTTCGGACACA.
EBER-2AS1 7075–7056 ACTTGCAAATGCTCTAGGCG.
2.21. Statistical analysis
All data are presented as the mean ± standard deviation, Significance was assessed using an unpaired two-tailed t-test. For survival analysis, Kaplan–Meier survival curves were constructed and differences between them were tested by the log-rank test using software in SPSS12. All tests of significance were set at P < 0.05. For analyzing the correlation between “EBVness” feature and the clinicopathologic characteristics, the Mann–Whitney U method was used. Fisher's exact test was used for analysis of the correlation of “EBVness” with EBV ISH, and a T-test to correlate EBVness with the absence or presence of APOBEC signatures.
2.21.1. Patient samples
The cohort of patient tumors analyzed retrospectively with in situ hybridization (ISH) consists of archival, paraffin-embedded breast cancer samples from patients diagnosed at the Jules Bordet Institute between 1995 and 2003 (Dedeurwaerder et al., 2011). The Ethics committee of the Jules Bordet Institute approved the present research project.
3. Results
3.1. EBV infection of primary and immortalized mammary epithelial cells, but not breast cancer cell lines is mediated by CD21
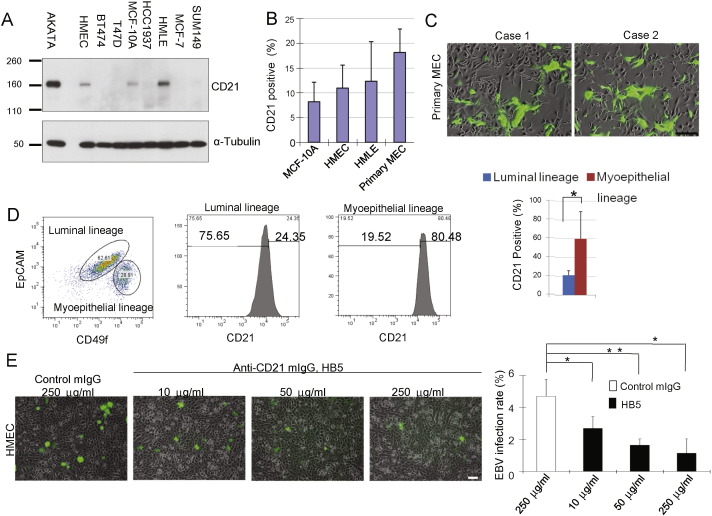
If EBV directly participates in breast tumorigenesis, then virus must gain access to MECs either as an initial event or at some later stage in the course of multi-step tumorigenesis. To determine the susceptibility of MECs to infection, we prepared primary mammary epithelial cells (PMECs) isolated from reduction mammoplasties, immortalized mammary epithelial cell lines (HMLE (Elenbaas et al., 2001), HMEC (Zhao et al., 2003) and MCF10A (Miller et al., 1993)) as well as established breast cancer cell lines and examined them for expression of the EBV-receptor, CD21, primarily expressed by B lymphocytes (Fingeroth et al., 1984, Fingeroth et al., 1988, Fingeroth, 1990, but also by a subset of epithelial cells (Fingeroth et al., 1999, Imai et al., 1998). We observed that CD21 was expressed on subpopulations of 9 to 25% of MECs (HMEC, HMLE, MCF10A and primary MECs (Fig. 1A, B, S1), but not on any of 5 breast cancer cell lines, representing ER + (MCF7, T47D), Her2 + (BT474) or triple-negative (HCC1937 and SUM149) breast cancers (Fig. 1A). Within the epithelial cell population, CD21 expression was highest in the myoepithelial subpopulation (Fig. 1D).
Fig. 1.
EBV infects human breast epithelial cells via CD21. (A) CD21 is expressed in immortalized normal breast epithelial cells, but not in breast cancer cells. MECs (HMLE, HMEC and MCF10A) were lysed and subjected to immunoblotting with anti-CD21 antibodies. An EBV-infected B-cell line (AKATA) served as positive control. (B) CD21 is expressed by subpopulations of MECs. (C) GFP-EBV infection of MECs. MECs were incubated with GFP-EBV for 2 h, images were taken 72 h after initial infection. (D) CD21 expression is highest in the myo-eoithelial subpopulation (CD49-high, EpCam-low). 60% of cultured primary MECs were of luminal lineage while 31% were of myoepithelial lineage. Counterstain with anti-CD21 antibodies showed significant enrichment of CD21 + cells in the myoepithelial compartment (bar graph, p < 0.05) (E) Competitive inhibition of EBV-infection of PMECs with anti-CR2 antibody HB5 (anti-CD21 moAb) and goat F(ab′)2 fragments to mouse IgG. MECs were incubated with increasing amounts HB5 followed by goat F(ab′)2 fragments (50 μg/ml)and then incubated with GFP-EBV for four hours. GFP-positive cells were counted 72 h later. Displayed are experimental triplicates ± SD. Scale bars represent 100 μm. * indicates p ≤ 0.05; ** indicates p ≤ 0.01 (two-sided T-test) (two-sided T-test).
When we incubated these cells with GFP-labeled EBV (Fig. 1, S2), we found that MECs, but not cancer cells, could be infected with EBV (Fig. 1C, E). The highest rate of infection was seen in primary MECs, isolated from reduction mammoplasties without prior passaging (Fig. 1C, S2A), followed by HMLE, HMEC and MCF10A (Fig. S2B). None of the cancer cell lines was susceptible to infection (data not shown). Significantly, competitive blockade of CD21 with the anti-CD21 antibody HB5 plus secondary immunoglobulin prevented the infection of MECs with EBV, confirming that EBV entered MECs via CD21 (Fig. 1 E). As GFP-EBV persisted in some MECs, suggesting a latent infection, we determined to test whether EBV promoted stepwise transformation into cancer as might occur in nature.
3.2. EBV infection induces expansion of a progenitor cell population
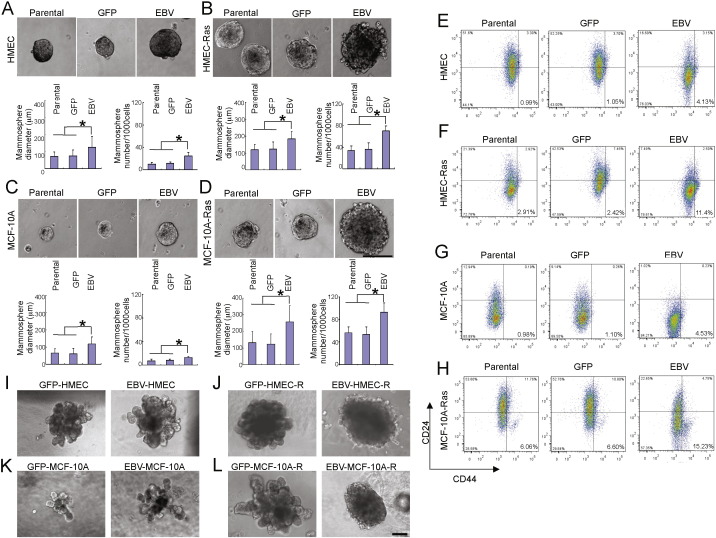
Latent EBV infection alters the transcriptional program and phenotype of host cells (Horikawa et al., 2007, Fingeroth et al., 1999) producing changes in growth and differentiation. This can include development of a stem cell-like signature and phenotype in EBV bearing NPC cells (Kondo et al., 2011, Kong et al., 2010). As EBV readily entered MECs, we asked whether EBV infection altered the self-renewal or differentiation properties of MECs. The HMEC line used to examine the effect of EBV infection on tumor formation in vivo was previously immortalized with telomerase (htert) and SV40 large T antigen (Hahn et al., 1999). In the mammosphere assay (Dontu et al., 2003) (Fig. 2A–D) we found a striking increase in both, the size and the number of spheres derived from EBV-infected MECs (Fig. 2A, C). Consistently, we detected expansion of CD24low/CD44 high expressing progenitor cells among the EBV-infected MECs (Fig. 2E–H).
Fig. 2.
EBV infection leads to de-differentiation and enhances proliferation of MECs. (A-D) Mammosphere cultures of EBV-infected and non-infected MECs after 2 weeks of culture. A: HMEC; B: HMEC-Ras: C: MCF-10A; D: MCF-10A-Ras. (E–H) MECs were stained with anti-CD24 anti-CD44 antibodies to visualize the stem cell compartment. E: HMEC; F: HMEC-Ras: G: MCF-10A; H: MCF-10A-Ras. (I–L) Differentiation cultures in 3D-matrigel. Mammospheres grown under stem cell conditions were transferred into differentiation medium (see Methods) and embedded in matrigel to allow for terminal differentiation. Scale bars represent 100 μm. Bar graphs display results of experimental triplicates ± SD. * indicates p ≤ 0.05.
To test if EBV-infection of MECs facilitated transformation, we infected immortalized HMECs with either p-EGFP-lentivirus or GFP-EBV, followed by Ras V12 or a matched retroviral vector control. The resulting cell lines (EBV-GFP/RasV12/SV40-HMEC called EBV-HMEC-R, and GFP/RasV12/SV40-HMEC called GFP-HMEC-R) were used for subsequent in vitro and xenotransplant experiments. Introduction of activated Ras increased the CD24low/CD44high progenitor cell population (Fig. 2F, H), and, correspondingly, the rate of mammosphere formation and their size (Fig. 2B, D), (Elenbaas et al., 2001), (Yu et al., 2007) Similarly, EBV infection led to an increase of the efficiency of mammosphere formation (Fig. 2A, C) and the percentage of CD24low/CD44high cells (Fig. 2E, G). Surprisingly, when both activated Ras and EBV were present (Fig. 2 F, H) the progenitor cell population increased fivefold, and mammosphere formation rate increased up to tenfold in HMEC (Fig. 2B) or MCF10A (Fig. 2D) that carried both activated Ras and EBV and their size was also increased. These data indicate that activated Ras and EBV cooperate to expand a MEC progenitor cell population with an increased proliferative capacity.
Having established an increased self-renewal capacity of EBV- and activated Ras carrying MECs, we examined their differentiation properties. When mammospheres were transferred into three-dimensional, matrigel-based cultures (Debnath et al., 2003, Gudjonsson et al., 2003), EBV-infected, GFP-control and MECs with activated Ras formed branching structures (Fig. 2I–L) indicative of terminal differentiation. EBV infection and activated Ras (V12D) combined on the other hand formed mulberry-shaped solid spheres, indicating a differentiation block (Fig. 2J, L). In summary, introduction of an activated oncogene, here Ras, into EBV-infected and immortalized MECs resulted in an expansion of progenitor cells with a differentiation block.
3.3. EBV Induces epithelial mesenchymal transition (EMT)
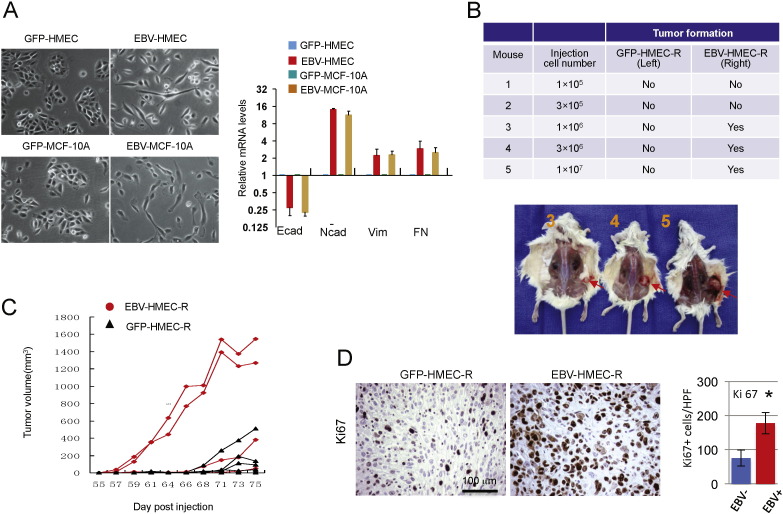
Given the altered progenitor cell properties, we asked if EBV-infected cells exhibited features of epithelial mesenchymal transition (EMT), often associated with early progenitor cell status (Mani et al., 2008, Yang et al., 2004). When grown on plastic, HMEC or MCF10A infected with EBV showed morphologic changes characteristic of EMT with loss of the epitheloid cobblestone pattern and increased cell mobility (Fig. 3A). Using RT-PCR we found a loss of E-cadherin expression and gain of Vimentin, N-cadherin and Fibronectin, all consistent with an EMT-like reprogramming of the EBV-infected MECs (Fig. 3A, right panel).
Fig. 3.
EBV infection leads to epithelial mesenchymal transition (EMT) and facilitates in vivo breast cancer formation. (A) Left: Two-dimensional morphology of EBV-infected (right in panel) versus GFP-control (left in panel). Right: Expression of Vimentin, E-cadherin, N-cadherin and Fibronectin as determined by quantitative RT-PCR of total RNA. Expression levels were normalized to GAPDH and displayed relative to the control cells. Bar graphs display results of experimental triplicates ± SD (B) Tumors generated from HMECs. Cells as indicated in the table were injected in the left (GFP-HMEC-R) or right (EBV-HMEC-R) flank of NOD/SCID mice. Tumors emerged only on the right (derived from EBV-HMEC-R) with a lag time of 12 weeks. (C) Growth curves of xenograft tumors generated from EBV-HMEC or GFP-HMEC. When injected at a dose of 2 × 107 in the left (GFP-HMEC-R) or right (EBV-HMEC-R) 4/5 injections resulted in tumors for each group. Displayed are the time of first palpation and the growth curves of the tumors. (D) Tumors were fixed, paraffin-embedded and stained with anti-Ki67 antibodies. The number of positive cells per high power field (HPF) was counted for each tumor in 3 different fields. * p ≤ 0.05 (two-sided T-test).
3.4. EBV infection facilitates breast tumor formation in vivo
When EBV-HMEC-R or GFP-HMEC-R were introduced as xeno-implants into NOD/SCID mice (Fig. 3B), EBV-infection decreased the lag time and increased the efficiency as well as the frequency of breast cancers: 1 × 106 to 107 EBV-HMEC-R were sufficient to induce tumor formation, while the corresponding number of GFP-HMEC-R did not induce tumors (Fig. 3B). The lag time of tumorigenesis in this experiment was 10–12 weeks, suggesting that the EBV-HMEC-R likely underwent additional transformational changes before cancers were formed. When EBV-HMEC-R or GFP-HMEC-R were injected at the highest dose of 2 × 107, tumors were derived from both EBV-HMEC and GFP-HMEC-R; however, the lag time for EBV-related tumors was much shorter than that for control tumors (Fig. 3C). The resulting EBV-related tumors had a high proliferative rate as determined by Ki67 (Fig. 3D). All tumors were characterized by the absence of ER, PR, Her2 and cytokeratin 8/18, and by the presence of cytokeratins CK5/6, CK14 and vimentin, indicative of a basal cell histology (Fig. S3).
3.5. EBV-infected HMECs display a type II latent pattern
EBV infection is typically classified as either lytic, latent 0, I, II and III or abortive (Kutok and Wang, 2006, Khanna et al., 1995). This holds true in particular for lymphoid tumors and NPC cell models. The infection patterns are not entirely precise in other systems, and primary tumor cells can show more variant expression (Kurokawa et al., 2005). We therefore determined EBV-gene expression in infected MECs and the tumors derived from these MECs (Fig. 4). Our in vivo tumors derived from human MECs in mice, thus contamination with EBV-carrying B-cells was excluded. We performed DNA PCR (Fig. 4A), to detect BZLF1, BNLF1 and EBER2 as markers EBV genome presence. We found that EBV-coding sequence was retained in all EBV-derived tumors (Fig. 4A). We found high expression of latent membrane protein 1 (LMP1) in EBV-MECs (Fig. 4B), while only weak expression of EBERS was detected in few tumor cells by in situ hybridization of EBV-derived tumors (Fig. 4C, lower panel), consistent with prior reports in breast cancer (Glaser et al., 2004, Bonnet et al., 1999a). A viral RNA expression array analysis (Wang et al., 2009, Kurokawa et al., 2005) clearly grouped EBV-infected MECs with the EBV-derived tumors, indicating a preserved EBV-gene expression pattern in the EBV-MEC-derived tumors (Fig. 4E, EBV-tumor 1–4). The gene expression pattern included high levels of EBNA-1, LMP1, LMP2A, LMP2B, BXLF2 and BFRF3, suggestive of a latent cycle of type II pattern accompanied by a significant lytic component, similar to NPC (Brooks et al., 1993, Yen et al., 2009). Interestingly, the CD21 antigen, consistently present on EBV-infected and uninfected MECs, was lost in the EBV-derived cancers (Fig. 4D), consistent with our earlier observations of absent of CD21 in breast cancer cell lines (Fig. 1A). In summary, we detected viral DNA, latency II accompanied by a lytic component and minimal EBERs expression in breast cancers derived from EBV-infected MECs.
3.6. EBV infection of MECs induces the MET-STAT3 pathway
In epithelial cells, EBV is known to induce EGFR expression (Miller et al., 1997) and activate the NFκb and STAT pathways (Kung and Raab-Traub, 2010, Kung and Raab-Traub, 2008, Miller et al., 1997). In an unbiased analysis of signal transduction pathways (Fig. 5A) using a receptor tyrosine kinase array (Fig. S4A) an increase in MET and STAT3 phosphorylation consistently distinguished EBV-infected MECs from GFP-control MECs (Fig. 5A), which was confirmed by Immunobloting analyses (Fig. 5B). Introduction of activated Ras also caused activation of the c-MET and STAT3 pathways, and presence of both, oncogenic Ras and EBV, led to a pronounced activation of MET and STAT3 signaling (Fig. 5B). Consistently phospho-MET- (Fig. 5C) and phospho-stat-3 (Fig. 5D) -positive cells were found in breast tumors that were derived from EBV-HMEC-R, but not in those derived from GFP-HMEC-R (Fig. 5C, D). LMP1 was abundantly expressed in all EBV-infected MECs (Fig. 4B), as expected from the viral expression array (Fig. 4E). Interestingly, METphosphorylation decreased in response to LMP1 depletion with siRNA (Fig. 5E), while STAT3 phosphorylation was not affected by LMP1 depletion, indicating that activation of MET, but not of STAT3, is mediated by LMP1. Phenotypically, depletion of LMP1 reduced the mammosphere-forming ability of EBV-infected MECs (Fig. 5F)), and treatments with the met-inhibitor XL-184 (Cabozatinib), a met-specific receptor-tyrosine kinase inhibitor, strongly reduced METphosphorylation (Fig. 5G), the mammosphere-forming ability of EBV-infected MECs (Fig. 5H. I) and the number of CD24low/CD44high cells (Fig. 5J). These shifts in the size of the progenitor cell population were not seen in GFP-HMEC-R treated with Cabozatinib (Fig. S4B–E). These results suggest that EBV induces METphosphorylation through expression of LMP1, which leads to an increase in the MEC progenitor cell compartment.
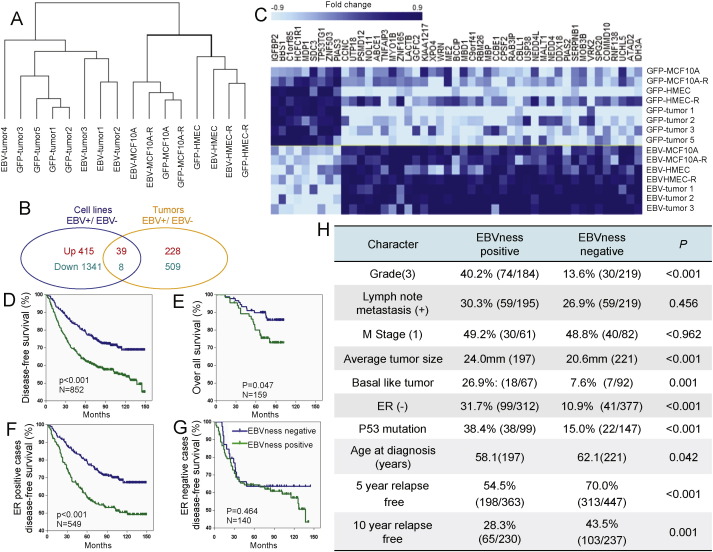
3.7. EBVness identifies a subset of breast cancers with features of poor prognosis
We obtained human gene expression arrays of our engineered cell lines (GFP-HMEC, EBV-HMECsand GFP-MEC-R, EBV-MEC-R) and the tumors derived from GFP-HMEC-R or EBV-HMEC-R. Within each category, cell lines or tumors, we then determined genes differentially expressed in EBV-infected samples compared to controls (Fig. 6A). As expected, new EBV infection in mammary epithelial cells led to a greater number of transcriptional changes than the presumably latent infection of established tumors (Fig. 6B). 47 alternately regulated genes were common to both groups, and formed the basis of our “EBVness” signature (Fig. 6C, Table S1). Of the 47 alternately regulated genes identified from the analysis of the U133A-Plus PM chip, 37 were represented in the archival datasets in GEO and thus used as an EBVness signature for further analysis (Table S4). The EBVness signature contains genes related to oncogenesis such as TNFα, Syndecan 3, insulin-like growth factor binding protein (IGFBP2) and inhibitors of STAT2 and STAT3 (PIAS2 and PIAS3), isocitrate dehydrogenase (IDH3) and DNA damage repair. An analysis of pathway activation in EBV-infected versus control MECs (Table S2) confirmed activation of the MET, EGFR and IL-6 pathways, all of which involve STAT3 phosphorylation (Fig. 5). Activation of the STAT3 pathway was also seen in the EBV-related versus control tumors (Table S3). Applying this signature to datasets of breast cancer patients in the publicly available GEO database (N = 852), we were able to identify a subset of breast cancers that carried the EBVness signature, though we make no claim that all of these contain EBV. EBVness-positive breast cancers tended to occur in younger patients (58.1 versus 62.1, p = 0.042), were more frequently of higher histological grade (40.2% versus 13.6%, p < 0.001), larger tumor size (average 24 mm versus 20.6 mm, p < 0.001), basal features (26.9% versus 7.6%, p = 0.001), ER-negative (31.7% versus 10.9%, p < 0.001), more frequently carried a p53 mutation (38.4% versus 15%, p < 0.001) (Fig. 6H), and, most importantly, were associated with a significantly lower disease free and overall survival rate (Fig. 6D,E, S5, Table S5).
Fig. 6.
An EBVness-signature for breast cancer correlates with poor clinical out come. (A) Cluster analysis of RNA expression profiles from 16 samples, including 4 EBV-infected MEC lines, 4 GFP-control cell lines, 4 EBV-MEC-derived breast cancers and 4 GFP-control tumors. EBV-tumor 4 was an outlier and is clustered singly, and was therefore not included in the following analysis. (B) Genes that were significantly different (P ≤ 0.001) between EBV-infected and GFP-control MECs (blue circle) and between EBV-infected BC and GFP-control BC (yellow circle) were determined separately. Differentially regulated genes were identified (39 up-regulated, and 8 down-regulated) as candidates for “EBVness” signature training. (C) A heatmap was generated displaying the 47 genes significantly and differentially expressed in EBV-infected samples both in vitro and in vivo. For further details see Table S1. (D–G) The “EBVness” signature is associated with decreased disease-free and overall survival. The identified gene signature was reduced from 47 to 37 to allow for analysis of archival datasets (HG-U133A chip) published in GEO. The data represent a pooled analysis of four data sets including GSE1456, GSE2990 GSE4922 and GSE2034 (HG-U133A platform). For further details and separate analysis see Fig. S4. (F) Clinicopathologic characteristics of breast cancers that carry the “EBVness” feature, again based on a pooled analysis of four data sets including GSE1456, GSE2990 GSE4922 and GSE2034 (HG-U133A platform). Also see Table S5.
The tumors that we derived from HMEC-R in mice were all ER-negative, regardless of their EBV status (Fig. S3). Because breast cancers, especially in the developed world, are more often ER-positive, we examined if the prognostic significance of EBVness was maintained in a subset analysis of ER-positive or negative breast cancers (Fig. 6F,G) and found that EBVness was significantly associated with shortened disease-free survival also in patients with ER-positive breast cancer.
3.8. EBVness correlates with the presence of EBV DNA in human breast cancers
To examine if the EBVness signature correlated with the absence or presence of EBV in tumor tissues, we examined 69 breast cancer specimen (Dedeurwaerder et al., 2011) for which U133 expression array data where available using fluorescent in-situ hybridization (FISH) with two probes that recognize EBV-LMP1 and EBV-BXLF2, respectively. The signal for both probes was RNAse-resistant, i.e. the probes detected EBV-DNA (Fig. S6). The FISH signal in breast cancer samples was strictly nuclear, consistent with a persistent EBV-episome at the nuclear envelope. EBV-FISH was typically positive for both probes and was only observed in epithelial cells, not in the architectural tissue or blood cells (Fig. 7A–E, S6). Breast cancers were considered EBV-FISH positive if a nuclear stain for both probes was seen in tumor cells as gauged by three independent investigators. There was considerable intra-tumoral heterogeneity in EBV-positive cases, with tumor cells staining positive in groups, while other areas of the tumor were negative. Therefore, the entire slide was scanned at high resolution for evaluation by virtual microscopy (Fig. 7A, B). 15/69 (22%) of the tumors were EBV-FISH positive, consistent with prior reports (Fina et al., 2001, Mazouni et al., 2011, Richardson et al., 2015). There was a significant correlation of tumors that were positive for EBV by FISH with the human gene expression signature EBVness”, Fig. 7F, p = 0.04).
EBV elicits and is edited by an APOBEC3 response in host cells (Suspene et al., 2011). APOBEC3 cytidine deaminases edit the single stranded viral genome resulting in loss of function. But more recently the APOBEC family of cytidine deaminases have also been found to leave characteristic mutational signatures, specifically in breast epithelium (Taylor et al., 2013, Nik-Zainal et al., 2014, Alexandrov et al., 2013) resulting in kataegis, clusters of hypermutation with C → T transitions, that potentially contribute to oncogenic transformation. To examine if EBVness had a correlation with the presence of the APOBEC signature in breast cancer, we translated and applied the EBVness signature to the expression data in TCGA. Despite the noise introduced by cross-platform translation of the signature and the considerable heterogeneity of the TCGA data set, there was a significant correlation between EBVness and presence of the APOBEC signature (Fig. S7), i.e. EBVness positive tumors were more likely to also be positive for the APOBEC signature. The correlation of the EBVness transcriptional signature with the presence of EBV-DNA in specific patients tumors as well as the correlation of EBVness with the APOBEC signature, are consistent with EBV infection of MECs being part of a multistep process of breast carcinogenesis in humans.
4. Discussion
4.1. EBV Infection of MECs provides a potential mechanism for oncogenesis
Our experimental data support a role for EBV in the early steps of malignant transformation. CD21 expression in epithelial cells had until now mostly been studied in cancer cell lines, of which only a small fraction, none of breast origin, was positive (Imai et al., 1998, Fingeroth et al., 1999, Birkenbach et al., 1992). Consistent with prior reports we found that breast cancer cells did not express CD21 (Speck and Longnecker, 2000). In contrast, we found that a subset of primary and immortalized MECs, expressed and exclusively utilized CD21 for EBV entry. The observations that CD21 was absent on all of the tumor cell lines, none of which became infected, and that analysis of the TCGA breast cancer RNAseq data revealed no active transcription of EBV (Khoury et al., 2013a, Tang et al., 2013), suggest that the EBV DNA that we detected in a subset of human breast cancers, is an inactive remnant of a previously active EBV infection that might have occurred in mammary epithelial cells years or even decades prior to cancer formation.
4.2. EBV induces a switch from virus-induced inflammation to transformation through activation of MET signaling
Infection of mammalian cells with DNA viruses, including EBV, leads to systematic perturbations of the host cell signaling networks initiated by the interaction of viral proteins with specific host proteins (Rozenblatt-Rosen et al., 2012). EBV infection of nasopharyngeal cells (NPCs) classically displays a latency II pattern that is characterized by restricted expression of a subset of virus genes, such as EBNA1, LMP1, LMP2A and LMP2B(Raab-Traub, 2002). In NPC, LMP1 has been shown to contribute to the development of genomic instability (Liu et al., 2004, Liu et al., 2005). The LMPs constitutively activate cell membrane receptors that generate signaling cascades, which lead to the activation of NF-κB (Gewurz et al., 2011, Gewurz et al., 2012). Notably, activation of NF-κB is essential for cell proliferation downstream from hepatocyte growth factor-stimulated c-MET (Muller et al., 2002), and both NF-kB and STAT3 activation have been shown to be required for a programmatic switch that occurs with the transition from inflammation to transformation (Iliopoulos et al., 2009). Our data suggested that LMP1 also induces this switch in infected MECs (Fig. 5E, F) and does so via activation of c-MET (Fig. 5G–J). Further evidence for this switch from a virus-induced inflammatory program to transformation are our observations that EBV infection induced EMT (Fig. 3A) and that the expansion of the progenitor cell population was dependent on activation of c-MET (Fig. 5H–J). MET activation has been observed in a fraction of breast cancers (reviewed in (Gastaldi et al., 2010)) and may dictate response to the met-inhibitor Cabozantinib (Yakes et al., 2011). Going forward it will be important to determine whether this same subset of breast tumors is EBV-related, and whether it is responsive to Cabozantinib.
4.3. EBVness correlates with adverse clinicopathological features, presence of EBV-DNA and an APOBEC mutational signature
We developed a signature for the transcriptional changes induced by EBV infection that we termed EBVness. This signature was associated with clinicopathological features of high grade tumors that tend to have a higher degree of genomic instability. While there was a positive correlation of EBVness with the finding of EBV DNA by ISH, only 33% of EBVness positive tumors were positive for EBV DNA, and in those cases there was marked tumoral heterogeneity with some areas of the tumor staining positive while others were negative (Fig. 7). Overall, these findings suggest that the presence of EBV is no longer required for tumor growth, most consistent with a “hit and-run” mechanism which would also explain why mining of the TCGA RNAseq data did not show active transcription of EBV (Tang et al., 2013, Khoury et al., 2013a).
The APOBEC family functions as an arm of the innate immune system through mutation followed by elimination of pathogen nucleic acids. The APOBEC3 cytidine deaminases can edit single stranded viral genomes such as EBV (Suspene et al., 2011) More recently, the APOBEC family of cytidine deaminases have also been implicated in causing kataegis, clusters of hypermutation with C -> T transitions that potentially contribute to breast cancer (Taylor et al., 2013, Nik-Zainal et al., 2014, Alexandrov et al., 2013). The correlative data presented here are consistent with a ‘hit-and-run’ model of carcinogenesis (Mcdougall, 2001) where the viral infection increases the odds of transformation of MECs into cancer cells by eliciting an APOBEC response that leads to a hypermutated phenotype.
4.4. Potential public health implications
Recent studies from Africa and Asia, where breast cancer occurs at a younger age, is typically more aggressive and of a basal type, suggested EBV might be associated with a particular subset of breast cancers. Epidemiologic data in support these observations include evidence of strong regional differences regarding association of EBV with breast cancer with higher rates being reported from Asia (Huo et al., 2012, Joshi et al., 2009) and North Africa (Hachana et al., 2011a, Fina et al., 2001) where EBV NPC is also endemic — which raises the specter of genetic predisposition. In addition to geography and genetics, the age at which primary EBV infection occurs may be important. Yasui found that “delayed” EBV-infection with mononucleosis in adolescence or early adulthood was an independent risk factor for the development of breast cancer (Yasui et al., 2001); this finding was not confirmed by an analysis of the Nurses Health Study (Massa et al., 2012), although that analysis was hampered by a low number of mononucleosis cases that could be followed prospectively and a the study population that is heavily skewed towards women of European descent.
It has been well documented that the incidence of breast cancer increases as societies develop. While this trend has long been recognized in Western Europe and North America, it is now also being observed in Asia, Africa and South America. A range of factors such as changes in diet, reproductive behavior, increased screening and longevity have all been linked to this trend. As EBV is frequently found in breast secretions including breast milk (Glenn et al., 2012a, Glenn et al., 2012b), MECs are naturally exposed to infectious virus, which could potentially constitute a risk factor. The tools now available, such as high sensitivity FISH and high-throughput DNA and RNA sequencing will allow, detection of the virus, even if it is an incomplete and inactive remnant of a much earlier infection.
The data presented here raise the possibility that delayed infection with EBV in late childhood or adolescence, when breast development in girls occurs, could lead to latent infection of MECs, increase mutational load and cooperate with other genetic or environmental factors to increase later breast cancer risk. If EBV infection indeed renders MECs vulnerable to malignant transformation into breast cancer cells, then a childhood immunization against EBV might have a potentially large public health impact.
Funding
This work was supported by funding from the AVON Foundation to GW and JF and through NIH grants R01AI063571 (JDF), CA019014, and DE018304 (DPD) and the University Cancer Research Fund (DPD).
Competing interests
The authors declare no financial conflicts.
Author contributions
Conceptualization: GMW, HH, and JDF; Methodology: GMW, HH, MLL, CD, SN, SY, PAK, EG, RHB, CS and DPD; Validation: HH, GP, XY and GMW; Formal analysis: HH, CD, PAK, SN, DPD,and GMW; Investigation HH, MLL, CD,SY, AH, RHB and GMW; Resources: EG, CS, CD, DPD, JDF, and GMW; Data curation: HH, CD, SN, CS, and DPD; Writing: HH, JDF, and GMW; Visualization HH and GMW; Supervision EG, CS, DPD, JDF, and GMW; Project administration: HH, JDF, and GMW; Funding acquisition JDF, EG, and GMW.
Acknowledgments
The authors thank Dr. Tom Roberts, Dana-Farber Cancer Institute and Dr. Lowell Schnipper, BIDMC, for their critical reading of the manuscript.
Footnotes
One sentence summary: Epstein–Barr Virus (EBV) can infect mammary epithelial cells and facilitate cancerous transformation consistent with a ‘hit-and-run’ mechanism.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.05.025.
Appendix A. Supplementary data
Fig. S1 CD21 is dominantly expressed in myoepithelial lineage of mammary epithelial cells. (A-C) Primary epithelial cells were stained with anti-EoCAM (FITC-labeled, vertical axis), anti-CD49f (APC, horizontal axis) antibodies and anti CD21. Quadrant gates were set according to isotype controls. (D) Myoepithelial lineage is minor population of mammary epithelial cells but contains major CD21 positive population.
Fig. S2 EBV infection of MECs. (A) Primary MECs isolated from reduction mammoplasties or (B) immortalized MEC cell lines as indicated were grown in MEGM, and incubated with EBV isolated from Akata cells for 4 h. Scale bars are 100 mm.
Fig. S3 Tumors generated from HMEC-htert are triple-negative have basal-like features. Upper panel: Absence of ER, PR and Her2 with positive controls (top row). Lower panel: Expression of cytokeratins 5/6, 14, p63 and Vimentin and absence of the luminal marker cytokeratin 8.
Fig. S4 (A) Image of RTK antibody array after incubation with cell lysates as indicated and scanning of the fluorescence intensity using an Odyssee reader. Red boxes highlight the position of p-Stat3, blue boxes p-Met. (B–E) Flow Cytometric determination of CD24 and CD44 in GFP-MECs (MCF10A or HMEC as indicated) treated with either vehicle control or the met-inhibitor Cabozatinib (XL 184). This is a control experiment for the data provided in Fig. 5J.
Fig. S5 The EBV-ness signature is a marker of poor prognosis in breast cancer. The plots show application of the “EBV-ness” signature to four different datasets accessed from GEO. Displayed is disease-free survival as a function of time in months. The merged results are shown in Fig. 6.
Fig. S6 Positive controls for the in situ hybridization of EBV: A. ISH for tumor xenografts derived from EBV-HMEC cells. B ISH for tumor xenografts from GFP-HMEC. (C, D) ISH stain for beta-actin and BfXL2 without (C) and with (D) RNAse treatment. (F, G) ISH stain for GAPDH and LMP1 without (F) and with (G) RNAse treatment. The ISH probes will hybridize with single stranded RNA or DNA. Only the LMP1 and BxLF2 signals were RNAse-resistant, indicating that the signal corresponds to EBV DNA.
Fig. S7 Correlation of the EBVness score with absence or presence of one or both APOBEC signatures in breast cancer. The EBVness score developed on the Affymetrix platform was transferred to the Agilent platform gene expression array as described and a continuous EBVness score was obtained for each sample. Significance of the correlation with the APOBEC signature was calculated using the t-test.
Table S1 List of genes differentially expressed at a significance level of p ≤ 0.001 in EBV-infected MECs and EBV-derived tumors versus GFP controls, according to the strategy outlined in Fig. 6B.
Table S2 Analysis of pathways differentially activated in EBV-infected MECs versus GFP controls MECs. The Hepatocyte Growth Factor Receptor (MET) pathway, and the IL6 related pathway, in which STAT3 is involved, are differentially activated in EBV-infected samples.
Table S3 Pathways differentially regulated between EBV-related BCs and GFP-control BCs.
Table S4 Training of the “EBVness” signature. 7 EBV positive and 8 EBV negative samples were included. 37 of the 47 significantly EBV regulated genes were used in the training, because these genes were represented in bot, the HT_HG-U133 + _PM array (used by us) as well as in the HG-U133A array (archival GEO data). Using BRB Array Tools, the signature was trained on the experimental tumors and cell lines. The diagonal linear discrimination analysis was applied and the weight of each gene is shown in the table.
Table S5 Correlation of the “EBVness” signature with clinicopathologic characteristics of the tumors in individual data sets.
References
- Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L., Boyault S., Burkhardt B., Butler A.P., Caldas C., Davies H.R., Desmedt C., Eils R., Eyfjord J.E., Foekens J.A., Greaves M., Hosoda F., Hutter B., Ilicic T., Imbeaud S., Imielinski M., Jager N., Jones D.T., Jones D., Knappskog S., Kool M., Lakhani S.R., Lopez-Otin C., Martin S., Munshi N.C., Nakamura H., Northcott P.A., Pajic M., Papaemmanuil E., Paradiso A., Pearson J.V., Puente X.S., Raine K., Ramakrishna M., Richardson A.L., Richter J., Rosenstiel P., Schlesner M., Schumacher T.N., Span P.N., Teague J.W., Totoki Y., Tutt A.N., Valdes-Mas R., Van Buuren M.M., Van ‘T Veer L., Vincent-Salomon A., Waddell N., Yates L.R., Australian Pancreatic Cancer Genome I., Consortium I.B.C., Consortium I.M.-S., Pedbrain I., Zucman-Rossi J., Futreal P.A., Mcdermott U., Lichter P., Meyerson M., Grimmond S.M., Siebert R., Campo E., Shibata T., Pfister S.M., Campbell P.J., Stratton M.R. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov L.B., Jones P.H., Wedge D.C., Sale J.E., Campbell P.J., Nik-Zainal S., Stratton M.R. Clock-like mutational processes in human somatic cells. Nat. Genet. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbach M., Tong X., Bradbury L.E., Tedder T.F., Kieff E. Characterization of an Epstein–Barr virus receptor on human epithelial cells. J. Exp. Med. 1992;176:1405–1414. doi: 10.1084/jem.176.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet M., Guinebretiere J.M., Kremmer E., Grunewald V., Benhamou E., Contesso G., Joab I. Detection of Epstein–Barr virus in invasive breast cancers. J. Natl. Cancer Inst. 1999;91:1376–1381. doi: 10.1093/jnci/91.16.1376. [DOI] [PubMed] [Google Scholar]
- Brooks L.A., Lear A.L., Young L.S., Rickinson A.B. Transcripts from the Epstein–Barr virus BamHI A fragment are detectable in all three forms of virus latency. J. Virol. 1993;67:3182–3190. doi: 10.1128/jvi.67.6.3182-3190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burga L.N., Tung N.M., Troyan S.L., Bostina M., Konstantinopoulos P.A., Fountzilas H., Spentzos D., Miron A., Yassin Y.A., Lee B.T., Wulf G.M. Altered proliferation and differentiation properties of primary mammary epithelial cells from BRCA1 mutation carriers. Cancer Res. 2009;69:1273–1278. doi: 10.1158/0008-5472.CAN-08-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burga L.N., Hu H., Juvekar A., Tung N.M., Troyan S.L., Hofstatter E.W., Wulf G.M. Loss of BRCA1 leads to an increase in epidermal growth factor receptor expression in mammary epithelial cells, and epidermal growth factor receptor inhibition prevents estrogen receptor-negative cancers in BRCA1-mutant mice. Breast Cancer Res. 2011;13:R30. doi: 10.1186/bcr2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J.I. Epstein–Barr virus infection. N. Engl. J. Med. 2000;343:481–492. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- Cohen J.I., Fauci A.S., Varmus H., Nabel G.J. Epstein–Barr virus: an important vaccine target for cancer prevention. Sci. Transl. Med. 2011;3 doi: 10.1126/scitranslmed.3002878. (107 fs7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S.K., Brugge J.S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Dedeurwaerder S., Desmedt C., Calonne E., Singhal S.K., Haibe-Kains B., Defrance M., Michiels S., Volkmar M., Deplus R., Luciani J., Lallemand F., Larsimont D., Toussaint J., Haussy S., Rothe F., Rouas G., Metzger O., Majjaj S., Saini K., Putmans P., Hames G., Van Baren N., Coulie P.G., Piccart M., Sotiriou C., Fuks F. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol. Med. 2011;3:726–741. doi: 10.1002/emmm.201100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G., Abdallah W.M., Foley J.M., Jackson K.W., Clarke M.F., Kawamura M.J., Wicha M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenbaas B., Spirio L., Koerner F., Fleming M.D., Zimonjic D.B., Donaher J.L., Popescu N.C., Hahn W.C., Weinberg R.A. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein M.A., Barr Y.M. Cultivation in vitro of human lymphoblasts from Burkitt's malignant lymphoma. Lancet. 1964;1:252–253. doi: 10.1016/s0140-6736(64)92354-2. [DOI] [PubMed] [Google Scholar]
- Epstein M.A., Achong B.G., Barr Y.M. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet. 1964;1:702–703. doi: 10.1016/s0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- Evans A.S. The spectrum of infections with Epstein–Barr virus: a hypothesis. J. Infect. Dis. 1971;124:330–337. doi: 10.1093/infdis/124.3.330. [DOI] [PubMed] [Google Scholar]
- Fina F., Romain S., Ouafik L., Palmari J., Ben Ayed F., Benharkat S., Bonnier P., Spyratos F., Foekens J.A., Rose C., Buisson M., Gerard H., Reymond M.O., Seigneurin J.M., Martin P.M. Frequency and genome load of Epstein–Barr virus in 509 breast cancers from different geographical areas. Br. J. Cancer. 2001;84:783–790. doi: 10.1054/bjoc.2000.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingeroth J.D. Comparative structure and evolution of murine CR2. The homolog of the human C3d/EBV receptor (CD21) J. Immunol. 1990;144:3458–3467. [PubMed] [Google Scholar]
- Fingeroth J.D., Weis J.J., Tedder T.F., Strominger J.L., Biro P.A., Fearon D.T. Epstein–Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc. Natl. Acad. Sci. U. S. A. 1984;81:4510–4514. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingeroth J.D., Clabby M.L., Strominger J.D. Characterization of a T-lymphocyte Epstein–Barr virus/C3d receptor (CD21) J. Virol. 1988;62:1442–1447. doi: 10.1128/jvi.62.4.1442-1447.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingeroth J.D., Diamond M.E., Sage D.R., Hayman J., Yates J.L. CD21-Dependent infection of an epithelial cell line, 293, by Epstein–Barr virus. J. Virol. 1999;73:2115–2125. doi: 10.1128/jvi.73.3.2115-2125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaldi S., Comoglio P.M., Trusolino L. The Met oncogene and basal-like breast cancer: another culprit to watch out for? Breast Cancer Res. 2010;12:208. doi: 10.1186/bcr2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewurz B.E., Mar J.C., Padi M., Zhao B., Shinners N.P., Takasaki K., Bedoya E., Zou J.Y., Cahir-Mcfarland E., Quackenbush J., Kieff E. Canonical NF-kappaB activation is essential for Epstein–Barr virus latent membrane protein 1 TES2/CTAR2 gene regulation. J. Virol. 2011;85:6764–6773. doi: 10.1128/JVI.00422-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewurz B.E., Towfic F., Mar J.C., Shinners N.P., Takasaki K., Zhao B., Cahir-Mcfarland E.D., Quackenbush J., Xavier R.J., Kieff E. Genome-wide siRNA screen for mediators of NF-kappaB activation. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2467–2472. doi: 10.1073/pnas.1120542109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S.L., Hsu J.L., Gulley M.L. Epstein–Barr virus and breast cancer: state of the evidence for viral carcinogenesis. Cancer Epidemiol. Biomark. Prev. 2004;13:688–697. [PubMed] [Google Scholar]
- Glenn W.K., Heng B., Delprado W., Iacopetta B., Whitaker N.J., Lawson J.S. Epstein–Barr virus, human papillomavirus and mouse mammary tumour virus as multiple viruses in breast cancer. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn W.K., Whitaker N.J., Lawson J.S. High risk human papillomavirus and Epstein Barr virus in human breast milk. BMC Res. Notes. 2012;5:477. doi: 10.1186/1756-0500-5-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasparri I., Bubman D., Cesarman E. EBV LMP2A affects LMP1-mediated NF-kappaB signaling and survival of lymphoma cells by regulating TRAF2 expression. Blood. 2008;111:3813–3820. doi: 10.1182/blood-2007-03-080309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson T., Ronnov-Jessen L., Villadsen R., Bissell M.J., Petersen O.W. To create the correct microenvironment: three-dimensional heterotypic collagen assays for human breast epithelial morphogenesis and neoplasia. Methods. 2003;30:247–255. doi: 10.1016/s1046-2023(03)00031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulley M.L. Molecular diagnosis of Epstein–Barr virus-related diseases. J. Mol. Diagn. 2001;3:1–10. doi: 10.1016/S1525-1578(10)60642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachana M., Amara K., Ziadi S., Romdhane E., Gacem R.B., Trimeche M. Investigation of Epstein–Barr virus in breast carcinomas in Tunisia. Pathol. Res. Pract. 2011;207:695–700. doi: 10.1016/j.prp.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Hachana M., Amara K., Ziadi S., Romdhane E., Gacem R.B., Trimeche M. Investigation of Epstein–Barr virus in breast carcinomas in Tunisia. Pathol. Res. Pract. 2011 doi: 10.1016/j.prp.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Hahn W.C., Counter C.M., Lundberg A.S., Beijersbergen R.L., Brooks M.W., Weinberg R.A. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- He J.R., Chen L.J., Su Y., Cen Y.L., Tang L.Y., Yu D.D., Chen W.Q., Wang S.M., Song E.W., Ren Z.F. Joint effects of Epstein–Barr virus and polymorphisms in interleukin-10 and interferon-gamma on breast cancer risk. J. Infect. dis. 2012;205:64–71. doi: 10.1093/infdis/jir710. [DOI] [PubMed] [Google Scholar]
- Horikawa T., Yang J., Kondo S., Yoshizaki T., Joab I., Furukawa M., Pagano J.S. Twist and epithelial-mesenchymal transition are induced by the EBV oncoprotein latent membrane protein 1 and are associated with metastatic nasopharyngeal carcinoma. Cancer Res. 2007;67:1970–1978. doi: 10.1158/0008-5472.CAN-06-3933. [DOI] [PubMed] [Google Scholar]
- Huang J., Chen H., Hutt-Fletcher L., Ambinder R.F., Hayward S.D. Lytic viral replication as a contributor to the detection of Epstein–Barr virus in breast cancer. J. Virol. 2003;77:13267–13274. doi: 10.1128/JVI.77.24.13267-13274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Q., Zhang N., Yang Q. Epstein–Barr virus infection and sporadic breast cancer risk: a meta-analysis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D., Hirsch H.A., Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Nishikawa J., Takada K. Cell-to-cell contact as an efficient mode of Epstein–Barr virus infection of diverse human epithelial cells. J. Virol. 1998;72:4371–4378. doi: 10.1128/jvi.72.5.4371-4378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Joshi D., Quadri M., Gangane N., Joshi R. Association of Epstein Barr virus infection (EBV) with breast cancer in rural Indian women. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Burrows S.R., Moss D.J. Immune regulation in Epstein–Barr virus-associated diseases. Microbiol. Rev. 1995;59:387–405. doi: 10.1128/mr.59.3.387-405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury J.D., Tannir N.M., Williams M.D., Chen Y., Yao H., Zhang J., Thompson E.J., Meric-Bernstam F., Medeiros L.J., Weinstein J.N., Su X. Landscape of DNA virus associations across human malignant cancers: analysis of 3,775 cases using RNA-Seq. J. Virol. 2013;87:8916–8926. doi: 10.1128/JVI.00340-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Wakisaka N., Muramatsu M., Zen Y., Endo K., Murono S., Sugimoto H., Yamaoka S., Pagano J.S., Yoshizaki T. Epstein–Barr virus latent membrane protein 1 induces cancer stem/progenitor-like cells in nasopharyngeal epithelial cell lines. J. Virol. 2011;85:11255–11264. doi: 10.1128/JVI.00188-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q.L., Hu L.J., Cao J.Y., Huang Y.J., Xu L.H., Liang Y., Xiong D., Guan S., Guo B.H., Mai H.Q., Chen Q.Y., Zhang X., Li M.Z., Shao J.Y., Qian C.N., Xia Y.F., Song L.B., Zeng Y.X., Zeng M.S. Epstein–Barr virus-encoded LMP2A induces an epithelial–mesenchymal transition and increases the number of side population stem-like cancer cells in nasopharyngeal carcinoma. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn-Hallek I., Sage D.R., Stein L., Groelle H., Fingeroth J.D. Expression of recombination activating genes (RAG-1 and RAG-2) in Epstein–Barr virus-bearing B cells. Blood. 1995;85:1289–1299. [PubMed] [Google Scholar]
- Kung C.P., Raab-Traub N. Epstein–Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor through effects on Bcl-3 and STAT3. J. Virol. 2008;82:5486–5493. doi: 10.1128/JVI.00125-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C.P., Raab-Traub N. Epstein–Barr virus latent membrane protein 1 modulates distinctive NF-kappaB pathways through C-terminus-activating region 1 to regulate epidermal growth factor receptor expression. J. Virol. 2010;84:6605–6614. doi: 10.1128/JVI.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa M., Ghosh S.K., Ramos J.C., Mian A.M., Toomey N.L., Cabral L., Whitby D., Barber G.N., Dittmer D.P., Harrington W.J., JR Azidothymidine inhibits NF-kappaB and induces Epstein–Barr virus gene expression in Burkitt lymphoma. Blood. 2005;106:235–240. doi: 10.1182/blood-2004-09-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutok J.L., Wang F. Spectrum of Epstein–Barr virus-associated diseases. Annu. Rev. Pathol. 2006;1:375–404. doi: 10.1146/annurev.pathol.1.110304.100209. [DOI] [PubMed] [Google Scholar]
- Labrecque L.G., Barnes D.M., Fentiman I.S., Griffin B.E. Epstein–Barr virus in epithelial cell tumors: a breast cancer study. Cancer Res. 1995;55:39–45. [PubMed] [Google Scholar]
- Liu M.T., Chen Y.R., Chen S.C., Hu C.Y., Lin C.S., Chang Y.T., Wang W.B., Chen J.Y. Epstein–Barr virus latent membrane protein 1 induces micronucleus formation, represses DNA repair and enhances sensitivity to DNA-damaging agents in human epithelial cells. Oncogene. 2004;23:2531–2539. doi: 10.1038/sj.onc.1207375. [DOI] [PubMed] [Google Scholar]
- Liu M.T., Chang Y.T., Chen S.C., Chuang Y.C., Chen Y.R., Lin C.S., Chen J.Y. Epstein–Barr virus latent membrane protein 1 represses p53-mediated DNA repair and transcriptional activity. Oncogene. 2005;24:2635–2646. doi: 10.1038/sj.onc.1208319. [DOI] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., Campbell L.L., Polyak K., Brisken C., Yang J., Weinberg R.A. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrao G., Habib M., Paiva A., Bicout D., Fallecker C., Franco S., Fafi-Kremer S., Simoes Da Silva T., Morand P., Freire de Oliveira C., Drouet E. Epstein–Barr virus infection and clinical outcome in breast cancer patients correlate with immune cell TNF-alpha/IFN-gamma response. BMC Cancer. 2014;14:665. doi: 10.1186/1471-2407-14-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa J., Hamdan A., Simon K.C., Bertrand K., Wulf G., Tamimi R.M., Ascherio A. Infectious mononucleosis and risk of breast cancer in a prospective study of women. Cancer Causes Control. 2012;23:1893–1898. doi: 10.1007/s10552-012-0064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazouni C., Fina F., Romain S., Ouafik L., Bonnier P., Brandone J.M., Martin P.M. Epstein–Barr virus as a marker of biological aggressiveness in breast cancer. Br. J. Cancer. 2011;104:332–337. doi: 10.1038/sj.bjc.6606048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdougall J.K. “Hit and run” transformation leading to carcinogenesis. Dev. Biol. (Basel) 2001;106:77–82. (discussion 82–3, 143–60) [PubMed] [Google Scholar]
- Miller F.R., Soule H.D., Tait L., Pauley R.J., Wolman S.R., Dawson P.J., Heppner G.H. Xenograft model of progressive human proliferative breast disease. J. Natl. Cancer Inst. 1993;85:1725–1732. doi: 10.1093/jnci/85.21.1725. [DOI] [PubMed] [Google Scholar]
- Miller W.E., Mosialos G., Kieff E., Raab-Traub N. Epstein–Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-kappaB activation. J. Virol. 1997;71:586–594. doi: 10.1128/jvi.71.1.586-594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M., Morotti A., Ponzetto C. Activation of NF-kappaB is essential for hepatocyte growth factor-mediated proliferation and tubulogenesis. Mol. Cell. Biol. 2002;22:1060–1072. doi: 10.1128/MCB.22.4.1060-1072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedobitek G., Meru N., Delecluse H.J. Epstein–Barr virus infection and human malignancies. Int. J. Exp. Pathol. 2001;82:149–170. doi: 10.1046/j.1365-2613.2001.iep0082-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S., Wedge D.C., Alexandrov L.B., Petljak M., Butler A.P., Bolli N., Davies H.R., Knappskog S., Martin S., Papaemmanuil E., Ramakrishna M., Shlien A., Simonic I., Xue Y., Tyler-Smith C., Campbell P.J., Stratton M.R. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet. 2014;46:487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin J., Vahrson W., Hines-Boykin R., Dittmer D.P. Real-time quantitative PCR analysis of viral transcription. Methods Mol. Biol. 2005;292:449–480. doi: 10.1385/1-59259-848-x:449. [DOI] [PubMed] [Google Scholar]
- Pavlidis P., Qin J., Arango V., Mann J.J., Sibille E. Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem. Res. 2004;29:1213–1222. doi: 10.1023/b:nere.0000023608.29741.45. [DOI] [PubMed] [Google Scholar]
- Peng J., Wang T., Zhu H., Guo J., Li K., Yao Q., Lv Y., Zhang J., He C., Chen J., Wang L., Jin Q. Multiplex PCR/mass spectrometry screening of biological carcinogenic agents in human mammary tumors. J. Clin. Virol. 2014;61:255–259. doi: 10.1016/j.jcv.2014.07.010. [DOI] [PubMed] [Google Scholar]
- Raab-Traub N. Epstein–Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 2002;12:431–441. doi: 10.1016/s1044579x0200086x. [DOI] [PubMed] [Google Scholar]
- Richardson A.K., Currie M.J., Robinson B.A., Morrin H., Phung Y., Pearson J.F., Anderson T.P., Potter J.D., Walker L.C. Cytomegalovirus and Epstein–Barr virus in breast cancer. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O., Deo R.C., Padi M., Adelmant G., Calderwood M.A., Rolland T., Grace M., Dricot A., Askenazi M., Tavares M., Pevzner S.J., Abderazzaq F., Byrdsong D., Carvunis A.R., Chen A.A., Cheng J., Correll M., Duarte M., Fan C., Feltkamp M.C., Ficarro S.B., Franchi R., Garg B.K., Gulbahce N., Hao T., Holthaus A.M., James R., Korkhin A., Litovchick L., Mar J.C., Pak T.R., Rabello S., Rubio R., Shen Y., Singh S., Spangle J.M., Tasan M., Wanamaker S., Webber J.T., Roecklein-Canfield J., Johannsen E., Barabasi A.L., Beroukhim R., Kieff E., Cusick M.E., Hill D.E., Munger K., Marto J.A., Quackenbush J., Roth F.P., Decaprio J.A., Vidal M. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A.I., Bhagabati N.K., Braisted J.C., Liang W., Sharov V., Howe E.A., Li J., Thiagarajan M., White J.A., Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Speck P., Longnecker R. Infection of breast epithelial cells with Epstein–Barr virus via cell-to-cell contact. J. Natl. Cancer Inst. 2000;92:1849–1851. doi: 10.1093/jnci/92.22.1849. [DOI] [PubMed] [Google Scholar]
- Stewart S.A., Dykxhoorn D.M., Palliser D., Mizuno H., Yu E.Y., An D.S., Sabatini D.M., Chen I.S., Hahn W.C., Sharp P.A., Weinberg R.A., Novina C.D. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspene R., Aynaud M.M., Koch S., Pasdeloup D., Labetoulle M., Gaertner B., Vartanian J.P., Meyerhans A., Wain-Hobson S. Genetic editing of herpes simplex virus 1 and Epstein–Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo. J. Virol. 2011;85:7594–7602. doi: 10.1128/JVI.00290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K.W., Alaei-Mahabadi B., Samuelsson T., Lindh M., Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat. Commun. 2013;4:2513. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B.J., Nik-Zainal S., Wu Y.L., Stebbings L.A., Raine K., Campbell P.J., Rada C., Stratton M.R., Neuberger M.S. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2 doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.Z., Roy D., Gershburg E., Whitehurst C.B., Dittmer D.P., Pagano J.S. Maribavir inhibits epstein–barr virus transcription in addition to viral DNA replication. J. Virol. 2009;83:12108–12117. doi: 10.1128/JVI.01575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakes F.M., Chen J., Tan J., Yamaguchi K., Shi Y., Yu P., Qian F., Chu F., Bentzien F., Cancilla B., Orf J., You A., Laird A.D., Engst S., Lee L., Lesch J., Chou Y.C., Joly A.H. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol. Cancer Ther. 2011;10:2298–2308. doi: 10.1158/1535-7163.MCT-11-0264. [DOI] [PubMed] [Google Scholar]
- Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yasui Y., Potter J.D., Stanford J.L., Rossing M.A., Winget M.D., Bronner M., Daling J. Breast cancer risk and “delayed” primary Epstein–Barr virus infection. Cancer Epidemiol. Biomark. Prev. 2001;10:9–16. [PubMed] [Google Scholar]
- Yen C.Y., Lu M.C., Tzeng C.C., Huang J.Y., Chang H.W., Chen R.S., Liu S.Y., Liu S.T., Shieh B., Li C. Detection of EBV infection and gene expression in oral cancer from patients in Taiwan by microarray analysis. J. Bioenerg. Biomembr. 2009;2009:904589. doi: 10.1155/2009/904589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J., Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Zhao J.J., Gjoerup O.V., Subramanian R.R., Cheng Y., Chen W., Roberts T.M., Hahn W.C. Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell. 2003;3:483–495. doi: 10.1016/s1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 CD21 is dominantly expressed in myoepithelial lineage of mammary epithelial cells. (A-C) Primary epithelial cells were stained with anti-EoCAM (FITC-labeled, vertical axis), anti-CD49f (APC, horizontal axis) antibodies and anti CD21. Quadrant gates were set according to isotype controls. (D) Myoepithelial lineage is minor population of mammary epithelial cells but contains major CD21 positive population.
Fig. S2 EBV infection of MECs. (A) Primary MECs isolated from reduction mammoplasties or (B) immortalized MEC cell lines as indicated were grown in MEGM, and incubated with EBV isolated from Akata cells for 4 h. Scale bars are 100 mm.
Fig. S3 Tumors generated from HMEC-htert are triple-negative have basal-like features. Upper panel: Absence of ER, PR and Her2 with positive controls (top row). Lower panel: Expression of cytokeratins 5/6, 14, p63 and Vimentin and absence of the luminal marker cytokeratin 8.
Fig. S4 (A) Image of RTK antibody array after incubation with cell lysates as indicated and scanning of the fluorescence intensity using an Odyssee reader. Red boxes highlight the position of p-Stat3, blue boxes p-Met. (B–E) Flow Cytometric determination of CD24 and CD44 in GFP-MECs (MCF10A or HMEC as indicated) treated with either vehicle control or the met-inhibitor Cabozatinib (XL 184). This is a control experiment for the data provided in Fig. 5J.
Fig. S5 The EBV-ness signature is a marker of poor prognosis in breast cancer. The plots show application of the “EBV-ness” signature to four different datasets accessed from GEO. Displayed is disease-free survival as a function of time in months. The merged results are shown in Fig. 6.
Fig. S6 Positive controls for the in situ hybridization of EBV: A. ISH for tumor xenografts derived from EBV-HMEC cells. B ISH for tumor xenografts from GFP-HMEC. (C, D) ISH stain for beta-actin and BfXL2 without (C) and with (D) RNAse treatment. (F, G) ISH stain for GAPDH and LMP1 without (F) and with (G) RNAse treatment. The ISH probes will hybridize with single stranded RNA or DNA. Only the LMP1 and BxLF2 signals were RNAse-resistant, indicating that the signal corresponds to EBV DNA.
Fig. S7 Correlation of the EBVness score with absence or presence of one or both APOBEC signatures in breast cancer. The EBVness score developed on the Affymetrix platform was transferred to the Agilent platform gene expression array as described and a continuous EBVness score was obtained for each sample. Significance of the correlation with the APOBEC signature was calculated using the t-test.
Table S1 List of genes differentially expressed at a significance level of p ≤ 0.001 in EBV-infected MECs and EBV-derived tumors versus GFP controls, according to the strategy outlined in Fig. 6B.
Table S2 Analysis of pathways differentially activated in EBV-infected MECs versus GFP controls MECs. The Hepatocyte Growth Factor Receptor (MET) pathway, and the IL6 related pathway, in which STAT3 is involved, are differentially activated in EBV-infected samples.
Table S3 Pathways differentially regulated between EBV-related BCs and GFP-control BCs.
Table S4 Training of the “EBVness” signature. 7 EBV positive and 8 EBV negative samples were included. 37 of the 47 significantly EBV regulated genes were used in the training, because these genes were represented in bot, the HT_HG-U133 + _PM array (used by us) as well as in the HG-U133A array (archival GEO data). Using BRB Array Tools, the signature was trained on the experimental tumors and cell lines. The diagonal linear discrimination analysis was applied and the weight of each gene is shown in the table.
Table S5 Correlation of the “EBVness” signature with clinicopathologic characteristics of the tumors in individual data sets.