Abstract
Mycobacterium tuberculosis (M. tuberculosis) is considered innately resistant to β-lactam antibiotics. However, there is evidence that susceptibility to β-lactam antibiotics in combination with β–lactamase inhibitors is variable among clinical isolates, and these may present therapeutic options for drug-resistant cases. Here we report our investigation of susceptibility to β-lactam/β–lactamase inhibitor combinations among clinical isolates of M. tuberculosis, and the use of comparative genomics to understand the observed heterogeneity in susceptibility. Eighty-nine South African clinical isolates of varying first and second-line drug susceptibility patterns and two reference strains of M. tuberculosis underwent minimum inhibitory concentration (MIC) determination to two β-lactams: amoxicillin and meropenem, both alone and in combination with clavulanate, a β–lactamase inhibitor. 41/91 (45%) of tested isolates were found to be hypersusceptible to amoxicillin/clavulanate relative to reference strains, including 14/24 (58%) of multiple drug-resistant (MDR) and 22/38 (58%) of extensively drug-resistant (XDR) isolates. Genome-wide polymorphisms identified using whole-genome sequencing were used in a phylogenetically-aware linear mixed model to identify polymorphisms associated with amoxicillin/clavulanate susceptibility. Susceptibility to amoxicillin/clavulanate was over-represented among isolates within a specific clade (LAM4), in particular among XDR strains. Twelve sets of polymorphisms were identified as putative markers of amoxicillin/clavulanate susceptibility, five of which were confined solely to LAM4. Within the LAM4 clade, ‘paradoxical hypersusceptibility’ to amoxicillin/clavulanate has evolved in parallel to first and second-line drug resistance. Given the high prevalence of LAM4 among XDR TB in South Africa, our data support an expanded role for β-lactam/β-lactamase inhibitor combinations for treatment of drug-resistant M. tuberculosis.
Abbreviations: XDR, extensively drug resistant; MIC, minimum inhibitory concentration; MDR, multidrug resistant; SNP, single nucleotide polymorphism; SNV, single nucleotide variant; WGS, whole-genome sequencing
Keywords: tuberculosis, Multi-drug resistant (MDR), Extensively drug resistant (XDR), Beta-lactam antibiotics, Antimicrobial chemotherapy, pks12, Recombination
Highlights
-
•
Paradoxical hypersusceptibility is observed drug susceptibility despite innate resistance in the wild type state.
-
•
Many MDR and XDR M. tuberculosis strains are susceptible to amoxicillin/clavulanate.
-
•
Whole-genome sequencing identified mutations associated with paradoxical hypersusceptibility.
-
•
An expanded role for β-lactams in drug-resistant M. tuberculosis is supported.
The global increase in drug-resistant tuberculosis has prompted a search for alternative therapies, including repurposing existing antibiotics. β-lactam antibiotics are safe drugs, however, they have previously been thought to be of limited use for tuberculosis due to innate resistance to this drug class. In this study, the authors found many drug-resistant tuberculosis isolates from South Africa to be susceptible to a β-lactam and β-lactamase combination, amoxicillin/clavulanate. With the use of comparative genomics, multiple genetic mutations were identified to be associated with this hypersusceptible phenotype. These findings support an expanded role of β-lactam/β-lactamase inhibitor combinations for treatment of drug-resistant TB.
1. Introduction
Approximately one third of the global population is infected with Mycobacterium tuberculosis (M. tuberculosis), the etiologic agent responsible for tuberculosis (TB). While many individuals who harbour M. tuberculosis never develop active disease, TB is often lethal and is estimated to have been responsible for 1.5 million deaths in the year 2013 (WHO, 2014). Multidrug-resistant (MDR) TB, which is defined as in vitro resistance to both rifampin and isoniazid, is a public health problem of increasing importance, with an estimated 480,000 incident cases in 2013 (WHO, 2014). Treatment of drug-resistant TB is more complex, lengthy, costly, toxic, and ultimately less successful at eradicating M. tuberculosis infection. With few novel antitubercular antibiotics in development, the emergence of extensively drug-resistant (XDR) M. tuberculosis strains (defined as MDR with additional resistance to both quinolones and second-line injectable agents) and the lack of effective treatment regimens have highlighted the potential to repurpose existing antibiotics in innovative ways (Wong et al., 2013).
M. tuberculosis has been considered innately resistant to β-lactam antibiotics, due to β–lactamase activity and the presence of non-classical transpeptidases in their cell wall (Hugonnet et al., 2009, Gupta et al., 2010). M. tuberculosis possesses a highly active β-lactamase, BlaC, that rapidly hydrolyzes many β-lactam drugs, rendering them ineffective (Hugonnet et al., 2009). Additionally, the existence of non-classical transpeptidases that crosslink the peptidoglycan cell wall of M. tuberculosis are thought to contribute to innate resistance to β-lactams (Gupta et al., 2010, Dubée et al., 2012).
Despite these barriers, there may be opportunities for clinical treatment of drug-resistant M. tuberculosis with β-lactam antibiotics (Payen et al., 2012, Keener, 2014). The addition of clavulanate, an oral β-lactamase inhibitor, irreversibly inhibits BlaC, and can enhance β-lactam activity against M. tuberculosis in vitro (Wang et al., 2006, Hugonnet and Blanchard, 2007). Furthermore, the carbapenem class of β-lactams is relatively resistant to hydrolysis by β-lactamases, and addition of clavulanate has been shown to further decrease the minimum inhibitory concentration (MIC) (Hugonnet et al., 2009). Meropenem/clavulanate has been shown to have high in vitro activity against XDR strains, but its high cost and intravenous dosing present challenges to its widespread use (Hugonnet et al., 2009, Gonzalo and Drobniewski, 2013). The laboratory strain of M. tuberculosis, H37Rv, and multiple drug-susceptible clinical isolates have been shown to be resistant to β-lactams as well as β-lactams plus β-lactamase inhibitors in vitro, but it is not yet clear to what extent the wider M. tuberculosis population may be susceptible to these antibiotics. If drug-resistant strains are generally more susceptible to this antibiotic class, then one might envision an expanded role for β-lactams in the treatment of drug-resistant TB, for which currently there are limited treatment options.
Here we describe an investigation of 89 South African clinical isolates and two reference strains of M. tuberculosis of varying drug susceptibility patterns. We determined the range of MICs to clinically available β-lactam antibiotics and used whole-genome sequencing (WGS) to understand the genetic basis of variability with respect to amoxicillin/clavulanate susceptibility. Our results provide insight into the clinical role of β-lactams in the treatment of drug-resistant TB and potential molecular markers of amoxicillin/clavulanate susceptibility.
2. Methods
2.1. Clinical Isolates of M. tuberculosis
We selected a subset of 86 clinical isolates of M. tuberculosis from our larger sequenced strain set (Cohen et al., 2015) for inclusion in this study. Briefly, sputum specimens were collected in KwaZulu-Natal, South Africa from 2008 to 2012 as part of a provincial drug-resistance surveillance study (Bantubani et al., 2014) and a prospective collection effort of patients newly initiating XDR regimens (OʼDonnell et al., 2014). Biomedical Research Ethics Council (BREC) approval from the University of KwaZulu-Natal was granted for whole genome sequencing of clinical strains. On all study isolates, drug susceptibility testing by critical concentration was performed prospectively for first and second-line TB drugs. The clinical isolate set represented diverse first and second-line drug susceptibility patterns, with 18 susceptible, three mono-drug resistant, five poly-drug resistant, 23 MDR and 37 XDR isolates.
2.2. Additional M. tuberculosis Strain
In addition to 86 clinical isolates recently isolated in South Africa, we also selected five additional strains for inclusion in this study, for a total of 91 unique isolates. These included reference strains H37Rv and CDC1551, which are both known to be fully susceptible to first and second-line TB drugs. Additionally, three clinical isolates that had previously been collected in KwaZulu-Natal (Koenig, 2007, Ioerger et al., 2009) were resequenced and included in this study. These were KZN4207, KZN1435 and KZN605, which have first/second-line drug susceptibility patterns of: susceptible, MDR and XDR, respectively.
2.3. β-lactam MIC Determination
The World Health Organization (WHO) does not recommend a standard methodology for β-lactam susceptibility or resistance testing in M. tuberculosis, thus we utilized the Microplate Alamar Blue Assay (MABA) (Collins and Franzblau, 1997) to determine the MIC of each strain to β-lactams alone, or β-lactams in combination with clavulanate, a β-lactamase inhibitor, as previously described (Lun et al., 2013). The β-lactams and concentrations utilized were amoxicillin (0.5 μg/mL to 32 μg/mL) and meropenem (0.25 μg/mL to 16 μg/mL). Briefly, 104 colony forming units (CFU) of a mid to late-log culture of each strain was plated onto a 96-well microplate in the presence of serial drug-dilutions with or without 2.5 μg/mL of clavulanate. Following seven days in culture, the lowest concentration of drug leading to at least a 90% reduction of bacterial growth signal by MABA was recorded as the MIC. Each assay was performed between one to 6 times on each isolate.
2.4. β-lactam Susceptibility Definitions
Based on favourable pharmacokinetics that would be necessary for clinical treatment (White et al., 2004), susceptibility to amoxicillin was defined as an MIC ≤ 4.0 μg/mL, and resistance as an MIC > 4.0 μg/mL; susceptibility to amoxicillin/clavulanate was defined as an MIC ≤ 4.0 μg/mL/2.5 μg/mL, and resistance as an MIC > 4.0 μg/mL/2.5 μg/mL. Susceptibility to meropenem was defined as an MIC ≤ 12.0 μg/mL, and resistance as an MIC > 12.0 μg/mL; susceptibility to meropenem/clavulanate was defined as an MIC ≤ 12.0 μg/mL/2.5 μg/mL, and resistance as an MIC > 12.0 μg/mL. We note that our association analysis used the amoxicillin/clavulanate MIC values directly, without dichotomization of resistant vs. susceptible.
2.5. β-lactamase Activity Testing
A subset of 10 M. tuberculosis isolates with varying average MICs to amoxicillin/clavulanate (range 0.5 μg/mL to > 32 μg/mL of amoxicillin in combination with 2.5 μg/mL clavulanate) underwent qualitative testing of their β-lactamase activity by nitrocefin hydrolysis. Nitrocefin (Calbiochem), a chromogenic β-lactamase substrate, was reconstituted in dimethyl sulfoxide (DMSO) to achieve 1 mg/mL. 60 μL of nitrocefin was diluted into 3 mL of phosphate buffered saline (PBS) nitrocefin to create a working solution. Mid to late log cultures of M. tuberculosis were diluted in PBS, and 105 colony forming units of each strain were plated into a 96-well plate. 100 μL of the nitrocefin working solution was added. Cultures were monitored for a color change from yellow to red that occurs with nitrocefin hydrolysis.
2.6. Whole-genome Sequencing
Whole-genome sequencing had been performed on all included strains as previously described and published in Cohen et al. (Cohen et al., 2015). Briefly, mycobacterial strains were single colony selected, and genomic DNA was extracted using published methods (Larsen et al., 2007). Library preparation and whole-genome sequencing were performed on the Illumina HiSeq 2000 at the Broad Institute in Boston, MA, as previously described (Walker et al., 2014). Both short (180 bp) and large (3–5 Kbp) insert paired-end libraries were created from each sample and sequenced using a read length of 101 bp. All sequencing data were retrieved from the Sequence Read Archive NCBI umbrella BioProject identifier PRJNA183624.
2.7. In silico Spoligotyping
In vitro spoligotypes are derived by a hybridization assay which determines whether or not each of 43 probe sequences (each 25 bp in length) hybridizes against a sample of DNA (Kamerbeek et al., 1997). Here spoligotypes were determined in silico from Illumina sequence reads by extraction of 25 bp sequences and their reverse complements from all possible positions, and subsequent search across this set for a match for each probe sequence. In the in vitro assay, probes may hybridize even if the sequences do not match exactly, and thus we allowed up to two nucleotide mismatches per probe. Hybridization patterns were assigned to known M. tuberculosis clades/lineages by comparison of exact pattern matches in the international SITVIT database (Demay et al., 2012) and interrogation of the TB Insight: TB-Lineage database (Shabbeer et al., 2012). Lineage designations were converted to the standardised nomenclature as presented in Gagneux et al. (Gagneux and Small, 2007).
2.8. Single nucleotide variant calling
Illumina reads for each of 91 strains were trimmed using seqtk (https://github.com/lh3/seqtk version 1.0-r31) with a Phred quality threshold of 0.01. Trimmed reads were mapped to the H37Rv reference genome (accession NC_000962.3) using BWA (Li and Durbin, 2009). Subsequent processing by SAMTools (Li et al., 2009) removed duplications, merged reads from different sequencing runs of the same isolate and removed unpaired reads. SNPs and small indels (SNVs) were called using SNVer (Wei et al., 2011) in a monoploid mode with default parameters, with the exception that the minimum base quality threshold was increased to q20. We also enforced a minimum frequency of five reads, in each of the forward and reverse orientations, supporting each SNV call. Variants for which the maximum a-posteriori genotype estimation as computed by SNVer did not support any allele, for which depth was less than 10, strand bias was smaller than 0.05, or p > 10− 10 were excluded from subsequent analyses.
2.9. Phylogenetic Reconstruction
Bayesian inference of phylogeny as implemented in MrBayes (Ronquist et al., 2012) was performed based on substitution patterns of single nucleotides at conserved genomic sites (i.e. for which high confidence SNP calls were obtained across all isolates). Genomic regions that are known to be prone to erroneous mapping of reads, namely repeat regions, mobile elements, phage proteins, PE and PPE proteins, were excluded (Casali et al., 2012). Mycobacterium canettii CIPT 140010059 (accession: NC_015848) was used as an outgroup for phylogenetic reconstruction. A generalized time-reversible (GTR) evolutionary model with gamma distributed rate variation across sites was applied (1,000,000 Markov chain Monte Carlo runs sampled every 1000 generations after a relative burn-in of 0.25, yielding a total of 750 replicates). As association tests require a phylogeny that estimates relatedness among isolates in terms of evolutionary time (Kang et al., 2008), a molecular clock constraint was imposed on branch lengths, enforcing all the leaves to be equidistant from the root. For visualization purposes, a tree with branch lengths that reflected the number of substitutions was also inferred (Fig. 2, Fig. 3).
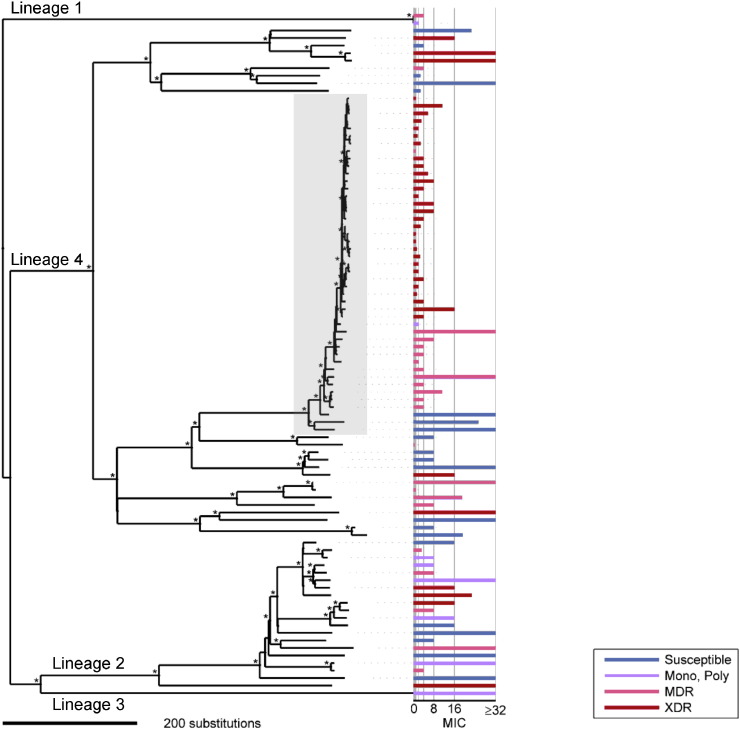
Fig. 2.
Amoxicillin/clavulanate hypersusceptible isolates were largely clustered within a monophyletic XDR group within the LAM4 clade. A Bayesian phylogeny of 91 M. tuberculosis isolates, rooted with the M. canettii outgroup, is shown. Bootstrap support values > 80% are marked by an asterisk. Globally recognised M. tuberculosis lineages are indicated. Amoxicillin/clavulanate MICs are represented in a horizontal bar graph to the right of the tree. MICs are reported by amoxicillin concentration, and clavulanate concentration was held constant at 2.5 μg/mL. As indicated in the figure key, the MIC bar graph is color-coded according to the first and second-line drug susceptibility patterns of the corresponding strain. The shaded box indicates LAM4 strains; a monophyletic cluster of spoligotype LAM4 strains with MDR and XDR level resistance is observed within lineage 4. While a significant number of isolates with hypersusceptibility to amoxicillin/clavulanate were concentrated within the LAM4 XDR clade, there are other notable examples of hypersusceptible strains that pertain to other global lineages and spoligotypes.
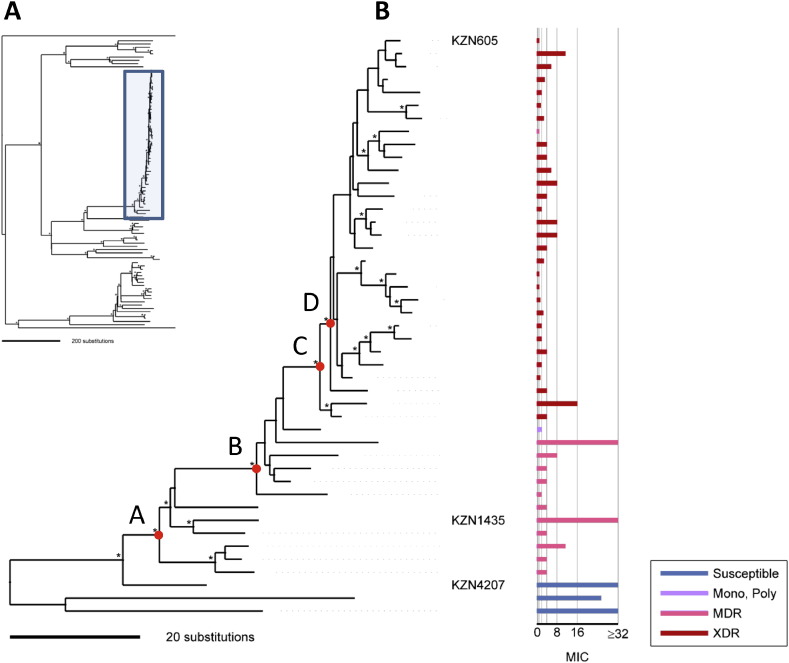
Fig. 3.
Tracing the emergence of amoxicillin/clavulanate susceptibility within the LAM4 clade. A. Bayesian phylogeny of 91 M. tuberculosis isolates as represented in Fig. 2. The LAM4 clade is marked by the blue box and is magnified in B. Nodes with bootstrap support values > 80% are marked by an asterisk. Nodes A-D define subclade divisions for which the corresponding pattern of genomic variants listed in Table 3 is conserved among all members of the subclade. Amoxicillin/clavulanate MICs (amoxicillin μg/mL, clavulanate concentration fixed at 2.5 μg/mL) are represented in a horizontal bar graph to the right of the tree. The bar graph is color-coded according to first and second-line drug susceptibility patterns as in Fig. 2. Historic LAM4 strains are identified by name: KZN605 (XDR), KZN1435 (MDR) and KZN4207 (first/second-line drug susceptible).
2.10. Gene-content Analysis
De novo sequence assemblies were annotated by Prokka (Seemann, 2014). Nucleotide sequences for all annotated coding sequence (CDS) regions were added to those annotated within 18 M. tuberculosis reference genomes retrieved from Genbank (accessions; NC_000962.3, NC_002755.2, NC_009565.1, NC_012943.1, NC_016768.1, NC_016934.1, NC_017522.1, NC_017523.1, NC_017524.1, NC_018078.1, NC_020089.1, NC_020559.1, NC_021054.1, NC_021192.1, NC_021193.1, NC_021194.1, NC_021740.1, NC_022350.1). A BLASTn all-against-all analysis was used to cluster CDS (percentage identity ≥ 90% and coverage ≥ 85%). Single representatives of each cluster were collated to form a non-redundant CDS list (preferentially selecting H37Rv reference CDS when present within a cluster). Non-H37Rv cluster representatives were screened against the H37Rv reference genome and removed if there was a BLASTn match with ≥ 90% identity and ≥ 85% coverage. (The H37Rv genome annotation has been well curated and thus we assume any sequence showing homology to regions of this genome, but which has not been annotated as a CDS in this genome, represents an erroneous annotation.) The presence of sequence regions representing each of the filtered, non-redundant CDS clusters (n = 3957) was estimated by BLASTn search against the de novo genome assemblies (percentage identity ≥ 90%, coverage ≥ 85%). Genes were considered putatively absent in cases where there was no BLAST match above the identity and coverage thresholds.
2.11. Association Tests
Genomic variants were tested for association with log MIC to amoxicillin/clavulanate. The association analysis used the amoxicillin/clavulanate MIC values directly rather than the dichotomous phenotype of resistant vs. susceptible. All SNVs for which base calls were available in all isolates were included in the analysis (including those in repeat regions, mobile elements, phage and PE/PPE genes). All genes represented by CDS in the filtered non-redundant list (see above) were also included in the analysis.
The initial sample consisted of 8580 SNVs and 8936 gene-content patterns. After excluding low frequency variants in which less than three isolates varied from all other isolates, 2555 SNVs and 228 gene-content patterns remained in the analysis. The close relatedness between many of the isolates in our sample leads to strong statistical dependencies between many pairs of variants, whose alleles have the same patterns across the sample. Such variants by definition comprise the same statistical hypothesis and result in the same p-value. We thus performed analysis of unique variant patterns, rather than of individual variants, and attached the p-value of each unique pattern to all SNVs having this pattern. Overall, 239 SNV patterns and 122 gene-content patterns (with an overlap of four patterns) were tested for association with drug resistance. FDR analysis was performed by considering the total number of tested hypotheses under both analyses.
A major challenge of genome-wide association testing is the presence of a population structure (Chen and Shapiro, 2015) in which sampled isolates are not identically and independently distributed. This can lead to spurious results due to confounding, e.g. in this case alleles that are common to a group of closely related susceptible isolates may falsely appear to be associated with ß-lactam susceptibility. To account for population structure in the setting of a quantitative phenotype (e.g. log-MIC), we applied a phylogenetically aware LMM (Kang et al., 2008). Given that M. tuberculosis is known to have a low rate of horizontal gene transfer (Liu et al., 2006), the effect of the population structure is assumed to be well-approximated by phylogeny. p-values were computed via simulation testing with one million simulations. See Supplemental Methods for more details.
3. Results
3.1. Clinical Isolates of M. tuberculosis Exhibit a Broad Range of MICs to β-lactams
We screened a set of 91 M. tuberculosis strains of varying drug susceptibility patterns by determining the MIC to β-lactam/β-lactamase inhibitor combinations (Table S1). Included clinical isolates of M. tuberculosis had been collected from 2008 to 2012 in KwaZulu-Natal, South Africa (Cohen et al., 2015). Phenotypic assessment of a strain's susceptibility to amoxicillin, amoxicillin/clavulanate, meropenem and meropenem/clavulanate was performed by the drug Microplate Alamar Blue Assay (MABA), as previously described (Collins and Franzblau, 1997, Lun et al., 2014).
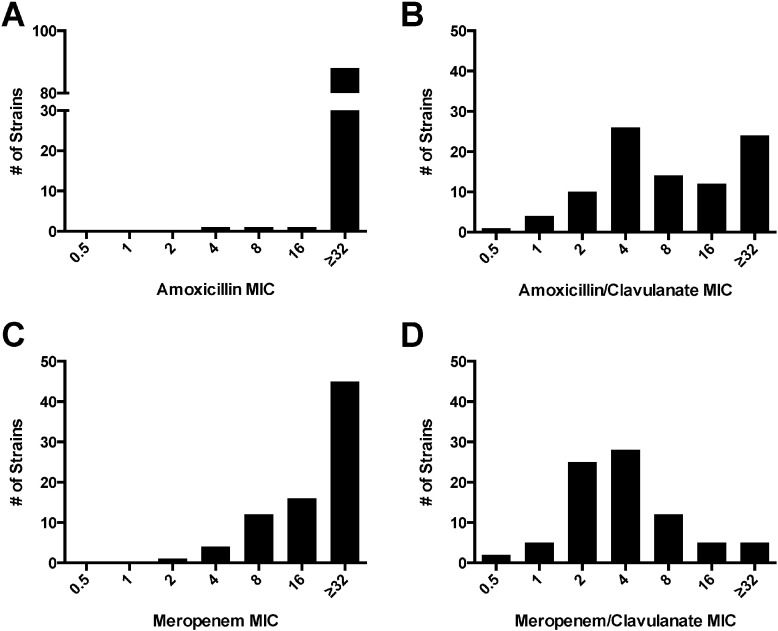
Phenotypic testing of strains revealed universally high MICs to amoxicillin by itself (Fig 1A), which confirms innate mycobacterial resistance to this antibiotic alone among these strains. However, with the addition of clavulanate the mean MIC to amoxicillin lowered from 55.4 to 15.2 (Table S2), as numerous isolates were observed to have very low MICs to amoxicillin/clavulanate (Fig. 1B). With drug combination amoxicillin/clavulanate, 41/91 (45%) isolates had an MIC ≤ 4 μg/mL/2.5 μg/mL and as such were classified as susceptible to this regimen (Table 1; Table S1). Given the wide range of MICs observed for amoxicillin/clavulanate, a significant number of M. tuberculosis isolates may be amenable to treatment with this orally active β-lactam/β-lactamase inhibitor combination.
Fig. 1A–1D.
Clinical isolates of M. tuberculosis exhibit a range of susceptibility to β-lactam/β-lactamase inhibitors. 91 isolates of M. tuberculosis underwent MIC determination to A) amoxicillin, B) amoxicillin/clavulanate, C) meropenem, and D) meropenem/clavulanate. MICs are reported by β-lactam concentration, and clavulanate concentration was held constant at 2.5 μg/mL.
Table 1.
A disproportionate number of MDR and XDR strains were susceptible to amoxicillin/clavulanate. Susceptibility to amoxicillin or amoxicillin/clavulanate was defined as MIC ≤ 4 μg/mL of the amoxicillin component. Susceptibility to meropenem or meropenem/clavulanate was defined as MIC ≤ 12 μg/mL of the meropenem component. Clavulanate was 2.5 μg/mL.
| First and second-line DST category | Amoxicillin susceptibility | Amoxicillin/Clavulanate susceptibility | Meropenem susceptibility | Meropenem/Clavulanate susceptibility |
|---|---|---|---|---|
| Susceptible | 0/21 (0%) | 3/21 (14%) | 3/16 (19%) | 14/16 (87.5%) |
| Mono-drug resistant | 0/3 (0%) | 0/3 (0%) | 0/3 (0%) | 3/3 (100%) |
| Poly-drug resistant | 0/5 (0%) | 2/5 (40%) | 1/5 (20%) | 5/5 (100%) |
| MDR | 0/24 (0%) | 14/24 (58%) | 5/18 (28%) | 19/21 (90%) |
| XDR | 1/38 (3%) | 22/38 (58%) | 11/36 (31%) | 36/37 (97%) |
| Total | 1/91 (1%) | 41/91 (45%) | 20/78 (26%) | 77/82 (94%) |
MIC determination was also performed for meropenem and meropenem/clavulanate. With meropenem alone, there was also a range of MICs observed among the tested isolates (Fig. 1C). With addition of clavulanate to meropenem (Fig. 1D), the mean observed MIC was lowered from 22.0 μg/mL for meropenem alone to 5.4 μg/mL/2.5 μg/mL for meropenem/clavulanate (Table S2). The resulting MIC's to meropenem and meropenem/clavulanate were consistent with those derived in a previous study (Hugonnet and Blanchard, 2007).
3.2. Paradoxical Hypersusceptibility to β-lactams among MDR and XDR Isolates
Interestingly, strains that had previously been classified as MDR or XDR on first and second-line TB drug susceptibility testing were more frequently associated with amoxicillin/clavulanate susceptibility than isolates that were fully susceptible to first and second-line drugs (Table 1). 14/24 (58%) MDR and 22/38 (58%) XDR isolates were classified as susceptible to amoxicillin/clavulanate, whereas only 3/21 (14%) first-line drug susceptible isolates were classified as susceptible to the same drug combination. Given that the overwhelming majority (18/21, 86%) of first-line susceptible strains including the reference laboratory strains H37Rv and CDC1551 were resistant to amoxicillin/clavulanate, it appeared paradoxical that MDR and XDR isolates would be susceptible to this drug combination. This was therefore termed “hypersusceptibility,” to indicate gain of susceptibility.
3.3. In vitro Assessment of β-lactamase Activity
To determine whether observed differences in the MIC to amoxicillin/clavulanate were due to differential β-lactamase activity, a phenotypic assessment was performed by the nitrocefin hydrolysis assay (see Section 2). A subset of 10 isolates with varying drug susceptibility patterns and a range of MICs to amoxicillin/clavulanate were selected for assessment (Table S3). All assayed strains, irrespective of drug susceptibility pattern or MIC to amoxicillin/clavulanate were able to hydrolyze nitrocefin, indicative of positive β-lactamase activity. This suggested that lack of β-lactamase activity did not account for low MICs to amoxicillin/clavulanate. These results are consistent with our observation that clavulanate addition is necessary to achieve a low MIC to amoxicillin.
3.4. Population Structure Reveals a Monophyletic Clade of LAM4 MDR and XDR Strains
We conducted a comparative genomic analysis to understand the evolution of hypersusceptibility to amoxicillin/clavulanate in our panel of clinical isolates. All 91 β-lactam phenotyped strains had whole-genome sequences available for analysis (Cohen et al., 2015). A Bayesian phylogeny (Fig. 2) was inferred from 7005 polymorphic sites identified through read-mapping and variant calling (Methods). In parallel, we performed an in silico spoligotype prediction (Methods) to contextualize the resulting population structure with respect to previously defined global M. tuberculosis lineages (Gagneux and Small, 2007, Hershberg et al., 2008) and SITVIT clades (Demay et al., 2012, Shabbeer et al., 2012). Strains represented 15 distinct shared types distributed across 11 SITVIT clades, each nesting within one of four global M. tuberculosis lineages (Table 2; Table S1).
Table 2.
Frequency and distribution of DST patterns and amoxicillin/clavulanate susceptibility determination across M. tuberculosis global lineages and SITVIT clades.
| M. tuberculosis lineage | Spoligotype designation | Total number of strains per spoligotype | First/second-line DST patterns |
Amoxicillin/clavulanate susceptibilities |
||||
|---|---|---|---|---|---|---|---|---|
| Susceptible | Mono/Poly | MDR | XDR | Susceptible | Resistant | |||
| 1 | EAI1-SOM | 2 | – | 1 (50%) | 1 (50%) | – | 2 (100%) | – |
| 2 | Beijing | 20 | 6 (30%) | 5 (25%) | 5 (25%) | 4 (20%) | 2 (10%) | 18 (90%) |
| 3 | CAS1-Kili | 1 | – | 1 (100%) | – | – | – | 1 (100%) |
| 4 | H1 | 1 | 1 (100%) | – | – | – | 1 (100%) | – |
| H37Rv | 2 | 2 (100%) | – | – | – | – | 2 (100%) | |
| LAM3 | 4 | 3 (75%) | – | – | 1 (25%) | – | 4 (100%) | |
| LAM4 | 47 | 4 (9%) | 1 (2%) | 13 (28%) | 29 (62%) | 32 (68%) | 15 (32%) | |
| S | 4 | – | – | 4 (100%) | – | 1 (25%) | 3 (75%) | |
| T1 | 3 | 2 (67%) | – | 1 (33%) | – | 1 (33%) | 2 (67%) | |
| T3 | 1 | – | – | – | 1 (100%) | – | 1 (100%) | |
| X3 | 5 | 2 (40%) | – | – | 3 (60%) | 1 (20%) | 4 (80%) | |
| UND | 1 | 1 (100%) | – | – | – | 1 (100%) | – | |
| Total | 91 | 21 (23%) | 8 (9%) | 24 (26%) | 37 (41%) | 41 (45%) | 50 (55%) | |
Lineages and SITVIT clades inferred from in silico spoligotype data (see Section 2). UND = undesignated spoligotype. First/second-line drug susceptibility (DST) results are abbreviated as follows: susceptible, mono or poly-drug resistant (Mono/Poly), multi-drug resistant (MDR) and extensively drug resistant (XDR). For amoxicillin/clavulanate susceptibility, strains were designated susceptible and resistant based on an MIC ≤ 4 μg/mL of the amoxicillin component.
There was an asymmetric distribution of MDR and XDR observed across the phylogeny (Fig. 2), as a disproportionate number of MDR and XDR isolates pertained to Lineage 4. More specifically, in silico spoligotyping revealed that these strains pertained to the LAM4 SITVIT clade and accounted for a great majority of MDR and XDR among the study isolates, with 68% and 85%, respectively. Of note, the LAM4 XDR strains clustered phylogenetically with reference strain KZN605 (Koenig, 2007, Ioerger et al., 2009), which was isolated in Tugela Ferry, KZN, South Africa in 2005 during the famed nosocomial XDR outbreak (Gandhi et al., 2006). A prior sequencing study by Cohen et al. (2015) has demonstrated that these same isolates are members of the clonal Tugela Ferry XDR epidemic in the region.
3.5. β-lactam Hypersusceptibility is Concentrated Within the Monophyletic Cluster of LAM4
While at least one strain susceptible to amoxicillin/clavulanate was observed in each of Lineages 1, 2 and 4, the vast majority (37/41 [90.2%]) of susceptible strains fell within Lineage 4 (Fig. 2 and Table 2). In fact, 32/41 (78%) of amoxicillin/clavulanate susceptible isolates pertained to the LAM4 clade, suggesting a monophyletic origin of hypersusceptibility.
In general, it can be difficult to disambiguate which genetic variants are responsible for a specific trait within a monophyletic clade as all genetic elements shared among the clade members will appear to be associated with that trait. However, the substantial phenotypic variability with respect to amoxicillin/clavulanate MIC despite limited genotypic variability in the LAM4 clade was suggestive that it may still be possible to identify key genomic variants responsible for or markers of the phenotype.
3.6. Comparative Genomic Analysis: Application of a Phylogenetically-aware Linear Mixed Model to Identify Candidate Variants
We next sought to test genomic markers of amoxicillin/clavulanate susceptibility using a phylogenetically-aware linear mixed model (LMM) (Kang et al., 2008) to identify candidate genomic variants. As our sample was dominated by a monophyletic phenotype of hypersusceptibility (Fig. 2), a phylogenetically-aware LMM was selected as this approach takes the phylogenetic structure into account and allows ranking of genomic phenotype-differentiating variants while remaining resilient to spurious variants (Chen and Shapiro, 2015). Single nucleotide polymorphisms (SNPs) and single nucleotide indels were considered together as single nucleotide variants (SNVs), and the goal of the analysis was focused on identifying genetic elements that differentiate closely related isolates that exhibit large phenotypic differences.
Rather than testing each independent SNV, variants were tested in groups defined by their pattern of representation across the set of genomes (i.e. variants shared by the same subset of genomes were grouped together). After excluding variants present in two or fewer genomes, we identified 239 distinct SNV groups. Due to the large number of tested hypotheses relative to the sample size, we controlled for the false discovery rate (FDR) rather than the family-wise error rate, using the Benjamini–Hochberg procedure (Benjamini and Hochberg, 1995). All p-values smaller than 1.68 × 10− 3 were determined to be significant at a 5% FDR, yielding 12 significant SNV groups. Of note, separately from the association analysis of SNVs, the LMM was also applied to a gene-content analysis (See Supplemental Results).
3.7. Identification of Twelve Significant Groups of Variants by LMM Analysis
The LMM analysis identified 12 significant groups of variants as highly associated with amoxicillin/clavulanate susceptibility, one of which included only a single synonymous variant (Table 3, Tables S4 and S5). These twelve groups with significant p-values comprised a total of 38 individual SNVs which included: 17 non-synonymous mutations within predicted coding sequences, 16 synonymous mutations (Table S5); four intergenic polymorphisms, and a single nucleotide deletion within a predicted pseudogene (PE_PGRS36). Coding sequences included those associated with the following functions as defined in the TubercuList database (Lew et al., 2011) or as otherwise cited: first and second-line drug resistance (rpoB, gyrA, ubiA (Safi et al., 2013)), virulence, transmembrane proteins (ctpB, drrA, Rv3921c) and cell wall synthesis (aftD, PE-PGRS genes (Brennan and Delogu, 2002) pks12 (Matsunaga and Sugita, 2012) and ubiA). As mapping and variant calling within the PE/PPE genes is notoriously difficult due to extent of sequence repeats within these genes, our findings regarding SNVs in these genes should be considered cautiously. Intergenic SNVs included one upstream of mmpL13a, a transmembrane transport protein, and one within two nucleotides of the espA transcriptional start site (Pang et al., 2013).
Table 3.
Single nucleotide variants (non-synonymous and intergenic) identified as putative genomic markers of amoxicillin/clavulanate susceptibility.
| Genomic loci | Rv number | Gene name | Effect* | Amino acid effect | # Isolates | p-Value | LAM4 sub-clade |
|---|---|---|---|---|---|---|---|
| 664929 | Rv0571c-Rv0572c | – | C ➜ A | Intergenic | 28 | 1.28E − 04 | D |
| 761161 | Rv0667 | rpoB | cTg ➜ cCg | L452P | 36 | 2.40E − 04 | B |
| 1272321 | Rv1144-Rv1145 | − mmpL13a | C ➜ A | Intergenic | 36 | 2.40E − 04 | B |
| 2,300,674 | Rv2048c | pks12 | Aac ➜ Gac | N2105D | 36 | 2.40E − 04 | B |
| 2300676 | Rv2048c | pks12 | tAc ➜ tTc | Y2104F | 36 | 2.40E − 04 | B |
| 3889150 | Rv3471c | – | gaC ➜ gaA | D64E | 36 | 2.40E − 04 | B |
| 761110 | Rv0667 | rpoB | gAc ➜ gGc | D435G | 30 | 2.88E − 04 | C |
| 763123 | Rv0667 | rpoB | aTc ➜ aCc | I1106T | 30 | 2.88E − 04 | C |
| 2246032 | Rv2000 | – | cTg ➜ cCg | L275P | 30 | 2.88E − 04 | C |
| 4056430 | Rv3616c-Rv3617 | espA-ephA | T ➜ C | Intergenic | 30 | 2.88E − 04 | C |
| 1212626 | Rv1087 | PE_PGRS21 | gAc ➜ gCc | D356A | 36 | 3.04E − 04 | – |
| 7570 | Rv0006 | gyrA | gCg ➜ gTg | A90 V | 32 | 4.96E − 04 | – |
| 4269271 | Rv3806c | ubiA | gTg ➜ gCg | V188 A | 31 | 6.08E − 04 | – |
| 1532777 | Rv1361c | PPE19 | cAg ➜ cGg | Q286R | 45 | 1.10E − 03 | – |
| 122107 | Rv0103c | ctpB | Ggt ➜ Agt | G23S | 42 | 1.17E − 03 | A |
| 3272997 | Rv2936 | drrA | Agg ➜ Ggg | R262G | 42 | 1.17E − 03 | A |
| 283614 | Rv0236c | aftD | Agc ➜ Ggc | S1080G | 8 | 1.25E − 03 | – |
| 340372 | Rv0280 | PPE3 | Tcg ➜ Ccg | S337P | 8 | 1.25E − 03 | – |
| 2357269 | Rv2098c | PE_PGRS36 (pseudogene) | G➜ | Deletion | 8 | 1.25E − 03 | – |
| 3462135 | Rv3093c | – | tgC ➜ tgG | C210W | 8 | 1.25E − 03 | – |
| 3942640 | Rv3512 | PE_PGRS56 | aTt ➜ aCt | I306T | 8 | 1.25E − 03 | – |
| 2074509 | Rv1829-Rv1830 | – | C ➜ G | Intergenic | 45 | 1.40E − 03 | – |
Genomic loci as defined in the H37Rv reference genome. For single nucleotide polymorphisms ‘Effect’ indicates the base differences (base in reference ➜ alternative base), polymorphisms within coding sequence regions are shown as capital letters in the context of the codons to which they belong (other positions shown in lower case). # Isolates indicates the number of M. tuberculosis strains that contained the corresponding polymorphism. LAM4 sub-clade indicates the sub-clade for which the SNP in question is both unique to and conserved across the sub-clade. All SNVs having the same p-value represent the same evolutionary pattern. Of note, our analyses did not identify any associations between amoxicillin/clavulanate susceptibility and variation in genes previously implicated for this phenotype e.g. mycobacterial l,d-transpeptidases, carboxypeptidases, penicillin-binding proteins and the BlaC beta-lactamase among others (Dubée et al., 2012, Datta et al., 2006, Kumar et al., 2012, Flores et al., 2005, Bhakta and Basu, 2002, Danilchanka et al., 2008, Dinesh et al., 2013, Fukuda et al., 2013, McDonough et al., 2005) (Table S6).
Genomic loci as defined in the H37Rv reference genome. For single nucleotide polymorphisms ‘Effect’ indicates the base differences (base in reference ➜ alternative base), polymorphisms within coding sequence regions are shown as capital letters in the context of the codons to which they belong (other positions shown in lower case). # Isolates indicates the number of M. tuberculosis strains that contained the corresponding polymorphism. LAM4 sub-clade indicates the sub-clade for which the SNP in question is both unique to and conserved across the sub-clade. All SNVs having the same p-value represent the same evolutionary pattern. Of note, our analyses did not identify any associations between amoxicillin/clavulanate susceptibility and variation in genes previously implicated for this phenotype e.g. mycobacterial l,d-transpeptidases, carboxypeptidases, penicillin-binding proteins and the BlaC beta-lactamase among others (Dubée et al., 2012, Datta et al., 2006, Kumar et al., 2012, Flores et al., 2005, Bhakta and Basu, 2002, Danilchanka et al., 2008, Dinesh et al., 2013, Fukuda et al., 2013, McDonough et al., 2005) (Table S6).
3.8. Clustering of Variants Within pks12 Likely Due to Recombination
Interestingly, eight of the identified variants were located within a 34 bp region of the 5′ pks12, which included two non-synonymous and six synonymous mutations. pks12 encodes a polyketide synthetase required for dimycocerosyl phthiocerol production, a major cell wall lipid that has been implicated in innate resistance to macrolides in M. avium (Philalay et al., 2004) and described in other M. tuberculosis drug-resistant strains (Farhat et al., 2013). This pattern of spatially clustered polymorphisms, previously observed in KZN-R506 (Ioerger et al., 2009), is highly unexpected, as TB has a low mutation rate. The longest open reading frame in M. tuberculosis, pks12 spans 12,456 nucleotides and is composed of two near identical sequences (Fig. S1). Eight of the SNVs were clustered at 5′ Beta-ketoacyl synthase (bks) domain, after which the surrounding DNA sequence is identical to the corresponding sequence at the 3′ bks domain. Taken together, these findings suggest a recombination event had occurred rather than multiple independent point mutations (Feil et al., 2000). pks12 has been identified by prior association studies in M. tuberculosis (Farhat et al., 2013); however, given the length of the gene, its association with amoxicillin/clavulanate susceptibility may well be spurious.
3.9. Amoxicillin/Clavulanate Susceptibility Within the LAM4 Phylogeny
In order to track the co-evolution of amoxicillin/clavulanate hypersusceptibility and first and second-line drug-resistance within LAM4, we investigated the significant SNVs in relationship to the LAM4 phylogeny (Fig. 3). A key observation was that 21/38 (55%) of the variants identified in our analyses were grouped into only four aggregates (indicated as A to D in Fig. 3), and each associated with a distinct division in the LAM4 sub-clade (Table 3). Division B coincided with rpoB L452, which confers rifampicin resistance and emergence of MDR level drug-resistance. This analysis suggests that within the LAM4 subclade, MDR level-resistance and amoxicillin/clavulanate susceptibility appear to have emerged in tandem.
All the SNVs associated with divisions A–D occurred solely within the LAM4 clade, thus further analysis in other populations will be required to determine whether these SNVs represent markers of amoxicillin/clavulanate susceptibility among the broader M. tuberculosis population.
4. Discussion
The global increase in MDR and XDR TB has prompted investigators to search for alternative treatment options, including repurposing of existing antibiotic therapies (Wong et al., 2013). While M. tuberculosis has traditionally been considered resistant to the β-lactams, there is emerging evidence that meropenem/clavulanate can have anti-tubercular activity relevant to the treatment of XDR-TB (Hugonnet et al., 2009). However, the extent to which meropenem/clavulanate or other β-lactam/β-lactamase inhibitor combinations are effective against the wider M. tuberculosis population is unclear.
We performed a phenotypic screen of 91 M. tuberculosis strains (Cohen et al., 2015) in order to determine the range of susceptibility to amoxicillin, amoxicillin/clavulanate, meropenem and meropenem/clavulanate. As expected, nearly all tested strains (94%) were susceptible to meropenem/clavulanate. Surprisingly, 45% of strains were also susceptible to amoxicillin/clavulanate, including a large proportion of MDR and XDR strains (58% each). As meropenem/clavulanate is intravenous, costly, and has multiple potential side effects (Linden, 2007), we chose to focus our investigation and discussion on amoxicillin/clavulanate, which is an inexpensive, oral, and globally accessible drug combination with an excellent safety profile (Salvo et al., 2009).
We sought to further investigate the phenomenon of amoxicillin/clavulanate susceptibility using comparative genomics. A phylogenetic analysis indicated that a large proportion of MDR and XDR isolates in our dataset were part of a LAM4 clade, which was responsible for the famed Tugela Ferry XDR outbreak in KwaZulu-Natal in 2005 (Gandhi et al., 2006), and still accounts for a considerable burden of XDR (Chihota et al., 2012, Müller et al., 2013, Gandhi et al., 2014). Additionally, a large proportion of amoxicillin/clavulanate susceptible isolates were also members of this clade, raising the possibility that this drug combination may be of benefit for individuals harboring these strains.
The pharmacodynamics of co-amoxiclav in the treatment of M. tuberculosis have not be formally studied. However for most bacteria the time above MIC (T > MIC) is considered the principal determinant of efficacy (Turnidge, 1998). The slow growth of M. tuberculosis means an extended T > MIC is likely to be critical for this organism. Despite this multiple and high dosing of co-amoxiclav could achieve drug levels expected to be active against M. tuberculosis strains with an MIC of ≤ 4 μg/mL/2.5 μg/mL amoxicillin/clavulanic acid (Chierakul et al., 2006).
Next we identified putative genomic markers of amoxicillin/clavulanate susceptibility through application of a phylogenetically-aware linear mixed model (LMM), which is suitable for samples wherein there is a large phenotypic variability within a group of closely related isolates (Supplementary material). The LMM identified 12 candidate variant patterns, comprising 38 unique variants (Table 3, Table S5). It should be remembered that despite the use of a phylogenetically-aware LMM, association analyses cannot separate the effects of genomic variants that show linkage disequilibrium (i.e. those that represent the same pattern), a factor which must be considered when interpreting our results. Consequently, we do not infer causality, but rather suggest that the variants identified here represent genomic markers of amoxicillin/clavulanate susceptibility, which warrant further investigation.
Although our analyses did identify a number of interesting variants, we did not identify any associations between amoxicillin/clavulanate susceptibility and variation in genes previously implicated for this phenotype e.g. mycobacterial l,d-transpeptidases, carboxypeptidases, penicillin-binding proteins (Dubée et al., 2012, Datta et al., 2006, Kumar et al., 2012, Flores et al., 2005, Bhakta and Basu, 2002, Danilchanka et al., 2008, Dinesh et al., 2013, Fukuda et al., 2013, McDonough et al., 2005). Furthermore, we found no evidence that altered β-lactamase genetics or activity accounts for observed differences in amoxicillin/clavulanate susceptibility. All 91 strains had a wild type blaC, the gene that encodes for the primary β-lactamase of M. tuberculosis. All strains in which β-lactamase activity was assessed—including strains with low MICs to amoxicillin/clavulanate—demonstrated the ability to hydrolyze nitrocefin, a β-lactam. Despite these findings, it is still conceivable that differences in BlaC expression may account for the observed range of β-lactam susceptibility, a hypothesis that will need to be tested in future experiments.
For M. tuberculosis, first and second-line drug susceptibility conforms to a classic model of drug resistance: wild type M. tuberculosis is susceptible to first and second-line antitubercular drugs (e.g. isoniazid, rifampin, ofloxacin, kanamycin etc.), and exposure to these same agents selects for resistant organisms. In contrast, our documentation of M. tuberculosis susceptibility to amoxicillin/clavulanate does not fit this classical model. Drug susceptible reference strains of M. tuberculosis and clinical isolates in basal phylogenetic groups (Fig. 2, Fig. 3) had innate resistance to amoxicillin/clavulanate, yet clinical isolates were observed to be susceptible, suggesting that they had acquired susceptibility to these agents. To distinguish this phenomenon from the classic model of drug resistance, we have named this concept “paradoxical hypersusceptibility,” which we define as phenotypic drug susceptibility observed in an organism in which its wild type state has innate resistance to that respective drug.
Further study will be necessary to fully elucidate the epidemiology of in vitro and in vivo susceptibility to β-lactam/β-lactamase inhibitor combinations globally and in other lineages of M. tuberculosis. The association between MDR and XDR clinical isolates and in vitro susceptibility to amoxicillin/clavulanate may provide effective treatment options for patients with XDR-TB, at least in regions of high LAM4 prevalence such as South Africa. However a recent study from Japan (Horita et al., 2014) suggests that drug-resistant strains with other genetic backgrounds also exhibit a similar phenotype. While the South African National TB program includes amoxicillin/clavulanate as part of a suggested drug regimen for XDR (Mangement of Drug-resistant Tuberculosis Policy Guidelines, 2012), only 61% of patients in a recently published long-term cohort of XDR in South Africa were prescribed this drug combination for at least some part of their treatment course (Pietersen et al., 2014). Given the poor treatment outcomes for XDR that have been reported, the fact that β-lactam drug susceptibility testing is not routinely performed, and relative safety of the drug, empiric addition of amoxicillin/clavulanate to patients with XDR-TB in South Africa may be advisable, although the optimal dosing regimen still needs to be established.
The following are the supplementary data related to this article.
Supplemental Methods and Results.
Study isolates are listed with sequencing identifiers, first/second-line DST pattern, MICs to amoxicillin, amoxicillin/clavulanate, meropenem and meropenem/clavulanate in addition to resistance or susceptibility classification to the respective antibiotics. “ND” indicates that phenotyping to that antibiotic was not performed for that isolate. Global M. tuberculosis lineage, SITVIT designation and spoligotype for each isolate were inferred in silico.
Association testing results.
List of candidate genes identified via a literature search that may be hypothesized to be associated with β-lactam susceptibility.
Author Contributions
KAC conceived of the project, designed the research, conducted experiments, analyzed data, and wrote the paper.
TEH contributed analytic tools, designed research, conducted experiments, analyzed data, and wrote the paper.
KLW designed research, conducted experiments, analyzed data, and wrote the paper.
OW contributed analytic tools, designed research, conducted experiments, analyzed data, and wrote the paper.
VM conducted experiments.
CY designed research, conducted experiments, analyzed data, and wrote the paper.
OS designed research, conducted experiments, and analyzed data.
RA designed research, conducted experiments, and analyzed data.
TC designed research, conducted experiments, and analyzed data.
YG designed research.
WRB designed the research.
ASP designed the research and wrote the paper.
All authors reviewed the manuscript.
Funding Information
KAC was supported by T32HL007633 of the National Heart, Lung and Blood Institute. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflicts of Interest
None declared.
Acknowledgements
We thank Eyal Privman for helpful discussions. We thank Anna Trigos for assistance with the gene-content analysis. We would like to thank Nonkqubela Bantubani of the Medical Research Council (MRC) in Durban, in addition to Drs. Max O′Donnell and Nesri Padayatchi for contributing clinical isolates of M. tuberculosis, which were used in this study. Lastly, we would like to thank Prof. Koleka P. Mlisana and Dr. Nomonde R. Mvelase of the KwaZulu-Natal National Health Laboratory Service and Dr. Gyanu Lamichhane for providing nitrocefin.
Contributor Information
Keira A. Cohen, Email: kacohen@partners.org.
Alexander S. Pym, Email: alex.pym@k-rith.org.
References
- Bantubani N., Kabera G., Connolly C. High rates of potentially infectious tuberculosis and multidrug-resistant tuberculosis (MDR-TB) among hospital inpatients in KwaZulu Natal, South Africa indicate risk of nosocomial transmission. PLoS One. 2014;9:e90868. doi: 10.1371/journal.pone.0090868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing on JSTOR. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- Bhakta S., Basu J. Overexpression, purification and biochemical characterization of a class A high-molecular-mass penicillin-binding protein (PBP), PBP1* and its soluble derivative from Mycobacterium tuberculosis. Biochem. J. 2002;361:635–639. doi: 10.1042/0264-6021:3610635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan M.J., Delogu G. The PE multigene family: a ‘molecular mantra’ for mycobacteria. Trends Microbiol. 2002;10:246–249. doi: 10.1016/s0966-842x(02)02335-1. [DOI] [PubMed] [Google Scholar]
- Casali N., Nikolayevskyy V., Balabanova Y. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 2012;22:735–745. doi: 10.1101/gr.128678.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.E., Shapiro B.J. The advent of genome-wide association studies for bacteria. Curr. Opin. Microbiol. 2015;25:17–24. doi: 10.1016/j.mib.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Chierakul W., Wangboonskul J., Singtoroj T. Pharmacokinetic and pharmacodynamic assessment of co-amoxiclav in the treatment of melioidosis. J. Antimicrob. Chemother. 2006;58:1215–1220. doi: 10.1093/jac/dkl389. [DOI] [PubMed] [Google Scholar]
- Chihota V.N., Müller B., Mlambo C.K. Population structure of multi- and extensively drug-resistant Mycobacterium tuberculosis strains in South Africa. J. Clin. Microbiol. 2012;50:995–1002. doi: 10.1128/JCM.05832-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen K.A., Abeel T., Manson McGuire A. Evolution of extensively drug-resistant tuberculosis over four decades: whole genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from KwaZulu-Natal. PLoS Med. 2015;12:e1001880. doi: 10.1371/journal.pmed.1001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins L., Franzblau S.G. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchanka O., Mailaender C., Niederweis M. Identification of a novel multidrug efflux pump of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2008;52:2503–2511. doi: 10.1128/AAC.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta P., Dasgupta A., Singh A.K., Mukherjee P., Kundu M., Basu J. Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation and mediates the formation of a trimeric complex involving FtsZ, FtsW and PBP3 in mycobacteria. Mol. Microbiol. 2006;62:1655–1673. doi: 10.1111/j.1365-2958.2006.05491.x. [DOI] [PubMed] [Google Scholar]
- Demay C., Liens B., Burguière T. SITVITWEB–a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect. Genet. Evol. 2012;12:755–766. doi: 10.1016/j.meegid.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Dinesh N., Sharma S., Balganesh M. Involvement of efflux pumps in the resistance to peptidoglycan synthesis inhibitors in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2013 doi: 10.1128/AAC.01957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubée V., Triboulet S., Mainardi J.-L. Inactivation of Mycobacterium tuberculosisl,d-transpeptidase LdtMt₁ by carbapenems and cephalosporins. Antimicrob. Agents Chemother. 2012;56:4189–4195. doi: 10.1128/AAC.00665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat M.R., Shapiro B.J., Kieser K.J. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet. 2013;45:1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil E.J., Smith J.M., Enright M.C., Spratt B.G. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics. 2000;154:1439–1450. doi: 10.1093/genetics/154.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores A.R., Parsons L.M., Pavelka M.S. Characterization of novel Mycobacterium tuberculosis and Mycobacterium smegmatis mutants hypersusceptible to beta-lactam antibiotics. J. Bacteriol. 2005;187:1892–1900. doi: 10.1128/JB.187.6.1892-1900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T., Matsumura T., Ato M. Critical roles for lipomannan and lipoarabinomannan in cell wall integrity of mycobacteria and pathogenesis of tuberculosis. MBio. 2013;4 doi: 10.1128/mBio.00472-12. (e00472–12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux S., Small P.M. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 2007;7:328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- Gandhi N.R., Moll A., Sturm A.W. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- Gandhi N.R., Brust J.C.M., Moodley P. Minimal diversity of drug-resistant Mycobacterium tuberculosis strains, South Africa. Emerg. Infect. Dis. 2014;20:394–401. doi: 10.3201/eid2003.131083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo X., Drobniewski F. Is there a place for β-lactams in the treatment of multidrug-resistant/extensively drug-resistant tuberculosis? Synergy between meropenem and amoxicillin/clavulanate. J. Antimicrob. Chemother. 2013;68:366–369. doi: 10.1093/jac/dks395. [DOI] [PubMed] [Google Scholar]
- Gupta R., Lavollay M., Mainardi J.-L., Arthur M., Bishai W.R., Lamichhane G. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat. Med. 2010;16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R., Lipatov M., Small P.M. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horita Y., Maeda S., Kazumi Y., Doi N. In vitro susceptibility of Mycobacterium tuberculosis isolates to an oral carbapenem alone or in combination with lactamase inhibitors. Antimicrob. Agents Chemother. 2014;58:7010–7014. doi: 10.1128/AAC.03539-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet J.-E., Blanchard J.S. Irreversible inhibition of the Mycobacterium tuberculosis beta-lactamase by clavulanate. Biochemistry. 2007;46:11998–12004. doi: 10.1021/bi701506h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet J., Tremblay L.W., Boshoff H.I., Barry C.E., Blanchard J.S. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science. 2009;323:1215–1218. doi: 10.1126/science.1167498. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger T.R., Koo S., No E.-G. Genome analysis of multi- and extensively-drug-resistant tuberculosis from KwaZulu-Natal, South Africa. PLoS One. 2009;4:e7778. doi: 10.1371/journal.pone.0007778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerbeek J., Schouls L., Kolk A. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.M., Zaitlen N.A., Wade C.M. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener A.B. Oldie but goodie: repurposing penicillin for tuberculosis. Nat. Med. 2014;20:976–978. doi: 10.1038/nm0914-976. [DOI] [PubMed] [Google Scholar]
- Koenig R. Tuberculosis. Few mutations divide some drug-resistant TB strains. Science. 2007;318:901–902. doi: 10.1126/science.318.5852.901a. [DOI] [PubMed] [Google Scholar]
- Kumar P., Arora K., Lloyd J.R. Meropenem inhibits d,d-carboxypeptidase activity in Mycobacterium tuberculosis. Mol. Microbiol. 2012:1–15. doi: 10.1111/j.1365-2958.2012.08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M.H., Biermann K., Tandberg S., Hsu T., Jacobs W.R. Genetic manipulation of Mycobacterium tuberculosis. Curr. Protoc. Microbiol. 2007 doi: 10.1002/9780471729259.mc10a02s6. (Chapter 10: Unit 10A.2) [DOI] [PubMed] [Google Scholar]
- Lew J.M., Kapopoulou A., Jones L.M., Cole S.T. TubercuList–10 years after. Tuberculosis (Edinb.) 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden P. Safety profile of meropenem: an updated review of over 6000 patients treated with meropenem. Drug Saf. 2007;30:657–668. doi: 10.2165/00002018-200730080-00002. [DOI] [PubMed] [Google Scholar]
- Liu X., Gutacker M.M., Musser J.M., Fu Y.-X. Evidence for recombination in Mycobacterium tuberculosis. J. Bacteriol. 2006;188:8169–8177. doi: 10.1128/JB.01062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun S., Guo H., Onajole O.K. Indoleamides are active against drug-resistant Mycobacterium tuberculosis. Nat. Commun. 2013;4:2907. doi: 10.1038/ncomms3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun S., Miranda D., Kubler A. Synthetic lethality reveals mechanisms of Mycobacterium tuberculosis resistance to β-lactams. MBio. 2014;5 doi: 10.1128/mBio.01767-14. (e01767–14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangement of Drug-resistant Tuberculosis Policy Guidelines. 2012. Department of Heatlh Republic of South Africa. [Google Scholar]
- Matsunaga I., Sugita M. Mycoketide: a CD1c-presented antigen with important implications in mycobacterial infection. Clin. Dev. Immunol. 2012;2012 doi: 10.1155/2012/981821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough J.A., Hacker K.E., Flores A.R., Pavelka M.S., Braunstein M. The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial β-lactamases. J. Bacteriol. 2005;187:7667–7679. doi: 10.1128/JB.187.22.7667-7679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B., Chihota V.N., Pillay M. Programmatically selected multidrug-resistant strains drive the emergence of extensively drug-resistant tuberculosis in South Africa. PLoS One. 2013;8:e70919. doi: 10.1371/journal.pone.0070919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OʼDonnell M.R., Wolf A., Werner L., Horsburgh C.R., Padayatchi N. Adherence in the treatment of patients with extensively drug-resistant tuberculosis and HIV in South Africa: a prospective cohort study. J. Acquir. Immune Defic. Syndr. 2014;67:22–29. doi: 10.1097/QAI.0000000000000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X., Samten B., Cao G. MprAB regulates the espA operon in Mycobacterium tuberculosis and modulates ESX-1 function and host cytokine response. J. Bacteriol. 2013;195:66–75. doi: 10.1128/JB.01067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen M.C., De Wit S., Martin C. Clinical use of the meropenem-clavulanate combination for extensively drug-resistant tuberculosis. Int. J. Tuberc. Lung Dis. 2012;16:558–560. doi: 10.5588/ijtld.11.0414. [DOI] [PubMed] [Google Scholar]
- Philalay J.S., Palermo C.O., Hauge K.A., Rustad T.R., Cangelosi G.A. Genes required for intrinsic multidrug resistance in Mycobacterium avium. Society. 2004;48:3412–3418. doi: 10.1128/AAC.48.9.3412-3418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen E., Ignatius E., Streicher E.M. Long-term outcomes of patients with extensively drug-resistant tuberculosis in South Africa: a cohort study. Lancet. 2014;383:1230–1239. doi: 10.1016/S0140-6736(13)62675-6. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., Van Der Mark P. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safi H., Lingaraju S., Amin A. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-d-arabinose biosynthetic and utilization pathway genes. Nat. Genet. 2013;45:1190–1197. doi: 10.1038/ng.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvo F., De Sarro A., Caputi A.P., Polimeni G. Amoxicillin and amoxicillin plus clavulanate: a safety review. Expert Opin. Drug Saf. 2009;8:111–118. doi: 10.1517/14740330802527984. [DOI] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Shabbeer A., Cowan L.S., Ozcaglar C. TB-lineage: an online tool for classification and analysis of strains of Mycobacterium tuberculosis complex. Infect. Genet. Evol. 2012;12:789–797. doi: 10.1016/j.meegid.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Turnidge J.D. The pharmacodynamics of beta-lactams. Clin. Infect. Dis. 1998;27:10–22. doi: 10.1086/514622. [DOI] [PubMed] [Google Scholar]
- Walker B.J., Abeel T., Shea T. Pilon : an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Cassidy C., Sacchettini J.C. Crystal structure and activity studies of the Mycobacterium tuberculosis beta-lactamase reveal its critical role in resistance to beta-lactam antibiotics. Antimicrob. Agents Chemother. 2006;50:2762–2771. doi: 10.1128/AAC.00320-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Wang W., Hu P., Lyon G.J., Hakonarson H. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011;39:e132. doi: 10.1093/nar/gkr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A.R., Kaye C., Poupard J., Pypstra R., Woodnutt G., Wynne B. Augmentin (amoxicillin/clavulanate) in the treatment of community-acquired respiratory tract infection: a review of the continuing development of an innovative antimicrobial agent. J. Antimicrob. Chemother. 2004;53(Suppl. 1):i3–20. doi: 10.1093/jac/dkh050. [DOI] [PubMed] [Google Scholar]
- WHO . 2014. Global Tuberculosis Report. [Google Scholar]
- Wong E.B., Cohen K.A., Bishai W.R. Rising to the challenge: new therapies for tuberculosis. Trends Microbiol. 2013;21:493–501. doi: 10.1016/j.tim.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Methods and Results.
Study isolates are listed with sequencing identifiers, first/second-line DST pattern, MICs to amoxicillin, amoxicillin/clavulanate, meropenem and meropenem/clavulanate in addition to resistance or susceptibility classification to the respective antibiotics. “ND” indicates that phenotyping to that antibiotic was not performed for that isolate. Global M. tuberculosis lineage, SITVIT designation and spoligotype for each isolate were inferred in silico.
Association testing results.
List of candidate genes identified via a literature search that may be hypothesized to be associated with β-lactam susceptibility.