Abstract
Although several ADAMs (A disintegrin-like and metalloproteases) have been shown to contribute to the amyloid precursor protein (APP) metabolism, the full spectrum of metalloproteases involved in this metabolism remains to be established. Transcriptomic analyses centred on metalloprotease genes unraveled a 50% decrease in ADAM30 expression that inversely correlates with amyloid load in Alzheimer's disease brains. Accordingly, in vitro down- or up-regulation of ADAM30 expression triggered an increase/decrease in Aβ peptides levels whereas expression of a biologically inactive ADAM30 (ADAM30mut) did not affect Aβ secretion. Proteomics/cell-based experiments showed that ADAM30-dependent regulation of APP metabolism required both cathepsin D (CTSD) activation and APP sorting to lysosomes. Accordingly, in Alzheimer-like transgenic mice, neuronal ADAM30 over-expression lowered Aβ42 secretion in neuron primary cultures, soluble Aβ42 and amyloid plaque load levels in the brain and concomitantly enhanced CTSD activity and finally rescued long term potentiation alterations. Our data thus indicate that lowering ADAM30 expression may favor Aβ production, thereby contributing to Alzheimer's disease development.
Abbreviations: ADAM, A Disintegrin and Metalloproteinase Domain; APP, amyloid precursor protein; BACE, Beta-site APP cleaving enzyme 1; BSA, bovine serum albumin; CamKIIα, Ca2 +/calmodulin-dependent protein kinase II alpha; COFRADIC, combined fractional diagonal chromatography; CTSD, cathepsin D; GKAP1, G kinase-anchoring protein 1; IRS4, insulin receptor substrate 4; LTP, long term potentiation; MMP, metalloproteinase; MRI, magnetic resonance imaging; PLA, proximity ligation assay; TD2, Type 2 diabetes
Keywords: Alzheimer, APP, ADAM30, Amyloid, Metabolism, LTP
Highlights
-
•
By transcriptomic analyses, low levels of ADAM30 expression was observed in the brain of AD cases (compared with controls).
-
•
Activation of cathepsin D by ADAM30 is required to modulate the APP metabolism in vitro, i.e. decrease in Ab secretion.
-
•
In AD-like mice, neuronal ADAM30 expression led to lower soluble Aβ42, amyloid plaques and to rescue long term potentiation
The amyloid precursor protein (APP) processing is central in the etiology of Alzheimer's disease (AD). Characterizing the actors of this metabolism is thus an important challenge.
We searched for proteins involved in the APP processing and we selected the A disintegrin-like and metalloprotease 30 (ADAM30). Our results revealed an axis in APP metabolism pathway through an ADAM30-dependent cathepsin D activation. When activated following ADAM30 expression, a decrease in Aβ secretion occurred in vitro and in vivo. ADAM30 under-expression observed in AD brains and thus the impairment of the ADAM30-dependent pathway may favor Aβ peptide production and, ultimately, AD development.
1. Introduction
Alzheimer's disease is a complex, multifactorial, neurodegenerative disease. It is the leading cause of dementia in elderly people. The main pathologic features of Alzheimer's disease are neurofibrillary tangles and senile plaque formation in the brain. The latter is caused by the progressive deposition of mainly 39- to 43-amino acid amyloid β (Aβ) peptides generated by proteolytic cleavage of the amyloid precursor protein (APP). The systematic observation of changes in APP metabolism in monogenic forms of Alzheimer's disease suggested that the Aβ/APP pathway is at the heart of the disease (Hardy and Selkoe, 2002). Even though the key role of APP processing in the etiology of Alzheimer's disease has been challenged in recent years, recent genetic and GWAS studies of sporadic forms of Alzheimer's disease seem to support the importance of dysfunctional APP metabolism and Aβ peptide production/degradation in the physiopathology of Alzheimer's disease (Lambert and Amouyel, 2011, Jonsson et al., 2012, Tian et al., 2013, Young et al., 2015).
The two major Aβ peptide species (Aβx-40 and Aβx-42) are produced by the sequential endoproteolysis of APP by β-secretase and γ-secretase complexes. APP can also undergo non-amyloidogenic cleavage by α-secretase within the Aβ sequence, which thereby precludes Aβ generation. The various enzymes and protein complexes involved in these secretase activities are increasingly well characterized: β-site APP cleaving enzyme 1 (BACE1) accounts for almost all the β-secretase activity, whereas a complex that includes presenilin 1 or 2 is responsible for the γ-secretase activity (De Strooper, 2003). More recently, an additional matrixine (MMP-MT5) has been shown to contribute to APP processing by acting upstream of the β-site (Willem et al., 2015, Baranger et al., 2016). Finally other N-terminal truncated Aβ peptide species can be generated by other proteases besides BACE1 (Wang et al., 2006, Schönherr et al., 2016).
In contrast, α-secretase activity is less well characterized even if several research groups have suggested that ADAM10 and ADAM17 are the major enzymes responsible for constitutive and regulated α-secretases-mediated pathways in the brain (Lammich et al., 1999, Kuhn et al., 2010). In addition to these direct cleavages taking place on APP, several additional proteins are likely to modulate APP levels by interfering with secretase activity, APP trafficking and/or APP degradation (Vincent and Checler, 2012). This complex network of protein-protein interactions is however poorly characterized and the identification of its components should improve our understanding of APP biology and fate, and might enable the delineation of therapeutic approaches.
Given this context, we decided to focus on ADAMs and related proteins. Our study was inspired by several observations besides our knowledge of the involvement of ADAMs as α-secretases (Rosenberg, 2009): (i) ADAMs and APP are involved in many different biological processes including brain development, plasticity and repair (Yang et al., 2006), and (ii) several matrix metalloproteases (MMP-2, -3 and -9) can degrade Aβ peptides (White et al., 2006, Reitz et al., 2010, Carson and Turner, 2002). We therefore performed a multi-angle screen for new components of APP metabolism, with a focus on MMPs, ADAMs and related proteins. We postulated that differentially expressed MMPs, ADAMs and related proteins (when comparing expression in Alzheimer's disease brains and control brains) might be clues for their involvement in APP physiology.
2. Materials and Methods
Written informed consent was obtained from study participants or, for those with substantial cognitive impairment, from a caregiver, legal guardian, or other proxy and the study protocols for all populations were reviewed and approved by the appropriate Institutional review boards of each country.
All animal experiments were approved by the local animal care and use committee (Comité d'Ethique en Experimentation Animale du Nord - Pas de Calais, Lille, France).
2.1. Study Design
We postulated that uncharacterized MMPS, ADAMs or related proteins may be involved in the APP metabolism. The purpose of this study was thus to explore this possibility. Potential candidates were selected from transcriptomic analyses targeting MMPs/ADAMs expression using total RNAs extracted from the brain of AD cases and controls. The strongest variations in expression were validated in an independent sample of brains using a different technology. Potential correlation between amyloid deposition in the brain of AD cases and expression of our genes of interest were examined. This work allowed us to select ADAM30 for further exploration.
We developed ADAM30 over- or under-expression experiments in different cellular models to assess ADAM30 impact on the APP metabolism. Potential α-, β- or γ-secretase activities of ADAM30 were examined. A without a priori research for ADAM30 substrates was performed using COFRADIC experiments. Impact of ADAM30 on APP metabolism through CTSD activation was tested using pharmacological or siRNA tools. All the experiments have been made at least in triplicates and by two independent manipulators for most of them. This work allowed us to demonstrate that ADAM30 modulates the APP metabolism through CTSD activity.
The in vitro observations were finally extended to an “Alzheimer-like” transgenic mouse model specifically over-expressing ADAM30 in neurons. Primary cultures of adult neurons were used to validate the results obtained from cell lines. Measurement of soluble Aβ42 and amyloid deposition were performed to corroborate the in vitro results in the mouse brains. Electrophysiological analyses were finally performed to extend the results to neuronal activity. All the analyses were performed in a blinded fashion. Our data demonstrated that ADAM30 over-expression led to a decrease in Aβ42 secretion in primary cultures, in soluble Aβ42 and amyloid deposition in the cerebral tissue and to a rescue of LTP in the Alzheimer-like mouse brain.
2.2. Microarray Analyses
The preparation of human brain samples is described in the Supplementary experimental procedures. Total RNA was extracted with a phenol/chloroform protocol (TRIzol® reagent, Invitrogen®, USA) from frozen frontal cortex brain tissue from one hundred fourteen Alzheimer's disease samples and one hundred sixty seven control samples. The quality of the total RNA extract was assessed with an Agilent 2100 Bioanalyzer (Agilent) and the ribosomal 28S/18S RNA ratio was estimated with the system's onboard biosizing software. Twelve Alzheimer's disease cases and twelve controls were selected among the initial samples by applying the following two criteria: (i) a ribosomal 28S/18S RNA ratio of 1.0 or more; (ii) a Braak stage below 2 (for control samples) (Table S1) (Bensemain et al., 2009).
Specific oligonucleotides for one hundred thirty two open reading frames (corresponding to MMPs, ADAMs, ADAMTSs and related proteins) were designed using OLIGOMER software (Mediagen) (Supplementary experimental procedures and Table S2; Bensemain et al., 2009). After synthesis, the oligonucleotides were purified to obtain a population that was homogeneous in terms of length (Sigma-Aldrich®, Germany). All oligonucleotides were 5′-functionalized with a C6H12NH2 arm.
In order to decrease the potential influence of inter-individual variability in the control population, we compared the genetic expression of each Alzheimer's disease case with the pool of control samples (see the Supplementary experimental procedures). Complementary RNA corresponding to 10 μg of the initial mRNA extract was produced by amplification and labeled with a Cy5 or Cy3 fluorophore using a Fluorescent Linear Amplification Kit® (Agilent), according to the manufacturer's instructions. After hybridization, microarrays were analyzed as described previously (Bensemain et al., 2009, see the Supplementary experimental procedures). It is important to note that the analyses of the one hundred thirty two ADAMs, MMPs and related proteins were performed with a one thousand seven hundred seventy one gene microarray used in a previous project in our laboratory (Bensemain et al., 2009, Chapuis et al., 2009). Accordingly, the threshold for statistical significance was set to p < 10− 5 (after Bonferroni correction), in order to select genes differentially expressed in the Alzheimer's disease brain.
2.3. Quantification of ADAM17, ADAM30, ADAM33 and ADAMTS16 mRNAs
Total RNA samples (from forty two controls and fifty one Alzheimer's disease cases) were randomly selected (see Supplementary material section) from among the samples not used in the transcriptomic experiments. Levels of ADAM17, ADAM30, ADAM33 and ADAMTS16 mRNAs were quantified as described by the supplier (Quantigene®, Panomics) (Canales et al., 2006), normalized against two different housekeeping genes (β-actin and β-glucuronidase: see the Supplementary material procedures) and were expressed in arbitrary units (AU). Comparisons were performed with a Mann-Whitney non-parametric test.
2.4. Immunohistochemistry Experiments
The brains used for immunohistochemistry (IHC) experiments were obtained at autopsy (at Lille University Medical Center) from Caucasian patients suffering from Alzheimer's disease and from Caucasian controls in whom the absence of Alzheimer's disease had been confirmed neuropathologically. These samples were not used in the transcriptomic experiments. In all patient samples, the neuropathologic features of Alzheimer's disease were confirmed by IHC and Western blot analysis of Tau, Aβ and α-synuclein (Delacourte et al., 2002). Brain samples were fixed in formalin for brightfield microscopy. Paraffin sections from the anterior frontal cortex (Brodmann area 10) were processed in an automatic Benchmark-XT system (Ventana, USA). Polyclonal antibodies against a sixteen-amino acid polypeptide within the human ADAM30 protein (Table S3) were raised according to a standard protocol (with three months of immunization, Interchim®, France). Anti-ADAM30 antibody and pre-immune rabbit serum (both diluted at 1:500) were applied after sample heating and were revealed using a standard immunoperoxidase technique.
2.5. Analyses of APP Metabolism as a Function of ADAM30WT or ADAM30mut Expression
Plasmid constructions are fully described in the Supplementary experimental procedures.
SKNSH-SY5Y-APP695WT and HEK293/HEK293-APP695WT cell lines were respectively cultured in DMEM and DEM/F12 supplemented with 10% serum FCS, 2 mM l-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin and (for SKNSH-SY5Y-APP695WT only) 1% MEM NEAA (Invitrogen®, USA) at 37 °C in a humidified atmosphere with 5% CO2.
For the analysis of APP metabolism, SKNSH-SY5Y-APP695WT and HEK-APP695WT cell lines (300,000 cells per well at seeding) were transfected with pcDNA-Mock, pcDNA-ADAM30WT, pcDNA-ADAM30mut, shRNA pU6/ADAM30 vectors (500 μg) or siADAM30 (ON-TARGETplus ADAM30 siRNA, GE Dharmacon). Transient transfection was performed using an Exgen500 protocol (Fermentas®, France) in SKNSH-SY5Y-APP695WT cells and a Fugene-HD protocol (Roche Diagnostics®, Switzerland) in HEK/HEK-APP695WT cells, according to the respective manufacturers' instructions. SiADAM30 were specifically transfected using Lipofectamine RNAi Max. Of note, because of the low endogenous level of ADAM30 expression, shRNA and siRNA targeting ADAM30 were first validated in SKNSH-SY5Y stably over-expressing ADAM30 (Fig. S12).
Forty-eight hours after transfection, supernatants were collected for the quantification of APP by-products and cells were lysed to assay for expression of ADAM30WT/mut or endogenous ADAM30 by Western blotting. Aβ1–40, Aβ1–42, sAPPα and sAPPβ were measured in sandwich ELISAs (INNOTEST® β-amyloid (1–42) and β-amyloid (1–40), Innogenetics, Ghent, Belgium), and the sAPPα and sAPPβ Assay kits (IBL-International®, Germany), according to the respective manufacturers' recommendations. Three independent, duplicate experiments were carried out for each mutant and measurements were performed twice for each sample.
For the BACE1 fluorometric assay, HEK293 cells were transfected with ADAM30WT/mut cDNAs. Forty-eight hours after transfection, cells were pipette-lysed with 50 μl of Tris-HCl (10 mM, pH 7.5) and monitored for BACE1 activity, as described previously (Andrau et al., 2003). For the γ-secretase activity assay, solubilized membranes were obtained from intact HEK293 cells transfected with ADAM30WT/mut cDNAs and analyzed as described previously (Sevalle et al., 2009).
For analyses of APPΔC8, APPF690S and APPE691V metabolism, HEK293 cell lines (300,000 cells per well at seeding) were transfected with pcDNA-Mock or pcDNA-ADAM30WT (250 μg) and pcDNA-APP695WT, pcDNA-APPΔC8, pcDNA-APPF690S or pcDNA-APPE691V (250 μg). Three independent, duplicate experiments were carried out for each mutant and all measurements were performed twice for each sample.
2.6. Identification of ADAM30 Substrates
Two HEK293-APP695WT cell lines stably overexpressing ADAM30WT or ADAM30mut (HEK293-ADAM30WT and HEK293-ADAM30mut) were generated (as mentioned above, one advantage of the HEK293 cell line is the absence of endogenous ADAM30 expression; see the Supplementary experimental procedures). These HEK293 cell lines were used to perform a targeted COFRADIC analysis of N-terminal peptides in a highly complex mixture, whereas all internal peptides were disregarded. The combination of COFRADIC, SILAC and N-terminal tagging chemistries generates quantitative data on the modification status of the N termini of the proteins present in the mixture. This approach provides an overall profile of enzyme activities and substrates (Staes et al., 2011; see the Supplementary experimental procedures).
For the CTSD assay, recombinant ADAM30WT and ADAM30mut were generated by Genscript (Piscataway, USA) (see the Supplementary experimental procedures). The activity of recombinant human CTSD (R&D Systems, Lille, France) was measured via cleavage of a fluorogenic peptide substrate (Mca-PLGL-Dpa-AR-NH2 from R&D Systems). CTSD (16 μg/ml) was pre-incubated at 37 °C for 30 min before being diluted eight-fold into 50 mM Tris-HCl, pH 7.5 buffer with 300 mM NaCl. Twenty-five microliter aliquots of enzyme sample were transferred into the wells of black 96-well microplates (Corning, Amsterdam, The Netherlands) and incubated with substrate at 37 °C for 40 min. The final volume of the incubation solution was 100 μl and the final reagent concentrations were 0.5 μg/ml of CTSD and 10 μM of substrate in assay buffer (0.1 M sodium acetate and 0.2 M NaCl, pH 3.5). Fluorescence was measured using a Victor3 V1420 plate reader (Perkin Elmer, Courtaboeuf, France) fitted with a 340 nm excitation filter and a 410 nm emission filter. The fluorescence of substrate blanks was subtracted. To evaluate the effect of ADAM30WT or ADAM30mut on CTSD activity, CTSD was pre-incubated with 84 μg/ml of each protein prior to dilution and substrate addition. The observed CTSD activities were compared with that measured when CTSD was pre-incubated with 84 μg/ml of bovine serum albumin (Sigma, Lyon, France).
For measuring intracellular CTSD activity, the SKNSH-SY5Y-APP695WT and HEK-APP695WT cell lines (300,000 cells per well at seeding) were transfected with pcDNA-Mock, pcDNA-ADAM30WT or pcDNA-ADAM30mut vectors. After 48 h, cells were lysed and the time course of CTSD activity (0, 5, 10 and 15 min) was monitored using a fluorometric SensoLyte® Cathepsin D assay (AnaSpec®, USA).
For the cathepsin inhibition experiments, HEK-APP695WT cell lines were transfected for 24 h with pcDNA-Mock, pcDNA-ADAM30WT or pcDNA-ADAM30mut vectors and then exposed for 24 h to pepstatin A (10 μM; Sigma-Aldrich, Germany), leupeptin (10 μM; Sigma-Aldrich, Germany) or E64-D (1 μM; Sigma-Aldrich).
Transient Co-transfection of CTSD siRNA (37.5 pmol ~ 0.75 μg, ON-TARGETplus CTSD siRNA, GE Dharmacon) and pCDNA3.1-ADAM30 vector (1.25 μg of DNA per well) was performed using Lipofectamine® 2000 Transfection Reagent (ThermoFisher Scientific) according to the manufacturer's recommendations. After 36 h of transfection and 24 h of secretion (wash after 12 h of transfection), cells were finally lysed and supernatants were recovered. Cell extracts were analyzed by western blot to validate both ADAM30WT over-expression and CSTD under-expression.
By-products of APP metabolism were then quantified as previously described. Three independent duplicate/triplicate experiments were carried out and measurements were performed twice for each sample.
2.7. Western Blot
Antibody against CTSD was diluted at 1/5000. The primary antibodies used to detect ADAM30 were diluted at 1/1000. The antibody 6E10 was used for APP detection at 1/5000. A monoclonal mouse antibody was used to detect β-actin at 1/10000. Antibody references are indicated in Table S3. Immunoreactive complexes were revealed using the ECL™ Western Blotting kit (Amersham®). Membranes were digitized using the ChemiDoc MP System (Bio-Rad, Marnes-la-Coquette, France).
2.8. Immunofluorescence and PLAs
The SKNSH-SY5Y-APP695WT cell line was cultured on poly-l-Lys-coated glass coverslips (Lab-Tek® Chamber Slide System 2 wells, Nunc, Roskilde, Denmark) for 24 h. The cells were then transfected with pcDNA-ADAM30WT-GFP, pcDNA-ADAM30mut-GFP, pcDNA-ADAM30WT or pcDNA-ADAM30mut vectors. After 48 h, cells were fixed in PBS containing 4% paraformaldehyde for 20 min at room temperature and further permeabilized with 0.25% (v/v) Triton X-100 in PBS. After blocking in 5% (w/v) BSA, fixed materials were incubated overnight at 4 °C with primary antibodies (diluted 1/500 in PBS supplemented with 5% (w/v) BSA and 0.25% Triton X-100.). After washing, appropriate secondary antibodies were used (diluted 1/400). Primary and secondary antibody combinations are described in Supplementary Table 3. The slides were read under a confocal microscope (Leica LSM 710) at the MICPAL microscopy facility at the Pasteur Institute of Lille (Lille, France).
For the PLAs, cells were washed with PBS and then fixed in PBS containing 4% paraformaldehyde for 30 min at room temperature. Cells were permeabilized with 0.25% Triton X-100 in PBS for 10 min. After blocking in 1% bovine serum albumin (BSA), cells were incubated for 2 h at room temperature with primary antibodies diluted in 1% BSA in PBS. The anti-CTSD antibody (Abcam, catalog number ab6313-100) was diluted 1/100 and the anti-ADAM30 antibody (Genetex, catalog number GTX117694) was diluted 1/50. Cells were then washed three times in PBS. All reagents used in the PLAs were purchased from Olink Bioscience (Uppsala, Sweden). The PLAs were performed according to the manufacturer's instructions by using anti-CTSD and anti-ADAM30 as primary antibodies.
2.9. Transgenic Mouse Experiments
2.9.1. Mouse Models
Two conditional ADAM30floxstopflox and ADAM30mutfloxstopflox transgenic mice were generated within a C57Bl6N background (Taconic, Germany). In brief, a construct containing the GAGGS promoter, a LoxP-NeomycineStop-LoxP cassette and the human Adam30WT or Adam30mut gene was introduced In the Rosa26 locus. CamKIIα/Cre mice (in which Cre gene expression is driven by the CamKIIα promoter (Tsien et al., 1996) and hAPPswe,Ind mice expressing a human APP gene bearing Swedish (670/671KM-NL) and Indiana (717 V-F) mutations were obtained from The Jackson Laboratory (Mucke et al., 2000). Both mice have a C57Bl6J background. The first cross was between hAPPswe,Ind mice and CamKIIα-Cre mice, yielding APP+/−/Cre+/− mice. The second cross was between APP+/− Cre+/− mice and hADAM30floxstopflox+/− or hADAM30mutfloxstopflox+/− mice, yielding triple transgenic hADAM30WTΔstop-hAPPswe,Ind-Cre or hADAM30mutΔstop-hAPP-Cre mice expressing ADAM30 or ADAM30mut in their brains. The mice data were systematically obtained from a crossing of a hADAM30floxstopflox+/− heterozygous C57BL6N female with an APP+/− Cre+/− heterozygous C57BL6J male without any backcross. So the genetic background has been systematically controlled and the phenotypes were analyzed only at the first generation.
All the mice were genotyped by PCR using genomic DNA isolated from tail tips, according to the provider's protocols (The Jackson Laboratory; Table S4 and Fig. S13), Mice were maintained on a standard diet with ad libitum access to water in a specific pathogen-free animal facility. The mice were studied at the age of 10 months ± 1 week. At sacrifice, the brains were removed. After sagittal section, one hemibrain was used for histological studies and the other was sectioned on brain matrices to isolate the cortices and hippocampi, which were then stored at − 80 °C.
2.9.2. Aβ42 Peptide Assays
Following the validation of gene expression in the brain (see the Supplementary experimental procedures), snap-frozen hippocampi were homogenized in guanidine buffer (Johnson-Wood et al., 1997). Hippocampal levels of Aβ42 were quantified with commercially available ELISA kits (Innogenetics kit for Aβ42). The protein concentration in hippocampal extracts was determined using a Bradford assay kit (Bio-Rad Laboratories).
2.9.3. Measurements of CTSD Activity Ex Vivo
Cortex samples were homogenized with dithiothreitol-containing assay buffer from the SensoLyte 520 Cathepsin D assay kit (AnaSpec®, USA), using a high-power homogenizer. After centrifugation for 10 min at 12,000 rpm and 4 °C, supernatants were assayed for CTSD with the SensoLyte® Cathepsin D assay kit (AnaSpec®), according to the manufacturer's instructions. The CTSD activity was measured independently in two different experiments: a time-course measurement every 5 min between 0 min and 30 min and an end-point measurement at 30 min (Fig. 4). Each measurement was performed at least twice for each cortex sample and the results were normalized against the sample weight and protein concentration. At least three independent, duplicate experiments were carried out and measurements were performed twice for each sample. Comparisons were performed using a Mann-Whitney non-parametric test (for the end-point experiment) or a two-tailed t-test (for the time-course experiment).
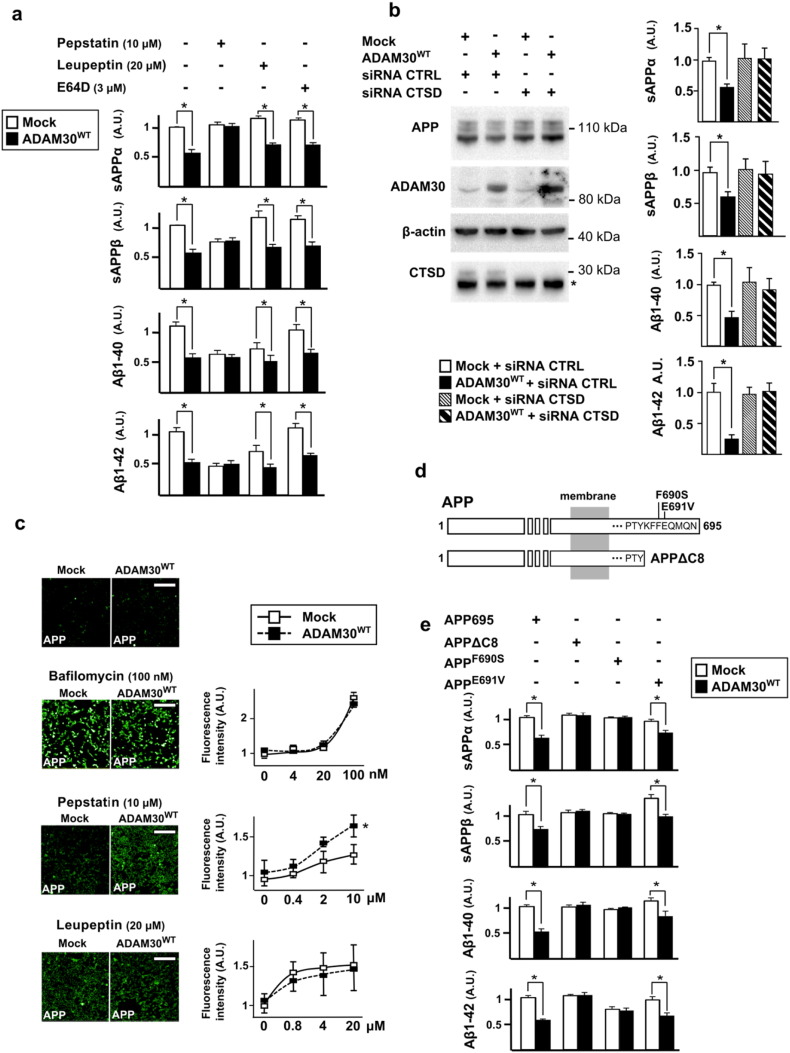
Fig. 4.
Assessment of the impact of ADAM30 on the APP metabolism through CTSD activation. (a) Impact of pepstatin, leupeptin or E64D treatment in HEK293-APP695WT cells 48 h after transfection with ADAM30WT or mock vector. Cells were treated for 24 h at the indicated concentration. Mean differences (± SEM) in the amounts of sAPPα, sAPPβ, Aβ1–40 and A-1β42 are shown. (b) Impact of CTSD inhibition by siRNA on APP metabolism according to ADAM30WT over-expression. Mean differences (± SEM) in the amounts of sAPPα, sAPPβ, Aβ1–40 and A1β-42 are shown. (c) The impact of inhibiting APP degradation in HEK293-APP695WT-ADAM30WT cells. APP degradation was blocked by 24 h of treatment with bafilomycin, pepstatin or leupeptin at the indicated concentration. Intracellular APP accumulation was assessed using immunofluorescence staining with LN27 antibody against the N-terminal part of APP. Scale bar: 100 μm. The graph represents the mean difference (± SEM) in fluorescence intensity per cell (n > 150) for the indicated concentration. (d) Schematic representation of the different mutation used in the C-ter APP. (e) The impact of ADAM30 overexpression on the metabolism of C-ter-mutated APP in HEK293 cells. Mean differences (± SEM) in the secreted amounts of sAPPα, sAPPβ, Aβ1–40, and Αβ1–42 are shown.
In all panels, three independent experiments were performed in duplicate after 24 h of transfection. *p < 0.05 (Mann-Whitney non-parametric test).
2.9.4. Histology
Hemibrains from APPsw,Ind, ADAM30WT/APPsw,Ind and ADAM30WT/APPsw,Ind/Cre mice were processed by a high-throughput neurohistological service (NeuroScience Associates). Briefly, hemibrains were embedded in a green-colored solid matrix (Multibrain technology) and cut along the rostrocaudal axis. For each brain, three sets of 30-μm-thick coronal sections (one hundred per set) were collected. One set was used for Aβ-peptide IHC experiments with a 6E10 monoclonal antibody and 3,3′-diaminobenzidine development. The other two series were stored for further analysis. Block-face photographs were taken (lateral resolution: 13 μm) before each section was prepared (Canon EOS 5D Mark III) (Dubois et al., 2007). Histology images were digitized using a flatbed scanner (ImageScanner III, GE Healthcare) with a lateral resolution of 5 μm.
2.9.5. 3D Reconstruction
All the image processing steps described below were performed using the BrainRAT pipeline 3D-HAPi (Dubois et al., 2007, Dubois et al., 2010, Vandenberghe et al., 2016) and a dedicated in-house image processing software package (BrainVISA, http://brainvisa.info). All image analyses were performed by operators blinded to the animal group assignments (Fig. S14). For each brain, block-face photographs and histological images were stacked. Brain tissue was automatically segmented on block-face photography volumes and masked to remove background. As block-face images were taken prior to sectioning at the same position section after section, reconstructed photographic volumes necessarily reflected the original shape of the frozen brains and were used as a 3D reference. Affine registrations between two-dimensional histological images and corresponding block-face photographs were estimated and used to (i) correct for deformations due to histological procedures and (ii) provide coherent histological volumes (xyz resolution: 5 × 5 × 90 μm3 for histological volumes stained with the 6E10 monoclonal antibody). For each animal, the block-face photography volume and the corresponding IHC volume stained with 6E10 monoclonal antibody were aligned within the same spatial reference framework.
2.9.6. Quantification of the Amyloid Load
Aβ peptide aggregates were segmented from the rest of the image by applying a supervised Bayesian classifier (Chubb et al., 2006) to reconstructed 6E10 IHC volumes. Each voxel was classified into either positively stained amyloid plaques, non-stained tissue or background as a function of the color intensities (red, green and blue) and the mean neighborhood intensity (the mean red, green and blue intensities within the voxel's four-connected neighbors). A mouse hemibrain atlas was derived from a publicly available, magnetic resonance imaging (MRI)-based mouse brain atlas (http://www.mouseimaging.ca/technologies/C57Bl6j_mouse_atlas.html) (Dorr et al., 2008) and registered on each photographic volume according to a protocol described and validated elsewhere (Lebenberg et al., 2010). Briefly, non-linear transformations were estimated for each brain (using MRI) and then applied to corresponding label atlas volumes. The percent volume occupied by the 6E10 staining was computed for the forebrain as a whole and the hippocampus in particular. Quality controls were performed visually for both registration and color segmentation processes. Comparisons between amyloid loads were performed with a Mann-Whitney non-parametric test.
2.9.7. Electrophysiology Experiments
Mice were shipped to E-PHY-SCIENCE (Sophia-Antipolis, France). In this study, twenty six 12-month-old transgenic mice (hADAM30WT-hAPPSw,Ind-Cre, hADAM30WT-hAPPSw,Ind, hAPPSw,Ind) and 8 WT mice were used. In total thirty four mice were scarified for the study. Of note, premature death is a phenotype of transgenic Alzheimer mice and no mice were lost in any of the groups during the course of the study. Acute slices (400 μm thick) were prepared with a vibratome (VT 1000S; Leica Microsystems, Bannockburn, IL) in ice-cold gassed aCSF enriched with sucrose. Sections were incubated in aCSF supplemented at 34 °C for 20 min and then kept at room temperature for at least 1 h before recording.
Recording was performed in a submerged chamber continuously perfused with aCSF, at 1–2 ml/min. A monopolar electrode was placed in the Schaffer collaterals, and stimulation was applied at 0.066 Hz (every 20 s) with stimulus intensity ranging from 0 to 100 μA, yielding evoked field EPSPs (fEPSPs) of 0–0.3 V.
fEPSPs were recorded in the stratum radiatum using a borosilicate micropipette filled with aCSF.
The signal was amplified with an Axopatch 200B amplifier (Molecular Devices, Union City, CA), digitized by a Digidata 1200 interface (Molecular Devices) and sampled at 10 kHz with Clampex 10 (Molecular Devices). aCSF, during incubation and recording, was composed of the following (in mM): 119 NaCl, 11 d-glucose, 1.3 MgCl2·6H2O, 1.3 NaH2PO4, 2.5 KCl, 2.5 CaCl2, 26 NaHCO3, gassed with O2/CO2 (95/5%) at least 20 min before use and throughout the experiment. Baseline was recorded for a minimum of 20 min or until stable. LTP was induced by stimulation with 100 Hz with four trains of a 1-s tetanus separated by 20 s.
Recordings were acquired using Clampex (Molecular Devices) and analyzed with Clampfit (Molecular Devices). Experimenters were blinded to treatment for all experiments. One or more slices from each mouse were used and were averaged, so that animals and not slices are considered biological replicates. Data were analyzed by measuring the slope of individual fEPSPs at 1–1.5 ms after the stimulus pulse by linear fitting using Clampfit (Molecular Devices).
3. Statistical Analyses
Statistical analysis was performed using SAS statistical software (version 9.1, SAS Institute Inc., Cary, NC, USA).
4. Results
4.1. Transcriptomic Analyses Show That ADAM30 is Under-expressed in AD Brains
Using total RNA from the frontal cortex of twelve Alzheimer's disease cases and twelve healthy controls, we performed a transcriptomic analysis of one hundred thirty two genes coding for MMPs, ADAMs and related proteins (the complete results are provided in Table S2). Four ADAMs were found to be differentially expressed in the brain of Alzheimer's disease cases compared with controls (ADAM17, ADAM33, ADAMTS16 over-expressed and ADAM30 underexpressed; p < 1 × 10− 5 for all comparisons after Bonferroni correction; see the Materials and Methods section and in Bensemain et al. (2009)). Of note, since the analyses of the 132 ADAMs, MMPs and related proteins were performed using a 2741-gene homemade microarray (see again the Materials and Methods section and in Bensemain et al. (2009), this design allowed us to determine whether the decrease in ADAM30 expression may be imputed to neuronal death. In order to evaluate this potential bias, we compared the expression of a subset of genes reported as only expressed in neuron (n = 113) to genes not exclusively (or not) expressed in neurons (n = 1789). Importantly, we did not observe any significant differences in terms of fold-change (Fig. S1). To bypass inherent risks of false positives in such systematic transcriptomic approaches despite of multiple testing corrections, the expression level of these four ADAMs was then analyzed in an independent, larger sample of AD cases (fifty one) and healthy controls (forty two). ADAM30 underexpression (Fig. 1a) and ADAM33 overexpression were confirmed whereas changes in the expression of ADAM17 and ADAMTS16 genes could not be validated in this larger sample (Fig. S2).
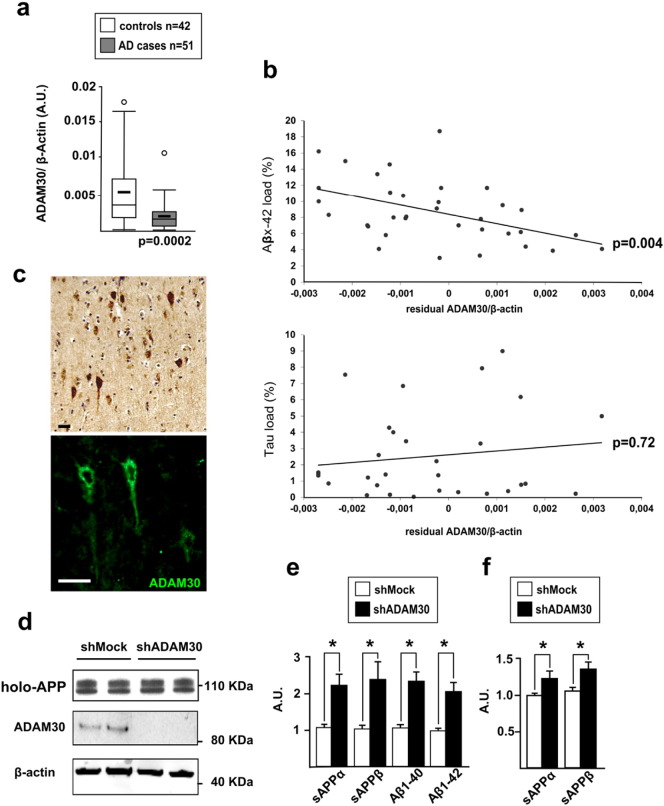
Fig. 1.
Characterization of ADAM30 expression in the brain. (a) Expression levels of ADAM30 in the brains of Alzheimer's disease cases (fifty one) and controls (forty two). All values are reported as arbitrary units (AU) following normalization against β-actin mRNA levels. All quantifications were carried out in triplicate in all individuals. The thick lines represent the median ADAM30 expression level in cases and controls. The midline represents the mean value and the upper and lower horizontal lines represent the first and third quartiles, respectively. Circles indicate individuals with extreme values (more than 2 SD above or below the mean value). p-Values refer to a Mann-Whitney non-parametric test. (b) Association of Aβx-42 and Tau loads in the brain of Alzheimer's disease cases with the expression of ADAM30 (residual correction), normalized against the expression of a-actin housekeeping gene. p values refer to Spearman's non-parametric test. (c) Immunohistochemistry experiments in human brain supporting ADAM30 expression in neurons. (d) A representative experiment measuring transfection of a shRNA vector against ADAM30 into SKNSH-5Y5Y-APP695WT cells (ADAM30, and β-actin). (e) Mean differences (± SEM) in the amounts of sAPPα, sAPPβ, A-1β40, A-1β42 in SKNSH-Sy5Y-APP695WT cells or (f) endogenous sAPPα, sAPPβ in SKNSH-SY5Y. Three independent experiments were performed in duplicate in SKNSH-5Y5Y-APP695WT and in triplicate in SKNSH-SY5Y. *p < 0.05 (Mann-Whitney non-parametric test).
We finally assessed whether the levels of ADAM30/ADAM33 expression may be correlated with AD hallmarks in the brain. The decrease in ADAM30 expression was significantly correlated with higher Aβ42 loads but not with Tau loads in Alzheimer's disease brain samples (Fig. 1b and Fig. S3) while ADAM33 expression did not correlate with these Alzheimer's disease markers (data not shown). Immunohistochemistry experiments in human brain tissue revealed a neuronal expression of ADAM30 (Fig. 1c and Fig. S4) as observed in a laser dissection transcriptomic analysis (GSE15222 dataset described in Liang et al. (2008)). We thus hypothesized that ADAM30 under-expression might be harmful by modulating Aβ peptide production and thus, we assessed the putative involvement of ADAM30 in APP processing.
4.2. ADAM30 Under-expression is Associated With Increased APP Catabolites In Vitro
We first investigated whether modulation of ADAM30 expression could be associated with an alteration of the APP metabolism in the SKNSH-SY5Y cell line stably expressing the wild-type (WT) APP695 isoform (SKNSH-SY5Y-APP695WT). This model allows measuring the production/secretion of all APP byproducts and quantifying separately Aβ1–40 and Aβ1–42 peptides. ADAM30 under-expression (transient transfection of SKNSH-SY5Y-APP695WT cells with a short hairpin RNA (shRNA) targeting ADAM30; Fig. 1d)) increased the levels of all APP products yielded by α-, β- and γ-secretases-mediated proteolysis and particularly Aβ1–40 and Aβ1–42 (Fig. 1e). Both endogenous sAPPα and sAPPβ secretion were also increased in the SKNSH-SY5Y cells after transient transfection of ADAM30-shRNA (Fig. 1f) or of an ADAM30-siRNAs (data not shown).
4.3. ADAM30 Catalytic Activity is Required for the Modulation of APP Metabolism
ADAM30 holds a unique zinc-binding motif HEXXHXXGXXHD, which is normally required for enzymatic activity (all metalloproteases harbour a HEXXH motif and half of the ADAM proteins present such a functional catalytic motif). We thus aimed at determining whether the catalytic function of ADAM30 accounts for the observed modulation of APP catabolites. To assess this possibility we generated mammalian expression vectors expressing either a wild-type ADAM30 (ADAM30WT) or an ADAM30 with a mutated catalytic site (ADAM30mut, see Supplementary information). These constructs were transfected either in the SKNSH-SY5Y-APP695WT cell line or a HEK293 cell line also stably expressing the wild-type APP695 isoform (HEK293-APP695WT).
Over-expression of ADAM30WT in both cell lines triggers the exact opposite effects observed when down-regulated ADAM30, i.e. decreased levels of all amyloidogenic and non-amyloidogenic APP catabolites (Fig. 2A and 2B). This effect was not observed after overexpression of catalytically silent ADAM30mut (Fig. 2a and b).
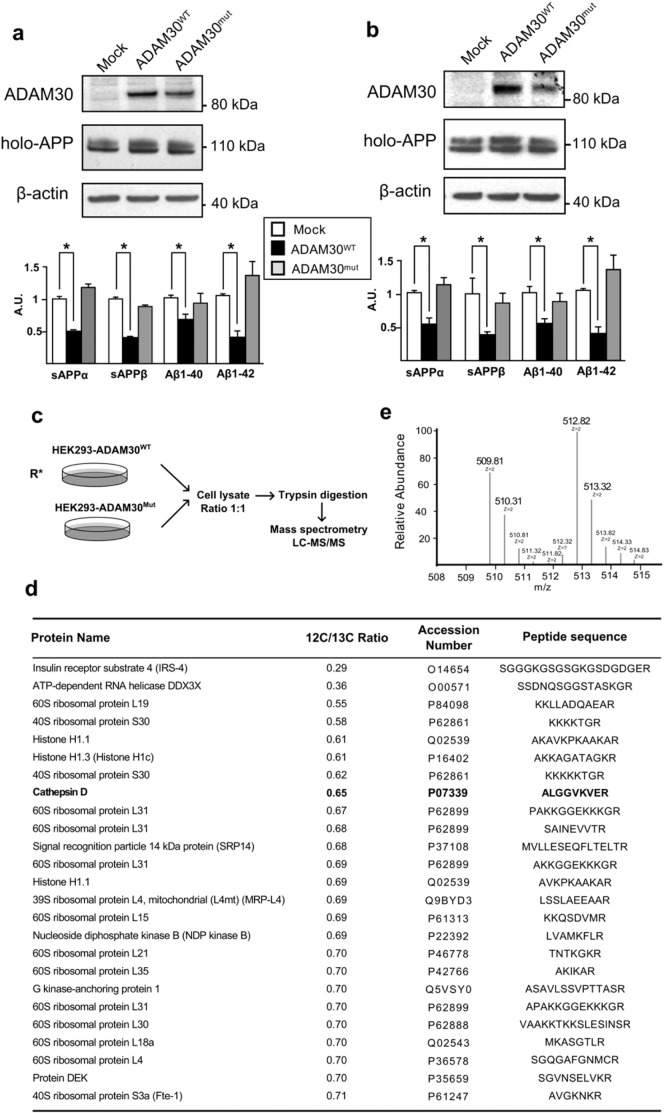
Fig. 2.
Representative experiment measuring ADAM30WT and ADAM30mut overexpression in (a) SKNSH-5Y5Y-APP695WT and (b) HEK-APP695WT and mean differences (± SEM) in the secreted amounts of sAPPα, sAPPβ, A1-β40 and A1-β42. Three independent experiments were performed in duplicate. *p < 0.05 (Mann-Whitney non-parametric test). (c) HEK293 cell lines stably over-expressing ADAM30WT or ADAM30mut and cultured them in SILAC medium supplemented with 12C- or 13C-labeled arginine, respectively. After lysis, equal amounts of proteins from the two samples were mixed, conditioned and then analyzed in a two-step HPLC process (see the Materials and Methods section). (c) Eighteen proteins (characterized by twenty five distinct peptide sequences in COFRADIC) were significantly over-represented in the HEK293-ADAM30WT cell line, relative to the HEK293-ADAM30mut cell line. (d) A representative mass spectrum of the neo N-terminal peptide of CTSD (ALGGVKVER). The peptide with mass m/z 509,81 Da originates from the 12C6 arginine-labeled cell line (mutant cell line), whereas the peptide with mass m/z 512,82 Da originates from the 13C6 arginine-labeled cell line (WT cell line).
We next started evaluating pathways that could link ADAM30 and APP catabolites. Strikingly, the fact that all APP catabolites were decreased by ADAM30 could have indicated that ADAM30 acted upstream of secretases cleavages. One possibility could be that ADAM30 directly modulates APP levels but we showed that APP mRNA levels were not affected by ADAM30 modulation (Fig. S5). An alternative explanation would be that ADAM30WT (but not ADAM30mut) modifies α-, β- and γ-secretase activities. Since several ADAMs has been proposed as genuine α-secretases, we first examined this possibility. However, ADAM30 did not exhibit α-secretase-like activity (Fig. S6a). Of note, ADAM30 is not present at the membrane (Fig. S6b, c). We also ruled out ADAM30 as a modulator of β- and γ-secretases, since overexpression of WT or mutated ADAM30 did not modify these activities (Fig. S7).
4.4. Identification of ADAM30 Substrates and Subsequent Targets
The above data suggest that ADAM30 might modulate an intracellular pathway or an intermediate effector contributing to APP physiology. Thus, we systematically searched for direct ADAM30 substrates or further downstream targets by applying N-terminal combined fractional diagonal chromatography (COFRADIC) (Staes et al., 2011) and comparing the “stable isotope labeling with amino acids in cell culture” (SILAC)-labeled N-terminones of HEK293 cells stably over-expressing either WT or catalytically inactive ADAM30 (Fig. 2b). We identified two thousand two hundred thirty eight proteins, eighteen of which (characterized by twenty five distinct peptide sequences) had neo-N-terminal peptides with higher intensities in cells over-expressing ADAM30WT, suggesting a putative susceptibility to ADAM30-mediated proteolysis (Fig. 2d). Most of these eighteen proteins were involved in transcription and translation (e.g. histones and ribosomal proteins). Even though the latter might well be substrates for ADAM30, we suspected that this observation might reflect potential differential extraction of proteins, e.g. potential differences in cell growth between HEK293-ADAM30WT and HEK293-ADAM30mut cell lines. After discarding all proteins linked to transcription and translation, only three peptides (and thus proteins) showed different ratios: insulin receptor substrate 4 (IRS4), cathepsin D (CTSD) and G kinase-anchoring protein 1 (GKAP1). We decided to focus on CTSD (see representative mass spectrum of the neo N-terminal peptide of CTSD, Fig. 2e). Indeed, the potential ADAM30 cleavage site identified in the COFRADIC experiment matched the CTSD endoproteolysis domain (residues 161–169) leading to a mature, active, lysosomal protease (Fig. 3A) (Benes et al., 2008). This latter observation suggests that ADAM30 acts as a CTSD maturation enzyme leading to enhanced cellular activity.
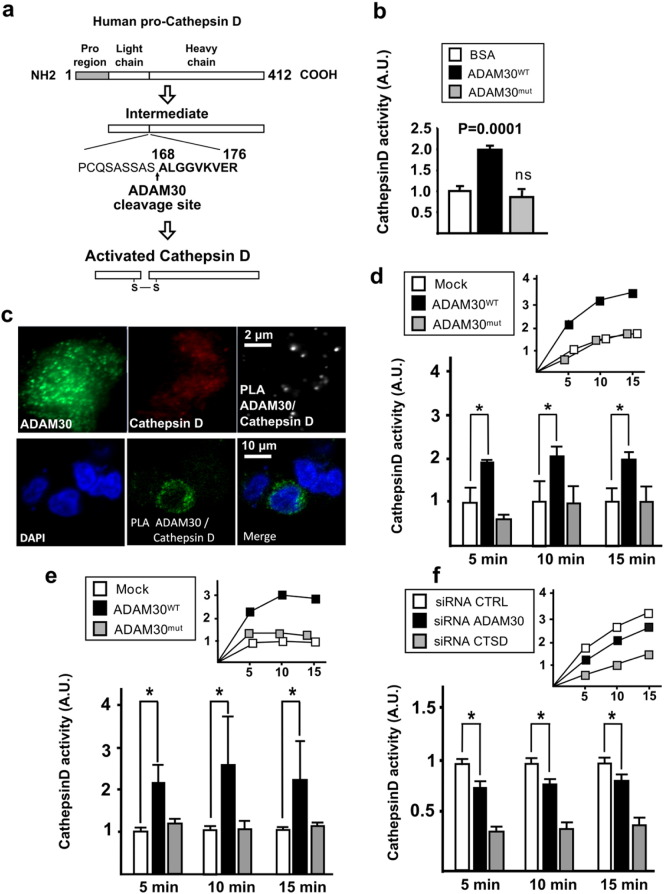
Fig. 3.
Characterization of CTSD activation by ADAM30. (a) Proteolysis of the pro-CTSD for CTSD activation. The black arrow shows the cleavage site for ADAM30 between amino acids 167 and 168 (corresponding to the activational processing of CTSD). (b) In vitro measurement of CTSD activity (mean ± SD) after incubation of recombinant BSA, ADAM30WT or ADAM30mut with a CTSD peptide substrate. (c) Proximity ligation assay with human endogenous ADAM30WT and CTSD proteins in native SKNSH-SY5Y and with overexpressed ADAM30WT and endogenous CTSD in HEK293 cells. CTSD activity in HEK293-APP695WT (d) or SKNSH-SY5Y-APP695WT (e) cells transfected with ADAM30WT, ADAM30mut or empty vector (Mock). The CTSD activity was assayed in three independent experiments, with a measurement every at 5, 10 and 15 min (means ± SEM). Top-right: representative kinetic experiment. (f) CTSD activity in SKNSH-SY5Y cells transfected with ADAM30-siRNA. The CTSD activity was assayed in three independent experiments, with a measurement every at 5, 10 and 15 min (means ± SEM). Top-right: representative kinetic experiment.
*p < 0.05 (Mann-Whitney non-parametric test). Three independent experiments were performed in duplicate.
4.5. CTSD Co-localizes With, and is Activated by ADAM30
To further investigate the hypothesis whereby ADAM30 targets CTSD, we set up a fluorescence-based assay of CTSD activity. A human, recombinant pro-CTSD was incubated with its substrate in the presence or absence of human recombinant ADAM30WT, ADAM30mut or bovine serum albumin (BSA). Incubation with ADAM30WT increased CTSD activity, whereas this effect was not detected after incubation with either ADAM30mut or BSA (Fig. 3b). In native SKNSH-SY5Y and transiently transfected HEK293-APP695WT cells, proximity ligand assay (PLA) revealed that respectively endogenous and overexpressed ADAM30WT co-localized with endogenous CTSD (Fig. 3c). Overexpression of ADAM30WT (but not ADAM30mut) was consistently associated with elevated CTSD activity in transiently transfected HEK293-APP695WT cells (Fig. 3d) or SKNSH-SY5Y-APP695WT cells (Fig. 3e). Inversely, transfection of ADAM30-siRNA led to a decrease in CTSD activity in native SKNSH-SY5Y cells (Fig. 3f). Of note, mutagenesis of ADAM30mut did not appear to alter the protein compartmentalization when compared with the main localization of ADAM30WT in late-endosome (Fig. S8).
4.6. CTSD Activity is Required for the Modulation of APP Metabolism by ADAM30
Both ADAM30's cellular localization and its ability to increase CTSD activity led us to postulate that ADAM30WT could contribute to the lysosomal CTSD maturation/activation and thereby, could favor APP degradation. This hypothesis may explain our observation of a general decrease in amyloidogenic and non amyloidogenic APP catabolites and agrees with our empirical observation that exposure to generic lysosome inhibitors (e.g. bafilomycin A1 and chloroquinone) abolished ADAM30WT-mediated impact on cellular APP metabolism (Fig. S9).
We further investigated this hypothesis by testing the impact of different pharmacological inhibitors: pepstatin (an aspartyl protease inhibitor with activity mainly directed towards CTSD), leupeptin (a serine and cysteine proteinase inhibitor with selective activity against cathepsin B) and E64D (a cysteine proteinase inhibitor that targets CTSB, cathepsin L and calpain). In the presence of pepstatin, the effect of ADAM30WT expression in HEK293 cells was no longer detected (Fig. 4a). In contrast, leupeptin (a serine and cysteine proteinase inhibitor with selective activity against CTSB) did not alter the impact of ADAM30WT expression on APP metabolism in HEK293-APP695WT cells (Fig. 4a). Similar results were obtained with E64D (Fig. 4a). Pepstatin could have elicited non-specific effect on the APP metabolism. In order to confirm the molecular cascade linking ADAM30, CTSD and APP, we finally down-regulated CTSD expression with a siRNA targeting CTSD in HEK293 cells over-expressing or not ADAM30WT. When CTSD was down-regulated, the effect of ADAM30WT expression in HEK293 cells was again no longer detected (Fig. 4b).
Altogether, these results strongly suggested that ADAM30 might be involved in APP degradation via selective activation of lysosomal CTSD. In line with our previous results, it is noteworthy that in a HEK293 cell line stably overexpressing APP695WT and ADAM30WT, exposure to pepstatin was also associated with a 51% increase in intracellular APP accumulation (relative to cells not overexpressing ADAM30WT; p < 0.01; Fig. 4c).
4.7. Lysosome Sorting of ADAM30 is Required for its Modulation of APP
We finally assessed whether the consensus sequences for lysosomal sorting in the APP C-terminal tail (Lai et al., 1995, Kouchi et al., 1998, Kouchi et al., 1999) was required for ADAM30WT impact on APP metabolism. In a first step, we mutated lysine 688 of APP695WT into a stop codon (APPΔC8) (Fig. 4d); this leads to deletion of the last eight C-terminal amino acids (the lysosome-addressing sequence YKFF, corresponding to YXXØ, where Ø is a highly hydrophobic residue) but does not modify the sequence controlling APP endocytosis. Fig. 4e shows that the transient co-expression of APPΔC8 and ADAM30WT in HEK293 cells abolished the decrease in APP catabolites associated with ADAM30WT expression. Further, we mutated phenylalanine at position 690 (APPF690S) into a serine residue within the lysosome-addressing sequence. Fig. 4e also shows that transient co-expression of APPF690S and ADAM30WT in HEK293 cells abolished the decrease in APP products associated with ADAM30WT expression. However, mutation of glutamic acid 691 (outside the lysosome-addressing sequence) into a valine (APPE691V), did not modify ADAM30 wild-type phenotype. These results thus indicate that only a mutation within the lysosome addressing sequence (APPF690S) abolished ADAM30-mediated impact on APP metabolism. Of note, we did not detect any modification on APPWT endocytosis in line with ADAM30WT over-expression (data not shown).
4.8. Overexpression of ADAM30 Modulates Aβ Production and Deposition In Vivo
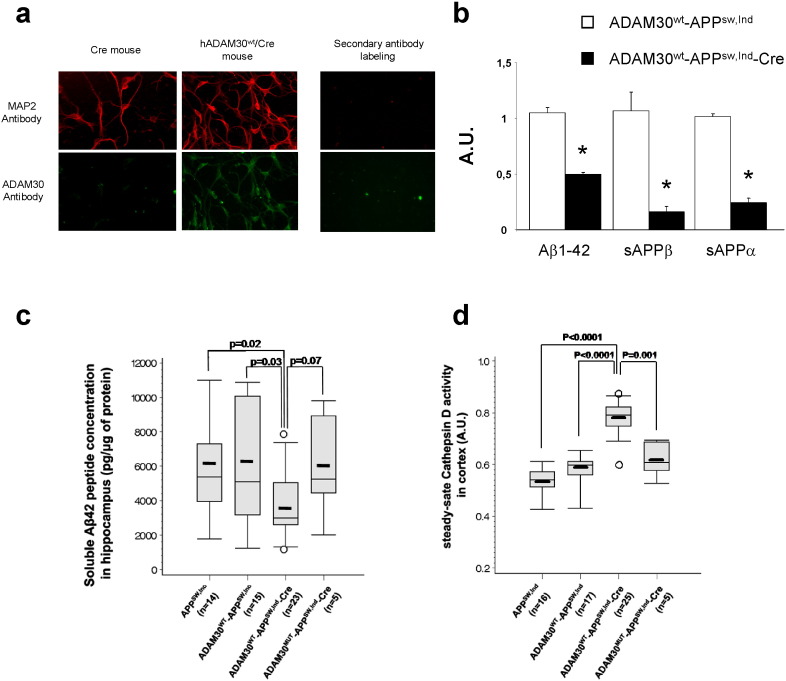
To corroborate our observations in vivo and further evaluate the impact of ADAM30 on Aβ peptide secretion and amyloidosis, we sought to generate a transgenic mouse model. It was not possible to develop ADAM30 knock-out mice (ADAM30 is not expressed in the mouse brain (Allen Brain Atlas, http://www.brain-map.org and our data, Fig. 5a and Supplementary Fig. 13) contrary to what we observed in humans. We thus generated APPSw,Ind mice that conditionally overexpressed hADAM30WT in the neurons of the forebrain (as previously mentioned, immunohistochemistry experiments were consistent with the expression of human ADAM30 in neurons in human brain tissue (Fig. 1c)). Primary cultures of adult neurons indicated that expression of ADAM30WT was only observed in neurons generated from hADAM30WT-hAPPSw,Ind-Cre mice (Fig. 5a). As observed in cell lines, a decrease in APP catabolites was also observed in primary cultures of adult neurons expressing ADAM30WT (Fig. 5b).
Fig. 5.
Impact of ADAM30 over-expression in neurons and AD-like models on Aβ secretion and CTSD activity. (a) ADAM30 expression at the protein level in adult neurons from 2 month mouse hippocampus (n = 3). (b) Mean differences (± SEM) in the secreted amounts of sAPPα, sAPPβ, and Aβ1–42 in adult neurons (Div 19) according to ADAM30WT over-expression. Three independent experiments were performed in duplicate. *p < 0.05 (Mann-Whitney non-parametric test). (c) Differences in levels of soluble Aβ1–42 peptide concentration in the hippocampi of different transgenic mice overexpressing ADAM30WT/mut or not. The thick lines represent the median Aβ1–42 level. The midline represents the mean value and the upper and lower horizontal lines represent the first and third quartiles, respectively. Circles indicate individuals with extreme values (more than 2 SD above or below the mean value). p-Values refer to a Mann-Whitney non-parametric test. (d) Differences in CTSD activity in the cortex of different transgenic mice overexpressing ADAM30WT/mut or not. The thick lines represent the median CTSD activity level. The midline represents the mean value and the upper and lower horizontal lines represent the first and third quartiles, respectively. Circles indicate individuals with extreme values (more than 2 SD above or below the mean value). p-Values refer to a Mann-Whitney non-parametric test.
We then extended our results by analyzing the level of soluble Aβ42 in the mouse hippocampus. We observed significantly lower hippocampal levels of soluble Aβ42 upon induction in mice overexpressing hADAM30WT (− 39%, relative to hADAM30WT-hAPPSw,Ind mice; p = 0.03). We also generated mice that conditionally overexpressed hADAM30mut in order to confirm that ADAM30-mediated potential effects are genuinely linked to its catalytic activity. Remarkably, we did not detect any lower levels of soluble Aβ42 in these mice overexpressing hADAM30mut (Fig. 5c). CTSD activity was also significantly higher in the cortex of mice overexpressing hADAM30WT (but not in those overexpressing hADAM30mut) than in hADAM30-hAPPSw,Ind mice (+ 34%, p < 0.0001, and + 7%, p = 0.82, respectively; Fig. 5D). Interestingly, we also observed a negative correlation between CTSD activity and soluble Aβ42 concentrations in hADAM30WT-hAPPSw,Ind-Cre mice (p < 0.05 in Spearman's correlation test, Fig. S10).
All these observations are in full accordance with our in vitro data and support the claim that our observed effects are indeed specifically due to ADAM30 enzymatic activity. Furthermore, our results support a direct link between ADAM30, CTSD activation and amyloid load.
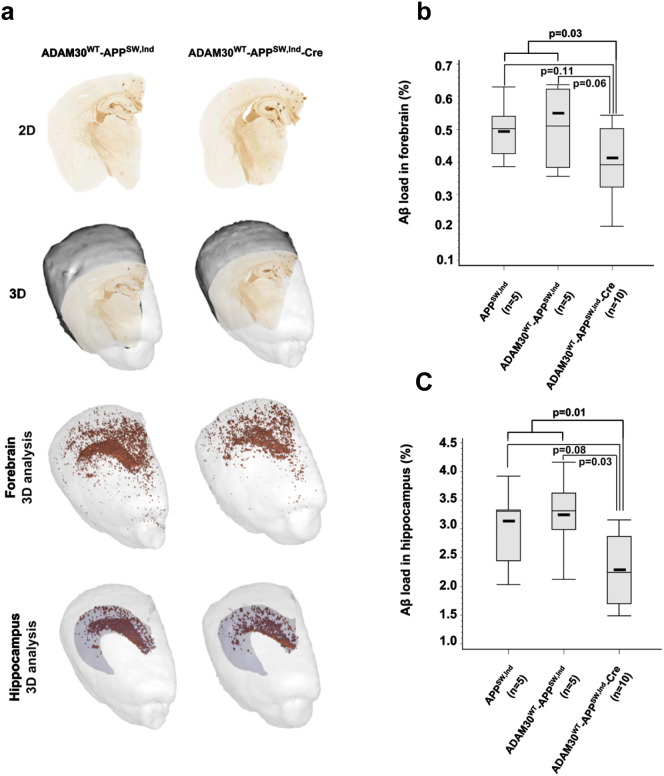
To assess this possibility, we used a reliable, automated, high-throughput, three-dimensional (3D) histology method to determine the Aβ load in mouse forebrain in general and the hippocampus in particular (see the Experimental procedures, Fig. 6a). Current standards for the analysis of brain histopathological markers heavily rely on manual intervention to delineate regions of interest and quantify the staining. Data collection is thus usually restricted to a few tissue sections. As a consequence, this approach allows avoiding eventual biases due to low section sampling rate as demonstrated elsewhere (Vandenberghe et al., 2016). By using this powerful approach, we observed that overexpression of hADAM30WT was associated with a 20% reduction in the Aβ load (p = 0.03; Fig. 6b) in the mice forebrain. As expected, this reduction was also observed in analysis restricted to the hippocampus (− 28%, p = 0.01; Fig. 6c).
Fig. 6.
Impact of ADAM30 over-expression in AD-like models on Aβ loads. (a) Representative 2D sections and 3D reconstruction of the brains in mice overexpressing ADAM30WT or not. Illustration in the same mice of the spatial distribution of 3D segmented Aβ peptide aggregates corresponding to forebrain and hippocampus regions (b) The Aβ load in the forebrain of APPsw,Ind transgenic mice overexpressing ADAM30WT or not. (c) The Aβ load in the hippocampus of APPsw,Ind transgenic mice overexpressing ADAM30WT or not. Box plots are as follows: The thick lines represent the median level. The midline represents the mean value and the upper and lower horizontal lines represent the first and third quartiles, respectively. Circles indicate individuals with extreme values (more than 2 SD above or below the mean value). p-Values refer to a Mann-Whitney non-parametric test.
4.9. Overexpression of ADAM30 Partially Rescues LTP Deficits
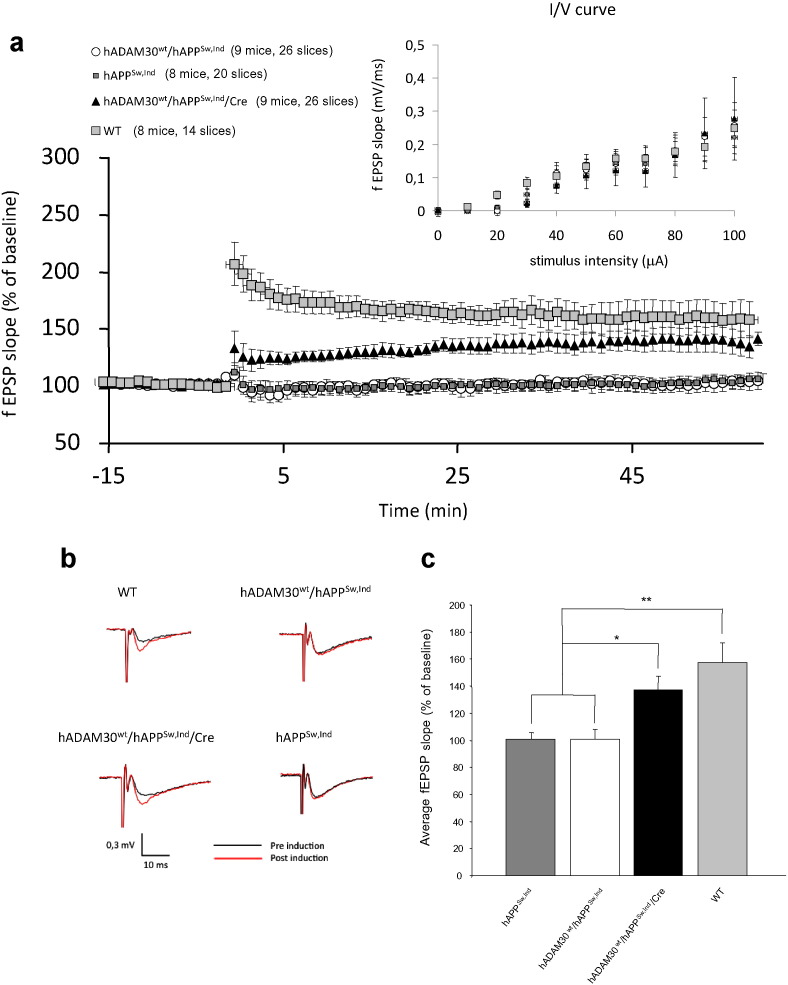
Since APP catabolites have been shown to block hippocampal long term potentiation (LTP) and impair learning function in mice (Nalbantoglu et al., 1997), we aimed at establishing the functional impact of hADAM30 overexpression on LTP in hAPPSw,Ind hippocampus. Several paradigms of basal synaptic transmission were analyzed: maximum fEPSP slope, Maximum fEPSP amplitude, maximum fiber volley amplitude and the function relating the stimulation intensity to fEPSP, which is an index of basal synaptic strength. None of these parameters were altered whatever the transgenic mice analyzed (Fig. 7a).
Fig. 7.
Impact of ADAM30WT over-expression on LTP in Alzheimer-like mouse model. (a) LTP at the CA3-CA1 synapses of hippocampal slices induced by 4 × 100-Hz stimulation is impaired in hADAM30WT-hAPPSw,Ind (nine mice) and hAPPSw,Ind ( eight mice) compared with WT mice (eight mice). Cre dependent gene expression of hADAM30WT in hADAM30WT-hAPPSw,Ind-Cre mice partially rescues LTP (nine mice). Inset graph shows input–output curves, which are comparable in hADAM30WT-hAPPSw,Ind (nine mice), hAPPSw,Ind (n = 8 mice), hADAM30WT-hAPPSw,Ind-Cre (n = 9 mice), and WT mice (eight mice). (b) Traces show the field excitatory postsynaptic potential (fEPSP) from a representative animal from each group, black traces represent the average of baseline recording before 4 × 100-Hz stimulation, red traces show the average over the last 20 min of post-stimulation recording. (c) Quantification indicating the percentage change in average fEPSP amplitude 40–60 min after 4 × 100-Hz stimulation. Data are mean ± SEM. **p = 0.0005, **p = 0.0015 (Mann-Whitney non-parametric test).
As expected, a LTP deficit was observed in hAPPSw,Ind and hADAM30WT-hAPPSw,Ind mice and this deficit was partially restored over time in hADAM30WT-hAPPSw,Ind-Cre mice (Fig. 7a and b). Mean fEPSP for the different groups was calculated over the last 20 min of recording post-high frequency stimulation. A significant difference in LTP was observed between the hADAM30WT-hAPPSw,Ind-Cre mice and the other transgenic mice in which no potentiation was observed. Even if ADAM30WT over-expression did not fully rescue potentiation, an about 60% recovery was observed (Fig. 7c).
5. Discussion
One of the main difficulties in differential expression analyses of postmortem tissues relates to whether the variations detected are causes or consequences of the disease process. By means of a multidisciplinary approach involving various state-of-the-art techniques and novel original transgenic mouse models, we identified ADAM30 as a player in APP metabolism and show that its underexpression likely directly contributes to Alzheimer's disease physiopathology.
Very little is known about the physiological role of ADAM30 (Cerretti et al., 1999, Hu et al., 2009, Ho et al., 2013, Almawi et al., 2013, Gupta et al., 2012, Ellis et al., 2014), its putative involvement in Alzheimer's disease and its protein partners. However, our results highlight a new axis in APP metabolism, with APP sorting to lysosomes, ADAM30-dependent CTSD activation and APP degradation. Overall, this set of data supports previous mechanistically unsolved observations of an important lysosomal contribution to the metabolism of APP (Funk and Kuret, 2012).
The potential APP degradation through ADAM30-dependent CTSD activation could be either linked to the CTSD activity by itself (CTSD was described to endoproteolyse the C100 APP fragment at multiple sites and to display β-secretase-like activity, in vitro) (Mackay et al., 1997, Chevallier et al., 1997) or to a CTSD-mediated subsequent activation of other hydrolases (Benes et al., 2008). In line with the recent description that PS1 mutations can be linked to defective lysosomal proteolysis (Lee et al., 2010), our data support the importance of the lysosome compartment in APP metabolism and as a consequence in the Alzheimer's disease pathophysiological processes.
We suspect that further investigation of other potential ADAM30 substrates might provide a better understanding of the ADAM30-dependent physiopathologic processes. Interestingly, all these targets can be directly or indirectly linked to insulino-resistance and/or APP metabolism. In one hand, GKAP1 was recently described to interact with IRS1 and to participate in insulino-resistance in adypocites (Ando et al., 2015) whereas the ADAM30 locus was reported as a genetic risk factor for Type 2 diabetes (T2D) (Cerretti et al., 1999, Hu et al., 2009, Ho et al., 2013, Almawi et al., 2013, Gupta et al., 2012, Ellis et al., 2014). On the other hand, IRs, insulino-resistance and TD2 were described as modulators of the Alzheimer's disease pathology though a potential modulation of the APP metabolism whereas CTSD was previously involved in APP processing in vitro (Dreyer et al., 1994, Mackay et al., 1997, Haque et al., 2008, Funk and Kuret, 2012). It is also noteworthy that IRS4 (also identified in our COFRADIC study) is able to directly interact with the Grb2 adaptor protein, a well-known actor of APP processing/sorting (Hinsby et al., 2004). Of note, since lysosomal proteolysis can represent a general mechanism for protein degradation/processing, we measured secreted catabolites of Met, a protein exhibiting a metabolism similar of APP (Lefebvre et al., 2012). We did not observe any modifications of the secreted Met products following co-transfection of Met and ADAM30 in HEK293 cells (Fig. S11). This observation suggests that the ADAM30-dependent CTSD-lysosome activation may be rather selective and that its metabolic consequences could be limited to a restricted number of proteins.
When considering the APP metabolism as a whole, our present findings (including the ADAM30-linked decreased secretions of APPs-α and -β and Aβ peptides) strongly suggest that either (i) ADAM30 is involved in a process that modifies the equilibrium between APP recycling (from early/late lysosomes to the membrane) and APP degradation in the lysosome or (ii) ADAM30 affects the cellular events that drives the dispatching of full-length membrane APP to either the endocytosis pathway or the lysosomal compartment, as was previously suggested (Lorenzen et al., 2010). However, it is noteworthy that we did not detect any changes in the internalization or endocytosis of full-length membrane APP in cells overexpressing ADAM30.
In conclusion, we hypothesize that ADAM30 is a key player in APP metabolism; it appears to be involved in APP degradation via sorting to lysosomes, targeted CTSD activation and then full-length APP recycling. The mechanisms that involve ADAM30 may restrict full-length APP recycling and therefore influence the pool of APP available for α-, β- and γ-secretase processing.
In conclusion, we characterized a new facet of the APP physiology which may provide new therapeutic targets to control amyloid peptide production in the brain.
The following are the supplementary data related to this article.
Supplementary material.
Transcriptomic data.
Conflict of Interest
The authors declare no conflict of interest.
Author contributions
F. Letronne., G.L., A-M.A., J.C., A.F, F.D, F.E and T.G. performed molecular and cellular biology experiments. G.L., A-M.A., J.C., F.D., N.M., E.W. and F. Lafont performed/supervised cellular imaging. F. Letronne., A-M.A., Y.S. performed mouse model breeding, brain preparation and soluble Aβ measurements. G.L., A.B., F.H. performed Quantigene assays. M.L., K.G. performed COFRADIC experiments and interpretation. M.E.V, A-S.H., M.D., N.S. and T.D. performed 3D brain amyloid load analyses and interpretation. F. Leroux., J.D., B.D., R.D. developed and performed recombinant CTSD/ADAM30 assays. M.L. and D.T. performed Met experiments. L.H., Y.L., D.H. performed microarray experiments and interpretation. L.C., C.B., F.C. performed β- and γ-secretase assays. F.P., C.B., J-J.H., D.M. provided human brain samples for immunohistochemistry and transcriptomic analyses. A-M.A., J.C., M.E.V, A.B., P.D., C.D., B.D., F. Lafont, M.D., D.M., P.A., F.C., D.H., T.D., J-C.L. participate to the writing and/or significantly revise the manuscript. A-M.A, J.C. M.L., R.D., D.H., F.C., T.D., J-C.L. conceived the experiments.
Acknowledgments
We thank the Lille Neurobank and UMR 1172 “Alzheimer & Tauopathies” for the brain immunohistochemistry experiments. We thank Florence Combes for her technical assistance. We thank Dr. Melissa Farinelli (E-Phy-Science) for her helpful discussion. F.L. was funded by the University of Lille II, the Nord-Pas de Calais Regional Council and the Fondation pour la Recherche Médicale (FRM). G.L. and F.E. was funded by the Institute Pasteur de Lille and the Nord-Pas de Calais Regional Council. J.C. and Y.S. were funded by the MEDIALZ Project (Grant 11001003) financed by the European Regional Development Fund and the Nord-Pas de Calais Regional Council. F.H. was funded by the Alzheimer's Association (grant IIRG-06-25487) and then the Ligue contre la Maladie d'Alzheimer (LECMA, grant 09705). C.D. was funded by Lille Metropole Communauté Urbaine. F.C. was supported by the Hospital University Federation (FHU OncoAge).
This work was also funded by the French government's CPER-Neuroscience (DN2M) program (Nord-Pas de Calais Regional Council and FEDER), INSERM (the ATC-vieillissement program), the Institut Pasteur de Lille, the Fondation pour la Recherche sur le Cerveau (FRC) and the French government's “Development of Innovative Strategies for a Transdisciplinary approach to Alzheimer's disease” Laboratory of Excellence (LABEX DISTALZ) program.
We acknowledge the support of Alzheimer's Research UK and Alzheimer's Society through their funding of the Manchester Brain Bank (from which samples were also obtained) under the Brains for Dementia Research (BDR) initiative.
References
- Almawi W.Y. A replication study of 19 GWAS-validated type 2 diabetes at-risk variants in the Lebanese population. Diabetes Res. Clin. Pract. 2013;102:117–122. doi: 10.1016/j.diabres.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Ando Y. Tumor Necrosis Factor (TNF)-α-induced repression of GKAP42 protein levels through cGMP-dependent kinase (CGK)-Iα causes insulin resistance in 3T3-L1 adipocytes. J. Biol. Chem. 2015;290:5881–5892. doi: 10.1074/jbc.M114.624759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrau D. BACE1- and BACE2-expressing human cells: characterization of β-amyloid precursor protein-derived catabolites, design of a novel fluorimetric assay, and identification of new in vitro inhibitors. J. Biol. Chem. 2003;278:25859–25866. doi: 10.1074/jbc.M302622200. [DOI] [PubMed] [Google Scholar]
- Baranger K. MT5-MMP is a new pro-amyloidogenic proteinase that promotes amyloid pathology and cognitive decline in a transgenic mouse model of Alzheimer's disease. Cell. Mol. Life Sci. 2016;73:217–236. doi: 10.1007/s00018-015-1992-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes P., Vetvicka V., Fusek M. Cathepsin D-many functions of one aspartic protease. Crit. Rev. Oncol. Hematol. 2008;68:12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensemain F. Evidence for induction of the ornithine transcarbamylase expression in Alzheimer's disease. Mol. Psychiatry. 2009;14:106–116. doi: 10.1038/sj.mp.4002089. [DOI] [PubMed] [Google Scholar]
- Canales R.D. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. (Available at: http://www.ncbi.nlm.nih.gov/pubmed/16964225) [DOI] [PubMed] [Google Scholar]
- Carson J.A., Turner A.J. Beta-amyloid catabolism: roles for neprilysin (NEP… [J Neurochem. 2002] - PubMed result. J. Neurochem. 2002;81:1–8. doi: 10.1046/j.1471-4159.2002.00855.x. (Available at: http://www.ncbi.nlm.nih.gov/pubmed/12067222) [DOI] [PubMed] [Google Scholar]
- Cerretti D.P. Isolation of two novel metalloproteinase-disintegrin (ADAM) cDNAs that show testis-specific gene expression. Biochem. Biophys. Res. Commun. 1999;263:810–815. doi: 10.1006/bbrc.1999.1322. [DOI] [PubMed] [Google Scholar]
- Chapuis J. Transcriptomic and genetic studies identify IL-33 as a candidate gene for Alzheimer's disease. Mol. Psychiatry. 2009;14:1004–1016. doi: 10.1038/mp.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier N. Cathepsin D displays in vitro beta-secretase-like specificity. Brain Res. 1997;750:11–19. doi: 10.1016/s0006-8993(96)01330-3. [DOI] [PubMed] [Google Scholar]
- Chubb C. BioVision: an application for the automated image analysis of histological sections. Neurobiol. Aging. 2006;27:1462–1476. doi: 10.1016/j.neurobiolaging.2005.08.023. [DOI] [PubMed] [Google Scholar]
- De Strooper B. Aph-1, Pen-2, and nicastrin with presenilin generate an active γ-secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- Delacourte A. Nonoverlapping but synergetic tau and APP pathologies in sporadic Alzheimer's disease. Neurology. 2002;59:398–407. doi: 10.1212/wnl.59.3.398. [DOI] [PubMed] [Google Scholar]
- Dorr A.E. High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. NeuroImage. 2008;42:60–69. doi: 10.1016/j.neuroimage.2008.03.037. [DOI] [PubMed] [Google Scholar]
- Dreyer R.N. Processing of the pre-beta-amyloid protein by cathepsin D is enhanced by a familial Alzheimer's disease mutation. Eur. J. Biochem./FEBS. 1994;224:265–271. doi: 10.1111/j.1432-1033.1994.00265.x. [DOI] [PubMed] [Google Scholar]
- Dubois A. Automated three-dimensional analysis of histological and autoradiographic rat brain sections: application to an activation study. J. Cereb. Blood Flow Metab. 2007;27:1742–1755. doi: 10.1038/sj.jcbfm.9600470. [DOI] [PubMed] [Google Scholar]
- Dubois A. Detection by voxel-wise statistical analysis of significant changes in regional cerebral glucose uptake in an APP/PS1 transgenic mouse model of Alzheimer's disease. NeuroImage. 2010;51:586–598. doi: 10.1016/j.neuroimage.2010.02.074. [DOI] [PubMed] [Google Scholar]
- Ellis K.L. Genetic variation at glucose and insulin trait loci and response to glucose–insulin–potassium (GIK) therapy: the IMMEDIATE trial. Pharmacogenomics J. 2014;15:55–62. doi: 10.1038/tpj.2014.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk K.E., Kuret J. Lysosomal fusion dysfunction as a unifying hypothesis for alzheimers disease pathology. Int. J. Alzheimers Dis. 2012 doi: 10.1155/2012/752894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V. Association analysis of 31 common polymorphisms with type 2 diabetes and its related traits in Indian sib pairs. Diabetologia. 2012;55:349–357. doi: 10.1007/s00125-011-2355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque A., Banik N.L., Ray S.K. New insights into the roles of endolysosomal cathepsins in the pathogenesis of Alzheimer's disease: cathepsin inhibitors as potential therapeutics. CNS Neurol. Disord. Drug Targets. 2008;7:270–277. doi: 10.2174/187152708784936653. [DOI] [PubMed] [Google Scholar]
- Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–357. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Hinsby A.M., Olsen J.V., Mann M. Tyrosine phosphoproteomics of fibroblast growth factor signaling: a role for insulin receptor substrate-4. J. Biol. Chem. 2004;279:46438–46447. doi: 10.1074/jbc.M404537200. [DOI] [PubMed] [Google Scholar]
- Ho M.M. Diabetes genes identified by genome-wide association studies are regulated in mice by nutritional factors in metabolically relevant tissues and by glucose concentrations in islets. BMC Genet. 2013;14:10. doi: 10.1186/1471-2156-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C. PPARG, KCNJ11, CDKAL1, CDKN2A-CDKN2B, IDE-KIF11- HHEX, IGF2BP2 and SLC30A8 are associated with type 2 diabetes in a Chinese population. PLoS ONE. 2009;4:7643. doi: 10.1371/journal.pone.0007643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Wood K. Amyloid precursor protein processing and A beta42 deposition in a transgenic mouse model of Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1550–1555. doi: 10.1073/pnas.94.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- Kouchi Z. The deletion of the C-terminal tail and addition of an endoplasmic reticulum targeting signal to Alzheimer's amyloid precursor protein change its localization, secretion, and intracellular proteolysis. Eur. J. Biochem. 1998;258:291–300. doi: 10.1046/j.1432-1327.1998.2580291.x. [DOI] [PubMed] [Google Scholar]
- Kouchi Z. Proteasome inhibitors induce the association of Alzheimer's amyloid precursor protein with Hsc73. Biochem. Biophys. Res. Commun. 1999;254:804–810. doi: 10.1006/bbrc.1998.9977. [DOI] [PubMed] [Google Scholar]
- Kuhn P.-H. ADAM10 is the physiologically relevant, constitutive α-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010;29:3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A., Sisodia S.S., Trowbridge I.S. Characterization of sorting signals in the β-amyloid precursor protein cytoplasmic domain. J. Biol. Chem. 1995;270:3565–3573. [PubMed] [Google Scholar]
- Lambert J.C., Amouyel P. Genetics of Alzheimer's disease: new evidences for an old hypothesis? Curr. Opin. Genet. Dev. 2011;21:295–301. doi: 10.1016/j.gde.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Lammich S. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebenberg J. Validation of MRI-based 3D digital atlas registration with histological and autoradiographic volumes: an anatomofunctional transgenic mouse brain imaging study. NeuroImage. 2010;51:1037–1046. doi: 10.1016/j.neuroimage.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Lee J.H. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre J. Met degradation: more than one stone to shoot a receptor down. FASEB J. 2012;26:1387–1399. doi: 10.1096/fj.11-197723. [DOI] [PubMed] [Google Scholar]
- Liang W.S. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen A. Rapid and direct transport of cell surface APP to the lysosome defines a novel selective pathway. Mol. Brain. 2010;3:11. doi: 10.1186/1756-6606-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay E.A. A possible role for cathepsins D, E, and B in the processing of beta-amyloid precursor protein in Alzheimer's disease. Eur. J. Biochem. 1997;244:414–425. doi: 10.1111/j.1432-1033.1997.00414.x. [DOI] [PubMed] [Google Scholar]
- Mucke L. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J. Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbantoglu J. Impaired learning and LTP in mice expressing the carboxy terminus of the Alzheimer amyloid precursor protein. Nature. 1997;387:500–505. doi: 10.1038/387500a0. [DOI] [PubMed] [Google Scholar]
- Reitz C. Matrix metalloproteinase 3 haplotypes and plasma amyloid beta levels: the Rotterdam Study. Neurobiol. Aging. 2010;31:715–718. doi: 10.1016/j.neurobiolaging.2008.05.033. [DOI] [PubMed] [Google Scholar]
- Rosenberg G.A. Matrix metalloproteinases and their multiple roles in neurodegenerative diseases. Lancet Neurol. 2009;8:205–216. doi: 10.1016/S1474-4422(09)70016-X. [DOI] [PubMed] [Google Scholar]
- Schönherr C. Generation of aggregation prone N-terminally truncated amyloid β peptides by meprin β depends on the sequence specificity at the cleavage site. Mol. Neurodegener. 2016;11:19. doi: 10.1186/s13024-016-0084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevalle J. Pharmacological evidences for DFK167-sensitive presenilin-independent γ-secretase-like activity. J. Neurochem. 2009;110:275–283. doi: 10.1111/j.1471-4159.2009.06131.x. [DOI] [PubMed] [Google Scholar]
- Staes A. Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat. Protoc. 2011;6:1130–1141. doi: 10.1038/nprot.2011.355. [DOI] [PubMed] [Google Scholar]
- Tian Y. Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer's APP-CTF for terminal degradation via autophagy. Proc. Natl. Acad. Sci. 2013;110:17071–17076. doi: 10.1073/pnas.1315110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien J.Z. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Vandenberghe M.E. High-throughput 3D whole-brain quantitative histopathology in rodents. Sci. Rep. 2016;6:20958. doi: 10.1038/srep20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent B., Checler F. α-Secretase in Alzheimer's disease and beyond: mechanistic, regulation and function in the shedding of membrane proteins. Curr. Alzheimer Res. 2012;9:140–156. doi: 10.2174/156720512799361646. [DOI] [PubMed] [Google Scholar]
- Wang D.S., Dickson D.W., Malter J.S. beta-Amyloid degradation and Alzheimer's disease. J. Biomed. Biotechnol. 2006;2006:58406. doi: 10.1155/JBB/2006/58406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A.R. Degradation of the Alzheimer disease amyloid β-peptide by metal-dependent up-regulation of metalloprotease activity. J. Biol. Chem. 2006;281:17670–17680. doi: 10.1074/jbc.M602487200. [DOI] [PubMed] [Google Scholar]
- Willem M. η-Secretase processing of APP inhibits neuronal activity in the hippocampus. Nature. 2015;526:443–447. doi: 10.1038/nature14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Baker K.A., Hagg T. The ADAMs family: coordinators of nervous system development, plasticity and repair. Prog. Neurobiol. 2006;79:73–94. doi: 10.1016/j.pneurobio.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Young J.E. Elucidating molecular phenotypes caused by the SORL1 Alzheimer's disease genetic risk factor using human induced pluripotent stem cells. Cell Stem Cell. 2015;16:373–385. doi: 10.1016/j.stem.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Transcriptomic data.