Abstract
Birth by Caesarian section is associated with short- and long-term respiratory morbidity. We hypothesized that mode of delivery affects the development of the respiratory microbiota, thereby altering its capacity to provide colonization resistance and consecutive pathobiont overgrowth and infections. Therefore, we longitudinally studied the impact of mode of delivery on the nasopharyngeal microbiota development from birth until six months of age in a healthy, unselected birth cohort of 102 children (n = 761 samples). Here, we show that the respiratory microbiota develops within one day from a variable mixed bacterial community towards a Streptococcus viridans-predominated profile, regardless of mode of delivery. Within the first week, rapid niche differentiation had occurred; initially with in most infants Staphylococcus aureus predominance, followed by differentiation towards Corynebacterium pseudodiphteriticum/propinquum, Dolosigranulum pigrum, Moraxella catarrhalis/nonliquefaciens, Streptococcus pneumoniae, and/or Haemophilus influenzae dominated communities. Infants born by Caesarian section showed a delay in overall development of respiratory microbiota profiles with specifically reduced colonization with health-associated commensals like Corynebacterium and Dolosigranulum, thereby possibly influencing respiratory health later in life.
Keywords: Microbiota, Microbiome, Mode of delivery, Caesarian section, Respiratory tract, Respiratory tract infection
Highlights
-
•
The respiratory microbiota is highly dynamic during the first six months of life.
-
•
The microbiota develops rapidly from a maternal/environmental-derived flora towards several niche-specific profiles.
-
•
Mode of delivery affects early respiratory microbiota development, especially colonization of potential protective commensals.
Birth by Caesarian section is associated with increased prevalence of respiratory diseases. We hypothesized that mode of delivery affects the micro-community of bacteria residing in the respiratory tract (microbiota), thereby influencing its ability to prevent invasion and outgrowth of potential pathogenic bacteria that can cause respiratory disease. We followed children during their first six months of life and show that the microbiota develops rapidly from a maternal/environmental-derived flora towards several niche-specific microbial profiles. Mode of delivery significantly affects respiratory microbiota development and interacts with breastfeeding, especially regarding the early presence of potential protective bacteria, which may contribute to respiratory health later in life.
1. Introduction
The total number of Caesarian sections (C-sections) has dramatically increased during the last decades; from 15% in 1990 to 27% in 2011 of all live births in industrialized countries (Mueller et al., 2015, OECD Publishing, 2013). This is worrisome as delivery by C-section has been associated with early life morbidity, including respiratory distress directly after birth (Karlström et al., 2013), hospitalization for respiratory syncytial virus infection (Kristensen et al., 2015), and long-term health problems, including development of asthma later in life (Guibas et al., 2013, Thavagnanam et al., 2008). One hypothesis that explains the association between the increase of infant disease and the mode of delivery is a disrupted mother-to-child bacterial transmission and thereby altered microbial colonization patterns in children born by C-section (Kristensen et al., 2015). Depending on mode of delivery, children are exposed to either the maternal vaginal and intestinal microbiota (vaginal delivery) or skin and environmental microbiota (C-section), leading to distinct microbial acquisition shortly after birth (Dominguez-Bello et al., 2010, Penders et al., 2013). This suggests that mode of delivery is likely to have profound impact on both the structure of early and late microbiota, as well as on processes depending on microbiota development, i.e. immune maturation, epithelial integrity, microbial tolerance, and pathogen resistance (Hooper et al., 2012).
The upper respiratory tract is the natural niche for respiratory bacterial and viral pathogens and the origin for consecutive respiratory tract infections (RTI). Potential pathogenic bacteria are embedded in a community of commensals, jointly forming the nasopharyngeal microbiota. Microbial colonization succession is highly influenced by environmental factors, host factors, and bacterial acquisition during the first years of life (Koppen et al., 2015). Recently, we published a first crude picture of microbiota development in children over the first two years of life (Biesbroek et al., 2014b). Despite large sampling intervals, the composition of the nasopharyngeal microbiota appeared highly dynamic, especially in the first six months of life. As we hypothesized that a balanced microbiota is more resilient to bacterial acquisition and overgrowth, we focused on the stability of the microbial profiles. Intriguingly, early microbiota composition (six weeks of life) predicted microbiota stability over the first two years of life: stable profiles were associated with (exclusive) breastfeeding and fewer respiratory tract infections in the consecutive period (Biesbroek et al., 2014a, Biesbroek et al., 2014b). These findings indicate that there is a window of opportunity during early life where a stable microbial signature is formed, that is associated with protection against respiratory symptoms. These data are supported by both epidemiological (Vissing et al., 2013) and murine (Gollwitzer et al., 2014) data. Although timing and order of exposure to specific groups of microbes, like mediated by mode of delivery, may have crucial consequences on development of the microbial profile, studies investigating the respiratory microbiota of healthy young children in detail in a longitudinal fashion are lacking.
We therefore decided to study the dynamics of the nasopharyngeal microbiota in relation to mode of delivery in detail, from birth until the age of six months in 62 vaginally delivered children and 40 children born by C-section.
2. Materials and Methods
Details can be found in the Supplemental data.
2.1. Study Population
Upper respiratory (nasopharyngeal) samples were obtained from 102 healthy children who participated in an ongoing prospective birth cohort study. The primary aim of this study is to characterize the development of the respiratory microbiota in term children born vaginally (n = 62) compared to children born by C-section (n = 40). Inclusion criteria at baseline were term birth (gestational age > 37 weeks). Exclusion criteria were major congenital anomalies, severe maternal or neonatal complications during birth, language barrier, intention to move outside the research area, or parents under 18 years of age. All participants were born between December 2012 and June 2014. Written informed consent was obtained from both parents before birth of the child. Participants did not receive any financial compensation. An acknowledged national ethics committee in the Netherlands (METC Noord-Holland, committee on research involving human subjects) approved the study (M012-015, NTR3986) and the study was conducted in accordance with the European Statements for Good Clinical Practice.
2.2. Data Collection
At baseline, data were collected on prenatal and perinatal characteristics. Follow-up of participants in the current study included visits directly post-partum, 24–36 h after delivery, and at 7 days, 14 days, and one, two, three, four, and six months of age. Deep nasopharyngeal swabs and a questionnaire on the health status of the child, including respiratory symptoms, were obtained during each visit by a trained research team of doctors and research nurses.
2.3. Construction of the Phylogenetic Library
Bacterial DNA of the nasopharyngeal samples was isolated using a mechanic disruption method as described previously (Biesbroek et al., 2012, Wyllie et al., 2014). Only samples with a bacterial density of at least 0.3 pg/μl above the background (DNA quantity of the negative controls) as measured with Real-Time PCR were analyzed to avoid interference of background DNA. After amplification of the hypervariable V4 region of the 16S rRNA gene, samples were sequenced by Illumina MiSeq (Illumina Inc., San Diego, CA, USA). Since the samples collected postpartum and at day one generally had very low DNA density, we further analysed 12 samples obtained at those time points that did not meet the inclusion criteria, however had shown to generate an amplicon in the16S qPCR and had generated > 200 reads in the sequence run. We used complete linkage hierarchical clustering based on the Bray-Curtis dissimilarity matrix including a broad set of negative controls (blanks) which had also generated > 200 reads (n = 16). We were able to make a distinction between samples that had significant different microbiota profiles (n = 9) and samples that did not (n = 3). Based on these results, we excluded the latter samples from further analyses. Sequences were processed using modules implemented in Btrim (Kong, 2011) and Mothur V1.31.2 ((Schloss et al., 2009), for modules see supplemental methods). The unsupervised method Minimum Entropy Decomposition (MED) was used to assemble the unique sequences into high resolution oligotypes ((Eren et al., 2014) and oligotyping.org; default settings were used except for the minimal substantive abundance which was set at 100). Taxonomic classification of oligotype node representatives was performed using a naive Bayesian RDP classifier. We calculated the relative abundance of oligotypes per sample and determined the Shannon diversity index to describe the microbial diversity.
2.4. Spectral Clustering
To analyze the trajectory of the microbiota composition of individuals over time, an unsupervised co-regularized spectral clustering algorithm was applied to all data of the 102 children according to previously described methods (Biesbroek et al., 2014b, Imangaliyev et al., 2015, Tsivtsivadze et al., 2013). In short, this multiview clustering algorithm allows for 1) the identification of clusters comprised of individuals with similar microbial profiles in an unbiased and robust manner and 2) the detection of bio-marker species by using an unsupervised feature selection approach (Tsivtsivadze et al., 2014). For further detail, see supplemental methods.
2.5. Statistical Analyses
For all analyses, we used normalized microbial abundance (Biesbroek et al., 2014b). Differences in baseline characteristics and metadata were statistically tested using the 2-sided Chi-square or Fisher's exact test, and Students T-test where appropriate (SPSS version 21). p-Values < 0.05 were considered significant. Calculations on Shannon diversity and observed oligotypes were performed using a Kruskal-Wallis test with Dunn's correction for multiple testing in GraphPad Prism (version 6).
Nonmetric multidimensional scaling plots based on Bray-Curtis dissimilarity of log-transformed relative abundances were used to visualize the differences between mode of delivery and time-dependent microbiota development. Statistical significance of the difference in the overall microbiota composition driven by mode of delivery was assessed using PERMANOVA.
To describe intra-individual changes in relative abundance and presence of microbial species over time, we calculated a relative change matrix (Harville, 1997) per oligotype for two-month timeframes. From this relative change matrix per timeframe, we calculated the magnitude of microbiota change (norm value) using L2 norm (Biesbroek et al., 2014b, Harville, 1997). The higher this norm value, the higher the magnitude of change.
To investigate whether there is a statistically significant association between colonization trajectory (e.g. the change of the clustering profile in time) and the mode of delivery, we conducted a randomization test (Biesbroek et al., 2014b).
To identify biomarker species that are associated with modus partus, we evaluated oligotypes that varied in relative abundance over time between vaginally and C-section born children, independent of feeding type (EDGE package R((Storey et al., 2005) and tutorial “Extraction of Differential Gene Expression Version 2.0.0” March 2015, R version 3.2.0)). We conducted the analysis described above for the total dataset (n = 102 children) and for a subset of children who were exclusively breast- or formula-fed during the first six months of life (respectively n = 19 and n = 28) to rule out confounding by differences in duration of breastfeeding between groups. Resulting p-values were corrected for multiple testing using the Benjamini-Hochberg method. q-Values < 0.05 for the whole dataset and q-values < 0.10 for the subset were considered significant and were interpreted to suggest that there is a difference in relative abundance of a given oligotype driven by birth mode as time goes on.
We used Excel (version 2011), Python (version 2.7.8, packages Numpy, Scipy, Scikits Learn, Matplotlib), R version 3.1.2 (packages ‘rChart’ (Sankey plot) and ‘ggplot2’), and Adobe Illustrator CS6 for visualization.
3. Results
3.1. Population Characteristics
Characteristics of the 102 children are depicted in Table S1. We observed a slight difference in gestational age (39.1 weeks (standard deviation 0.9) versus 39.7 weeks (standard deviation 1.1)) between children born by C-section and vaginally born children respectively. Moreover, vaginally born children were almost twice as likely to receive breastfeeding for longer than three months compared to children born by C-section (59.7% of the vaginally born were breastfed versus 32.5% of the C-section born children, p = 0.009 (Chi-square)). Otherwise, there were no significant differences between the two populations.
3.2. Sequence Characteristics
We evaluated nine sample moments per child, for 102 children in total. We had to exclude in total 149 samples from further analyses because of bacterial DNA concentrations that were below the threshold level or provided unreliable microbiota profiles in comparison to negative control samples (91 of the 555 collected samples of vaginally born children and 58/355 for C-section delivered children and table S2). Most of these samples were collected shortly (< 2 h and/or at 24 h) after birth. We found no significant association between the DNA concentration and mode of delivery overall or for the excluded samples (Fig. S1). The remaining 761 samples were sequenced in five Miseq runs, resulting in 18,140,708 high-quality sequences (mean 23,838 and range 9973–91,376 sequences per sample; average coverage 97%, range 94–99%), which could be assigned to 1354 oligotypes. The number of oligotypes per sample increased during the first months of life from a mean of 103 oligotypes (95% confidence interval (CI) 95–111) at one week of age to 129 oligotypes (95% CI 121–138) at three months of age (p < 0.001, Kruskal-Wallis with Dunn's correction for multiple testing). Simultaneously, the Shannon diversity increased from 2.2 (95% CI 2.1–2.3) at week one to 2.5 (95% CI 2.4–2.6) at month three (p < 0.01, Kruskal-Wallis with Dunn's correction for multiple testing) (Fig. S2). Although we observed no significant difference in diversity between the children born by C-section compared to the vaginally born children, we found significant differences in the overall microbiota composition over time between C-section and vaginally delivered infants (PERMANOVA unadjusted R2 = 0.004, p-value = 0.007, Fig. S3).
3.3. Dynamics in Nasopharyngeal Microbial Communities
Overall, the most abundant genera in the first six months of life were Corynebacterium (25%), Moraxella (21%), Staphylococcus (19%), Streptococcus (11%), Dolosigranulum (11%), and Haemophilus (4%). The dominant oligotypes were Staphylococcus sp. es13 (19%), Moraxella catarrhalis/nonliquefaciens (18%), and Corynebacterium pseudodiphteriticum/propinquum (17%), followed by Dolosigranulum pigrum (11%), Streptococcus viridans (6%), Corynebacterium sp. (4%), Moraxella lincolnii (3%), Streptococcus pneumoniae (2%), and two types of Haemophilus influenzae (2% and 2% respectively). By culture and whole genome shotgun analysis (data not shown), we were able to assign Staphylococcus sp. es13 to Staphylococcus aureus, therefore this annotation will be used hereafter.
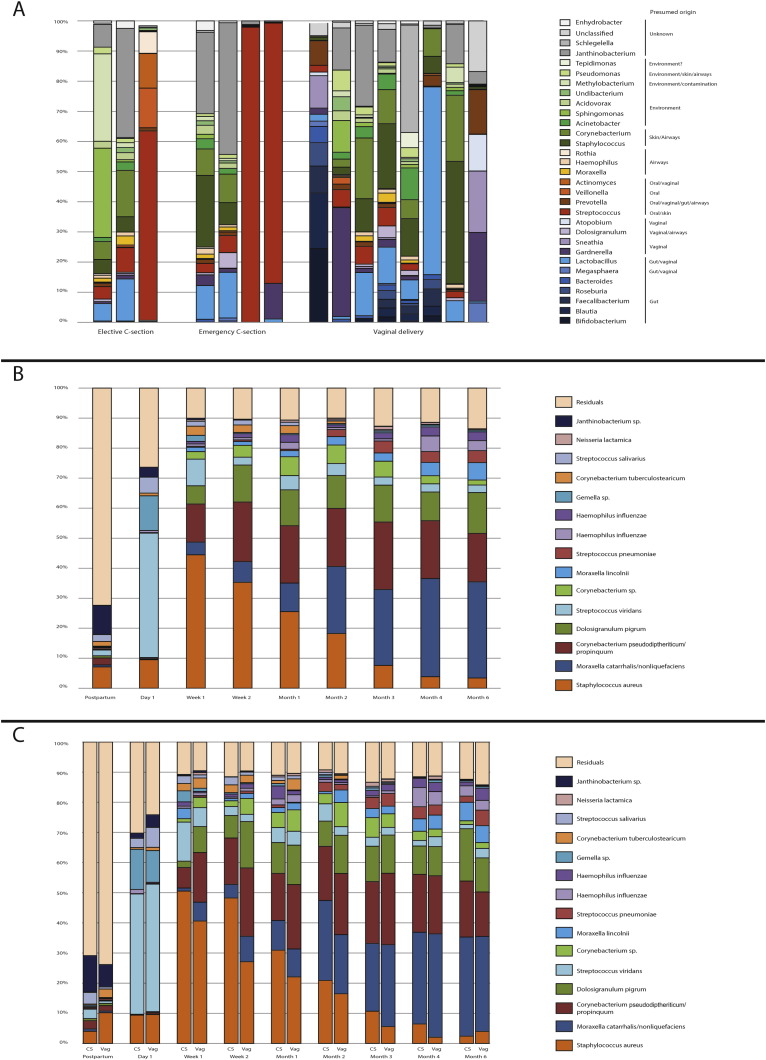
Directly after birth, we detected DNA from a mixture of bacteria, presumably from fecal, vaginal, skin, and/or environmental origin (Fig. 1a, color code represent presumed origin based on literature references). Bacterial DNA from fecal origin was exclusively found in children born by vaginal delivery, with DNA from Faecalibacterium and Blautia being detected in five out of eight samples available from vaginally born children. Furthermore, bacterial DNA from presumed vaginal origin was observed in vaginally delivered children but also in children born by C-section, especially when born by secondary C-section. Although duration of ruptured membranes differed between C-section groups, with 0 h for elective C-sections and a median of 9 h (range 0–48) for secondary C-sections (p < 0.0001, Kruskal-Wallis), we found no direct correlation between the duration of ruptured membranes and the microbial profile postpartum, which is most likely due to a lack of power.
Fig. 1.
Microbial profiles from birth till 6 months of age.
(a) Origin of samples collected postpartum. Genera present in the samples collected postpartum (n = 16) were coloured based on the presumed niches of origin (as known in literature), i.e. intestinal (blue), vaginal (purple), airways or oral (orange), skin or environmental sources (green) and unknown (grey). Samples were divided by the mode of delivery.
(b) Relative abundances of the 15 most abundant oligotypes are depicted for all samples per sampling moment.
(c) Relative abundance of the 15 most abundant oligotypes for either children born by C-section (left) or vaginal birth (right) per sampling moment.
Abbreviation: CS = C-section born children, Vag = vaginally born children. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Around 24 h after birth we observed outgrowth of especially S. viridans (41% of all reads) and to a lesser extend Gemella spp. (12% of all reads) in almost all children born vaginally and by C-section (Fig. 1b). Subsequently, at one week of age S. viridans/Gemella profiles were replaced by niche-specific bacterial profiles containing mostly S. aureus, followed by Corynebacterium spp. and D. pigrum. Between the ages of two weeks and six months, S. aureus (45% of all reads at two weeks of age) colonization gradually declined and M. catarrhalis/nonliquefaciens (32% of all reads at six months of age), Corynebacterium, and D. pigrum emerged. In addition, bacterial colonization with H. influenzae and S. pneumoniae started to emerge over the first 6 months of life (Fig. 1c).
3.4. Individual Microbial Succession Patterns
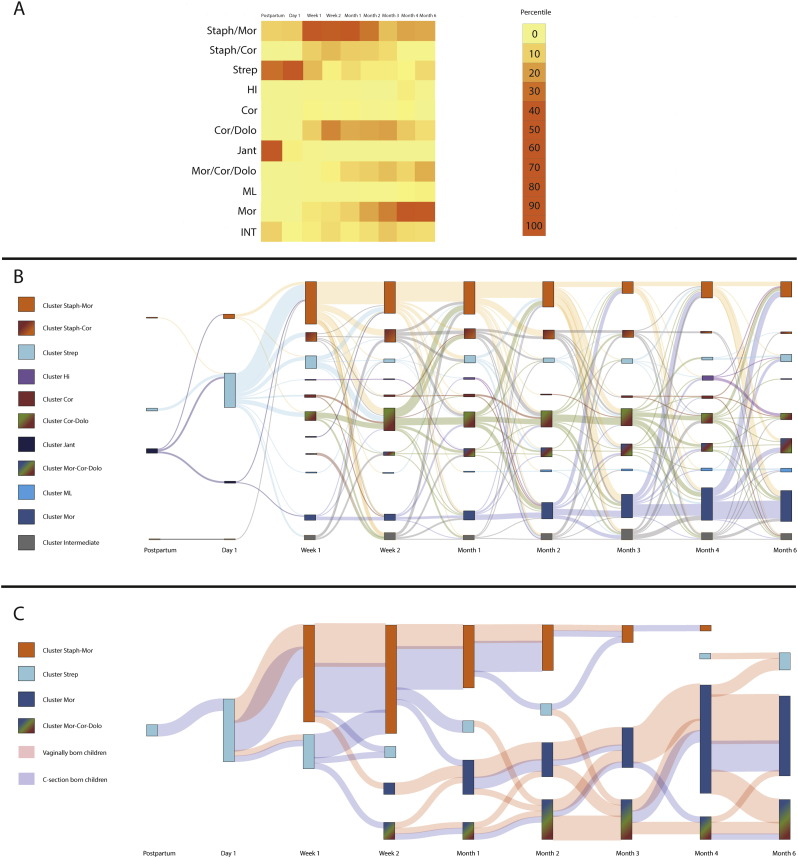
To gain more insight into microbial succession patterns within the study population, we used co-occurrence spectral clustering algorithms to define clusters of samples with similar bacterial co-occurrence patterns. Using this method, we identified 11 distinct clusters (Fig. S4, Fig. S5). The probabilistic assignment to a particular cluster varies between zero and one. The higher the probability, the higher the assignment of a child to a cluster. Based on biomarker analyses, we named each cluster after the most discriminative, abundant biomarker specie(s) (Fig. S6). Most samples collected postpartum and on day one belonged to three of the clusters, i.e. the S. aureus, S. viridans and the Janthinobacterium-dominated profiles of which the latter profile was only present directly after birth and disappeared during follow-up. From week one on, nine distinct profiles were observed: (1) M. lincolnii (ML), (2) M. catarrhalis/nonliquefaciens (Mor), (3) H. influenzae (HI), (4) C. pseudodiphteriticum (Cor) (5) S. viridans (Strep), (6) a cluster typified by high abundance of S. aureus at the early time points and M. catarrhalis/nonliquefaciens later on (Staph-Mor); (7) a combination of Staphylococcus-Corynebacterium (Staph-Cor), (8) a mix of Moraxella-Corynebacterium-Dolosigranulum (Mor-Cor-Dolo), and (9) a combination of Corynebacterium-Dolosigranulum (Cor-Dolo) (Fig. 2a). In line with the above, we observed strong age-related dynamics of the clusters over time (Fig. 2). Clusters with S. aureus emerged early in life and were gradually replaced by clusters dominated by Moraxella, Corynebacterium, Dolosigranulum, and/or Haemophilus species.
Fig. 2.
Microbial succession patterns during the first six months of life.
In order to gain more insight into microbial succession patterns, we tracked the children over time to follow their time path from one cluster at a certain time point to the same or another cluster at the consecutive time point.
(a) Heatmap showing per time point the percentile of children that belonged to each of the 11 clusters found.
(b) and (c) Graphic representation of the flow of children between clusters over time. By using co-regularized spectral clustering, we obtained per cluster a probabilistic likelihood (between zero and one) that a child belonged to this cluster. The higher the probability score, the higher the assignment of a child to a cluster. By depicting this for every child per time point, we were able to track cluster switching over time. The bars depict the clusters per time point (e.g. postpartum, day one), and the size of the bar represents the number of children belonging to that cluster. The surface between one cluster at a certain time point and the adjacent time point indicate the movements of children from that cluster to another. Panel b shows the movement of children over time when using a cut-off of the probabilistic score of 0.5. In panel c the cut-off of 0.8 is used and additionally the mode of delivery is depicted (pink surface between clusters = vaginally born children, blue = C-section born children).
Children that did not meet a value above the cut-off were put in the intermediate cluster (in panel b) or for convenience were left out of the figure (in panel c). Please note that since two adjacent time points fulfilling the criteria with a likelihood of > 0.5 (b) or > 0.8 (c) were required to track the switching between clusters, some cluster have a mismatch between “input”(left surface, going into a bar) and “output” (right side going to the next bars).
Abbreviations; Staph, Staphylococcus; Jant, Janthinobacterium; Strep, Streptococcus viridans; ML, Moraxella lincolnii; Cor, Corynebacterium; Dolo, Dolosigranulum; Mor, Moraxella; HI, Haemophilus influenzae; INT, Intermediate cluster (children with a probabilistic likelihood < 0.5). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
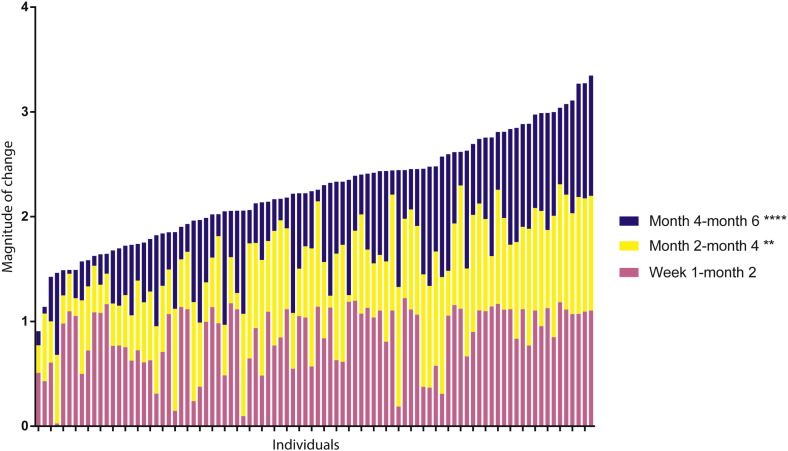
We calculated microbiota change over three periods, i.e. 0–2 months, 2–4 months and 4–6 months. The microbiota changed most during the first two months of life when compared to the other two intervals (p < 0.01 and p < 0.0001 respectively, Friedman test with Dunn's correction for multiple testing) (Fig. 3). Especially cluster HI was associated with a high magnitude of change (Fig. S7), and with a temporary colonization state (observed in individual children for a short period of time, i.e. only one time point). Moreover, the HI cluster was highly associated with respiratory infections as parents reported respiratory tract infection symptoms in five out of ten children in the HI cluster, (50.0%) vs 152/539 (22.0%) in the other clusters (p = 0.03, Chi-square). This association was observed independent of mode of delivery, although power was limited for this analysis.
Fig. 3.
Magnitude of relative change of the microbial profiles during the first six months of life.
The L2 norm value is depicted for the change between the samples at age one week and two months (pink), two and four months (yellow) and four and six months of age (blue), respectively. A higher norm value indicates a higher magnitude of change between the time points. The children were sorted from a lower magnitude of change (left) to a higher magnitude of total change (right). Although there are inter-individual differences in change over time, the highest magnitude of change occurred in general during the first two months of life, followed by the two-four months of age time frame and the four-six months of age time frame (p < 0.01 and p < 0.0001 respectively as compared to week one-two months of age). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
For identifying more stringent clusters, we used a probability assignment score of > 0.8/1.0 as a cut-off for assigning an individual sample to a specific cluster (Fig. 2c). Now, we were able to identify four very stringent clusters, dominated by Streptococcus, Moraxella-Corynebacterium-Dolosigranulum, Staphylococcus-Moraxella, and M. catarrhalis/nonliquefaciens. With respect to mode of delivery, we observed that children born by vaginal delivery tend to switch to the Moraxella- and Corynebacterium/Dolosigranulum-dominated profiles in an earlier stage compared to the children born by C-section, while the C-section born children stayed longer in the S. aureus-dominated profile (Fig. 2b). Moreover, time-related dynamics of microbiota development between C-section and vaginally born children appeared to be different (p = 0.015, randomization test). More in-depth analyses showed mode of delivery to be the strongest driver of S. aureus colonization and duration, and we found no significant other characteristics explaining duration of Staphylococcus-dominated colonization.
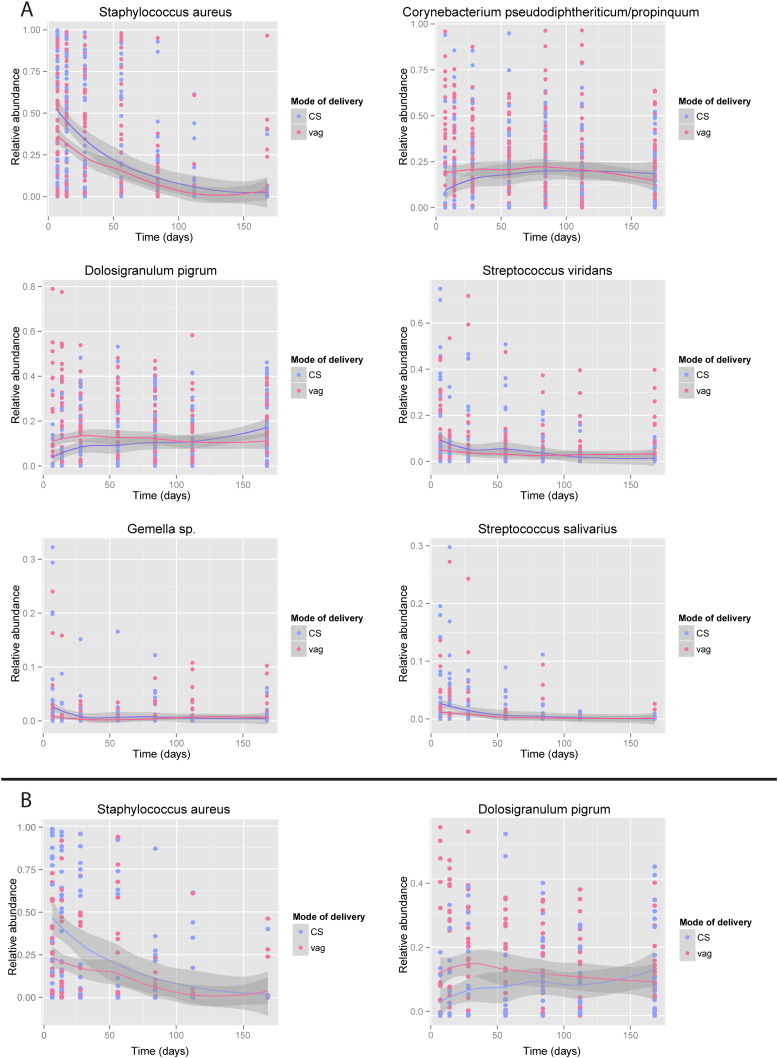
Additionally, we performed time-dependent analyses of oligotype abundance and dynamics to identify the biomarkers that are associated with modus partus, simultaneously correcting for feeding type, which is a known confounder. Over time, C-section born children had a higher abundance of S. aureus, S. viridans, Gemella sp., S. salivarius (q = 0.036, 0.036, 0.024, 0.036 respectively, calculated by natural spline regression models with Benjamini Hochberg correction for multiple testing) and a lower abundance of C. pseudodiphteriticum/propinquum and D. pigrum compared to vaginally born children (respectively q = 0.044, 0.007, natural spline regression models with Benjamini Hochberg correction for multiple testing). These patterns were most prominent at the early time points (Fig. 4a). The biomarkers identified using this approach were similar to the biomarkers of the clusters that were identified using co-occurrence spectral clustering algorithms, implicating that vaginally delivered children are more likely to develop towards the Corynebacterium and Dolosigranulum dominated profiles and less likely to stay within the Staphylococcus dominated profiles. Since vaginally born children in our study were more likely to be breastfed and many children switched feeding type over the duration of the study (in both mode of delivery groups), we repeated the analyses for children exclusively breast- (n = 19) or formula fed (n = 28) during the full six months of follow-up. Also in this subgroup analyses, we were still able to confirm lower abundance of S. aureus (q = 0.08) and higher abundance of D. pigrum (q = 0.08) in vaginally delivered children compared to C-section born children when corrected for feeding type (Fig. 4b).
Fig. 4.
Time course analyses of biomarker species.
Time-course behavior of (a) S. aureus, (b) C. pseudodiphteriticum/propinquum, (c) D. pigrum, (d) S. viridans, (e) Gemella sp., (f) S. salivarius based on mode of delivery in the overall cohort (panel a).
Time-course behavior of (a) S. aureus and (b) D. pigrum in a subset of children comparing exclusively breastfed with exclusively formula-fed children (respectively n = 19 and n = 28 children) (panel b).
4. Discussion
In this prospective birth cohort study, we show the dynamics of nasopharyngeal microbiota development with special emphasis on the mode of delivery in detail, from directly after birth until the age of six months in a large group of healthy term infants.
4.1. From Initial Colonization to Niche Differentiation
Directly after birth, different niches of the human body highly resemble each other (Dominguez-Bello et al., 2010). We confirm this to also be true for the respiratory tract at several hours after birth, showing a mixed profile of (presumed) fecal, vaginal, skin, oral, and environmental species. However, for the nasopharyngeal niche we observed niche-differentiation as early as at one week of age, with initial high incidence and abundance of S. aureus colonization. Furthermore, we found a dynamic process characterized by a rapid reduction in S. aureus and simultaneous emergence of Corynebacterium species and D. pigrum, followed by M. catarrhalis/nonliquefaciens, S. pneumoniae, and H. influenzae during the first six months of life. These profiles were also observed in other studies in young children (Biesbroek et al., 2014a, Biesbroek et al., 2014b, Teo et al., 2015). As almost all children were colonized by S. aureus at early age before switching to a profile characterized by Corynebacterium, Dolosigranulum and/or Moraxella, we speculate that S. aureus represents a so-called keystone species in early life, functioning as a backbone for a healthy development of the nasopharyngeal microbiota. S. aureus may be transferred from the skin of the parents or caregivers to the neonate. As S. aureus is known for its immune-evasive capacities (Johannessen et al., 2012) and induction of a low inflammatory profile of epithelium (Følsgaard et al., 2013), colonization of S. aureus may create a milieu that is more receptive to colonization by other commensal bacteria, such as Corynebacterium, Dolosigranulum, and/or Moraxella spp., which in turn are associated with niche stability over time and reduced risk of respiratory symptoms (Biesbroek et al., 2014b, Teo et al., 2015).
In contrast, it was found that early colonization by H. influenzae leads to a more pro-inflammatory profile (Følsgaard et al., 2013) and is associated with an increased risk of mild (Biesbroek et al., 2014b) and severe respiratory symptoms, including bronchiolitis (Vissing et al., 2013), pneumonia (Vissing et al., 2013), and otitis media (Leibovitz et al., 2004). Additionally, in our study H. influenzae is associated with parental-reported respiratory symptoms, which is accompanied by an unstable transient profile. Interestingly, the profile that is dominated by H. influenzae significantly differs from the profiles that preceded or followed a respiratory infection, suggesting that H. influenzae accompanies temporary dysbiosis and susceptibility to respiratory symptoms.
Although S. pneumoniae emerged between the ages of two and six months, in our study population it mostly showed to be a low-abundant community member of microbiota cluster dominated by other species in these first 6 months of life, which is in contrast to what we and others observed later in life where S. pneumoniae predominance is more commonly found (Bosch et al., 2015, Miller et al., 2011).
4.2. Impact of Mode of Delivery
Mode of delivery appears to be an important driver for respiratory health (Karlström et al., 2013, Kristensen et al., 2015). Birth by C-section has been associated with differences in innate and adaptive immunity such as lower levels of Th1-related cytokines (Jakobsson et al., 2014) and non-specific humoral immune responsiveness of the mucosa (Huurre et al., 2008). These differences in immunological responses may be a consequence of differences in microbial composition, since microbial colonization has shown to affect mucosal tolerance and homeostasis by microbiota-mediated signaling (Beck et al., 2012, Cernadas, 2011, Clarke, 2014). Although our results may suggest a limited direct impact of mode of delivery on nasopharyngeal microbiota composition directly after birth, we still observed subtle though significant differences in respiratory microbial development between children born by vaginal delivery and C-section over time. This suggests that the impact may not be mediated directly by the maternal inoculum, but rather by other indirect mechanisms such as low abundant bacteria that promote the outgrowth of new species, or by the indirect influences of microbial communities in other niches like the gut. Alternatively, lack of power to identify potential microbial drivers at the earliest time points might be a plausible explanation as well since we were only capable of profiling 15% of samples at day 0. Nevertheless, differences found in development of upper respiratory tract microbiota over the first six months of life may contribute to infant health, as C-section born children were 1) more likely to switch between clusters, indicating instability of the microbial profile, and 2) had a lower abundance of the potentially beneficial Corynebacterium and Dolosigranulum, especially in the first months of life. Both of these phenomena were related to an increase in respiratory infections over time in a previously published study (Biesbroek et al., 2014b). Moreover, vaginal birth as well as breastfeeding (Biesbroek et al., 2014a) seem to stimulate a microbial profile with (presumed) beneficial bacteria. Considering that C-section born children tend to more often receive formula feeding, which is presumed to lead to a less beneficial profile, it may be an interesting option to actively promote breastfeeding for C-section born children.
4.3. Window of Opportunity
Not only the exposure to microbes, but also the timing of this exposure is likely to have important clinical implications, as an early microbial profile seems to predict increased susceptibility to disease later in life (Biesbroek et al., 2014b, Martinez, 2014, Teo et al., 2015) and early infections may predispose to recurrent infections later on in life (Kvaerner et al., 1997). A window of opportunity was proposed in which establishing microbiota may be more dynamic and can therefore more easily be restored towards a healthy microbial profile. Although it is still unknown how long this period spans and to what extent the microbiota can be restored, results of a recently published pilot study showed that the microbiota of C-section born children partially reverted to the microbiota profiles of vaginally born children when the child was rubbed with the maternal vaginal microbiota directly after birth. The microbiota of the gut, skin, and oral niche of these children were found to be especially enriched with vaginal bacteria, which was most pronounced during the first week of life (Dominguez-Bello et al., 2016). Our data also show that the microbial succession pattern differs based on delivery mode between C-section born and vaginally delivered children beyond the first weeks of life; this could implicate that mode of delivery has a direct effect on the microbiota development and may have important clinical consequences to set an individual on a trajectory towards respiratory health or disease further in life.
4.4. Strengths and Limitations
The longitudinal study design with short time intervals allowed us to determine the microbial trajectory directly after birth until the age of six months. Strengths of our study include the consistency of data and the deep nasopharyngeal sample collection in a large, well defined and unselected birth cohort conducted by a trained research team. Therefore, we were able to describe reliably and in detail the very early dynamics in the nasopharyngeal microbiota. Enrollment took place over two consecutive years, thereby limiting potential confounding by environmental factors such as seasonal changes. Due to the prospective nature of the study, we were able to reliably correlate metadata of each time point to the corresponding microbial composition. An additional strength in this study is that direct perinatal effects of antibiotics will be minimal, since only one dose of antibiotic prophylaxis was administered during C-sections and only after the mother had delivered the child. In addition, perinatal use of antibiotics was very limited in our population with only 4/102 (4%) mothers receiving antibiotics in the first week of the infant's life. Therefore, we expect minimal direct influence of prophylactic/perinatal antibiotic use on the development of the neonatal microbiota. Moreover, in this study we used the more advanced oligotyping technique allowing us to annotate bacteria more commonly up to species level. Lastly, we used robust, unbiased, machine learning methods to determine clusters of similarity and the change in microbial succession over time.
Our study has some limitations; first, despite the short sampling intervals, the microbial composition at any given sample moment provides only a snapshot of a constantly changing ecosystem. Therefore, we can only make assumptions on the true dynamical changes that have occurred over time. Second, we were able to analyze 84% of samples, and only 15% of samples from the earliest time point limiting the statistical power to detect potential bacterial drivers of the observed differences between respiratory microbiota development of C-section and vaginally delivered infants.
5. Conclusion
In conclusion, the respiratory microbiota in neonatal life develops from an initially maternally or environmentally transmitted mixed flora via S. viridans to S. aureus dominance, followed by differentiation into one of several niche-specific microbiota profiles after one week of age. Mode of delivery affected early respiratory microbiota development significantly, especially the timing of seeding and the abundance of potentially protective commensals. These effects on early microbiota development may be an important clue for the role of the microbiota in respiratory health later in life, which therefore deserves further investigation.
Author Contributions
Author contributions: D.B., M.A. van H., and E.A.M.S. conceived and designed the experiments. A.A.T.M.B., G.B., P.C.M. de G. and P.P. included the patients, A.A.T.M.B. and G.B. executed the study follow up. R.H. performed the DNA isolation of the samples. A.A.T.M.B, E.L., G.K., W.A.A. de S.P., B.K., E.A.M.S., and D.B. analysed the data. A.A.T.M.B., E.L., B.K., and D.B. wrote the paper. All authors significantly contributed to interpreting the results, critically revised the manuscript for important intellectual content, and approved the final manuscript.
Funding
This work was supported by the Spaarne Gasthuis Hoofddorp and University Medical Center Utrecht, the Dutch Organization for Scientific Research through NWO-Vidi grant (91715359), and Top consortia for Knowledge and Innovation, Agri & Food (TKI-AF-12190). The funding sources had no role in the design, execution, analyses, or interpretation of the data of this study.
Conflict of Interest
EAMS declares to have received unrestricted research support from Pfizer, grant support for vaccine studies from Pfizer and GSK and fees paid to the institution for advisory boards or participation in independent data monitoring committees for Pfizer and GSK. None of the fees or grants listed here was received for the research described in this paper. All other authors report no competing interests.
Acknowledgments
We wish to thank the department of Obstetrics and Gynecology of the Spaarne Gasthuis and participating midwifery clinics for recruiting participants for this study; the research team of the Spaarne Gasthuis Academy for their outstanding dedication in executing the study; the laboratory staff of the University Medical Center Utrecht and TNO for their excellent lab work, and the cooperating institutes for their commitment to the project. Most of all, we are indebted to all the participating children and their families.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.05.031.
Appendix A. Supplementary Data
Supplementary material.
References
- Beck J.M., Young V.B., Huffnagle G.B. The microbiome of the lung. Transl. Res. 2012;160:258–266. doi: 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesbroek G., Sanders E.A.M., Roeselers G., Wang X., Caspers M.P.M., Trzciński K., Bogaert D., Keijser B.J.F. Deep sequencing analyses of low density microbial communities: working at the boundary of accurate microbiota detection. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesbroek G., Bosch A.A.T.M., Wang X., Keijser B.J.F., Veenhoven R.H., Sanders E.A.M., Bogaert D. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am. J. Respir. Crit. Care Med. 2014;190:298–308. doi: 10.1164/rccm.201401-0073OC. [DOI] [PubMed] [Google Scholar]
- Biesbroek G., Tsivtsivadze E., Sanders E.A.M., Montijn R., Veenhoven R.H., Keijser B.J.F., Bogaert D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 2014 doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- Bosch A.A.T.M., van Houten M.A., Bruin J.P., Wijmenga-Monsuur A.J., Trzciński K., Bogaert D., Rots N.Y., Sanders E.A.M. Nasopharyngeal carriage of Streptococcus pneumoniae and other bacteria in the 7th year after implementation of the pneumococcal conjugate vaccine in the Netherlands. Vaccine. 2015;34:531–539. doi: 10.1016/j.vaccine.2015.11.060. [DOI] [PubMed] [Google Scholar]
- Cernadas M. It takes a microbiome: commensals, immune regulation, and allergy. Am. J. Respir. Crit. Care Med. 2011;184:149–150. doi: 10.1164/rccm.201105-0828ED. [DOI] [PubMed] [Google Scholar]
- Clarke T.B. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect. Immun. 2014;82:4596–4606. doi: 10.1128/IAI.02212-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello M.G., Costello E.K., Contreras M., Magris M., Hidalgo G., Fierer N., Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello M.G., De Jesus-Laboy K.M., Shen N., Cox L.M., Amir A., Gonzalez A., Bokulich N.A., Song S.J., Hoashi M., Rivera-Vinas J.I., Mendez K., Knight R., Clemente J.C. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016;22:250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren A.M., Morrison H.G., Lescault P.J., Reveillaud J., Vineis J.H., Sogin M.L. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 2014;9:968–979. doi: 10.1038/ismej.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Følsgaard N.V., Schjørring S., Chawes B.L., Rasmussen M.A., Krogfelt K.A., Brix S., Bisgaard H. Pathogenic bacteria colonizing the airways in asymptomatic neonates stimulates topical inflammatory mediator release. Am. J. Respir. Crit. Care Med. 2013;187:589–595. doi: 10.1164/rccm.201207-1297OC. [DOI] [PubMed] [Google Scholar]
- Gollwitzer E.S., Saglani S., Trompette A., Yadava K., Sherburn R., McCoy K.D., Nicod L.P., Lloyd C.M., Marsland B.J. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 2014;20:642–647. doi: 10.1038/nm.3568. [DOI] [PubMed] [Google Scholar]
- Guibas G.V., Moschonis G., Xepapadaki P., Roumpedaki E., Androutsos O., Manios Y., Papadopoulos N.G. Conception via in vitro fertilization and delivery by Caesarean section are associated with paediatric asthma incidence. Clin. Exp. Allergy. 2013;43:1058–1066. doi: 10.1111/cea.12152. [DOI] [PubMed] [Google Scholar]
- Harville D.A. Springer; New York: 1997. Matrix Algebra From a Statistician's Perspective | Springer [WWW Document] [Google Scholar]
- Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huurre A., Kalliomäki M., Rautava S., Rinne M., Salminen S., Isolauri E. Mode of delivery - effects on gut microbiota and humoral immunity. Neonatology. 2008;93:236–240. doi: 10.1159/000111102. [DOI] [PubMed] [Google Scholar]
- Imangaliyev S., Keijser B., Crielaard W., Tsivtsivadze E. Personalized microbial network inference via co-regularized spectral clustering. Methods. 2015;83:28–35. doi: 10.1016/j.ymeth.2015.03.017. [DOI] [PubMed] [Google Scholar]
- Jakobsson H.E., Abrahamsson T.R., Jenmalm M.C., Harris K., Quince C., Jernberg C., Björkstén B., Engstrand L., Andersson A.F. Decreased gut microbiota diversity, delayed bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- Johannessen M., Sollid J.E., Hanssen A.-M. Host- and microbe determinants that may influence the success of S. aureus colonization. Front. Cell. Infect. Microbiol. 2012;2:56. doi: 10.3389/fcimb.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlström A., Lindgren H., Hildingsson I. Maternal and infant outcome after caesarean section without recorded medical indication: findings from a Swedish case-control study. BJOG. 2013;120:479–486. doi: 10.1111/1471-0528.12129. (discussion 486) [DOI] [PubMed] [Google Scholar]
- Kong Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics. 2011;98:152–153. doi: 10.1016/j.ygeno.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Koppen I.J.N., Bosch A.A.T.M., Sanders E.A.M., van Houten M.A., Bogaert D. The respiratory microbiome during health and disease; a paediatric perspective. Pneumonia. 2015;6:90–100. doi: 10.15172/pneu.2015.6/656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen K., Fisker N., Haerskjold A., Ravn H., Simões E.A.F., Stensballe L. Caesarean section and hospitalization for respiratory syncytial virus infection: a population-based study. Pediatr. Infect. Dis. J. 2015;34:145–148. doi: 10.1097/INF.0000000000000552. [DOI] [PubMed] [Google Scholar]
- Kvaerner K.J., Nafstad P., Hagen J.A., Mair I.W., Jaakkola J.J. Recurrent acute otitis media: the significance of age at onset. Acta Otolaryngol. 1997;117:578–584. doi: 10.3109/00016489709113441. [DOI] [PubMed] [Google Scholar]
- Leibovitz E., Jacobs M.R., Dagan R. Haemophilus influenzae: a significant pathogen in acute otitis media. Pediatr. Infect. Dis. J. 2004;23:1142–1152. [PubMed] [Google Scholar]
- Martinez F.D. The human microbiome. Early life determinant of health outcomes. Ann. Am. Thorac. Soc. 2014;11(Suppl. 1):S7–12. doi: 10.1513/AnnalsATS.201306-186MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E., Andrews N.J., Waight P.A., Slack M.P.E., George R.C. Herd immunity and serotype replacement 4 years after seven-valent pneumococcal conjugate vaccination in England and Wales: an observational cohort study. Lancet Infect. Dis. 2011;11:760–768. doi: 10.1016/S1473-3099(11)70090-1. [DOI] [PubMed] [Google Scholar]
- Mueller N.T., Bakacs E., Combellick J., Grigoryan Z., Dominguez-Bello M.G. The infant microbiome development: mom matters. Trends Mol. Med. 2015;21:109–117. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD Publishing . 2013. “Caesarean Sections” [WWW Document]. Heal. A Glance 2013 OECD Indic. [Google Scholar]
- Penders J., Gerhold K., Stobberingh E.E., Thijs C., Zimmermann K., Lau S., Hamelmann E. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J. Allergy Clin. Immunol. 2013;132:601–607. doi: 10.1016/j.jaci.2013.05.043. [DOI] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., Sahl J.W., Stres B., Thallinger G.G., Van Horn D.J., Weber C.F. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Xiao W., Leek J.T., Tompkins R.G., Davis R.W. Significance analysis of time course microarray experiments. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo S.M., Mok D., Pham K., Kusel M., Serralha M., Troy N., Holt B.J., Hales B.J., Walker M.L., Hollams E., Bochkov Y.A., Grindle K., Johnston S.L., Gern J.E., Sly P.D., Holt P.G., Holt K.E., Inouye M. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015 doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thavagnanam S., Fleming J., Bromley A., Shields M.D., Cardwell C.R. A meta-analysis of the association between Caesarean section and childhood asthma. Clin. Exp. Allergy. 2008;38:629–633. doi: 10.1111/j.1365-2222.2007.02780.x. [DOI] [PubMed] [Google Scholar]
- Tsivtsivadze E., Borgdorff H., Wijgert J.v.d., Schuren F., Verhelst R., Heskes T. Springer; Berlin Heidelberg, Berlin, Heidelberg: 2013. Partially Supervised Learning, 2nd IAPR International Workshop on Partially Supervised Learning, PSL 2013, 13 May 2013 through 14 May 2013, Nanjing, 8183 LNAI, 80–90, Lecture Notes in Computer Science. [Google Scholar]
- Tsivtsivadze E., Imangaliyev S., Keijser B.J.F. Workshop on Machine Learning in Computational Biology (MLCB) Annual Conference on Neural Information Processing Systems (NIPS); Montréal, Canada: 2014. Unsupervised multi-view feature selection via co-regularization. [Google Scholar]
- Vissing N.H., Chawes B.L.K., Bisgaard H. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. Am. J. Respir. Crit. Care Med. 2013;188:1246–1252. doi: 10.1164/rccm.201302-0215OC. [DOI] [PubMed] [Google Scholar]
- Wyllie A.L., Chu M.L.J.N., Schellens M.H.B., van Engelsdorp Gastelaars J., Jansen M.D., van der Ende A., Bogaert D., Sanders E.A.M., Trzciński K. Streptococcus pneumoniae in saliva of Dutch primary school children. PLoS One. 2014;9 doi: 10.1371/journal.pone.0102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.