Abstract
The innate immune protein Stimulator of interferon genes (STING) promotes the induction of interferon beta (IFN-β) production via the phosphorylation of its C-terminal tail (CTT) by TANK-binding kinase 1 (TBK1). Potent ligands of STING are, therefore, promising candidates for novel anti-cancer drugs or vaccine adjuvants. However, the intrinsically flexible CTT poses serious problems in in silico drug discovery. Here, we performed molecular dynamics simulations of the STING fragment containing the CTT in ligand-bound and unbound forms and observed that the binding of a potent ligand cyclic GMP-AMP (cGAMP) induced a local structure in the CTT, reminiscent of the known structure of a TBK1 substrate. The subsequent molecular biological experiments confirmed the observed dynamics of the CTT and identified essential residues for the activation of the IFN-β promoter, leading us to propose a new mechanism of STING activation.
Keywords: STING C-terminal tail, Molecular dynamics simulation, Type I IFNs, TBK1; cGAMP.
Highlights
-
•
The binding of the potent ligand cGAMP to STING induces local structural ordering in the flexible C-terminal tail of STING.
-
•
Site-directed mutagenesis studies, designed based on this observation, elucidated residues essential for STING function.
-
•
The local structure formation in the CTT was shown to be essential for the induction of IFN-β production.
1. Introduction
Stimulator of interferon genes (STING) is an essential adaptor protein in innate immunity, which plays an important role in inducing interferon beta (IFN-β) production when a cell is infected by pathogens (Ishikawa and Barber, 2008, Ishikawa et al., 2009). In the cytosol, cyclic GMP-AMP synthase (cGAS) binds to dsDNA derived from pathogen or host-damaged cells, and produces a small molecule ligand, cyclic GMP-AMP (cGAMP) (Ablasser et al., 2013, Diner et al., 2013, Sun et al., 2013). After the binding of cGAMP to STING, STING is phosphorylated by a SER/THR kinase, TANK-binding kinase 1 (TBK1) (Gao et al., 2013, Tanaka and Chen, 2012, Zhang et al., 2013). Phosphorylated STING recruits a transcription factor, interferon regulatory factor 3 (IRF3) and facilitates the phosphorylation of IRF3 by TBK1, which leads to the dimerization of IRF3 (Larabi et al., 2013, Liu et al., 2015, Qin et al., 2003, Shu et al., 2013, Tanaka and Chen, 2012). The IRF3 dimer then is translocated to the nucleus and binds to the IFN-β promoter, resulting in its production (Tanaka and Chen, 2012). Small compounds that can bind to STING, therefore, are promising candidates for anti-cancer drugs or vaccine adjuvants.
STING consists of 379 residues and forms a homo-dimer. STING is divided into three parts, an N-terminal domain that includes four transmembrane regions (1–154), a dimerization and ligand-binding domain (155–342), and a C-terminal tail (CTT, 343–379) that includes residues phosphorylated by TBK1. More than 20 crystal structures of mouse and human STING have been determined to date. These structures include complex structures with cGAMP and cyclic-di-GMP (c-di-GMP). The latter is also a natural ligand and shows lower activity for IFN-β production than cGAMP. All of the known STING structures contain only the dimerization and ligand binding domain, probably due to the structural flexibility of the N- and C-terminal domains (Gao et al., 2013, Zhang et al., 2013). These structures show that ligand binding leads to a conformational change in a segment (224–244, hereafter referred to as the Lid) proximal to the ligand-binding site (Huang et al., 2012, Zhang et al., 2013). In the ligand-bound structures, the Lid segment from each subunit form a closed conformation by forming a β-hairpin and putting the two β-hairpin together to form a four-stranded antiparallel β-sheet. This β-sheet covers the ligand-binding pocket located in the interface of the two subunits, hence the name “lid”. On the other hand, in the ligand-unbound form, the Lid has an open conformation and the ligand-binding site is exposed to the solvent. In c-di-GMP bound human STING, however, four of the five crystal structures have open Lid conformations, suggesting that low-activity ligands cannot fully maintain the closed Lid conformation (Huang et al., 2012). The small-molecule compound 5,6-dimethylxanthenone-4-acetic acid (DMXAA) has been shown to bind and activate mouse, but not human, STING (Gao et al., 2013). Gao et al. (2014) have demonstrated that the mutation of G230I in the Lid of human STING leads to the binding of DMXAA and the activation of IFN-β production. This mutation induces the closed-Lid conformation, even with the supposedly low affinity of DMXAA for human STING, suggesting that the closure of the Lid by ligand binding is essential for IFN-β activity.
Molecular features of STING have been investigated by many research groups, and a number of amino acid residues implicated for STING function have been identified. Those residues include phosphosites to induce the activation of the IFN-β promoter (Konno et al., 2013, Tanaka and Chen, 2012, Zhong et al., 2008); multiple studies using molecular biological techniques have identified both common and unique amino acids, such as S324, S326, S358 and S366 in human STING, to be phosphorylated on STING activation. In addition, genetic approaches with human SNP analysis or genome analysis of goldenticket mice have identified other amino acid residues important for ligand binding (Sauer et al., 2011). However, no correlation has been established so far between the IFN-β inducing activity of a ligand and its binding affinity (Gao et al., 2013). One possible explanation is the differences in the ability of these ligands to induce structural changes in STING. For example, changes in the dynamics of the CTT may lead to the phosphorylation of SER and THR residues by TBK1.
In the current work, we adopted an interdisciplinary approach and predicted the dynamics of STING-CTT by in silico simulations, followed by the characterization of candidate residues by molecular biological techniques. We first constructed a structural model of the CTT in human STING and performed molecular dynamics (MD) simulations in ligand-bound and unbound forms. The simulation of cGAMP-bound human STING-CTT showed a behavior completely different from those in the c-di-GMP-bound and ligand-unbound forms. Site-directed mutagenesis studies, designed based on these observations, elucidated a phosphosite S354 in mouse (S355 in human) and three residues important for STING function, L362, L363 and M223 in mouse (L363, L364, and K224 in human, respectively), all of which have not been reported before. Our results suggest that a combination of computer simulations and molecular biological analysis is a powerful approach to investigating complex molecular features.
2. Materials and Methods
2.1. Computation
2.1.1. The Construction of STING-CTT Models and the Execution of MD Simulations
We constructed a structural model of the CTT manually by using SYBYL-X (Tripos International, 1699), and performed energy minimization (50,000 steps), equilibration (500 ps NVT and NPT) and short MD simulation (3 ns) at 300 K in the cubic box (protein + 1.0 nm) filled with tip3p solvents, using the amber99sb force field by Gromacs 4.6.2 (Abraham et al., 2015) on Fujitsu PRIMEHPC FX10. The structurally stabilized CTT model was added to the end of both subunits in crystal structures of human STING (PDBIDs 4ksy, 4f5d and 4f5e). We performed energy minimization (50,000 steps) and equilibration (1 ns) of the model of STING with the CTT (STING-CTT), where the topologies of ligands were calculated by using amber12 antechamber (Wang et al., 2006, Wang et al., 2004). The structurally stabilized STING-CTT models were used as the initial structures of three MD simulations, as described in Results. The MD simulations of STING-CTT models were performed at 300 K for 700 ns in the same conditions as described above. The simulation of the cGAMP-bound form was extended to 1 μs, to observe the formation and stability of the local structure in the CTT.
2.1.2. Calculation of Electrostatic Potential on Molecular Surfaces
The electrostatic potential on the molecular surfaces of proteins and ligands were calculated by SCB (Nakamura and Nishida, 1987). The program assigns an electrostatic potential value to each vertex of triangles that construct a molecular surface calculated by the program MSP (Connolly, 1983). All the figures were drawn by the interactive molecular viewer jV (Kinoshita and Nakamura, 2004).
2.1.3. Calculation of Hydrophobic Interactions
The hydrophobic interactions involving L374 were estimated at every 50 ns from the interaction network calculated by RINerator (Doncheva et al., 2011), where the programs Reduce (Word et al., 1999b) and Probe (Word et al., 1999a) are implemented.
2.2. Experiments
2.2.1. Cell Culture
HEK293 were purchased from the American Type Culture Collection (ATCC), and were maintained in DMEM supplemented with FCS and 50 mg/ml each of penicillin and streptomycin. The sting knockout MEFs and its counterpart MEFs were kindly gifted by G. Barber (University of Miami Miller School of Medicine). All MEFs used in this study were maintained in DMEM supplemented with FCS and 50 mg/ml each of penicillin and streptomycin.
2.2.2. Plasmids Construction
STING expression plasmids were generated by PCR amplification with template of a mouse spleen cDNA library. The cDNA fragments were introduced into pCIneo (Promega) with HA-tag at the carboxyl terminus. The expression plasmids of STING point mutants were constructed as shown previously (Jounai et al., 2007).
2.2.3. Generation of Stably Transformed MEFs
Recombinant lentiviruses expressing STING or STING mutants were generated by transfection with pCAG-HIVgp, pCMV-VSV-G-RSV-Rev. and either CS-STING-IRES-puro or other CS-based expression plasmids into HEK293 cells. Forty-eight to seventy-two hours following transfection, the culture supernatants containing recombinant lentiviruses were recovered, and transferred to the fresh MEFs. The MEFs expressing STING or various STING mutants were selected in the presence of puromycin (2 mg/ml).
2.2.4. Luciferase Assay
HEK293 cells seeded on 24-well plates (2 × 105 cells/well) were transiently transfected with 25 ng of firefly luciferase reporter plasmid encoding IFN-β promoter, 25 ng of Renilla luciferase plasmid, and 450 ng of expression plasmid for wild type of STING or STING point mutants. Forty-eight hours later from transfection, the luciferase expression level was measured with the Dual-luciferase Reporter Assay System (Promega, Madison, WI). The firefly luciferase activity was normalized to Renilla luciferase activity in each sample.
2.2.5. Immunoprecipitation and Immunoblotting
Immunoprecipitation and immunoblotting analyses were performed using anti-phosphorylated SER/THR (Cell signaling technology), anti-IRF3 (Cell signaling technology), anti-p-IRF3 (Ser396) (Cell signaling technology), or anti-HA (Roche Diagnostics) antibodies, as described previously (Jounai et al., 2007). To observe STING phosphorylation in immunoblotting, 7.5% poly-acrylamide gel was prepared. For evaluating the STING expression levels in the luciferase assay, NuPAGE gradient gel (4–12%, Thermo Fisher Scientific) was used.
2.2.6. In vitro Kination Assay
HA-tagged STING (STING-HA) was immunoprecipitated from MEFs stably expressing STING-HA with an anti-HA antibody (COVANCE). STING-HA substrates were mixed with TBK1 kinase (MILLIPORE) in the phosphorylation buffer (20 mM HEPES buffer (pH 7.4) with 100 mM NaCl, 0.1% NP-40, 10% glycerol, 5 mM MgCl2, 5 mM MnCl2, 20 mM β-glycerophosphate, and 100 μM Na3VO4) on ice. Some samples were additionally mixed with 100 μM ATP with/without 50 nM staurosporine (Calbiochem). The kination reaction was performed at 30 °C for 20 min, and immunoblotting was performed immediately.
3. Results
3.1. MD Simulations of Human STING-CTT
We generated a random structure as a model of the CTT and attached it to the C-terminal end of both subunits in selected crystal structures of human STING (see Materials and Methods for details. These structures are herein referred to as STING-CTTs). The energy-minimized STING-CTTs were used as the initial structures of three MD simulations (Supplementary Fig. S1a, see Materials and Methods): 1) cGAMP-bound, 2) c-di-GMP-bound and 3) ligand-unbound forms (with simulation times of 1 μs, 700 ns and 700 ns, respectively). Snapshots at the end of the simulations are shown in Fig. 1 and Supplementary Fig. S1b. The c-di-GMP-bound and ligand-unbound forms produced stable but open Lid structures with no major movements of the CTTs after about 300 ns, while the cGAMP-bound form produced stable structures after 600 ns (Supplementary Fig. S1c). We will describe three major observations from the simulations below.
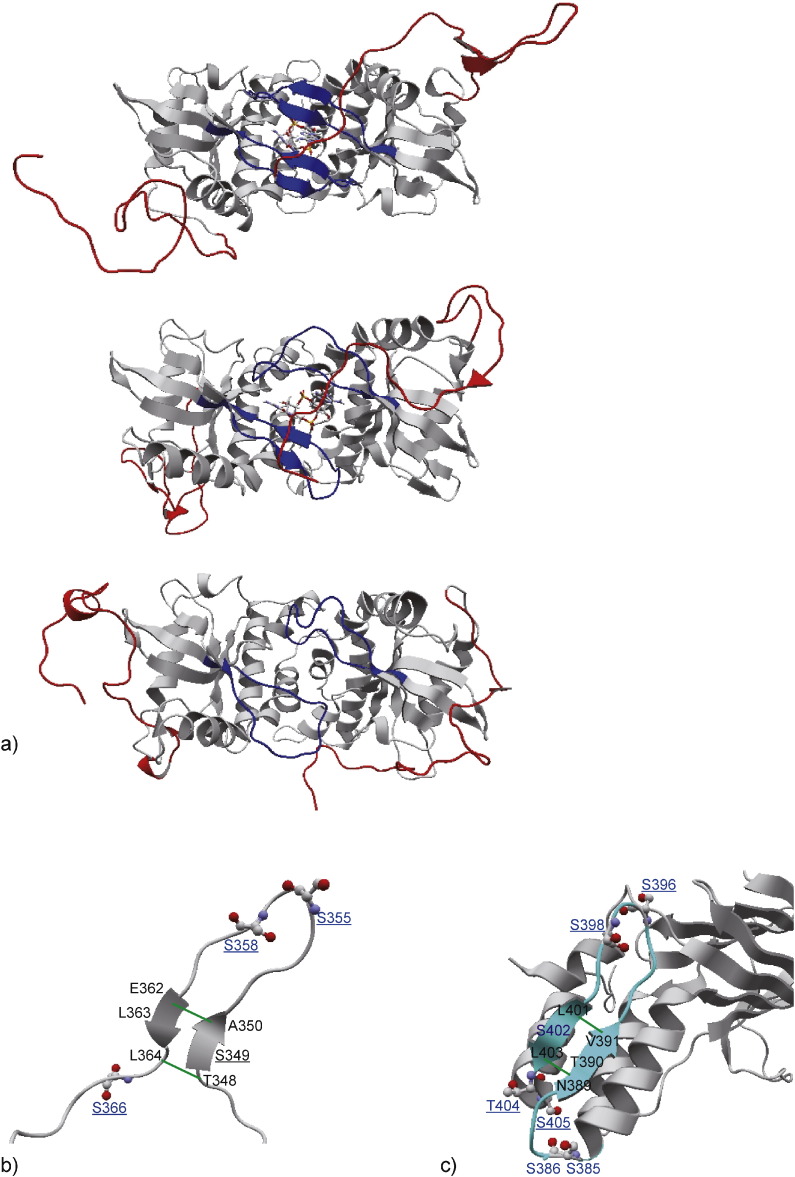
Fig. 1.
The results of the MD simulations of STING-CTT. a) Snapshots at the end of the following three MD simulations, 1) cGAMP-bound human STING-CTT (simulation time: 1 μs), 2) c-di-GMP-bound form (700 ns) and 3) ligand-unbound form (700 ns). The Lid and the CTT are colored blue and red, respectively, where the CTT of the right subunit showed a remarkable movement. The ligands cGAMP and c-di-GMP are shown in ball and stick model. b) The observed CTT structure in the cGAMP-bound form at 780 ns. The hydrogen bonds in the β-sheet are formed between main-chain atoms in T348 and L364 and those in A350 and E362, which were estimated by HBPLUS (McDonald and Thornton, 1994) and shown by green lines. The conserved SERs in mouse and human STING are underlined, where putative phosphosites are colored blue. c) The structure of the SER-rich region in the recognition domain of IRF3 (PDBID 3a77). The SER-rich region that contains phosphosites around the β-sheet is colored cyan, where hydrogen bonds between main-chain atoms in the β-sheet are shown by green lines. Blue colored residues are identified as phosphosites by several experimental studies (Lin et al., 1998, Yoneyama et al., 1998). The residues corresponding to the putative phosphosites in STING-CTT are underlined.
3.1.1. The cGAMP-bound Form Maintained the Closed Lid Conformation
The Lid retained the closed form in the cGAMP-bound form throughout the simulation, while it became open in the c-di-GMP-bound form even though the initial structure had a closed Lid conformation (Fig. 1a, where the Lid structures are colored blue). In the c-di-GMP-bound form, the Lid started to open at around 30 ns and the distance between the Lid regions in both subunits increased to about 2.5 times the initial length (Supplementary Fig. S2a). These observations indicate that only cGAMP has the ability to maintain the closed Lid conformation.
3.1.2. The End of the CTT Moved Onto the Lid in the Ligand-bound Forms
The end of the CTT moved onto the Lid and stayed there in the ligand-bound forms, while it kept away from the center of the Lid in the ligand-unbound form (Fig. 1a, where the CTT structures are colored red). We quantified this observation by the number of CTT atoms in contact with at least one atom in the Lid from either subunit, where the contacting atoms were defined to be within 6.0 Å of each other (Supplementary Fig. S2b). In all three simulations, the CTT from one subunit (the right subunit in Fig. 1a) came closer to the Lid and stayed there. We will describe only these CTTs hereafter. In the ligand-bound forms, the “right” CTT atoms (in subunit B) frequently contacted both the “right” and “left” Lid atoms (subunits B and A, respectively) after the movement of the end of the CTT onto the Lid (around 29 ns and 38 ns in the cGAMP- and c-di-GMP-bound forms, respectively). On the other hand, in the ligand-unbound form, the “right” CTT atoms (subunit B) started to approach the Lid from the other direction and started contacting only the “left” Lid (subunit A) after 250 ns (Fig. 1a and Supplementary Fig. S2b). Thus, only in the ligand-bound forms, the end of the CTT can cover the entire portion of the Lid.
3.1.3. A Local Structure in the CTT was Formed Only in the cGAMP Bound Form
Only in the cGAMP-bound form, a β-sheet was formed in the CTT (Fig. 1b). The β-sheet was formed between regions 348–350 and 362–364, because of the formation of hydrogen bonds between the main-chain atoms of residues 348 and 364 and those between residues 350 and 362. The β-sheet was formed at around 547 ns, and it was not broken until the end of the simulation. The prediction of disordered regions (Shimizu et al., 2007) and secondary structures (Jones, 1999) showed partial order and β-structures with moderate confidence around the β-sheet observed in the simulation (Supplementary Fig. S2c), which supports the simulation result. Note that the transient secondary structure formation during the MD simulation is compatible with no electron densities observed for the CTT in the existing crystal structures; only those structures stable for the time-scale of crystallographic analysis should be visible.
TBK1 also phosphorylates IRF3 in a SER-rich region of its recognition domain (Lin et al., 1998, Yoneyama et al., 1998). We compared the local structure observed in STING-CTT to the crystal structure of the SER-rich region in IRF3 and found that the known phosphorylated SER residues were placed in similar positions with respect to the β-sheet (Fig. 1c). These observations suggest that the β-sheet in the CTT is an appropriate conformation for the substrate recognition by TBK1.
3.2. Putative Phosphosites in the CTT
There are four SERs and two THRs in the CTT in mouse STING (seven and four in human STING, respectively), and all the four SERs and two THRs in mouse are conserved between the two species (Fig. 1b). After the β-sheet formation (at 547 ns), T348 and S349 formed new hydrogen bonds with the residues in the regions other than the CTT, while T376 formed hydrogen bonds before the β-sheet formation and retained them until the end of the simulation (shown as dots in Supplementary Fig. S2d). The other three SER residues, S355, S358 and S366, formed no hydrogen bonds after the β-sheet formation and had large accessible surface areas of the O-γ atoms (Supplementary Fig. S2e), suggesting that these three SERs have a high possibility to be phosphorylated by TBK1.
3.3. Site-directed Mutagenesis Study of Conserved SERs and THRs in the CTT
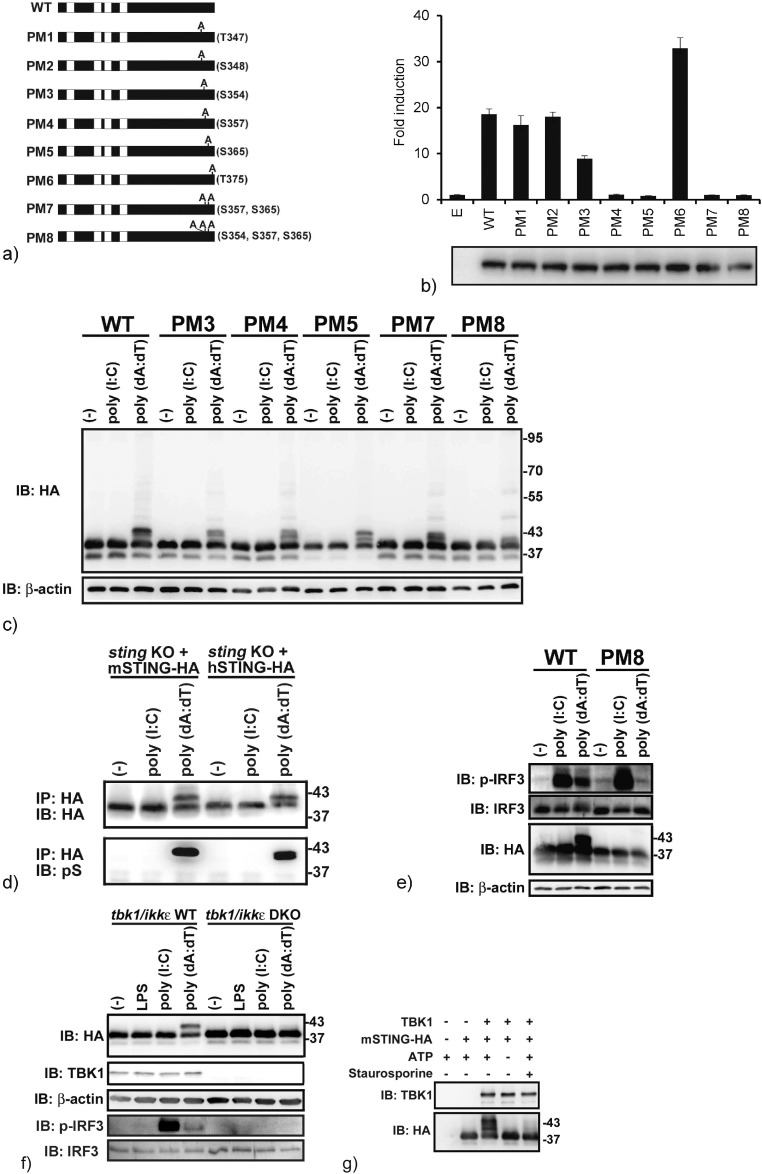
Previous reports showed that the CTT of STING included several phosphosites related to its function of sensing cytosolic DNA. Since our simulation study showed three of the four conserved SERs as candidate phosphosites, all the SER and THR residues conserved between human and mouse were mutated to ALA, and then all point mutants of mouse STING were expressed in HEK293 cells to evaluate IFN-β promoter activation (Fig. 2a and b). The point mutation at T347, S348 and T375 (human T348, S349 and T376, respectively) induced the IFN-β promoter activity comparable to that of the wild type mouse STING (Fig. 2b). However, the point mutation at S354 (human S355) reduced IFN-β promoter activity, and that at S357 (human S358) or S365 (human S366) completely abolished the ability to activate the IFN-β promoter, indicating that S354, S357 and S365 of mouse STING are important for the activation of the IFN-β promoter (Fig. 2b).
Fig. 2.
Experimental verification of the simulation results. a) A schematic representation of the STING point mutants generated. b) IFN-β promoter activity. HEK293 cells were co-transfected with pIFN-β Luc, pRG-TK and indicated plasmids. Forty-eight hours post-transfection, the reporter assay was performed. The expression levels of Renilla luciferase were used as internal control. The bottom image shows expression levels of STING-HA by immunoblotting with an anti-HA antibody. The experiments were performed twice, and representative figure was shown. The data were expressed the means ± SD from triplicates. c–e) The sting KO MEFs stably expressing point mutants of STING were treated with dsRNA or dsDNA for 3 h, and then immunoprecipitation and/or immunoblotting analyses were performed with the antibodies indicated. f) The tbk1/ikke WT or DKO MEFs, stably expressing HA-tagged STING, were stimulated with indicated stimuli for 3 h, and then immunoblotting was performed. g) HA-tagged STING was immunoprecipitated from sting KO MEFs stably expressing HA-tagged STING, and precipitates were subjected to an in vitro kination assay.
To confirm whether S354, S357 and S365 of mouse STING are actually phosphorylated in response to intracellular dsDNA, point mutants of STING were stably expressed into sting knockout murine embryonic fibroblasts (MEFs), and then those MEFs were stimulated by dsRNA or dsDNA to observe STING phosphorylation (Fig. 2a and c). Single point mutations at S354, S357 or S365 showed a slight decrease in STING phosphorylation (size of nearly 43 kDa) by dsDNA stimulation compared with the wild type (Fig. 2c). However, double mutations at S357 and S365 changed the band mobility of phosphorylated STING (PM7 in Fig. 2c), and the triple mutation dramatically decreased the phosphorylated STING bands upon dsDNA stimulation (PM8 in Fig. 2c), suggesting that at least S354, S357 and S365 of mouse STING (S355, S358 and S366 of human STING) are phosphorylated in response to cytosolic dsDNA. To prove dsDNA-mediated phosphorylation of STING, anti-pSER/THR antibodies were used for immunoblotting (Fig. 2d). As expected, both mouse and human STING showed the STING protein bands containing phosphorylated SER and THR residues upon dsDNA stimulation (Fig. 2d).
3.4. The Importance of the Phosphorylation of STING for IRF3 Activation
To evaluate the importance of STING phosphorylation at S355, S358 and S366 for IRF3 phosphorylation, MEFs stably expressing the wild type or the triple mutant mouse STING were stimulated with dsRNA or dsDNA, and then immunoblotting was performed (Fig. 2e). The treatment of dsRNA led to IRF3 phosphorylation in both transformants (Fig. 2e). However, the MEFs with the triple mutant mouse STING showed decreased ability to phosphorylate IRF3 in response to cytosolic dsDNA, indicating that STING phosphorylation is essential for IRF3 activation upon dsDNA stimulation.
3.5. TBK1 Phosphorylates STING Directly in Response to dsDNA Stimulation
To confirm whether the phosphorylation of STING upon intracellular dsDNA-sensing is also regulated by TBK1, tbk1/ikke double knockout MEFs that stably expressed HA-tagged mouse STING were constructed (Fig. 2f). The treatment with dsDNA, but not LPS or dsRNA, resulted in the phosphorylation of mouse STING in the tbk1/ikke wild-type MEFs (Fig. 2f). However, in the tbk1/ikke double knockout MEFs, the phosphorylation of mouse STING in response to dsDNA was abolished completely. In addition, IRF3 was phosphorylated by stimuli in the tbk1/ikke wild-type MEFs but not the tbk1/ikke knockout MEFs, suggesting that TBK1 is required for the phosphorylation of STING in response to intracellular dsDNA (Fig. 2f). To demonstrate that TBK1 directly phosphorylates mouse STING, an in vitro kination experiment was performed. As shown in Fig. 2g, TBK1 phosphorylated mouse STING in an ATP dependent manner, and the addition of staurosporine, a pan-kinase inhibitor, reduced the phosphorylated STING bands, suggesting that TBK1 directly phosphorylates STING.
3.6. Site-directed Mutagenesis Study of the Residues Forming the β-Sheet in the CTT
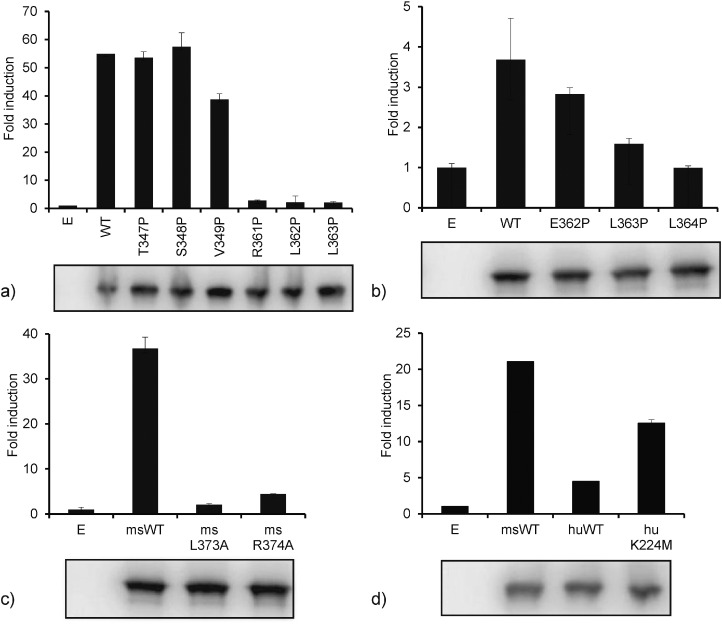
We replaced the six residues in mouse STING that corresponded to the residues forming the β-sheet in human STING-CTT with PRO to confirm the significance of the β-sheet formation. Of these six mutations, R361P, L362P and L363P in mouse STING almost entirely abolished the activation of the IFN-β promoter (Fig. 3a). The corresponding E362, L363 and L364 in human STING were also replaced with PRO, and L363P and L364P showed a decrease in IFN-β activity (Fig. 3b), suggesting that conserved LEU residues (L362 and L363 of mouse STING, and L363 and L364 of human STING) are important for IFN-β promoter activation mediated by STING phosphorylation.
Fig. 3.
Identification of amino acids in STING required for the activation of IFN-β promoter. a–d) IFN-β promoter activities. HEK293 cells were co-transfected with pIFN-β Luc, pRG-TK and indicated plasmids. Forty-eight hours post-transfection, the reporter assay was performed. The expression levels of Renilla luciferase were used as the internal control. The experiments were performed twice, and representative figures were shown. The data were expressed as means from duplicates.
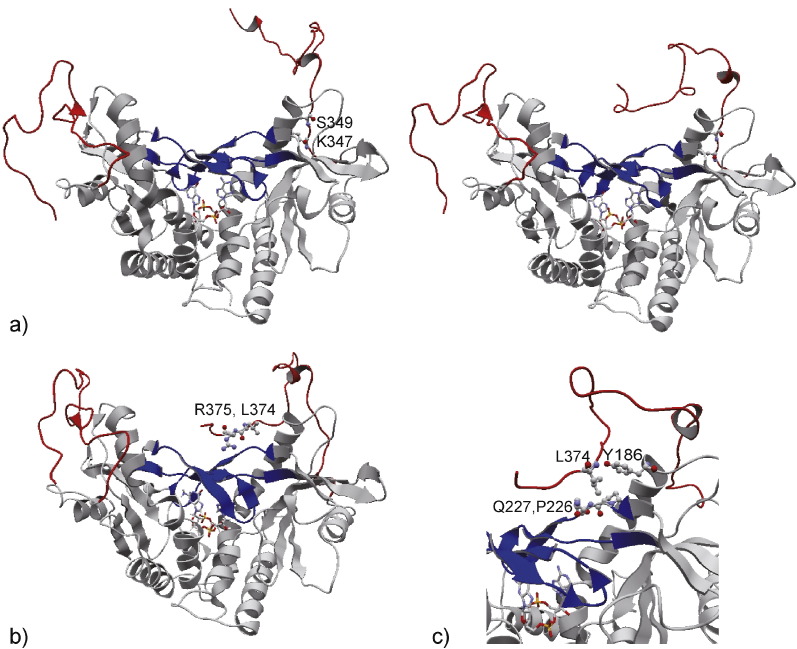
We observed the residues at the end of the CTT, L374 and R375 in human STING, to be involved in the formation of the β-sheet indirectly (see Discussions). L373 and R374 in mouse STING correspond to these two residues in human and they were also mutated to ALA, and the expression plasmids were subjected to evaluation of IFN-β promoter activity. Both point mutants showed a significant decrease in IFN-β promoter activity compared with wild-type STING, confirming the importance of the putative β-sheet formation for STING activation (Fig. 3c).
3.7. Site-directed Mutagenesis Study of a Non-conserved Residue in the Lid
Finally, we wished to identify candidate residues to explain the differences in STING activity between mouse and human, because mouse STING-mediated IFN-β promoter activity is higher than human STING-mediated one (Fig. 3a and b). Our simulation hinted us a site, in which a basic residue in human (K224) is replaced with a non-polar one in mouse (M223), as will be described in detail later (Fig. 4). Interestingly, human wild-type STING showed lower activity of the IFN-β promoter than mouse wild-type STING, whereas a human mutant STING (K224M) showed recovered activity of the IFN-β promoter, suggesting that a non-polar residue at this position (224 in human and 223 in mouse) is key to STING function via the CTT movement and IFN-β activation (Fig. 3d).
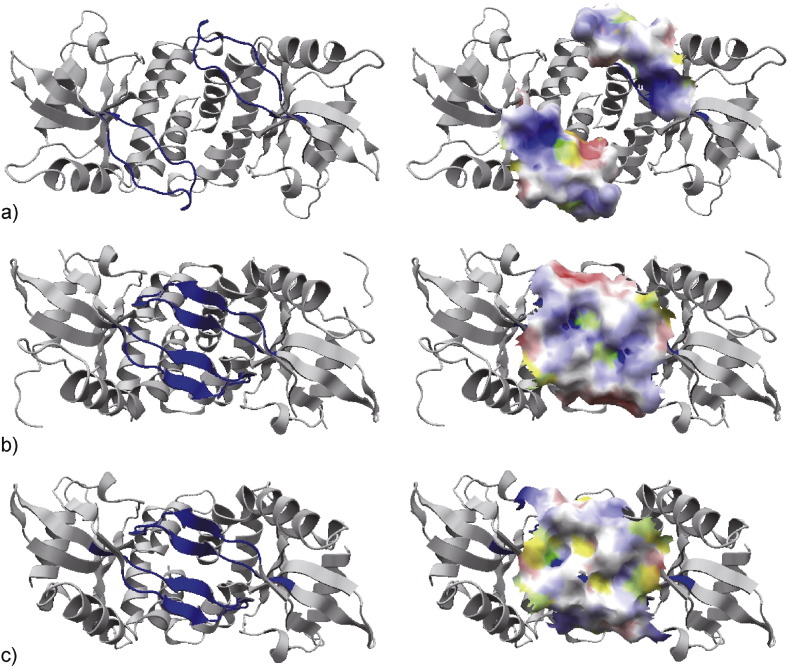
Fig. 4.
The electrostatic properties on the Lid. a) The electrostatic potential on the molecular surface of the Lid in ligand-unbound human STING (PDBID 4f5e), b) that in cGAMP-bound human STING (4ksy), and c) that in cGAMP-bound mouse STING (4loj). The molecular surfaces of the Lid are superimposed onto the respective known structures (that are identical to the structures on the left, where the Lid is colored blue). The electrostatically negative and positive regions are colored red and blue, respectively. The hydrophobic region is colored yellow. The region colored orange or green shows hydrophobic and electrostatically negative or hydrophobic and electrostatically positive, respectively.
4. Discussions
In both mouse and human STING, ligand binding causes a conformational change in the Lid, from open to close, and the natural ligands, cGAMP and c-di-GMP, with their binding affinity to STING, possess the ability to close the Lid (Huang et al., 2012, Zhang et al., 2013). However, several experimental studies showed that only cGAMP-bound human STING had sufficiently high IFN-β activity. In our simulation study of human STING, only the cGAMP-bound STING-CTT maintained the closed Lid conformation throughout the simulation (Fig. 1a), suggesting that the maintenance of the closed Lid conformation for a sustained period of time is required for the phosphorylation by TBK1 and the production of IFN-β.
Only in the ligand-bound forms, the ends of the CTTs moved onto the Lid at an early stage of the simulations (during 28–29 ns and 37–38 ns in the cGAMP- and c-di-GMP-bound forms, respectively), and stayed on the Lid throughout the simulations (Fig. 1a), indicating that the conformational change in the Lid by ligand binding affects the behavior of the CTT. The conformational change in the Lid alters its electrostatic environment; it is highly positive in the ligand-unbound form, due to free K224, R232 and R238 pointing outward (Fig. 4a and see Materials and Methods for the calculation of the electrostatic potential), while it is lowly positive in the ligand-bound forms, because K224 is hydrogen-bonded to other Lid residues, and R232 and R238 interact with the ligand, making the positively charged side-chains in the ARG residues pointing inward (Fig. 4b). It suggests that changes in the electrostatic environments (weakened positive charges) on the Lid contributed to the movement of the CTT onto the Lid. The two ARGs above are conserved in mouse and human STING, while K224 in human corresponds to M223 in mouse. Therefore, the electrostatic property on the Lid in ligand-bound mouse STING is weaker than that in human STING (Fig. 4c). The site-directed mutagenesis study of K224M in human STING showed that this mutation enhanced IFN-β production in human STING (Fig. 3d). In general, IFN-β activity is higher in mouse than in human STING (Fig. 3d), suggesting that the weakening of the positive charges on the Lid leads to the enhanced IFN-β production. These observations support the hypothesis that the movement of the end of the CTT onto the Lid leads to phosphorylation by TBK1.
This movement may be facilitated by the formation of hydrogen bonds in the CTT as shown in Fig. 5 and Supplementary Fig. S3a. In the cGAMP-bound form, the hydrogen bonds in L374 and R375 appeared to have anchored the end of the CTT to the Lid (Fig. 5b and Supplementary Fig. S3a). L374 is especially important, because no other residue pairs between the CTT and Lid formed main-chain hydrogen bonds (Supplementary Fig. S3a), and L374 also formed hydrophobic interactions with several residues in the Lid throughout the simulation (Fig. 5c, see Materials and Methods). These observations suggest that L374 is the main contributor to anchoring the end of the CTT to the Lid. The site-directed mutagenesis study showed that L373A and R374A in mouse STING, corresponding to L374A and R375A in human STING, decreased IFN-β production (Fig. 3c), which is consistent with a previous study (Tanaka and Chen, 2012) showing that S366 and L374 in human STING are important for IRF3 activation. L374A and R375A cannot form the above-described hydrophobic interactions and hydrogen bonds to anchor the CTT, suggesting the important roles of these residues in IFN-β production. These observations support our hypothesis that anchoring the end of the CTT to the Lid facilitated β-sheet formation in the CTT. On the other hand, in the c-di-GMP-bound form, the end of the CTT moved onto the Lid but the end of the CTT continued to move from side to side on the Lid during the simulation (Supplementary Fig. S3b); Lid opening led to no continuous formation of hydrogen bonds between the residues in the Lid and the end of the CTT (Supplementary Fig. S3c). These observations suggest that without anchoring the end of the CTT to the Lid, no local structures were formed in the CTT for a duration sufficient for its phosphorylation. Therefore, we hypothesize that anchoring the end of the CTT is necessary for the formation of a local structure in the CTT, such as the β-sheet observed in the cGAMP-bound form.
Fig. 5.
Anchoring the end of the CTT in the cGAMP-bound form. a) The hydrogen bonds in K347 and S349, which are involved in the movement of the end of the CTT onto the Lid during 28–29 ns. These residues and the ligand cGAMP are shown in ball and stick model and stick model, respectively. The Lid and the CTT are colored blue and red, respectively, where the right CTT showed a remarkable movement. b) The hydrogen bonds observed in L374 and R375 at 125 ns, which anchored the end of the CTT to the Lid. c) The hydrophobic interactions observed in L374 with Y186 in loop1, and P226 and Q227 in the Lid at 50 ns, which were formed after moving the end of the CTT onto the Lid and retained until the end of the simulation.
Among the six residues involved in the formation of the β-sheet in the CTT in the cGAMP-bound form, the mutations L363P and L364P in human STING and the corresponding L362P and L363P in mouse STING showed a decrease in IFN-β production (Fig. 3a and b). The PRO replacement is considered to break secondary structures, both α-helix and β-sheet, suggesting that the break of the β-sheet by the mutation L363P and L364P results in the decreased IFN-β activity. The secondary structure prediction in L363P and L364P in human STING showed lower probability in forming a β-sheet around these PRO residues (Supplementary Fig. S4a) compared to that in the wild type (Supplementary Fig. S2c). These observations indicate that the formation of a β-sheet in the CTT is essential for phosphorylation by TBK1 and IFN-β activity.
The phosphorylation of STING by TBK1 is required for the phosphorylation of IRF3, as described above (Fig. 2e), indicating that both STING and IRF3 are substrates of TBK1. IRF3 consists of two domains, DNA-binding and recognition domains. The recognition domain contains a SER-rich region, where several SER and THRs exist and some of them have been identified as phosphosites, such as 2S (S385, S386) and 5ST (S396, S398, S402, T404 and S405) (Lin et al., 1998, Qin et al., 2003, Yoneyama et al., 1998). The known structures of the recognition domain of IRF3 form a β-sheet in the SER-rich region (Fig. 1c), which is similar to the conformation of the CTT observed in the simulation of the cGAMP-bound form (Fig. 1b). The putative phosphosites, S355, S358 and S366 in STING-CTT, are located in the positions corresponding to S396, S398 and T404 or S405 in IRF3, respectively. It suggests that the observed CTT structure adopts a conformation appropriate for phosphorylation by TBK1.
Phosphorylated STING recruits IRF3 and facilitates the phosphorylation of IRF3 by TBK1 (Tanaka and Chen, 2012). It has been shown that the recruitment of IRF3 by phosphorylated STING is through the binding of STING to an electrostatically positive region in the recognition domain of IRF3 (Supplementary Fig. S4b) (Liu et al., 2015). Our results suggest that the phosphorylation of the CTT would create an electrostatically negative β-sheet (Supplementary Fig. S4c), which may preferentially bind to the electrostatically positive region in IRF3. Based on these observations, we speculate that the β-sheet formation in the CTT leads to the phosphorylation of both STING and IRF3.
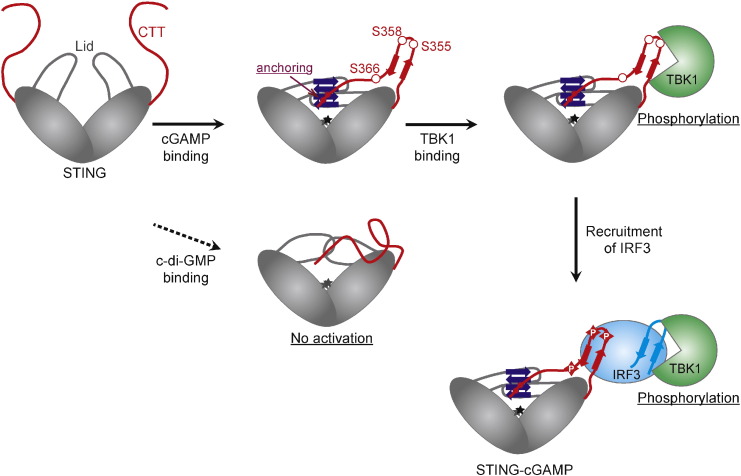
In conclusion, our MD simulations of STING-CTT and experimental verification revealed the dynamics of the CTT involved in ligand binding and phosphorylation. The site-directed mutagenesis studies confirmed the importance of the formation of the local structure in the CTT, and further identified several residues essential for IFN-β activity, such as K224, L363 and L364. Based on the results obtained in this study, we propose a putative mechanism of STING phosphorylation upon binding of a potent ligand as shown in Fig. 6. Structure-based in silico screening is an attractive strategy for searching for novel STING ligands as candidates for anti-cancer drugs or vaccine adjuvants. However, methods relying solely on the structure of the ligand-binding domain (without the CTT) may fall short in evaluating the IFN-β activity, because of the complex relationship between the ligand-binding and IFN-β activity. We believe that considering the dynamics of the CTT is essential for a complete understanding of STING function, and plan to devise a new screening system based on our STING-CTT MD simulations.
Fig. 6.
A putative mechanism of STING phosphorylation. Based on the simulation results and experimental verification, we propose that the following sequence of events must occur for STING activation upon binding of a potent ligand such as cGAMP; the binding of only a potent ligand cGAMP leads to the formation of the closed Lid conformation and the subsequent movement and anchoring of the end of the CTT to Lid, which facilitates the formation of a β-sheet in the CTT. The β-sheet formation presents the conserved SERs to TBK1 and leads to the phosphorylation of these residues by TBK1. The phosphorylated CTT recruits IRF3 by binding to the recognition domain of IRF3, which facilitates the phosphorylation of IRF3 by TBK1. On the other hand, the binding of the low-activity ligand c-di-GMP does not maintain the closed Lid conformation and thus, the end of the CTT is not anchored to the Lid and no local structures are formed in the CTT.
Author contributions
Y.T. designed and performed the molecular dynamics simulations and analyzed the data. N.J. and F.T. designed and performed the experiments. Y.T. and N.J. wrote the manuscript. K.J.I. and K.M. revised the manuscript and supervised all of the research.
Acknowledgements
This work was in part supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (Grant Numbers 25430186 and 25293079; to K.M.) and from Japan Agency for Medical Research and Development (“The adjuvant database project”; to K.J.I. and K.M.). This study was partially supported by MEXT SPIRE Supercomputational Life Science. We thank Y. Namiuchi and Y. Eguchi for advice on supercomputing.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.05.039.
Contributor Information
Yuko Tsuchiya, Email: tsuchiya@protein.osaka-u.ac.jp.
Ken J. Ishii, Email: kenishii@nibiohn.go.jp.
Kenji Mizuguchi, Email: kenji@nibiohn.go.jp.
Appendix A. Supplementary data
Supplementary material.
References
- Ablasser A., Goldeck M. cGAS produces a 2′–5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham M.J., Murtola T. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- Connolly M.L. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221(4612):709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Diner E.J., Burdette D.L. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3(5):1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncheva N.T., Klein K. Analyzing and visualizing residue networks of protein structures. Trends Biochem. Sci. 2011;36(4):179–182. doi: 10.1016/j.tibs.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Gao P., Ascano M. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013;154(4):748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Zillinger T. Binding-pocket and lid-region substitutions render human STING sensitive to the species-specific drug DMXAA. Cell Rep. 2014;8(6):1668–1676. doi: 10.1016/j.celrep.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.H., Liu X.Y. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat. Struct. Mol. Biol. 2012;19(7):728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Ma Z. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Jounai N., Takeshita F. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. U. S. A. 2007;104(35):14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K., Nakamura H. eF-site and PDBjViewer: database and viewer for protein functional sites. Bioinformatics. 2004;20(8):1329–1330. doi: 10.1093/bioinformatics/bth073. [DOI] [PubMed] [Google Scholar]
- Konno H., Konno K. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155(3):688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larabi A., Devos J.M. Crystal structure and mechanism of activation of TANK-binding kinase 1. Cell Rep. 2013;3(3):734–746. doi: 10.1016/j.celrep.2013.01.034. [DOI] [PubMed] [Google Scholar]
- Lin R., Heylbroeck C. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 1998;18(5):2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Cai X. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347(6227) doi: 10.1126/science.aaa2630. (aaa2630) [DOI] [PubMed] [Google Scholar]
- McDonald I.K., Thornton J.M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238(5):777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Nakamura H., Nishida S. Numerical calculations of electrostatic potentials of protein-solvent systems by the self consistent boundary method. J. Phys. Soc. Jpn. 1987;56:1609–1622. [Google Scholar]
- Qin B.Y., Liu C. Crystal structure of IRF-3 reveals mechanism of autoinhibition and virus-induced phosphoactivation. Nat. Struct. Biol. 2003;10(11):913–921. doi: 10.1038/nsb1002. [DOI] [PubMed] [Google Scholar]
- Sauer J.D., Sotelo-Troha K. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 2011;79(2):688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K., Hirose S. POODLE-S: web application for predicting protein disorder by using physicochemical features and reduced amino acid set of a position-specific scoring matrix. Bioinformatics. 2007;23(17):2337–2338. doi: 10.1093/bioinformatics/btm330. [DOI] [PubMed] [Google Scholar]
- Shu C., Sankaran B. Structural insights into the functions of TBK1 in innate antimicrobial immunity. Structure. 2013;21(7):1137–1148. doi: 10.1016/j.str.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wu J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Chen Z.J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 2012;5(214):ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripos International . 1699. South Hanley Rd, St. Louis, Missouri, 63144, USA. [Google Scholar]
- Wang J., Wolf R.M. Development and testing of a general amber force field. J. Comput. Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang W. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25(2):247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Word J.M., Lovell S.C. Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J. Mol. Biol. 1999;285(4):1711–1733. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]
- Word J.M., Lovell S.C. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999;285(4):1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Suhara W. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17(4):1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shi H. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell. 2013;51(2):226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B., Yang Y. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29(4):538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.