Abstract
The accumulation of abnormal prion protein (PrPSc) converted from the normal cellular isoform of PrP (PrPC) is assumed to induce pathogenesis in prion diseases. Therefore, drug discovery studies for these diseases have focused on the protein conversion process. We used a structure-based drug discovery algorithm (termed Nagasaki University Docking Engine: NUDE) that ran on an intensive supercomputer with a graphic-processing unit to identify several compounds with anti-prion effects. Among the candidates showing a high-binding score, the compounds exhibited direct interaction with recombinant PrP in vitro, and drastically reduced PrPSc and protein-aggresomes in the prion-infected cells. The fragment molecular orbital calculation showed that the van der Waals interaction played a key role in PrPC binding as the intermolecular interaction mode. Furthermore, PrPSc accumulation and microgliosis were significantly reduced in the brains of treated mice, suggesting that the drug candidates provided protection from prion disease, although further in vivo tests are needed to confirm these findings. This NUDE-based structure-based drug discovery for normal protein structures is likely useful for the development of drugs to treat other conformational disorders, such as Alzheimer's disease.
Keywords: Prion, Drug discovery, In silico screening, Small chemical compounds, Conformational disorders
Highlights
-
•
NPRs are hunted by a structure-based drug discovery (SBDD) algorithm carried on an intensive supercomputer “DEGIMA”.
-
•
NPRs bind to PrPC using surface plasmon resonance analysis and thermal shift assay.
-
•
NPRs bind to PrPC by van der Waals interaction using the fragment molecular orbital calculation.
-
•
NPRs suppress PrPSc levels and aggresomes in persistently prion-infected cells.
-
•
NPRs inhibit PrPSc levels and microglia activation in prion-infected mice brain.
-
•
In silico drug discovery using SBDD enable to develop the available therapeutic agents for prion disease.
None of effective and efficient therapeutic agents for prion disease is available. In this manuscript, we describe the drug discovery against prion disease using novel screening system. In this system, we performed docking simulation using original calculation system, termed DEGIMA supercomputer, and searched candidate compounds as anti-prion drug from among the many chemical compounds library. The compounds directly interacted with recombinant prion protein, and drastically reduced abnormal form prion protein in the prion-infected cells and the brain of illness mice. This drug discovery may be useful for the therapeutic development to cure conformational disorders.
1. Introduction
Conformational diseases, which include Alzheimer's disease and Parkinson's disease, are the result of the accumulation of intracellular dysfunctional proteins, such as amyloid-beta and alpha-synuclein (Jucker and Walker, 2011). Abnormal prion protein (PrPSc) has also been shown to be a pathogenic protein, which is formed by conformational changes to the native cellular prion protein (PrPC) (Weissmann et al., 2002). The molecular mechanisms of conversion remain poorly understood, although drug discovery studies have focused on the conversion process from PrPC and PrPSc. A variety of drugs have been reported to reduce PrPSc levels by halting the conversion process as described below: acridines including quinacrine (Vogtherr et al., 2003); anti-PrP antibodies including D18 (Peretz et al., 2001), 6H4 (Enari et al., 2001) and ICSM38 (White et al., 2003); polyanions including pentosane polysulfate (PPS) (Doh-ura et al., 2004, Priola and Caughey, 1994); dextran sulfate (Caughey and Raymond, 1993) and HM2602 (Adjou et al., 2003); the polyene antibiotics including amphotericin B (Mange et al., 2000) and filipin (Marella et al., 2002); the others including suramin (Gilch et al., 2001), Congo-Red (Caughey and Race, 1992), Cpd B (Kawasaki et al., 2007), GN8 (Kuwata et al., 2007) and luminescent-conjugated polythiopherenes (LCPs) (Herrmann et al., 2015). Other studies have focused on the intracellular proteolytic system, such as autophagy of insoluble proteins, because the PrPSc complex and the PrP oligomer may have toxic effects on the cell (Aguzzi and Calella, 2009). In vitro and in vivo studies using compounds such as lithium (Heiseke et al., 2009), trehalose (Aguib et al., 2009), rapamycin (Ishibashi et al., 2015), tamoxifen (Marzo et al., 2013), FK506 (Nakagaki et al., 2013), IU-1 (Homma et al., 2015), have reported anti-prion effects. Among them, PPS, Cpd B, LCPs, and FK506 significantly prolong survival periods in mice inoculated with RML or FK-1 prion strains (Doh-ura et al., 2004, Herrmann et al., 2015, Kawasaki et al., 2007, Nakagaki et al., 2013). Recently, it especially has been reported that Anle138b has potent and broad spectrum activity for different protein aggregation disease models (Wagner et al., 2013). Studies have continued to identify suitable compounds for treating the diseases, although none have provided any evidence of benefits against human prion disease, even though some were tested in clinical trials (Tsuboi et al., 2009, Haik et al., 2014).
The structure-based drug discovery (SBDD) using computer simulation was recently facilitated to develop effective chemical compounds. This novel approach is based on virtual screening for drug discovery and has successfully identified compounds for treating several diseases, such as nelfinavir (Kaldor et al., 1997) and amprenavir (Highleyman, 1999) for AIDS; zanamivir for influenza (McCauley, 1999); celecoxib (Stratton and Alberts, 2002) and rofecoxib (Mardini and FitzGerald, 2001) as cyclooxygenase 2 inhibitors; antibacterial agents (Simmons et al., 2010); Ras inhibitor for human cancer (Shima et al., 2013). SBDD has also been used in prion disease, showing that Cp-60, − 62 compounds that mimic the dominant negative PrPC mutant inhibit PrPSc formation (Perrier et al., 2000) and that GN8 strongly stabilises normal conformation by binding to a specific region in PrPC, which suppresses PrPSc production and prolongs survival of prion-infected mice (Kuwata et al., 2007). Furthermore, other small compounds that target the same position as the interaction between GN8 and PrPC have been discovered by virtual screening which used original docking simulation, and those compounds reduced PrPSc levels in RML prion-infected cells (Hyeon et al., 2015). In this study, we performed original docking simulations, termed Nagasaki University Docking Engine (NUDE) for PrPC conformation and small compounds in a large chemical compound database using the DEGIMA supercomputer system. Binding interactions were analysed using the fragment molecular orbital (FMO) method to identify novel anti-prion drugs. Following virtual screening, we tested the ability of candidate compounds to bind to PrPC using surface plasmon resonance (SPR) analysis. The thermal shift assay (TSA) was used to determine whether the compounds influenced thermal change-dependent PrPC stabilisation. We also evaluated the anti-prion effect of compounds using persistently prion-infected cells and mice, which revealed novel therapeutic candidates.
2. Materials and methods
2.1. Regents and antibodies
All candidate compounds (NPR-015, − 050, − 053, − 056 and − 065) identified by in silico calculation were purchased from ASINEX (Supplementary Fig. S8). GN8 was gifted from Prof. Kuwata (Gifu University) and served as the positive anti-prion control drug (Kuwata et al., 2007). These compounds were completely dissolved in 100% dimethyl sulfoxide (DMSO) and adjusted to 10 mM as stock solution. Stock solutions of compound were diluted in sterile water, culture medium or phosphate buffered saline (PBS) to perform the several assay in this study. Antibodies specific for PrP (Santa Cruz Biotechnology, M20; SPI-Bio, SAF61), Iba-1 (WAKO, 019-19741 for IHC and 016-20001 for WB) and β-actin (MBL) were purchased from the indicated vendors. Horseradish peroxidase-conjugated anti-goat (Jackson ImmunoResearch) and anti-mouse (GE Healthcare Life Sciences) IgG antibodies were used for immunoblotting.
2.2. In silico screening
To obtain potential candidate anti-prion compounds, we performed a docking simulation using an original chemical compound library that included approximately 210,000 compounds. The three-dimensional structure of each compound was produced by Open Babel software (O'Boyle et al., 2011), which was followed by energy minimisation using GAFF force field (Wang et al., 2004). In our docking simulation, a structure of the globular domain of human PrPC (124–230th amino acid residues) was used as a receptor, whose atomic coordinates were prepared from the Nuclear Magnetic Resonance (NMR) structure (Biljan et al., 2012) (Protein Data Band code: 2LSB). A cubic space (64 × 64 × 64 Å) was used as the search region for the docking simulation, and binding with the whole surface of the target protein was examined. We employed an original docking simulation program designed as a Graphics Processing Unit (GPU). In this study, the DEGIMA (DEstination for Gpu Intensive MAchine) supercomputer from the Nagasaki Advanced Computing Center, which was constructed with more than 100 GPUs, was used for the docking simulation.
2.3. Fragment molecular orbital calculation
To analyse binding conformations obtained from the docking simulation, FMO calculations (Kitaura et al., 1999) were performed. To prepare reliable atomic coordinates for the FMO calculations, hydrogen atoms were added to the docking structure and the N-terminal of the G124 was capped by —COCH3. An energy minimisation with classical force fields (AMBER99SB (Hornak et al., 2006) and GAFF (Wang et al., 2004)) was performed using the AMBER 10 program (Case, 2008), and the energy minimised structure was used for the FMO calculations. Amino acid residues and compounds were treated as a single fragment, except for C179 and C214, which were merged into one fragment, because they have a disulfide bond. Interaction energies were calculated at the Hartree–Fock (HF) level and second-order Møller–Plesset perturbation (MP2) level with resolution of the identity approximation (Ishikawa and Kuwata, 2009) using cc-pVDZ basis set (Dunning, 1989). In this study, the FMO calculations were performed using the PAICS program (Ishikawa et al., 2009).
2.4. Surface plasmon resonance (SPR) analysis
Interactions between recombinant PrP and compounds were evaluated using a Biacore T200 system (GE Healthcare) as previously described (Nakagaki et al., 2013). Human or mouse PrP (23 − 231) was synthesised by Escherichia coli in a protein expression system using the pET vector, and the protein was purified by imidazole in a NTA column as previously described (Atarashi et al., 2011). The recombinant PrP solutions were diluted to 10 μg/mL with running buffer (10 mM HEPES, pH 7.4 containing 150 mM NaCl, 0.05% Tween20 [Sigma Aldrich] and 5% DMSO), and the ligands were immobilised on a CM5 sensor chip (GE, BR-100,530) using an amine coupling kit (GE, BR-1000-50). Immobilisation of PrP was performed with an average of 2000 RUs. Compounds of various concentrations were diluted with the same running buffer and evaluated by injecting them sequentially for 2 min at a flow rate of 30 mL/min, after which the running buffer alone was injected for a further 20 min at the same flow rate to wash out the bound compounds. Data were corrected using a blank sensor chip as a control. Each compound was dissolved in DMSO and diluted to 5% with running buffer (Nakagaki et al., 2013).
2.5. Cell cultures
Mouse neuroblastoma Neuro 2a cells were obtained from the American Type Culture Collection (CCL 131). N2a-58 cells are established from N2a cells overexpressing PrPC and integrating mouse prnp gene in Neuro 2a cells. N2a-FK cells established from N2a-58 cells were infected with a mouse-adapted Gerstmann–Sträussler–Scheinker strain, Fukuoka-1, as previously described (Ishibashi et al., 2015, Ishibashi et al., 2011). N2a-22L cells were established from N2a-58 cells infected with a mouse-adapted scrapie 22L strain as previously described (Homma et al., 2015, Homma et al., 2014a, Nishida et al., 2000). The above cells were grown at 37 °C in 5% CO2 in Dulbecco's-modified Eagle's medium (Wako) containing 4500 mg/mL glucose, 10% heat-inactivated fetal bovine serum, 100 units/mL penicillin and 100 μg/mL streptomycin (Nakarai Tesque). Cell viability was determined by counting the living cells with Luna automatic cell counter instrument (Logos Biosystems) and cell form was visualised by a microscope.
2.6. Immunobloting
Immunoblotting was performed as previously described (Homma et al., 2014b). For PrPSc detection, the lysates were digested with 20 μg/mL proteinase K (PK; Nakarai Tesuque) at 37 °C for 30 min. After addition of SDS-sample buffer, the samples were applied to 15% SDS-PAGE gel and were subsequently transferred to a PVDF membrane. To detect PrPSc, M20 served as the primary antibody (1:1000) and anti-goat IgG-HRP as the secondary antibody (1:5000), respectively. Bands were visualised using the Chemi-Lumi One (Nakarai Tesque) or ECL prime Western Blotting Detection Kit (GE Healthcare). Band intensities were quantified using ImageJ software (National Institutes of Health).
2.7. Immunofluorescence analysis
In the prion-infected cells, immunofluorescence staining was performed as previously described (Homma et al., 2014b, Ishibashi et al., 2015). Cells were treated with DMSO or 10 μM GN8 and NPRs for 48 h in a chamber slide (BD Falcon). The cells were then washed twice in PBS followed by fixation with 4% formaldehyde for 30 min at room temperature. After permeabilisation using 0.5% Triton X-100, the slides were incubated in 5% skim milk for 1 h at room temperature. For PrPSc detection, the slides were incubated with 3 M guanidine thiocyanate for 5 min at room temperature. The cells were then incubated overnight at 4 °C in a SAF61-specific primary antibody (1:200) to detect PrPSc, followed by Alexa Fluor® 488-conjugated anti-mouse IgG (Invitrogen) (1:500) for 1 h at 37 °C. Nuclei were stained with Vectashield mounting medium containing DAPI (Vector Laboratories). All images were visualised using a confocal laser-scanning microscope LSM 700 (Carl Zeiss).
2.8. Fluorescence-based thermal shift assay (F-TSA)
Fluorescence-based thermal shift assay was used to measure interactions between recombinant human PrP and compounds, which were revealed as temperature changes. These changes were detected using a LightCycler® 480 system (Roche Diagnostics) and SYPRO orange (Life Technologies), which has a maximum excitation at 470 nm and maximum emission at 570 nm. The reaction mixture was as follows: 100 μg/mL recombinant PrP, 150 mM NaCl, 25 mM PIPES (pH 7.0) and 1000-fold diluted SYPRO orange with or without 10 μM compounds in a 96-well white plate (Roche Diagnostics). PrP stabilisation was evaluated by measuring optical density at each temperature. The temperature was gradually increased from 37 °C to 90 °C and fluorescence scanning was continuously measured in 1-°C increments using the LightCycler® 480 instrument with a wavelength filter of 483/610 nm. The fluorescence intensity from each protein dissociation curve was plotted for each thermal point and fitted to the Boltzmann sigmoidal equation using Origin 8.5 software. Each Tm (melting temperature) value was calculated from the maximum value of the first derivative curve of the melting curve, respectively. The mean ± standard deviation (SD) obtained from results of at least three experiments was considered for data analysis.
2.9. Protein aggregation assay
Protein aggregation in cells was evaluated using the ProteoStat® Aggresome Detection Kit (Enzo Life Science). Proteostat selectively interacts with proteins that have a β-sheet-rich structure similar to an aggresome formation and has a maximum excitation at 500 nm and maximum emission at 600 nm as fluorescence dye. All kit reagents and components were prepared according to the manufacturer's instructions. After treating cells with 10-μM GN8 or NPRs compounds for 48 h, the cells were washed with PBS and permeabilised with 0.5% Triton-X-100 containing 3 mM EDTA for 30 min on ice. After washing with PBS, the cells were incubated for 30 min at room temperature in the dark with a dual detection reagent (1:2000 Proteostat, 1:1000 Hoechst 33342 for nuclei staining). The proteostat-positive cells were then observed using a LSM 700 confocal microscope.
2.10. In vivo infection experiments
Four-week-old ddY male mice were purchased from SLC (Hamamatsu, Japan) and were intracerebrally inoculated with 20 μL of a 10− 1 dilution of brain homogenate prepared from mice terminally infected with the FK-1 strain. The compounds were dissolved in normal saline containing 0.25% DMSO, and the mice were intraperitoneally administered 1.0 mg compound/kg/day every other day. As a control, age- and strain-matched mice were intraperitoneally inoculated with normal saline containing 0.25% DMSO. Mice were monitored every other day until the terminal stage of disease or until sacrificed. Clinical onset was defined as the presence of two or more symptoms: greasy and/or yellowish hair, hunchback, weight loss, yellow pubic hair, ataxic gait and non-parallel hind limbs. The mice were sacrificed at clinical onset (110 d.p.i.) or terminal stage and the brains and spleens were removed. The right hemisphere of the brains and a section of spleens were immediately frozen and homogenised at 20% (weight/volume) in PBS. For immunoblotting analysis, total proteins were extracted by mixing with an equal amount of 2 × lysis buffer (300 mM NaCl, 1% Triton-X-100, 1% sodium deoxycholate and 100 mM Tris-HCl; pH 7.5). The remaining brains and spleens were fixed in 10% neutral-buffered formalin.
2.11. Ethics statement
All animal experiments were approved by the Committee on the Animal Care and Use Committees of the Nagasaki University. All animals were cared for in accordance with the Guidelines for Animal Experimentation of Nagasaki University.
2.12. Pathological analysis
The fixed tissues were embedded in paraffin and sectioned into 3 μm-thick sections. To evaluate spongiform changes, the sections were stained with hematoxylin (WAKO, 131-09665) and eosin (WAKO, 056-06722). For Iba-1 staining, after deparaffinisation and rehydration, the sections were boiled in Target Retrieval Solution, Citrate pH 6 (DAKO, S2369) for 20 min for antigen retrieval. The blocked sections were treated with 0.3% hydrogen peroxidase (WAKO, 086-07445) in methanol (Hayashi Pure Chemical, 130-02069) for 30 min to inactivate endogenous peroxidase and then incubated with 3% nonfat dry milk (Megmilk Snow Brand, FA-08) in Tris-buffered saline containing 0.1% Tween 20 (TBST) for 60 min at room temperature. The blocked sections were incubated with a primary antibody (anti-Iba-1 antibody, WAKO, 019-19741) overnight at room temperature, followed by envision polymer horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (DAKO, K4002) for 60 min at room temperature. Immunostaining was visualised using 3, 3-diaminobenzidine (DAB; Dojindo Lab, D006). The hydrolytic autoclaving and formic acid method for PrPSc staining have been previously described (Ishibashi et al., 2012).
2.13. Statistical analysis
Results in the graph represent mean ± SD from at least three independent experiments. Statistical analysis of all data was performed using Statcel 2 from Excel and GraphPad Prism software. The Student's t-test was used to compare between two groups, and one- or two-way analysis of variance followed by the Tukey–Kramer test was used for multiple comparisons. The log-rank test was used to analyse the mortality in prion-infected mice.
3. Results
3.1. In silico screening
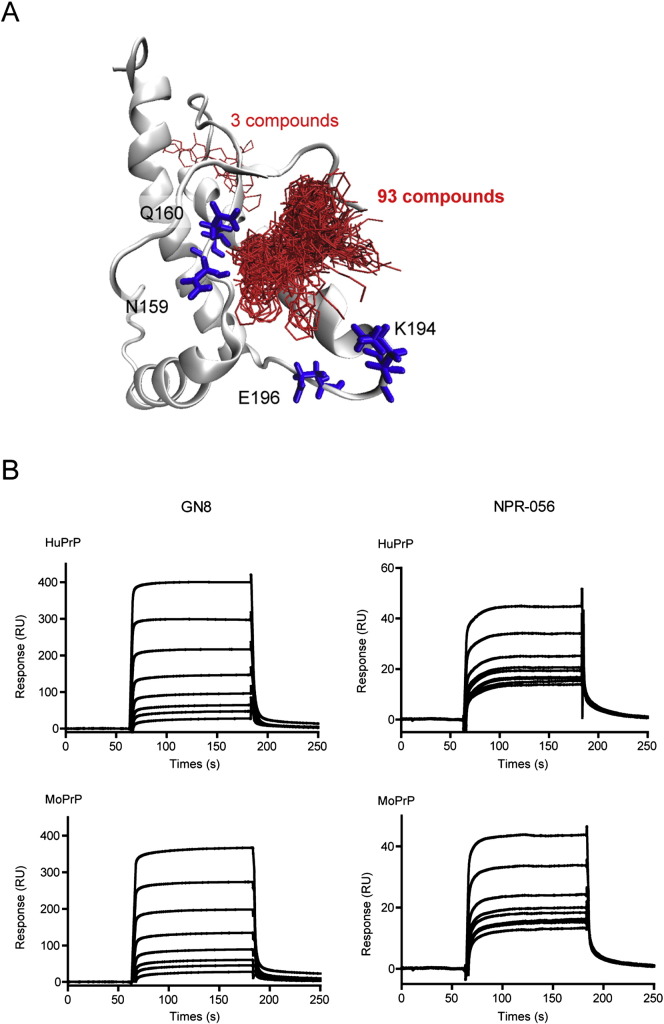
Docking scores, which are related to the binding affinity with human PrPC, were obtained from simulations for all compounds in the chemical compound library. We selected 96 compounds as candidates for anti-prion agents by analysing docking scores and similarities of chemical structures. In Fig. 1A, the docking conformations of the 96 selected compounds are shown together with the four amino acid residues (N159, Q160, K194 and E196), which were reported to be important for binding of GN8 (Ishikawa et al., 2009, Kuwata et al., 2007). A total of 93 compounds were located near N159 and Q160, but slightly away from K194 and E196. The other three compounds were located in a position completely different from the pocket. We investigated whether these candidate compounds, termed ‘NPR,’ exhibited anti-prion effects.
Fig. 1.
Interaction of candidate compounds by in silico calculation with PrP.
(A) Binding conformations of the 96 compounds selected by docking simulation. The red sticks represent compound conformation and the blue sticks represent the four amino acid residues reported to have a strong interaction with GN8. (B) Direct binding of compounds with PrP23–231. Sensorgrams of GN8 and NPR-056 for binding with mouse or human PrP23–231. The GN8 concentrations were 0, 0.98, 1.95, 3.91, 7.81, 15.6, 31.3 and 62.5 μM, and the NPR-056 concentrations were 0, 0.2, 0.39, 0.78, 1.56, 3.13, 6.25, 12.5, 25 and 50 μM (from bottom to top). (For interpretation of the references to color in this figure legend, the reader is referred to the online version of this chapter.)
3.2. Evaluation of binding affinity between PrPC and candidate compounds using surface plasmon resonance (SPR) analysis
To evaluate the interactions between NPRs and PrPC, we analysed the direct binding affinity of NPRs to refolded recombinant PrP. High-throughput screening was used to quantify binding affinity and 10-μM NPRs were sequentially subjected to SPR analysis using sensor chips with solid-phase human or mouse PrPC (Biacore™). Results confirmed that a region within the NPRs had a high-binding ability for human and mouse PrPC. In particular, the binding response (RU) of NPR-056 was estimated to be at least 3-fold greater than others (Supplementary Fig. S1). Kinetic analysis, which revealed the sensorgram slopes of NPR-056, showed that the dose-dependent response was similar to GN8 (positive control). The KD, which shows as dissociation constant, values of NPR-056 were significantly less compared with GN8 (Fig. 1B. and Supplementary Fig. S2). These results suggest that a useful compound binding to PrPC was appropriately selected by SPR analysis from among the putative NPRs compounds which calculated by the Nagasaki docking system on the DEGIMA supercomputer.
3.3. The reduction of PrPSc in persistently prion-infected cells by candidate compounds
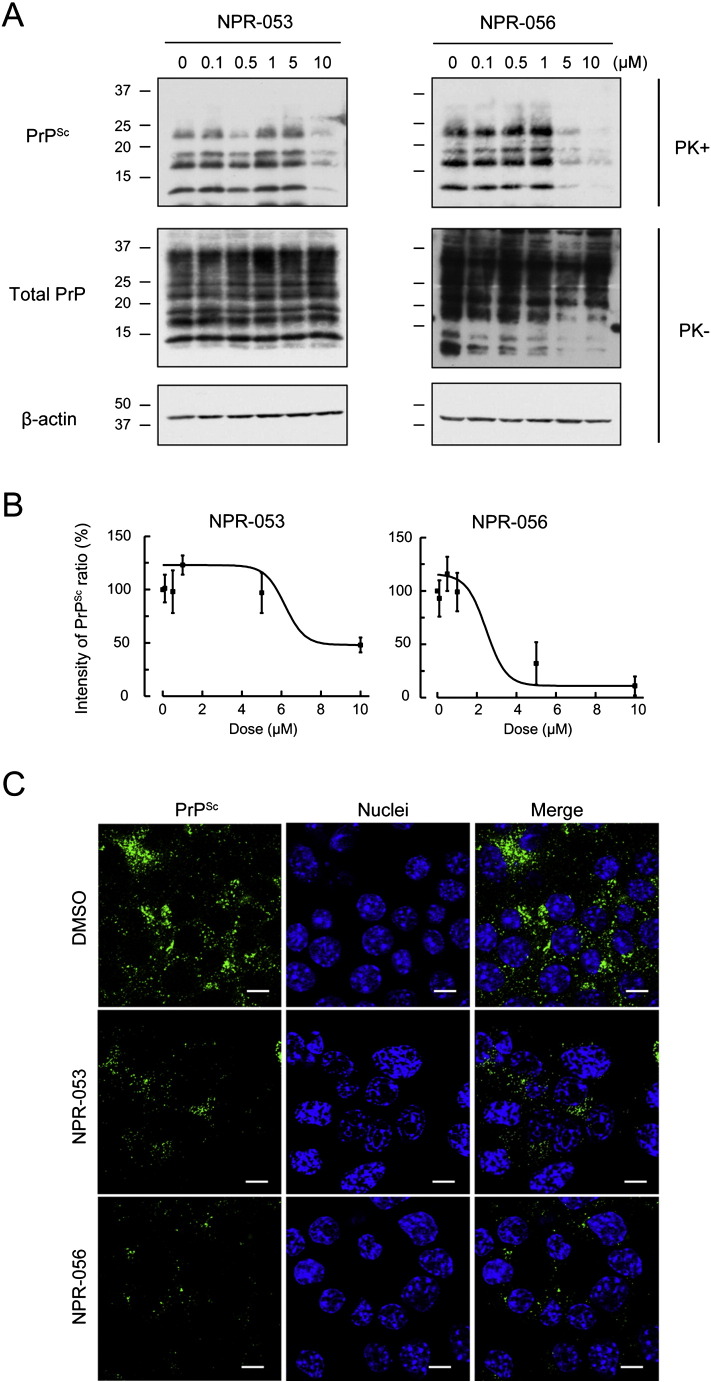
To examine the effect of NPRs on PrPSc, we performed Western Blotting (WB) analysis using 10-μM NPR-treated persistently prion-infected cells. Results confirmed significantly reduced PrPSc protein levels in persistently Gerstmann–Sträussler–Scheinker syndrome-derived Fukuoka-1 prion-infected cells (N2a-FK) that were incubated for 48 h after NPR treatment compared with cells treated with DMSO (Supplementary Fig. S3A: NPR-005, - 014, -015, -020, -030, -050, -053, -056 and -065). These NPRs also had a similar effect on scrapie-derived 22L prion-infected cells (N2a-22L) (Supplementary Fig. S3B). To determine the respective IC50 concentrations, PrPSc levels in cells treated with NPRs were reduced in a dose-dependent manner (0.1-, 0.5-, 1-, 5- and 10-μM NPRs) (Fig. 2A and B) (Supplementary Fig. S4A and B). The effective concentration for 50% reduction of PrPSc (IC50) in NPR-053, -056, -015, -050 and -065 were between 3.72 and 7.68 μM. In particular, NPR-056 and -065 resulted in significantly lower concentrations compared with GN8 (Table 1). These results suggest that the candidate NPRs compounds, which were calculated by the docking system and confirmed by PrPC-binding affinity, effectively reduced PrPSc in persistently prion-infected cells in a manner similar to GN8. Intriguingly, the low PrPC-binding affinity groups (NPR-015, -050 and -065) also exhibited anti-prion effects, although the detailed mechanisms remain to be shown. To support these results, we subsequently visualised PrPSc using immunofluorescence analysis. To selectively detect PrPSc, persistently prion-infected cells were incubated with an anti-PrP monoclonal antibody following treatment with guanidine thiocyanate to reveal aggregated PrPSc in the cells (Homma et al., 2015, Ishibashi et al., 2015). Results confirmed significant PrPSc reduction in the N2a-FK cells following treatment with NPR-053 and -056 (Fig. 2C). NPR-015, one of the low PrPC-binding affinity groups, also resulted in a similar PrPSc reduction in the cells (Supplementary Fig. S4C).
Fig. 2.
Dose responses of anti-prion compounds in prion-infected cells.
(A) N2a-FK cells were treated with different concentrations (0, 0.1, 0.5, 1, 5 and 10 μM) of NPR-053 and -056 compounds for 48 h. PrPSc, total PrP and β-actin as loading control were detected by immunoblotting. (B) PrPSc band intensities treated by individual NPR concentrations are shown as a percentage of the negative control. Results in the graph present mean ± SD from at least three to five independent experiments. The dose-dependent PrPSc reduction curves were plotted using Boltzmann curve fitting. (C) Localisation of PrPSc in 10 μM NPR-053 or − 056-treated N2a-FK cells using immunofluorescent staining. PrPSc and nuclei were detected using SAF61 antibody (green) and DAPI (blue), respectively. Cells were visualised by confocal laser-scanning microscopy. Scale bars represent 10 μm.
Table 1.
The anti-prion effects of NPRs in N2a-FK prion-infected cells (IC50).
| Compounds | IC50 (μM)a | Statistics (p value)b |
|---|---|---|
| GN8 | 6.72 ± 1.13 | – |
| NPR-053 | 7.68 ± 2.64 | 0.5279 |
| NPR-056 | 3.72 ± 1.57* | 0.0115 |
| NPR-015 | 4.46 ± 3.43 | 0.2513 |
| NPR-050 | 5.94 ± 3.43 | 0.6796 |
| NPR-065 | 3.73 ± 2.13* | 0.0486 |
This anti-prion assay have been at least performed 5 times-trial, independently.
Statistics have been calculated using student's t-test. ⁎p < 0.05 vs GN8.
3.4. Analysis of PrPC–NPR interaction using fragment molecular orbital calculation
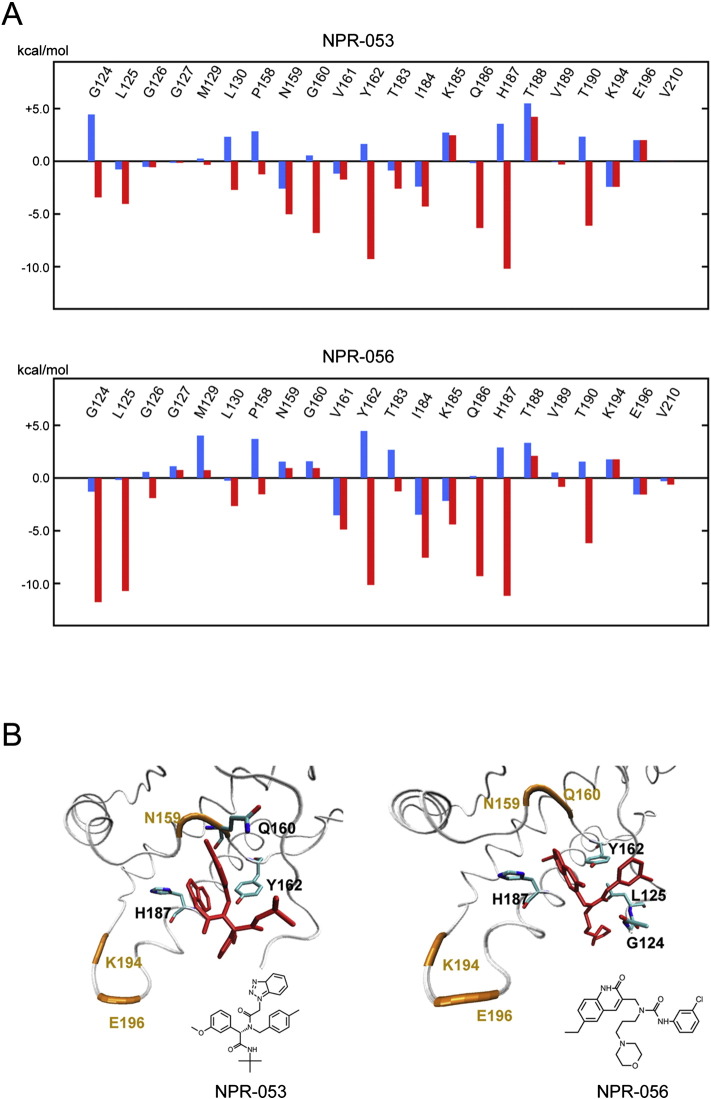
The interaction energies between amino acid residues and NPR-053 and − 056 were calculated using the FMO method, which used binding structures from the docking simulation. In Fig. 3, the calculated energies are provided for residues located within 4.0 Å of the compounds. As a reference, interaction energies of the four residues that are important for GN8 binding are also listed. Negative interaction energy represents an attractive interaction with the compounds. The energies obtained by the HF method mainly included a polar interaction (i.e., electrostatic and charge-transfer interaction), which is a major component of a hydrogen-bonding interaction. Conversely, the energies obtained using the MP2 method included a van der Waals interaction (or non-polar interaction). Thus, a difference between HF and MP2 energies was considered to be van der Waals energy. Residues with large negative interaction energy were not found in the HF results, indicating that an important hydrogen-binding interaction does not exist. Conversely, MP2 energies of several residues result in a large negative value. These results suggested that the van der Waals interaction plays a major role in the binding of NPR-053 and -056 with PrPC.
Fig. 3.
Interaction energy of each amino acid residue with NPR-053 and -056.
(A) Energies between amino acid residues and NPRs were calculated using the FMO method. Interaction energies of residues located within 4.0 Å are provided. Electrostatic and charge-transfer interactions caluculated by the HF (blue); van der Waals interaction calculated by MP2 (red). (B) Binding conformation of NPR-053 and -056 were obtained from docking simulation. Human PrPC (white tube) and compounds (red stick). Several residues exhibiting large interaction energy with the compound are represented by a stick model. For the purpose of reference, positions of the four residues largely interacting with GN8 are represented (orange).
3.5. Effect of NPRs on stabilising recombinant PrP with thermal changes
Generally, high-throughput screening is performed using the TSA to determine whether low-molecular-weight ligands, such as chemical compounds, might interact with target proteins. TSA allows for the quantification of a direct interaction between compounds and progressive protein denaturation. Fluorescence-based TSA (F-TSA) uses a fluorescence dye, such as Sypro® orange, to bind to hydrophobic regions within the protein; this low-cost, simple and readily measurable method is widely used to rapidly acquire data. In this study, we examined whether the compounds were able to interrupt PrPC denaturation depending on incremental temperature increases. Fluorescence intensity levels of samples containing recombinant PrPC as the substrate were revealed as thermal melting curves, which gradually increased in temperature from 37 °C to approximately 70 °C. Eventually, fluorescence intensity levels gently became reduced by the time of measurable maximum degree (approximately 90 °C), but not no substrate line (Fig. 4). The protein stability profiles of F-TSA following GN8 and NPR treatment revealed distinctive Tm values that represented responsive degrees to the maximum value in the first derivative curve of the thermal melting curve. The difference between compound treatment and no treatment was calculated as ∆ Tm. NPR-053 treatment revealed a ∆ Tm of 2.692 ± 0.189 degrees, which was significantly greater than with GN8 treatment, suggesting that NPR-053 might stabilise PrPC by binding to PrPC. These results were not observed with NPR-056 (Table 2). To confirm the effect of NPRs on PrPC expression and cellular cytotoxicity, we treated mouse neuroblastoma cells overexpressing PrPC (N2a-58) with different concentrations of NPR-015, − 053 and − 056 for 48 h, and determined PrPC levels and cell viability. Results showed that none of the NPRs influenced PrPC levels and cell viability in the N2a-58 cells (Supplementary Fig. S5).
Fig. 4.
Recombinant PrP stabilisation by NPRs using fluorescence-based thermal shift assay.
The temperature-dependent recombinant PrP denaturation profile shows a two-phase transition. PrP-binding affinity of several compounds can be assessed from the shift of unfolding temperature to easily obtain potential compounds with a stabilising influence on PrP. Recombinant PrP was analysed by F-TSA in the presence of 0.5% DMSO, 10 μM GN8, NPRs, or buffer only (no PrP). Figures show each representative denaturation profile, respectively. Solid lines: no compounds (PrP only), grey lines: buffer only.
Table 2.
The effects of NPRs against recombinant prion protein stability using fluorescence-based thermal shift assay.
| Compounds | ∆ Tm (°C)a | Statistics (p value)b |
|---|---|---|
| GN8 | 0.680 ± 1.037 | 0.3198 |
| NPR-053 | 2.692 ± 0.189 | 1.596E − 05 |
| NPR-056 | − 0.005 ± 0.631 | 0.9907 |
The F-TSA have been at least performed 3 times-trial, independently.
Melting temperature (Tm) values indicate maximum responsive value in first derivative curve from the thermal melting curve. Each ∆ Tm value shows difference of respective samples compared with no-compound.
Statistics have been calculated using student's t-test. ⁎⁎⁎p < 0.001 vs without compound.
3.6. Bioassay using prion disease model mice treated by NPRs
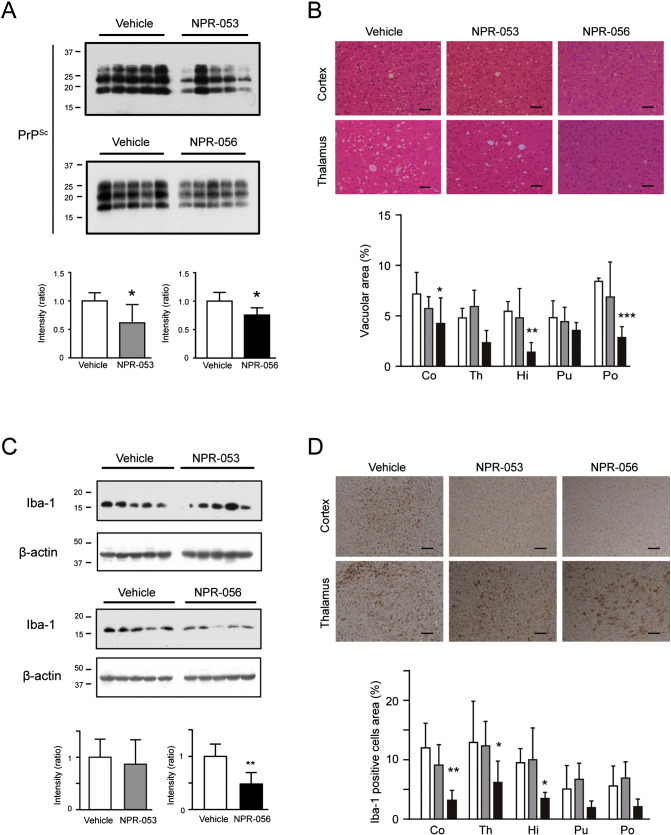
To elucidate whether the NPRs were effective in animal models of prion diseases, CD-1 mice inoculated with Fukuoka-1 brain homogenate were administered either NPR-053 or NPR-056 three times in a week from 2 days post-inoculation (d.p.i.) until sacrifice. We analysed PrPSc and histological changes in the brains of mice at 110 d.p.i., which was the time point at which the mice developed symptoms. PrPSc levels in the NPR-treated brains were 20–30% less than in the vehicle-treated brains at disease onset (Fig. 5A). NPR-056 treatment significantly reduced the vacuolar area in the cortex compared with vehicle treatment. However, NPR-053 treatment resulted in no obvious changes in the cortex (Fig. 5B). We also evaluated gliosis levels in the brains of NPR-treated mice at onset. At 110 d.p.i., NPR-056 treatment significantly reduced expression of Iba-1 protein (also known as allograft inflammatory factor 1: AIF1), which is an EF-hand protein and a marker of activated microglia (Ito et al., 1998, Sakai et al., 2013) (Fig. 5C). Immunohistochemical staining revealed that the area occupied by Iba-1-positive cells was significantly decreased in the cortex, thalamus and hippocampus (Fig. 5D). However, the survival periods of the treated groups remained unchanged compared with the mice treated with vehicle (Supplementary Fig. S6). These results showed that NPR-056 inhibited PrPSc accumulation and the associated pathological changes, including neuronal cell death and microgliosis, suggesting that this compound could serve as a candidate drug for prion diseases.
Fig. 5.
NPR-053 and -056 suppress PrPSc, vacuolation and microgliosis in mouse brains at 110 d.p.i.
(A) Mice were sacrificed at 110 d.p.i. and PrPSc levels were measured in the brains. PrPSc accumulation (band intensity) was analysed by western blotting using M20 antibody as a primary antibody. (B) Degree of vacuolation change was compared by HE staining. The vacuole-occupied area in each section was calculated. (C) Iba-1 expression in the mouse brains was analysed by western blotting and immunohistochemical staining. The ratio of Iba-1/β-actin was calculated from band intensities by western blotting. (D) Microglial activation was analysed by quantifying Iba-1-positive cells following immunohistochemistry. In all bar graphs, white, grey and black bars represent vehicle, NPR-053 and − 056 groups, respectively. In histological analysis, all scale bars represent 50 μm. Statistical significances in immunoblotting were determined using the two-tailed Student's t-test, and two-way analysis of variance followed by the Tukey–Kramer test for multiple comparisons was used for histological analysis in brain regions (cortex: Co, thalamus: Th, hippocampus: Hi, putamen: Pu and pons: Po). *p < 0.05 and **p < 0.01.
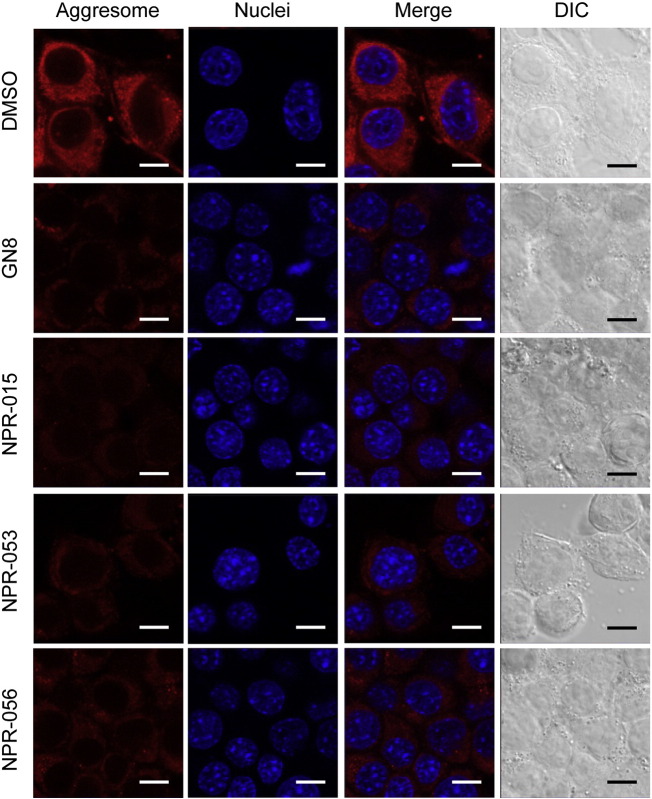
3.7. NPR effect on aggresomes in N2a-FK cells
Aggresomes are established by the aggregation of a variety of inessential proteins and intracellular inclusion bodies co-localising with the accumulation of phosphorylated tau and alpha-synuclein in pathological tissues and cell culture models of conformational diseases (Kothawala et al., 2012, Shen et al., 2011). Aggresomes are large complexes of aberrant proteins that exhibit resistance to degradation and are, therefore, sequestered as inclusion bodies. It is thought that sequestrated aggresomes can be degraded by the autophagy system (Doi et al., 2013, Matsumoto et al., 2011, Watanabe et al., 2012). Recent reports have shown that aggresomes containing p62 and PrPSc form in persistently prion-infected cells; p62 interacts closely with PrPSc and is critical in promoting degradation via the autophagy system (Homma et al., 2014b).
In this study, we used the aggresome detection probe as Proteostat to determine whether NPRs affect aggresome levels in persistently prion-infected cells. Many aggresomes were detected in the cytoplasm of N2a-FK cells treated by DMSO only. The anti-prion compounds that were shown to suppress PrPSc (GN8, NPR-015, -053 and -056) also significantly reduced the number of intracellular aggresomes in the N2a-FK cells after treatment for 48 h with 10 μM of the compounds, respectively (Fig. 6). These results indicated that NPRs might facilitate clearance of PrPSc, as well as dysfunctional and unnecessary proteins in the cytoplasm.
Fig. 6.
Aggresome detection in prion-infected cells after NPR treatment.
N2a-FK cells treated with 10 μM GN8, NPR-015, -053, or -056 compounds for 48 h were subjected to protein aggregation assays. Aggresomes in cells were detected using ProteoStat® (red). Cell nuclei were counterstained with Hoechst 33342 (blue). Cells were visualised by confocal laser-scanning microscopy, and whole cells were visualised by differential interference contrast (DIC) with transmitted light. Scale bars represent 10 μm.
4. Discussion
In the search for drugs for human prion diseases, we conducted docking simulation using a DEGIMA supercomputer. Results from the in vitro and ex vivo drug screening revealed that NPR-053 and -056 significantly reduced PrPSc levels (Fig. 2, Fig. 5). Although the affinity of NPR-056 for PrPC was approximately 2-fold greater than NPR-053 (Supplementary Fig. S1), thermal stabilisation of the NPR-053–PrPC complex was significantly stronger than the GN8–PrPC complex (Fig. 4 and Table 2), indicating that bonding varies among NPRs. Intriguingly, although NPR-015 and -065 did not bind with PrPC in the SPR assay, all NPRs (-015, -050, -053, -056 and -065) exhibited anti-prion effects in FK-1 and 22L strains (Supplementary Fig. S3B). These results suggested that NPRs might exhibit suppressant effects against PrPSc in universal prion strains depending on the binding of NPRs to PrPC, although these results differed from drugs with a strain-dependent function such as Cpd B (Kawasaki et al., 2007). Conversely, NPRs had no effect on PrPC levels and cellular toxicity (Supplementary Fig. S5), suggesting that NPRs do not promote PrPC degradation, compared with activation of the ubiquitin–proteasome system by IU-1 (Homma et al., 2015) and macroautophagy by lithium (Heiseke et al., 2009) and FK506 (Nakagaki et al., 2013). Results from the bioassay showed that NPR-053 and -056 treatments markedly suppressed PrPSc and gliosis levels in the brains of mice at onset, but had no effect on survival (Fig. 5 and Supplementary Fig. S6), indicating that the effective drugs which is left in brain after NPR-treatment might be very low levels. The pharmacokinetics, such as transportation to the brain and drug stabilisation, remains poorly understood, so future studies should focus on the routes, periods, frequency and dosages of NPRs.
The intracellular aggregations of dysfunctional misfolded proteins are removed by a variety of homeostatic mechanisms, including endoplasmic reticulum-associated degradation, the ubiquitin–proteasome system and the autophagy system. PrPSc has been shown to co-localise with p62 and form aggresomes in prion-infected cells (Homma et al., 2014b). Intriguingly, results from this study shown that some NPRs significantly reduced the development of aggresomes (Fig. 6). Previous reports have shown that aggresomes are degraded by a selective autophagy system that recognises LC3-tagged protein complexes (Bjorkoy et al., 2005, Ichimura et al., 2008, Komatsu et al., 2007, Pankiv et al., 2007). The location of senile plaques in the Alzheimer's disease brain, which is the result of Aβ and neurofibrillary tangles caused by hyper-phosphorylated tau and inclusion bodies, which are formed by alpha-synuclein in Parkinson's disease cell culture models, is identical to the location of aggresomes (Kothawala et al., 2012, Shen et al., 2011). These results suggest that NPRs could protect against prion disease as well as other conformational diseases that accumulate aggresomes.
Molecular simulation and analysis of atomic level interactions between NPRs and PrPC showed that the binding sites of NPR-053 and -056 were located around the four amino acid residues — N159, Q160, K194 and E196 (Fig. 1A). High-pressure NMR shows that this position is less stable (Kuwata et al., 2002) and is considered to be a “hot spot” for the pathogenic conversion of prion diseases (Kuwata et al., 2007, Yamamoto, 2014). Indeed, GN8 can stabilise PrPC and inhibit conversion by binding to this position (Ishikawa et al., 2009, Kuwata et al., 2007). Furthermore, several different structure compounds were reported, which have anti-prion activity by binding to the similar position (Ferreira et al., 2014). Our simulation results show that the overall binding characters of our compounds are consistent with these previous studies, suggesting that the hydrophobic pocket might be important for binding of other anti-prion compounds with heterogeneous PrP. However, the FMO calculations clearly demonstrate that the detailed interaction mechanisms of NPR-053 and -056 are different than GN8. In the case of GN8, polar interactions, including hydrogen-bonding interactions with N159, Q160, K194 and E196 were critical (Ishikawa et al., 2009). Conversely, HF level calculations revealed no large negative interaction energy (Fig. 3A), suggesting that polar interactions are not important for the binding of NPR-053 and -056. Instead, MP2 energies of several residues were negatively large, indicating that van der Waals interactions play an important role in complex stability. In particular, G124, L125, Y162, Q186 and H187 exhibit key interactions with the compounds. The docking structures of NPR-053 and -056 are shown in Fig. 3B. We note that the aromatic rings of these compounds are in contact with the amino acid residues, which are capable of generating a large attractive interaction with PrPC. Thus, our analysis of interaction energies using the FMO method was consistent with binding conformations from the docking simulation.
It must be taken into account that the compounds may also interact with the flexible N-terminal domain because NPRs have been derived by molecular simulation from human PrP (124–230 a.a.). To address this point, an in vitro-binding experiment (i.e. SPR) with purified PrPs (121–231 and 90–231 a.a.) will be needed. In addition, interaction with abnormal form is also another possible action of anti-prion drugs and we cannot exclude this possibility regarding with the NPRs. Continuously, conformation of fibril formed PrP has been analysed using solid-state NMR, X-ray fiber diffraction, Hydrogen exchange mass spectrometry or electron micrograph with high resolution (Laws et al., 2001, Helmus et al., 2008, Wille et al., 2009, Damo et al., 2010, Smirnovas et al., 2011), thereby high content of beta sheet and their stacking structure were suggested (Wille et al., 2002, Groveman et al., 2014). However, there are still some obligations to overcome before using the structural information in docking simulation on DEGIMA at present, because some of them are hypothetical and not sufficient. In the future, specific anti-prion drugs that can directly interact with abnormal PrP structure will be necessary for treatment of prion disease, because normal cellular PrP has a critical physiological function on maintenance of myelin and axon (Bremer et al., 2010).
In conclusion, results from in silico screening and targeting of the PrPC structure showed that this method could be useful for drug discovery against prion disease. Future studies should evaluate these effects in animal models of disease, in in vitro screening with post-translational modifications of PrPC and in the structure analysis using NMR of the compounds–PrPC complexes. Results also showed that NPRs could be made widely available as candidate drugs for conformational disease.
Competing interests
The authors declare that they have no competing interests.
Author contribution statement
D.I., T.N., T.I., F.C. and T.H. coordinated and performed the entire project. R.A., K.W. and N.N. supervised and discussed the data. D.I., T.N. and T.I wrote the manuscript. D.I. and N.N. revised the manuscript.
Acknowledgments
We thank Dr. Yoshimasa Tanaka and Prof. Katsuya Satoh from Nagasaki University and Dr. Yuji O. Kamatari from Gifu University for helpful discussions and critical assessment of the manuscript, and Hanako Nakayama, Ayako Nakazaki, Atsuko Matsuo and Megumi Tanaka for technical assistance. This work was supported by a Grant-in-Aid of the Research Committee of Prion Disease and Slow Virus Infection from the Ministry of Health, Labor and Welfare of Japan; a Grant-in-Aid of the Research Committee of Molecular Pathogenesis and Therapies for Prion Disease and Slow Virus Infection, the Practical Research Project for Rare and Intractable Disease from the Japan Agency for Medical Research and Development, AMED; a grant from the Takeda Science Foundation; a grant from the Japan Intractable Disease Research Foundation; a Grant-in-Aid from the Tokyo Biochemical Research Foundation; and a grant provided by the YOKOYAMA Foundation for Clinical Pharmacology (Grant No. YRY1502).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.06.010.
Appendix A. Supplementary data
Supplementary material
References
- Adjou K.T., Simoneau S., Sales N., Lamoury F., Dormont D., Papy-Garcia D., Barritault D., Deslys J.P., Lasmezas C.I. A novel generation of heparan sulfate mimetics for the treatment of prion diseases. J. Gen. Virol. 2003;84:2595–2603. doi: 10.1099/vir.0.19073-0. [DOI] [PubMed] [Google Scholar]
- Aguib Y., Heiseke A., Gilch S., Riemer C., Baier M., Schatzl H.M., Ertmer A. Autophagy induction by trehalose counteracts cellular prion infection. Autophagy. 2009;5:361–369. doi: 10.4161/auto.5.3.7662. [DOI] [PubMed] [Google Scholar]
- Aguzzi A., Calella A.M. Prions: protein aggregation and infectious diseases. Physiol. Rev. 2009;89:1105–1152. doi: 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- Atarashi R., Satoh K., Sano K., Fuse T., Yamaguchi N., Ishibashi D., Matsubara T., Nakagaki T., Yamanaka H., Shirabe S. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat. Med. 2011;17:175–178. doi: 10.1038/nm.2294. [DOI] [PubMed] [Google Scholar]
- Biljan I., Giachin G., Ilc G., Zhukov I., Plavec J., Legname G. Structural basis for the protective effect of the human prion protein carrying the dominant-negative E219K polymorphism. Biochem. J. 2012;446:243–251. doi: 10.1042/BJ20111940. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on Huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer J., Baumann F., Tiberi C., Wessig C., Fischer H., Schwarz P., Steele A.D., Toyka K.V., Nave K.A., Weis J. Axonal prion protein is required for peripheral myelin maintenance. Nat. Neurosci. 2010;13:310–318. doi: 10.1038/nn.2483. [DOI] [PubMed] [Google Scholar]
- Case D. University of California; Sanfrancisco: 2008. AMBER 10. [Google Scholar]
- Caughey B., Race R.E. Potent inhibition of scrapie-associated PrP accumulation by congo red. J. Neurochem. 1992;59:768–771. doi: 10.1111/j.1471-4159.1992.tb09437.x. [DOI] [PubMed] [Google Scholar]
- Caughey B., Raymond G.J. Sulfated polyanion inhibition of scrapie-associated PrP accumulation in cultured cells. J. Virol. 1993;67:643–650. doi: 10.1128/jvi.67.2.643-650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damo S.M., Phillips A.H., Young A.L., Li S., Woods V.L., Jr., Wemmer D.E. Probing the conformation of a prion protein fibril with hydrogen exchange. J. Biol. Chem. 2010;285:32303–32311. doi: 10.1074/jbc.M110.114504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doh-ura K., Ishikawa K., Murakami-Kubo I., Sasaki K., Mohri S., Race R., Iwaki T. Treatment of transmissible spongiform encephalopathy by intraventricular drug infusion in animal models. J. Virol. 2004;78:4999–5006. doi: 10.1128/JVI.78.10.4999-5006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi H., Adachi H., Katsuno M., Minamiyama M., Matsumoto S., Kondo N., Miyazaki Y., Iida M., Tohnai G., Qiang Q. p62/SQSTM1 differentially removes the toxic mutant androgen receptor via autophagy and inclusion formation in a spinal and bulbar muscular atrophy mouse model. J. Neurosci. 2013;33:7710–7727. doi: 10.1523/JNEUROSCI.3021-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning T.H. Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. J. Chem. Phys. 1989;90:1007–1023. [Google Scholar]
- Enari M., Flechsig E., Weissmann C. Scrapie prion protein accumulation by scrapie-infected neuroblastoma cells abrogated by exposure to a prion protein antibody. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9295–9299. doi: 10.1073/pnas.151242598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira N.C., Marques I.A., Conceicao W.A., Macedo B., Machado C.S., Mascarello A., Chiaradia-Delatorre L.D., Yunes R.A., Nunes R.J., Hughson A.G. Anti-prion activity of a panel of aromatic chemical compounds: in vitro and in silico approaches. PLoS One. 2014;9 doi: 10.1371/journal.pone.0084531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilch S., Winklhofer K.F., Groschup M.H., Nunziante M., Lucassen R., Spielhaupter C., Muranyi W., Riesner D., Tatzelt J., Schatzl H.M. Intracellular re-routing of prion protein prevents propagation of PrP(Sc) and delays onset of prion disease. EMBO J. 2001;20:3957–3966. doi: 10.1093/emboj/20.15.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groveman B.R., Dolan M.A., Taubner L.M., Kraus A., Wickner R.B., Caughey B. Parallel in-register intermolecular beta-sheet architectures for prion-seeded prion protein (PrP) amyloids. J. Biol. Chem. 2014;289:24129–24142. doi: 10.1074/jbc.M114.578344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haik S., Marcon G., Mallet A., Tettamanti M., Welaratne A., Giaccone G., Azimi S., Pietrini V., Fabreguettes J.R., Imperiale D. Doxycycline in Creutzfeldt–Jakob disease: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2014;13:150–158. doi: 10.1016/S1474-4422(13)70307-7. [DOI] [PubMed] [Google Scholar]
- Heiseke A., Aguib Y., Riemer C., Baier M., Schatzl H.M. Lithium induces clearance of protease resistant prion protein in prion-infected cells by induction of autophagy. J. Neurochem. 2009;109:25–34. doi: 10.1111/j.1471-4159.2009.05906.x. [DOI] [PubMed] [Google Scholar]
- Helmus J.J., Surewicz K., Nadaud P.S., Surewicz W.K., Jaroniec C.P. Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6284–6289. doi: 10.1073/pnas.0711716105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann U.S., Schutz A.K., Shirani H., Huang D., Saban D., Nuvolone M., Li B., Ballmer B., Aslund A.K., Mason J.J. ). Structure-based drug design identifies polythiophenes as antiprion compounds. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aab1923. (299ra123) [DOI] [PubMed] [Google Scholar]
- Highleyman L. New anti-HIV drugs in development. Beta. 1999;12:7–8. [PubMed] [Google Scholar]
- Homma T., Ishibashi D., Nakagaki T., Fuse T., Sano K., Satoh K., Atarashi R., Nishida N. Persistent prion infection disturbs the function of Oct-1, resulting in the down-regulation of murine interferon regulatory factor-3. Sci. Rep. 2014;4:6006. doi: 10.1038/srep06006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma T., Ishibashi D., Nakagaki T., Satoh K., Sano K., Atarashi R., Nishida N. Increased expression of p62/SQSTM1 in prion diseases and its association with pathogenic prion protein. Sci. Rep. 2014;4:4504. doi: 10.1038/srep04504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma T., Ishibashi D., Nakagaki T., Fuse T., Mori T., Satoh K., Atarashi R., Nishida N. Ubiquitin-specific protease 14 modulates degradation of cellular prion protein. Sci. Rep. 2015;5:11028. doi: 10.1038/srep11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon J.W., Choi J., Kim S.Y., Govindaraj R.G., Jam Hwang K., Lee Y.S., An S.S., Lee M.K., Joung J.Y., No K.T. Discovery of novel anti-prion compounds using in silico and in vitro approaches. Sci. Rep. 2015;5:14944. doi: 10.1038/srep14944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kumanomidou T., Sou Y.S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- Ishibashi D., Yamanaka H., Mori T., Yamaguchi N., Yamaguchi Y., Nishida N., Sakaguchi S. Antigenic mimicry-mediated anti-prion effects induced by bacterial enzyme succinylarginine dihydrolase in mice. Vaccine. 2011;29:9321–9328. doi: 10.1016/j.vaccine.2011.10.017. [DOI] [PubMed] [Google Scholar]
- Ishibashi D., Atarashi R., Fuse T., Nakagaki T., Yamaguchi N., Satoh K., Honda K., Nishida N. Protective role of interferon regulatory factor 3-mediated signaling against prion infection. J. Virol. 2012;86:4947–4955. doi: 10.1128/JVI.06326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi D., Homma T., Nakagaki T., Fuse T., Sano K., Takatsuki H., Atarashi R., Nishida N. Strain-dependent effect of macroautophagy on abnormally folded prion protein degradation in infected neuronal cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Kuwata K. Fragment molecular orbital calculation using the RI-MP2 method. Chem. Phys. Lett. 2009;474:195–198. doi: 10.1021/jz201697x. [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Ishikura T., Kuwata K. Theoretical study of the prion protein based on the fragment molecular orbital method. J. Comput. Chem. 2009;30:2594–2601. doi: 10.1002/jcc.21265. [DOI] [PubMed] [Google Scholar]
- Ito D., Imai Y., Ohsawa K., Nakajima K., Fukuuchi Y., Kohsaka S. Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res. Mol. Brain Res. 1998;57:1–9. doi: 10.1016/s0169-328x(98)00040-0. [DOI] [PubMed] [Google Scholar]
- Jucker M., Walker L.C. Pathogenic protein seeding in Alzheimer disease and other neurodegenerative disorders. Ann. Neurol. 2011;70:532–540. doi: 10.1002/ana.22615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldor S.W., Kalish V.J., Davies J.F., 2nd, Shetty B.V., Fritz J.E., Appelt K., Burgess J.A., Campanale K.M., Chirgadze N.Y., Clawson D.K. Viracept (nelfinavir mesylate, AG1343): a potent, orally bioavailable inhibitor of HIV-1 protease. J. Med. Chem. 1997;40:3979–3985. doi: 10.1021/jm9704098. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y., Kawagoe K., Chen C.J., Teruya K., Sakasegawa Y., Doh-ura K. Orally administered amyloidophilic compound is effective in prolonging the incubation periods of animals cerebrally infected with prion diseases in a prion strain-dependent manner. J. Virol. 2007;81:12889–12898. doi: 10.1128/JVI.01563-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaura K., Sawai T., Asada Y., Nakano T., Uebayasi M. Pair interaction molecular orbital method: an approximate computational method for molecular interactions. Chem. Phys. Lett. 1999;312:319–324. [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Kothawala A., Kilpatrick K., Novoa J.A., Segatori L. Quantitative analysis of alpha-synuclein solubility in living cells using split GFP complementation. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwata K., Li H., Yamada H., Legname G., Prusiner S.B., Akasaka K., James T.L. Locally disordered conformer of the hamster prion protein: a crucial intermediate to PrPSc? Biochemistry. 2002;41:12277–12283. doi: 10.1021/bi026129y. [DOI] [PubMed] [Google Scholar]
- Kuwata K., Nishida N., Matsumoto T., Kamatari Y.O., Hosokawa-Muto J., Kodama K., Nakamura H.K., Kimura K., Kawasaki M., Takakura Y. Hot spots in prion protein for pathogenic conversion. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11921–11926. doi: 10.1073/pnas.0702671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws D.D., Bitter H.M., Liu K., Ball H.L., Kaneko K., Wille H., Cohen F.E., Prusiner S.B., Pines A., Wemmer D.E. Solid-state NMR studies of the secondary structure of a mutant prion protein fragment of 55 residues that induces neurodegeneration. Proc. Natl. Acad. Sci. U. S. A. 2001;98:11686–11690. doi: 10.1073/pnas.201404298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mange A., Nishida N., Milhavet O., McMahon H.E., Casanova D., Lehmann S. Amphotericin B inhibits the generation of the scrapie isoform of the prion protein in infected cultures. J. Virol. 2000;74:3135–3140. doi: 10.1128/jvi.74.7.3135-3140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardini I.A., FitzGerald G.A. Selective inhibitors of cyclooxygenase-2: a growing class of anti-inflammatory drugs. Mol. Interv. 2001;1:30–38. [PubMed] [Google Scholar]
- Marella M., Lehmann S., Grassi J., Chabry J. Filipin prevents pathological prion protein accumulation by reducing endocytosis and inducing cellular PrP release. J. Biol. Chem. 2002;277:25457–25464. doi: 10.1074/jbc.M203248200. [DOI] [PubMed] [Google Scholar]
- Marzo L., Marijanovic Z., Browman D., Chamoun Z., Caputo A., Zurzolo C. 4-Hydroxytamoxifen leads to PrPSc clearance by conveying both PrPC and PrPSc to lysosomes independently of autophagy. J. Cell Sci. 2013;126:1345–1354. doi: 10.1242/jcs.114801. [DOI] [PubMed] [Google Scholar]
- Matsumoto G., Wada K., Okuno M., Kurosawa M., Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- McCauley J. Relenza. Curr. Biol. 1999;9:R796. doi: 10.1016/s0960-9822(99)80491-6. [DOI] [PubMed] [Google Scholar]
- Nakagaki T., Satoh K., Ishibashi D., Fuse T., Sano K., Kamatari Y.O., Kuwata K., Shigematsu K., Iwamaru Y., Takenouchi T. FK506 reduces abnormal prion protein through the activation of autolysosomal degradation and prolongs survival in prion-infected mice. Autophagy. 2013;9:1386–1394. doi: 10.4161/auto.25381. [DOI] [PubMed] [Google Scholar]
- Nishida N., Harris D.A., Vilette D., Laude H., Frobert Y., Grassi J., Casanova D., Milhavet O., Lehmann S. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J. Virol. 2000;74:320–325. doi: 10.1128/jvi.74.1.320-325.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open babel: an open chemical toolbox. J. Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Overvatn A., Bjorkoy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Peretz D., Williamson R.A., Kaneko K., Vergara J., Leclerc E., Schmitt-Ulms G., Mehlhorn I.R., Legname G., Wormald M.R., Rudd P.M. Antibodies inhibit prion propagation and clear cell cultures of prion infectivity. Nature. 2001;412:739–743. doi: 10.1038/35089090. [DOI] [PubMed] [Google Scholar]
- Perrier V., Wallace A.C., Kaneko K., Safar J., Prusiner S.B., Cohen F.E. Mimicking dominant negative inhibition of prion replication through structure-based drug design. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6073–6078. doi: 10.1073/pnas.97.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priola S.A., Caughey B. Inhibition of scrapie-associated PrP accumulation. Probing the role of glycosaminoglycans in amyloidogenesis. Mol. Neurobiol. 1994;8:113–120. doi: 10.1007/BF02780661. [DOI] [PubMed] [Google Scholar]
- Sakai K., Hasebe R., Takahashi Y., Song C.H., Suzuki A., Yamasaki T., Horiuchi M. Absence of CD14 delays progression of prion diseases accompanied by increased microglial activation. J. Virol. 2013;87:13433–13445. doi: 10.1128/JVI.02072-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D., Coleman J., Chan E., Nicholson T.P., Dai L., Sheppard P.W., Patton W.F. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem. Biophys. 2011;60:173–185. doi: 10.1007/s12013-010-9138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima F., Yoshikawa Y., Ye M., Araki M., Matsumoto S., Liao J., Hu L., Sugimoto T., Ijiri Y., Takeda A. In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8182–8187. doi: 10.1073/pnas.1217730110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons K.J., Chopra I., Fishwick C.W. Structure-based discovery of antibacterial drugs. Nat. Rev. Microbiol. 2010;8:501–510. doi: 10.1038/nrmicro2349. [DOI] [PubMed] [Google Scholar]
- Smirnovas V., Baron G.S., Offerdahl D.K., Raymond G.J., Caughey B., Surewicz W.K. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 2011;18:504–506. doi: 10.1038/nsmb.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton M.S., Alberts D.S. Current application of selective COX-2 inhibitors in cancer prevention and treatment. Oncology (Williston Park) 2002;16:37–51. [PubMed] [Google Scholar]
- Tsuboi Y., Doh-Ura K., Yamada T. Continuous intraventricular infusion of pentosan polysulfate: clinical trial against prion diseases. Neuropathology. 2009;29:632–636. doi: 10.1111/j.1440-1789.2009.01058.x. [DOI] [PubMed] [Google Scholar]
- Vogtherr M., Grimme S., Elshorst B., Jacobs D.M., Fiebig K., Griesinger C., Zahn R. Antimalarial drug quinacrine binds to C-terminal helix of cellular prion protein. J. Med. Chem. 2003;46:3563–3564. doi: 10.1021/jm034093h. [DOI] [PubMed] [Google Scholar]
- Wagner J., Ryazanov S., Leonov A., Levin J., Shi S., Schmidt F., Prix C., Pan-Montojo F., Bertsch U., Mitteregger-Kretzschmar G. Anle138b: a novel oligomer modulator for disease-modifying therapy of neurodegenerative diseases such as prion and Parkinson's disease. Acta Neuropathol. 2013;125:795–813. doi: 10.1007/s00401-013-1114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Tatebe H., Taguchi K., Endo Y., Tokuda T., Mizuno T., Nakagawa M., Tanaka M. p62/SQSTM1-dependent autophagy of Lewy body-like alpha-synuclein inclusions. PLoS One. 2012;7 doi: 10.1371/journal.pone.0052868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C., Enari M., Klohn P.C., Rossi D., Flechsig E. Molecular biology of prions. Acta Neurobiol. Exp. (Wars) 2002;62:153–166. doi: 10.55782/ane-2002-1434. [DOI] [PubMed] [Google Scholar]
- White A.R., Enever P., Tayebi M., Mushens R., Linehan J., Brandner S., Anstee D., Collinge J., Hawke S. Monoclonal antibodies inhibit prion replication and delay the development of prion disease. Nature. 2003;422:80–83. doi: 10.1038/nature01457. [DOI] [PubMed] [Google Scholar]
- Wille H., Michelitsch M.D., Guenebaut V., Supattapone S., Serban A., Cohen F.E., Agard D.A., Prusiner S.B. Structural studies of the scrapie prion protein by electron crystallography. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3563–3568. doi: 10.1073/pnas.052703499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H., Bian W., McDonald M., Kendall A., Colby D.W., Bloch L., Ollesch J., Borovinskiy A.L., Cohen F.E., Prusiner S.B. Natural and synthetic prion structure from X-ray fiber diffraction. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16990–16995. doi: 10.1073/pnas.0909006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N. Hot spot of structural ambivalence in prion protein revealed by secondary structure principal component analysis. J. Phys. Chem. B. 2014;118:9826–9833. doi: 10.1021/jp5034245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material