Abstract
Retinoic acid inducible gene-I (RIG-I) is a cytosolic pathogen recognition receptor that initiates the immune response against many RNA viruses. Upon RNA ligand binding, RIG-I undergoes a conformational change facilitating its homo-oligomerization and activation that results in its translocation from the cytosol to intracellular membranes to bind its signaling adaptor protein, mitochondrial antiviral-signaling protein (MAVS). Here we show that RIG-I activation is regulated by reversible acetylation. Acetyl-mimetic mutants of RIG-I do not form virus-induced homo-oligomers, revealing that acetyl-lysine residues of the RIG-I repressor domain prevent assembly to active homo-oligomers. During acute infection, deacetylation of RIG-I promotes its oligomerization upon ligand binding. We identify histone deacetylase 6 (HDAC6) as the deacetylase that promotes RIG-I activation and innate antiviral immunity to recognize and restrict RNA virus infection.
Keywords: RIG-I, HDAC6, Deacetylation, HCV, West Nile virus, Interferon, Innate immunity
Graphical Abstract
Highlights
-
•
RIG-I undergoes reversible deacetylation during acute virus infection.
-
•
Acetylation of RIG-I Repressor domain controls RIG-I activation by restricting dimerization.
-
•
HDAC6 is the cellular deacetylase essential for RIG-I deacetylation to induce antiviral innate immunity.
RIG-I is a cytosolic pathogen recognition receptor at the frontline of immune response against RNA virus infection. RIG-I is expressed in most cells of the body and becomes activated after sensing and binding to pathogen associated molecular pattern (PAMP) RNA motifs within products of virus infection. The RIG-I activation process involves multiple regulatory events including PAMP binding and ATP hydrolysis, protein conformational change and intracellular redistribution, and specific post-translational modifications. Our results define reversible acetylation of RIG-I and its deacetylation by HDAC6 as key to controlling innate antiviral immune induction in response to RNA virus infection.
1. Introduction
Retinoic acid inducible gene-I (RIG-I) plays a major role in pathogen recognition to trigger innate immunity and signal the initiation of the immune response against RNA virus infection. RIG-I is the prototypical member of the RIG-I-like receptor (RLR) family of cytosolic RNA helicases includes melanoma differentiation-associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2) proteins. RIG-I functions as a cytosolic pathogen recognition receptor (PRR) (Loo and Gale, 2011) and drives immune signaling after binding to pathogen associated molecular pattern (PAMP) motifs within viral RNA that accumulate during acute infection of many RNA viruses (Kell et al., 2015). RIG-I recognizes 5′-triphosphate (5′-ppp) in conjunction with double-stranded (ds) RNA as non-self PAMPs. Our studies have shown that during acute hepatitis C virus (HCV) infection RIG-I recognizes and binds to the poly-uridine/cytosine (poly-U/UC) motif of the HCV RNA, thus signaling an innate immune response that restricts hepatocyte permissiveness for infection (Schnell et al., 2012, Saito et al., 2008). Active RIG-I then mediates downstream signaling by binding to the adaptor protein, mitochondrial antiviral-signaling protein (MAVS), located on the outer membrane of the mitochondria, peroxisomes, and on mitochondria-associated membrane (MAM) (Dixit et al., 2010, Horner et al., 2011). This process results in establishment of a MAVS signalosome that drives the activation of interferon regulatory factor 3 (IRF3) and NF-κB, leading to the expression of interferon (IFN)-β and a variety of antiviral and immunomodulatory genes that confer antiviral defense and that regulate immunity to infection (Loo and Gale, 2011). Viral evasion and control of these processes supports chronic HCV infection in nearly 200 million people, thus marking RIG-I regulation of hepatic defenses as a critical determinant impacting HCV infection and immunity (Liu and Gale, 2010, Sumpter et al., 2005).
RIG-I has tandem amino-terminal caspase activation and recruitment domains (CARDs) that mediate downstream signaling. RIG-I signaling is governed through inhibitory intramolecular interactions of the CARDs with its helicase domain and the carboxyl-terminal repressor domain (RD) to hold RIG-I in an autorepressed state until its activation mediated by binding to RNA PAMP ligand (Saito et al., 2007, Cui et al., 2008, Jiang et al., 2011). RD engagement of 5′-ppp RNA induces homotypic RD interaction with a partner RIG-I molecule bound to the RNA ligand. Through this process the RD is shown to release the autorepression of RIG-I, thereby facilitating RIG-I oligomerization and ATP hydrolysis and ultimately leading to a signaling-on conformation in which it mediates interaction with cognate RD on the RIG-I partner (Cui et al., 2008). This process then facilitates the formation of a RIG-I translocon consisting of RIG-I/14-3-3ε/TRIM25 complex that mediates the redistribution or “translocation” of RIG-I from cytosol to intracellular membrane compartments to bind MAVS through homotypic CARD-CARD interactions (Liu et al., 2012). The E3 ubiquitin ligase TRIM25 interacts with RIG-I at amino acid T55 and conjugates K63-linked ubiquitin to RIG-I in a process that is required to release RIG-I from autorepression (Gack et al., 2007). Introducing a T55I mutation into RIG-I abrogates its interaction with TRIM25 and interrupts RIG-I immune signaling, thus the cells become highly permissive to infection by HCV and other RNA viruses (Sumpter et al., 2005, Gack et al., 2007). RIG-I signaling is further regulated by Riplet-mediated K63-linked polyubiquitination of the RIG-I RD, which also serves to release the RIG-I CARDs from auto-inhibition for signaling activation (Oshiumi et al., 2013), and by reversible phosphorylation of the CARDs that fine-tunes its activation (Wies et al., 2013). Following ligand-induced ATP hydrolysis, RIG-I is placed in a signaling-on conformation (Guo et al., 2011, Civril et al., 2011, Lu et al., 2011). Thus, in the current model of RIG-I activation, the RNA PAMP ligand, the translocon, oligomeric assemblies containing RIG-I, and polyubiquitination are the prerequisites for RIG-I to signal through downstream adaptor MAVS to initiate innate immune responses.
The reversible acetylation of cytoplasmic protein complexes has been shown to regulate major cellular processes, including signal transduction and protein localization (Choudhary et al., 2009). Several histone acetyl-transferases (HATs) and histone deacetylases (HDACs) have been found to play critical roles in regulating type I interferon production and response (Nusinzon and Horvath, 2006, Suh et al., 2010, Zhu et al., 2011, Masumi, 2011). For example, a study identified that HDAC6, a cytosolic HDAC, could enhance β-catenin nuclear translocation to facilitate IRF3 activation and type I IFN production during virus infection (Zhu et al., 2011). Interestingly, a high-resolution mass spectrometry study identified lysine acetylation of RIG-I at residues K858 and K909 located within the RD (Choudhary et al., 2009). However, whether lysine acetylation participates in RIG-I activation and innate antiviral immunity is not known. Here, we define a role for the reversible acetylation of RIG-I in modulating innate immune signaling during RNA virus infection. We found that RIG-I is acetylated at adjacent sites within the RD in resting cells, and this posttranslational modification prevents RIG-I oligomerization. During acute virus infection, RIG-I acetylation is removed by HDAC6, which directly binds to RIG-I. This process facilitates PAMP RNA-induced RIG-I signaling activation. Our studies reveal that HDAC6 activity is crucial for RIG-I-dependent signaling to innate antiviral immunity.
2. Materials & Methods
2.1. Cells
Huh7 and HEK293 cells have been described (Saito et al., 2008). RIG-I-knock-down cells were produced through transduction and selection of cells with gene-specific pLKO.1-puro vector-based shRNA expression cassettes (Sigma).
2.2. Plasmids and Constructs
Flag-tagged RIG-I constructs have been described (Saito et al., 2007). HA-tagged and Myc-tagged RIG-I expression constructs were generated by cloning specific PCR products into pcDNA3.1 or pEF-Tak (Saito et al., 2007). Acetyl-mimetic mutants of RIG-I were generated by using QuikChange® Site-Directed Mutagenesis Kit (Stratagene). Flag-tagged HDAC or SIRT constructs were obtained from Addgene (Addgene plasmid 13812, 13813, 13819, 13821, 13822, 13823, 13824, and 13825).
2.3. RNA ChIP Assay
Huh7 cells were transfected with pEFBos-Flag-RIG-I and RIG-I acetyl-mimetic mutants. Following 48 h post-transfection, in vitro transcribed biotinyl-HCV PAMP RNA was transfected into these cells for 1 h. After two PBS washes, formaldehyde was added to the media to crosslink the RNA and protein, and 0.125 M of glycine was added to stop the reaction. The lysates were then used for anti-Flag immunoprecipitation and eluted by Flag peptide. The recovered products were directly dotted to nitrocellulose membrane for later immunoblotting to detect Flag-tagged RIG-I by anti-Flag antibodies and biotinyl-PAMP RNA by HRP-conjugated Strepavidin.
2.4. Virus Infection
For virus infection, 105 cells were cultured in wells of a 48-well plate and were infected with HCV (strain HCV 2a/JFH1) in serum-free media at 37 °C. One hour later, the cells were rinsed in PBS and incubated in media containing 10% FBS. At the time of harvest, cells were washed twice with PBS and fixed in 3% Paraformadehyde:PBS solution. Cells were immunostained with anti-HCV serum as described.
2.5. Immunoprecipitation
Cells were lysed in ice-cold RIPA buffer (50 mM Tris-Cl pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) in the presence of Protease Inhibitor Cocktail (Roche) for 10 min. Lysates were clarified by centrifugation and incubated with 2 μg of antibodies for 16 h followed by Protein A/G agarose for 1 h at 4 °C. The immunocomplexes were washed 3 times with cold RIPA buffer and resuspended in 15 μl of 2 × SDS sample buffer for SDS-PAGE. Commercial antibodies used in this study were: Anti-FLAG monoclonal antibody, Sigma-Aldrich Cat# P2983 RRID:AB_439685; anti-HA onoclonal antibody, Antibodies-Online.com - The Marketplace for Antibodies Cat# ABIN289443 RRID:AB_10775823; anti-ubiquitin polyclonal antibody, Antibodies-Online.com - The Marketplace for Antibodies Cat# ABIN361830 RRID:AB_10789886; anti-HDAC6 polylconal antibody, Fitzgerald Industries International Cat# 20R-1669 RRID:AB_11191264.
2.6. ATPase Assay
WT and mutant RIG-I were expressed and purified from Escherichia coli. 10 nM of purified RIG-I protein was incubated with 125 nM of RNA ligands, including 18-bp dsRNA, 14-bp dsRNA, 24-mer hairpin RNA and 5′-ppp hairpin RNA. The initial ATP concentration was 1 mM, and spiked with [γ-32P]ATP at 37 °C. A time course (0–30 min) of the ATPase reaction was used to determine the rates of ATP hydrolysis.
2.7. Gel Shift Assay
[γ-32P] end-labeled 18-bp RNA was incubated with purified WT or mutant RIG-I protein at vrious concentration from 0.2 pmol to 10 pmol in 20 μl reactions. The protein-RNA complex was then run in a 6% Native PAGE followed by radioautography.
2.8. Luciferase Reporter Assay
Dual luciferase assays to measure Interferon β promoter activity were conducted as described (Saito et al., 2008).
2.9. In Vivo Mouse Infections
C57BL/6 (WT) mice were purchased from Jackson Laboratories, Bar Harbor, ME. HDAc6 deficient mice were generously provided by Dr. Eduardo M. Sotomayor (H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33612) (Cheng et al., 2014). All mice were genotyped for positive identification and were bred and housed in in specific pathogen-free conditions in the animal facility at the University of Washington. All experiments were performed in accordance with the University of Washington Institutional Animal Care and Use Committee guidelines. Age-matched 7–8 week old mice were inoculated subcutaneously in the rear footpad with 100 PFU of WNV TX-02 strain (Keller et al., 2006) in a 20 μl inoculum diluted in phosphate buffered saline (PBS) supplemented with 1% heat-inactivated fetal bovine serum. Mice were monitored daily for morbidity and mortality. For clinical scoring, infected mice were monitored daily for signs of hind limb dysfunction and paresis. Mice were scored using the following scale of 1–6: 1, ruffled fur/lethargy, no paresis; 2, very mild to mild paresis; 3, frank paresis involving at least one hind limb and/or conjunctivitis; 4, severe paresis; 5, true paresis; 6, moribund. Each experiment included cohorts of 10 each WT and HDAC6−/− mice either mock-infected or infected with WNV. Three experiments were performed. Statistical significance for weight and clinical scores were determined using the Holm-Sidak method of multiple t-tests, with alpha = 5.000%. *p value < 0.005.
2.10. Statistical Analysis
Data were compared using Student's t-test.
3. Results
3.1. RIG-I is Acetylated in Resting Cells but Deacetylated During Acute Virus Infection
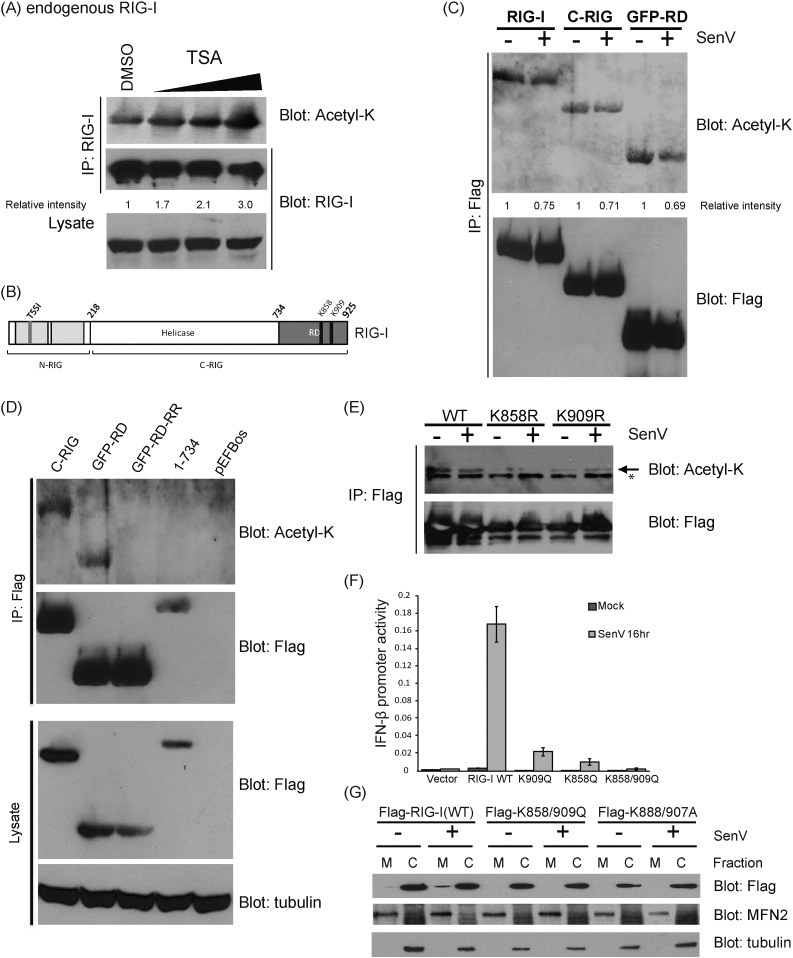
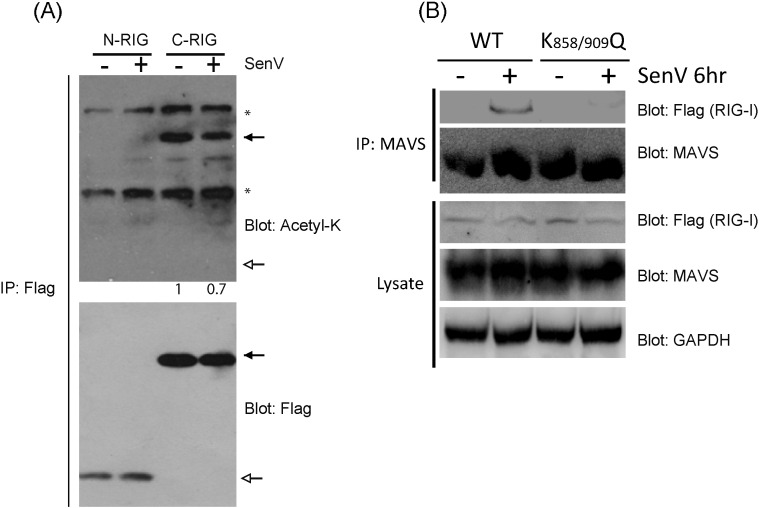
To determine if RIG-I is acetylated in resting cells, endogenous, full-length RIG-I was recovered by immunoprecipitation (IP) from HEK293 cells that were treated with increasing doses of a pan-deacetylase inhibitor, Trichostatin A (TSA). The recovered RIG-I was assessed for acetylation by immunoblot analysis using an anti-acetyl-lysine antibody. We found that RIG-I was basally acetylated in resting, non-infected cells and that increasing doses of TSA resulted in increased levels of acetyl-RIG-I (Fig. 1A). To identify the potential sites of lysine acetylation of RIG-I, we ectopically expressed Flag-tagged RIG-I and RIG-I truncation mutants containing C-terminal domain of RIG-I (C-RIG) or the green fluorescence protein fused to the RIG-I RD (GFP-RD) (Fig. 1B) and used immunoblot assay to probe for levels of acetyl-lysine on RIG-I following immunoprecipitation of each construct. These experiments were conducted in the presence or absence of infection with Sendai virus (SenV), an RNA virus that triggers RLR signaling and accurately models hepatitis C virus (HCV) activation of RIG-I (Saito et al., 2008). Full-length RIG-I, C-RIG, and GFP-RD were acetylated in resting, non-infected cells but during SenV infection the acetylation levels decreased (Fig. 1C). Notably, a construct encoding the Flag-tagged N-terminal CARDs of RIG-I (N-RIG) was not acetylated (Fig. S1A). Taken together, these results localize acetylation to the RIG-I RD inclusive of K858 and K909. To assess whether K858 and K909 were indeed the sites for RIG-I acetylation, Flag-tagged GFP-RD K858-909R mutant (GFP-RD-RR) was generated by site-direct mutagenesis. We then assessed the level of acetyl-lysine within the GFP-RD-RR mutant when expressed in 293 cells. GFP-RD-RR protein was recovered by anti-FLAG IP and subjected to anti-acetyl-lysine immunoblot assay in parallel analyses with IP products recovered from cells expressing C-RIG, GFP-RD, RIG-I 1-734, or vector alone (pEFBos). We found that while both C-RIG and GFP-RD were acetylated in resting cells, neither RIG-I 1-734 nor GFP-RD-RR was acetylated (Fig. 1D). Moreover, full-length RIG-I mutants containing individual point mutation of K858R or K909R each exhibited reduced acetylation levels compared to RIG-I WT (Fig. 1E). These results verify that residues K858 and K909 of RIG-I are acetylation targets in resting cells.
Fig. 1.
RIG-I is acetylated in resting cells. (a) Endogenous RIG-I was immunoprecipitated (IP) from mock or TSA treated cell lystate. Acetyl-RIG-I level was detected by anti-acetyl-lysine antibody. (b) Illustration of RIG-I mutants used in (c) to (g). (c) Ectopically expressed Flag-tagged RIG-I and mutants were recovered by IP from mock or SenV-infected cell lystate. Acetyl-lysine levels were detected by immunoblotting. Relative intensities of acetyl-RIG-I to total Flag-RIG-I was measured and calculated by Image J software and expressed as a value relative to the corresponding uninfected control. (d) Ectopically expressed Flag-tagged RIG-I and mutants were recovered by IP and analyzed for acetylation levels by immunoblot (blot) assay. GFP-RD-RR referred to Flag-tagged GFP-fused RIG-I RD with K858-909R mutations. (e) Ectopically expressed Flag-tagged RIG-I and mutants were recovered by IP from mock or SenV-infected cell lystate. Acetyl-lysine levels were detected by immunoblot assay. (f) SenV-induced IFN-β promoter induction in RIG-I knock-down Huh7 cells complemented with WT RIG-I or RIG-I acetyl-mimetic mutants. (g) Membrane (M) localization of RIG-I to the MAM from a resting, cyosolic (C) state during SenV infection. RIG-I acetyl-mimetic mutants were not detected in the membrane fractions, whereas WT RIG-I was redistributed to the MAM as marked by mitofusin 2, a MAM protein during SenV infection. Tubulin marks the cytosolic fraction.
Supplementary Fig. 1.
(a) RIG-I acetylation at K858 and K909 of the C-terminal RD. Ectopically expressed Flag-tagged RIG-I and indicated mutants were immunoprecipitated (IP) from mock or SenV-infected cell lystate. Acetyl-lysine levels were detected by immunoblot assay. C-RIG and N-RIG positions are indicated by filled or open arrows respectively. Non-specific bands are indicated by stars. (b) Virus infection-inducible interaction between MAVS and WT RIG-I but not the acetyl-mimetic RIG-I mutant. Flag-tagged WT RIG-I or RIG-I K858-909Q were ectopically expressed in Huh7 cells then infected with SenV for 6 h. Cell lysates were subjected to immunoprecipitation with rabbit anti-MAVS antibodies. Flag-tagged WT RIG-I was co-immunoprecipitated with MAVS during SenV infection, whereas RIG-I K858-909Q could not form significant amout of virus-inducible complex with MAVS.
To assess how the acetylation of RIG-I regulates its signaling activities, we introduced acetyl-mimetic mutations into a RIG-I expression construct by site-directed mutagenesis, whereby the lysine at residues K858 and K909 were mutated to glutamine to mimic the steric and charge characteristics of acetylated lysine (Xiong and Guan, 2012). The resultant K858Q or K909Q single-mutant or the K858-909Q double mutant in comparison to WT RIG-I were assessed for their ability to signal IFN-β promoter activation in response to SenV infection. For these experiments RIG-I knockdown Huh7 cells that stably express an shRNA targeting RIG-I were reconstituted with either WT or mutant RIG-I constructs in the presence of an IFN-β-luciferase reporter construct, and either mock-infected or infected with SenV. As compared to WT RIG-I, cells expressing any of the three acetyl-mimetic mutants (K858Q, K909Q, and K858-909Q) of RIG-I were significantly impaired in their ability to signal IFN-β promoter activation in response to SenV infection (Fig. 1F). We previously found that RIG-I redistributes to membrane fractions in response to virus infection, and this translocation is a measure of RIG-I signaling activation wherein the RIG-I translocon facilitates RIG-I binding to MAVS (Liu et al., 2012). We evaluated whether acetylation played a role in regulating the relocalization process by comparing WT RIG-I and RIG-I acetyl-mimetic mutants for their ability to redistribute from the cytosol to membrane fractions in response to SenV infection. As a control we included a RIG-I K888-907A mutant that cannot bind PAMP RNA and therefore precludes the formation of the RIG-I translocon (Liu et al., 2012). In cell fractionation studies, we found that a portion of WT RIG-I redistributed to the membrane (M) fraction that also contains the mitochondria associated membranous protein mitofusin 2 (MFN2) in response to SenV infection, as expected (Fig. 1G). In contrast, the RIG-I K858-909Q and the RIG-I K888-907A mutants remained in the cytosolic (C) fraction along with control protein tubulin. Neither mutant could be detected in the membrane fractions regardless of whether the cells were infected with SenV, similar to RIG-I in resting cells. Consistent with these observations, RIG-I K858-909Q could not undergo virus-induced interaction with MAVS but WT RIG-I formed a complex with MAVS upon SenV infection (Fig. S1B). These results reveal RIG-I to be acetylated within the RD at lysine residues K858 and K909 wherein acetylation at these residues restricts RIG-I translocation to intracellular membranes for MAVS interaction in response to virus infection.
3.2. Acetylation Controls RIG-I Oligomer Formation
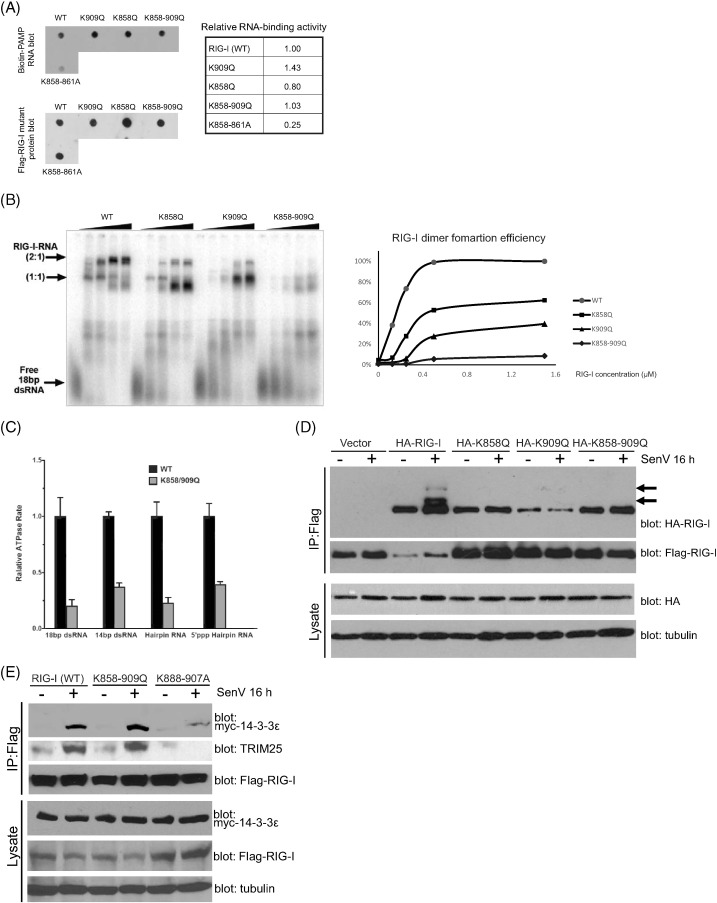
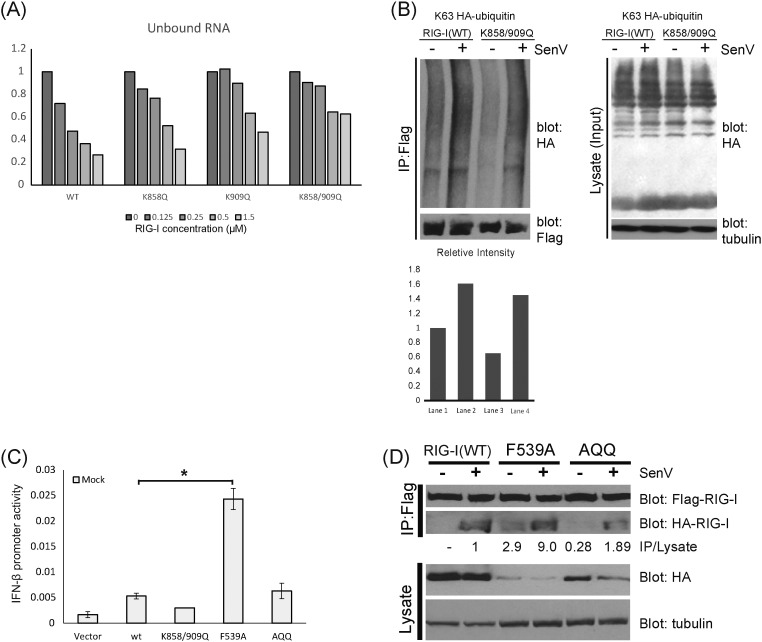
To define the molecular mechanism by which acetylation of RIG-I at K858 and K909 restricts RIG-I activation, we further assessed the acetyl-mimetic RIG-I mutants for their ability to bind PAMP RNA, to become polyubiquitinated, and to oligomerize, all of which are step-wise processes essential for RIG-I signaling activation (Chan and Gack, 2015). In particular, K858 of RIG-I has been implicated in participating in binding to RNA ligand cooperatively with K861 (Shigemoto et al., 2009). We first evaluated WT RIG-I and the acetyl-mimetic mutants, RIG-I K858Q, RIG-I K909Q, and RIG-I K858-909Q for their ability to bind to synthetic 5′ppp-containing ssRNA PAMP ligands. As a further control, we included for comparison a RNA-binding defective RIG-I K858-861A mutant (Shigemoto et al., 2009). We transfected in vitro transcribed biotinylated HCV 5′-ppp poly-U/UC PAMP RNA (Schnell et al., 2012, Saito et al., 2008) into Huh7 cells that express Flag-tagged WT RIG-I, RIG-I acetyl-mimetic mutant, or RIG-I K858-861A, and assessed the level of PAMP RNA that was recovered in a complex with Flag RIG-I by IP and immunoblot analysis. PAMP RNA that is recovered as part of the RIG-I complex was detected using alkaline phosphatase-conjugated streptavidin. We found that each acetyl-mimetic mutant of RIG-I (K858Q, K909Q, and K858-909Q) retained PAMP RNA-binding activity similar to WT RIG-I (Fig. 2A). In contrast, RNA binding activity of the RIG-I K858-861A mutant was attenuated in comparison to WT RIG-I. We further evaluated WT and mutant RIG-I for their ability to bind to a dsRNA PAMP ligand in electrophoretic mobility shift assays (EMSA) using purified recombinant proteins. The acetyl-mimetic mutants of RIG-I all retained the ability to bind to an 18-bp dsRNA PAMP ligand in a concentration-dependent manner, which is measured by the decrease in signal from the free 18-bp dsRNA band and the concentration-dependent formation of slower migrating bands of RIG-I:RNA complexes (Figs. 2B and S2A). Among the mutants, RIG-I K858-909Q exhibited comparably reduced dsRNA-binding efficiency in this assay wherein lower levels of the 1:1 RIG-I:dsRNA complex were detected with the K858-909Q mutant compared to WT, and this mutant formed little, if any, of the higher 2:1 RIG-I:dsRNA complexes (Fig. 2B). More importantly, all three RIG-I acetyl-mimetic mutants were attenuated in their ability to oligomerize in the presence of dsRNA, as observed by the reduction in signals of the 2:1 RIG-I:dsRNA bands in comparison to WT RIG-I, with the K858-909Q mutant having almost a complete loss of the 2:1 oligomer product (Fig. 2B). These results suggest that although they retain the ability to recognize and bind PAMP RNA, acetyl-mimetic RIG-I is restricted in its ability for forming RNA-induced oligomers. Thus, PAMP RNA-induced oligomerization of RIG-I is regulated by acetylation of RIG-I at K858 and K909 sites.
Fig. 2.
Oligomerization difficiency of RIG-I acetyl-mimetic mutants. (a) in vitro transcribed biotinylated-PAMP RNA from HCV 3′UTR was transfected into cells expressing the indicated Flag-tagged RIG-I mutants. After crosslinking, PAMP RNA/RIG-I complexes were recovered by anti-Flag IP and blotted onto nitracellulose membrane. RNA is visualized using Streptavidin-HRP. Relative RNA binding activities were quantified using Image J software, and expressed as a value relative to the corresponding WT control. (b) Representative gel-shift assays for WT and RIG-I mutants in the presence of 18-bp dsRNA. The oligomer formation efficiency of RIG-I-RNA (2:1) complex by band intensities was quantified and plotted. (c) Relative RNA-stimulated ATP hydrolysis of WT RIG-I and three RIG-I mutants. Data were normalized to WT RIG-I and shown as the relative level ATPase rate. Error bars represent the standard deviation of triplicate measurements. (d) Impaired oligomerization of RIG-I acetyl-mimetic mutants during SenV infection. Flag-tagged wt RIG-I and HA-tagged RIG-I acetyl-mimetic mutants were co-transfected into HEK293 cells. After SenV infection, cell extracts were subjected to IP by anti-Flag antibody. RIG-I oligomerization activities were shown by the levels of co-IP HA-tagged RIG-I. Arrows indicate Flag-RIG-I oligomers. (e) RIG-I translocon formation. HEK293 cells expressing Flag-tagged WT or mutant RIG-I were co-expressed with Myc-14-3-3ε and cells were infected with 100 HAU SenV for 16 h followed by anti-Flag IP of cell extracts. The products were analyzed by immunoblot with anti-TRIM25 antibody or anti-Myc antibody.
Binding to PAMP RNA induces RIG-I hydrolysis of ATP to facilitate the conformational change that places RIG-I into a signaling-on conformation that promotes oligomerization (Chan and Gack, 2015). To directly assess how RIG-I oligomerization is regulated by acetylation at K858 and K909, and how this process might impact RIG-I interactions with TRIM25 and 14-3-3ε to assemble the translocon, we conducted ATPase and co-IP studies to assess the state of RIG-I activation. We assessed ATPase activity of RIG-I WT and RIG-I K858-909Q acetyl mimetic mutant induced by different PAMP RNA ligand including 18-bp or 14-bp dsRNA, and 24-mer hairpin RNA with or without 5′-ppp. Compared to WT RIG-I, the ATPase activity of RIG-I K858-909Q induced by each RNA ligand was attenuated (Fig. 2C), indicating that RD acetylation of RIG-I impedes its ability to hydrolyze ATP and thus may impair oligomerization of RIG-I. We next co-expressed Flag-tagged RIG-I WT and acetyl-mimetic mutants of HA-tagged RIG-I, and evaluated their capacity in forming virus-induced interaction by co-IP assays. SenV infection induced robust interaction of HA-RIG-I WT with FLAG-RIG-I WT, but interaction of RIG-I WT with either HA-RIG-I K858Q, K909Q or K858-909Q mutant failed to occur in response to SenV infection (Fig. 2D). We then assessed virus-induced complex formation of RIG-I with TRIM25 and 14-3-3ε, the essential components of the RIG-I translocon (Liu et al., 2012). For these analyses we assessed the partner binding activity of the RIG-I K858-909Q double mutant compared to RIG-I WT and a RIG-I K888-907A double mutant control that is known to completely abolish RNA-binding activity (Wang et al., 2010). We found that the RIG-I K858-909Q acetyl mimetic double mutant was still able to form a virus-induced complex to interact with both 14-3-3ε and TRIM25 during SenV infection at a similar level to RIG-I WT (Fig. 2E). However, the RIG-I K888-907A mutant was defective in its ability to undergo virus-induced interaction with TRIM25 and 14-3-3ε, thus defining PAMP RNA binding as a critical step in promoting RIG-I translocon formation. We further assessed the extent of K63-linked RIG-I ubiquitination as mediated by TRIM25 (Gack et al., 2007) and found that the ubiquitination levels of the RIG-I WT and K858-909Q acetyl mimetic mutant were similar following induction by SenV infection (Fig. S2B). Taken together, these data reveal that K858-909 acetylation restricts RNA-induced RIG-I ATPase activity and oligomer formation but not induction of translocon formation nor ubiquitination of RIG-I, and that this control occurs at a step following RIG-I binding to PAMP RNA ligand.
Supplementary Fig. 2.
(a) Graphical representation of specific RNA binding by WT RIG-I or RIG-I mutants over a dose range of RIG-I concentration. (b) K63-linked ubquitination of the RIG-I CARDs in RIG-I acetyl-mimetic mutants remains intact. WT or mutant Flag-RIG were co-expressed with HA-tagged K63ubiquitin in Huh7 cells. Cell extracts were subject to immunoprecipitation with anti-Flag antibody. Ubiquitination level of RIG-I products was evaluated by anti-HA immunoblot. Immnoblotting of input lysates is shown in the right panel, respective ubiquitin product for each lane of upper left panel is presented in the lower left panel. (c) Constiutive IFN-β promoter activity in resting/noninfected RIG-I knock-down Huh7 cells complemented with RIG-I WT and RIG-I mutants. *: p value < 0.005. (d) Flag-tagged WT or HA-tagged RIG-I mutants were co-trasnfected with WT HA-RIG-I in Huh7 cells. Cells mock-infected (−) or infeted with SenV (+) for 16 h, extracts prepared and subjected to co-immunoprecipitation with anti-Flag antibody. Input (lysate) and IP products were assessed by immunoblot assay with anti-HA or anti-Flag antibodfy.
To determine how acetylation might serve to control RIG-I from spurious activation in non-infected, resting cells, we evaluated RIG-I oligomerization and activation of innate immune signaling among acetyl mimetic RIG-I mutants designed to disrupt interdomain interactions that otherwise regulate RIG-I signaling. In particular, we compared the signaling actions of WT and RIG-I K858-909Q to RIG-I F539A. In resting cells RIG-I is maintained in an autorepressed state through the RD which imparts helicase (Hel2i) domain interactions of F539 with the CARDs wherein F539A mutation of RIG-I releases this autorepression resulting in a level of constitutive RIG-I signaling (Kowalinski et al., 2011). We expressed WT RIG-I, RIG-I F539A, or the acetyl mimics of RIG-I, in Huh7 RIG-I knockdown cells that lacked expression of endogenous RIG-I. We found that RIG-I F539A exhibited a level of constitutive signaling activity but this constitutive activity was reduced in the context of the K858-909Q acetyl-mimetic mutations (RIG-I AQQ triple mutant) (Fig. S2C). Thus, even when RIG-I is placed in a “signaling-on” state through disruption of inhibitory domain interactions, as is the case of the F539A mutant, acetyl mimetic mutation of K858-909Q restricts RIG-I signaling. To test how these acetyl-mutations impacted RIG-I protein-protein interactions in resting cells we evaluated the interaction of HA-RIG-I WT with Flag-tagged RIG-I WT, RIG-I F539A, and RIG-I AQQ mutants following co-expression in Huh7 RIG-I knockdown cells. Cells were harvested at 24 h post-transfection, and proteins were co-immunoprecipitated with anti-Flag antibody then subjected to anti-HA immunoblot analysis. As expected, in noninfected, resting cells, WT RIG-I was present only as a monomer and subsequently formed a RIG-I complex upon SenV infection. However, RIG-I F539A formed a constitutive complex with RIG-I WT, consistent with its constitutive signaling activity (Fig. S2D). In contrast, this constitutive RIG-I binding activity of the F539A mutant exhibited a > 4-fold reduction when this mutation was placed in context of the RIG-I AQQ triple mutant. Taken together, these observations show that K858-909Q mutation regulates intramolecular interactions of RIG-I to control its signaling activity. These results imply that acetylation governs RIG-I oligomerization actions to thus prevent aberrant activation in the absence of virus infection. Deacetylation of RIG-I is therefore required to facilitate the RIG-I oligomerization/activation process.
3.3. HDAC6 is the RIG-I Deacetylase
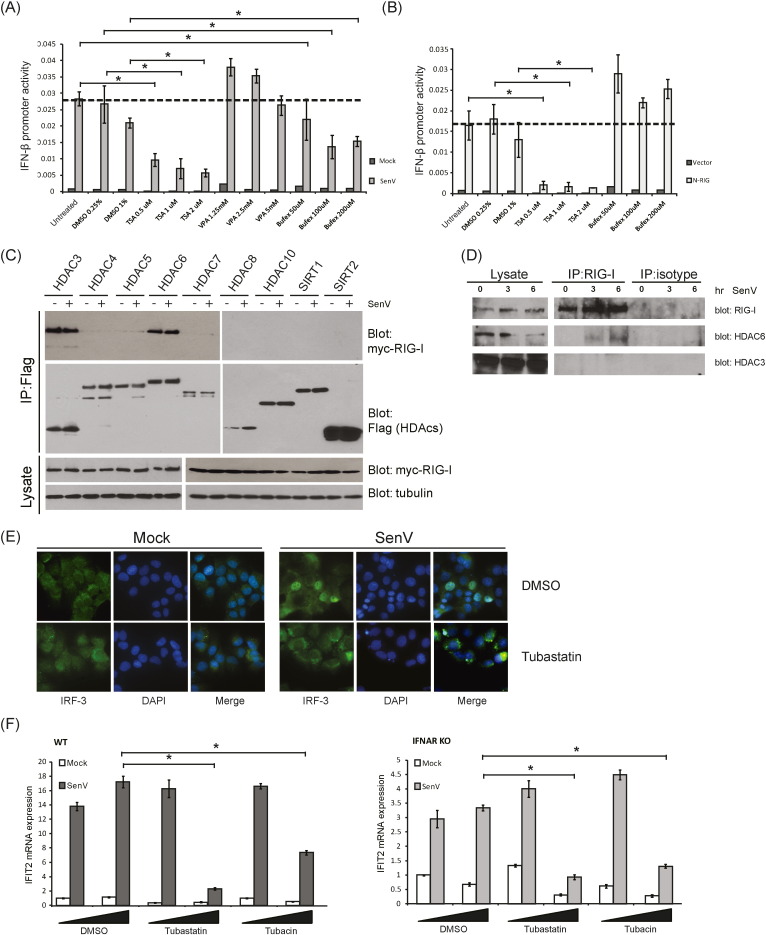
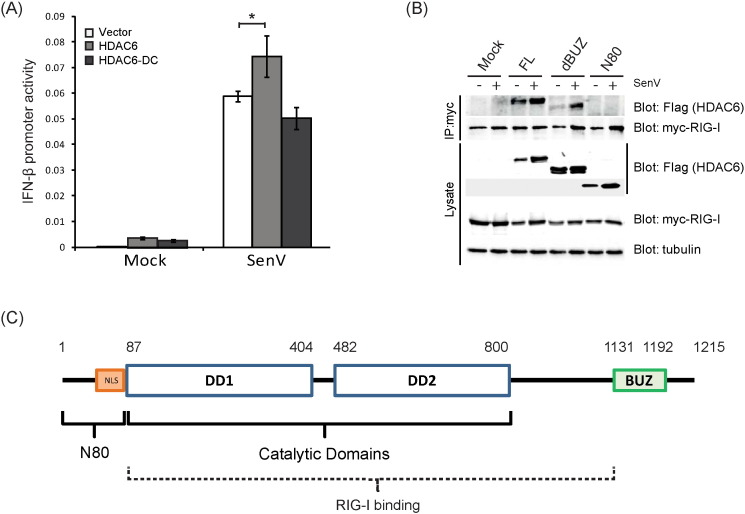
To identify the specific deacetylase that mediates RIG-I deacetylation, we examined RIG-I signaling to the IFN-β promoter in cells treated with HDAC inhibitors. HEK293 cells were treated with a pan-HDAC inhibitor (Trichostatin A, TSA), nuclear HDAC inhibitor (valproate, VPA), or cytoplasmic HDAC inhibitor (Bufexamac) (Bantscheff et al., 2011) followed by SenV infection to induce RIG-I signaling and activation an IFN-β promoter luciferase construct. TSA treatment has been shown previously to block IFN-β production (Nusinzon and Horvath, 2006). While both Bufexamac and TSA treatment blocked IFN-β promoter induction by SenV in a dose-dependent manner, treatment of cells with VPA did not decrease IFN-β promoter induction as compared to nontreated cells (Fig. 3A). TSA also blocked IFN-β promoter induction by the constitutively active N-RIG construct but importantly, Bufexamac treatment did not block IFN-β promoter induction by N-RIG. Thus, RIG-I lacking its C-terminal region containing the acetylation sites is not subject to regulation by the cytoplasmic HDACs (Fig. 3B). To identify the HDACs that target RIG-I, we co-expressed Myc-tagged RIG-I and Flag-tagged cytoplasmic HDACs in HEK293 cells and subjected lysates to a co-IP analysis. Both HDAC3 and HDAC6 were recovered with RIG-I in anti-Flag IP reactions, regardless of virus infection (Fig. 3C). We therefore assessed endogenous RIG-I interaction with endogenous HDAC6 or HDAC3. Intriguingly, we found that endogenous RIG-I and HDAC6 formed a virus-inducible complex within three hours after SenV infection but endogenous HDAC3 did not interact with RIG-I (Fig. 3D). Thus, HDAC6 forms a virus-induced complex with RIG-I during acute virus infection.
Fig. 3.
RIG-I deacetylation by the cytoplasmic deacetylase HDAC6 during viral infection. (a) and (b) RIG-I signaling to the IFN-β promoter was measured by promoter-luciferase assay in mock-infected or SenV-infected Huh7 cells that were treated with DMSO control or HDAC inhibitor. (a) cells were treated with increasing dose of Tricostatin A (TSA), Valproate (VPA), or Bufexamac (Bufex). (b) Huh7 cells were transfected with vector lone or the constitutively active N-RIG mutant and cultured in the presence of treatment with DMSO (control), TSA, or Bufexamac. 12 h later the cells were harvested and assessed for IFN-β-luciferase activity. *: p value < 0.05. (c) Screening of RIG-I interacting HDACs. Flag-tagged HDACs and Myc-tagged RIG-I were co-transfected into HEK293 cells. Cells were mock or SenV infected and extracts prepared 16 h later. Proteins were recovered by anti-Myc co-IP and identified by anti-Flag or anti-Myc immunoblot analysis. (d) Endogenous RIG-I and HDAC6 form virus-inducible complexes. Mock or SenV-infected Huh7 cell lysates were used for IP by anti-RIG-I antibody. Recovered proteins were identified by immunoblot assay for RIG-I, HDAC6 and HDAC 3. (e) Huh7 cells were mock infected or infected with SenV for 16 h in the presence of DMSO (control) or Tubastatin A treatment. 16 h later the cells were co-stained with DAPI to detect the nucleus and anti-IRF3 antibody followed by anti-rabbit flourescein secondary antibody for immunostain analysis of IRF3 subcellular distribution. (f) WT and IFNAR KO MEFs were treated with DMSO control or with increasing levels of HDAC6 specific inhibitor, Tubastatin A or Tubacin, during mock infection or SenV infection. 16 h later the cells were harvested and the level of IFIT2 mRNA was quantified by RT-qPCR assay. *: p value < 0.05.
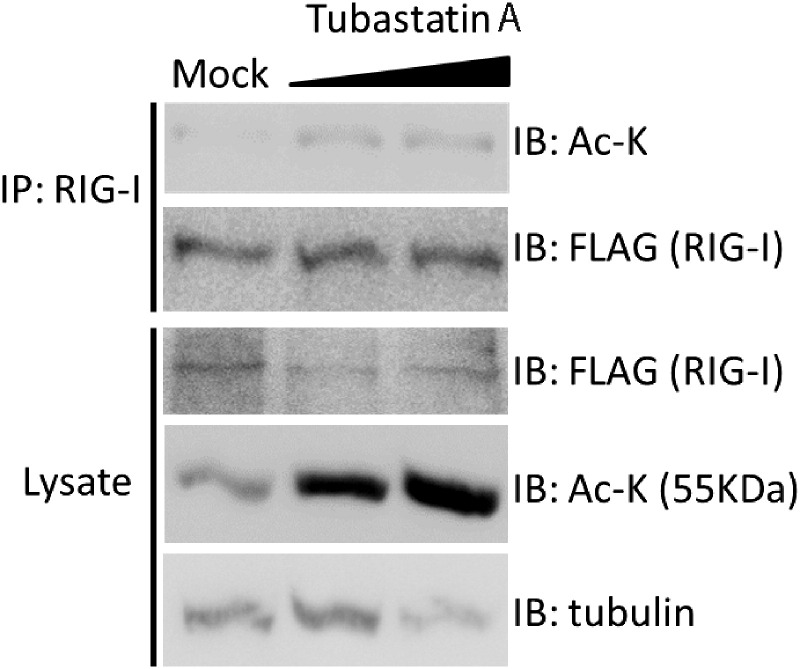
To assess the role of HDAC6 as a RIG-I deacetylase, we examined the level of acetyl RIG-I in cells treated with increasing amounts of the HDAC6-specific inhibitor, Tubastatin A (Haggarty et al., 2003). Tubastatin A treatment resulted in increased abundance of acetyl-RIG-I (Fig. S3). We therefore tested whether or not Tubastatin A could suppress RIG-I-dependent signaling of IRF3 activation in Huh7 cells. Cells were pretreated with Tubastatin A for 2 h and then infected with SenV for 16 h to induce RIG-I-dependent IRF3 activation, which can be measured by assessing IRF3 nuclear translocation. SenV infection induced the nuclear accumulation of IRF3 but this was blocked by treatment of cells with Tubastatin A (Fig. 3E). To determine if HDAC6 was required for induction of direct IRF3-target genes, we assessed the expression of interferon-induced protein with tetratricopeptide repeats (IFIT)2 gene in WT mouse embryo fibroblasts (MEFs) and in MEFs from mice with a targeted deletion in the type-1 interferon receptor (IFNAR KO). Cells were treated with HDAC6 specific inhibitors, Tubastatin A or Tubacin, infected with SenV, and harvested for RT-qPCR assay to measure IFIT2 mRNA level (Fig. 3F). IFIT gene expression can be induced after virus infection directly upon IRF3 activation as well as upon IFN signaling because of the presence of both IRF3 and ISGF3 binding sites in the promoter (Fensterl and Sen, 2011, Daffis et al., 2007). Virus-induced IFIT2 mRNA levels in both WT and IFNAR−/− MEFs were suppressed in cells treated with Tubastatin A or Tubacin (Fig. 3F, left and right panels respectively). Similar results but with a higher level of SenV-induced IFIT2 mRNA expression was observed in WT MEFs, likely reflecting IFN receptor-dependent signaling amplification. It is notable that HDAC6 has been reported to act as a coactivator of the IFN-β promoter-enhancer wherein HDAC6 was shown to operate upstream of IRF3 (Nusinzon and Horvath, 2006). Taken together, these observations indicate that HDAC6 imparts control of RIG-I-dependent signaling of IRF3 activation and target gene expression. Thus, HDAC is the RIG-I deacetylase.
Supplementary Fig. 3.
Increaseing RIG-I acetylation levels in cells treated Tubastatin A in a dose-dependent manner. Huh7 cells were treated with increasing dose of Tubastatin A for 6 h. Cells were harvested and subjected to immunoblot assay with anti-RIG-I or anti-acetyl-lysine antibodies. A dominant band in the lysate detected in the acetyl-lyine blot at the molecular weight around 55 kDa, overlapping with endogenous tubulin as detected by anti-tubulin antibodiesy. Tubastatin A treatment increases the levels of acyl RIG-I and acetyl-tubulin (control).
3.4. HDAC6 is Essential for RIG-I-dependent Antiviral Innate Immunity
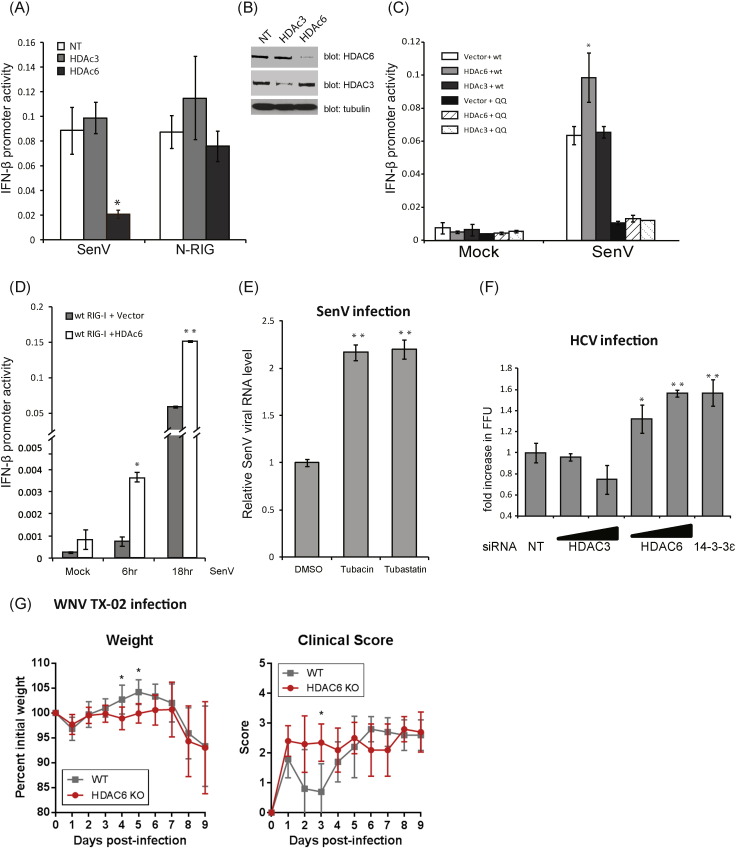
To determine the role of HDAC6 in RIG-I signaling of innate antiviral immunity, we examined innate immune signaling and virus infection in the context of HDAC knockdown in human cells. Huh7 cells were transfected with non-targeting (NT) siRNA, or with siRNA targeting HDAC3 (control) or HDAC6, followed by SenV infection or transfected with N-RIG expression construct to trigger RIG-I signaling (Fig. 4A and B). Knockdown of HDAC6 but not HDAC3 resulted in reduced IFN-β promoter induction upon SenV infection, as compared to NT siRNA transfected control cells (Fig. 4A). However, ectopic expression of N-RIG in NT control cells or HDAC3- or HDAC6- knockdown cells induced IFN-β promoter activity (Fig. 4A), indicating that HDAC6 requires the RIG-I C-terminal region to negatively regulate RIG-I signaling. We also assessed whether ectopic HDAC6 expression could enhance RIG-I signaling. HEK293 cells were co-transfected with vector, HDAC3, or HDAC6 in combination with an over-expression construct encoding RIG-I WT or the RIG-I K858-909Q double mutant. As shown in Fig. 4C, HDAC6 but not HDAC3 enhanced signaling by RIG-I WT after SenV infection but failed to enhance signaling by RIG-I K858-909Q. Furthermore, ectopic HDAC6 supported more rapid RIG-I signaling after SenV infection (Fig. 4D) but an HDAC6 active site mutant, HDAC6-DC, lacking acetyl-transferase activity, did not enhance RIG-I signaling (Fig. S4A). We also conducted parallel studies to assess MDA5 signaling in the presence of ectopic HDAC6 expression or in HDAC6 knockdown cells during mock-infection or infection with encephalomyocarditis virus (EMCV), a known MDA5-specific agonist (Loo and Gale, 2011). EMCV-induced signaling of MDA5 to the IFN-β promoter was neither altered by HDAC6 inhibitor treatment nor HDAC6 knockdown (data not shown), indicating that HDAC6 specifically regulates RIG-I and not MDA5. We therefore assessed HDAC6 interaction with RIG-I to identify the domain of HDAC6 that mediates binding to RIG-I. We found that binding to RIG-I occurred exclusive of the N-terminal 80 a.a. of HDAC6 and independently of the ubiquitin-binding domain (BUZ domain; Fig. S4B), suggesting that HDAC6/RIG-I interaction is mediated by the HDAC6 C-terminal region that encodes the catalytic domain (Fig. S4C).
Fig. 4.
HDAC6 deacetylase activity is required for RIG-I signaling induction. (a) Huh7 cells were transfected with nontargeting siRNA control (NT) or siRNA specific for knockdown of HDAC3 or HDAC6. Cells were then infected with SenV or transfected with vector or N-RIG to induce signaling in the presence of the IFN-β-luciferase promoter construct. IFN-β promoter activity was measured 16 h later. (b) HDAC6 and HDAC3 protein levels from siRNA treated cells in A. (c) WT or mutant RIG-I was ectopically expressed with vector control, HDAC3 or HDAC6 in combination in the presence of the IFN-β-luciferase promoter in Huh7 cells. Cells were mock infected or infected with SenV and harvested for assessment of IFN-β promoter activity 16 h later. (d) WT RIG-I was ectopically expressed with vector control or HDAC6 in combination in the presence of the IFN-β-luciferase promoter in Huh7 cells. Cells were mock infected or infected with SenV and harvested at the indicated time for assessment of IFN-β promoter activity. (e) Huh7 cells were treated with DMSo control, Tubastatin A or Tubacin and mock infected or infected with SenV. Cells were harvested and viral RNA measured by RT-qPCR assay 24 h later. (f) Huh7 cells were treated with NT siRNA or increasing levels of siRNA for knockdown of HDAC3 or HDAC6. Cells were infected with HCV. Infectious virus in supernatant was determined at 48 h post-infection by viral focus forming unit assay. *: p value < 0.05; **: p value < 0.01. (G) WT and HDAC KO mice were challenged with WNV and monitored for clinical scores and weight loss. Statistical significance was determined using the Holm-Sidak method of multiple t-tests, with alpha = 5.000%. *: p value < 0.05.
Supplementary Fig. 4.
(a) The deacetylase activity of HDAC6 was required to enhance RIG-I dependent IFN-β promoter activity. HDAC6-DC, the catalytic site defective mutant of HDAC6, could not boost RIG-I signaling through IFN-β promoter above level sof cells expressing vector alone. *: p value < 0.05. (b) Mapping of the RIG-I interaction domain of HDAC6. Cells were cotransfected with Myc-RIG-I and Flag vector control or the indicated Flag-HDAC6 WT or mutant construct. FL, full-length HDAC6; dBUZ, HDAC6 with BUZ domain deletion; N80, the first 80 amino acid of HDAC6 from the N-terminus. IP products were assesed by immunoblot assay.
To determine if HDAC6 was required to protect the cells from RNA virus infection, we further evaluated viral RNA level or replication of distinct RNA viruses including SenV (a paramyxovirus), HCV (a hepacivirus), and West Nile Virus (WNV, a flavivirus) infection. Tubastatin A or Tubacin treatment of Huh7 cells resulted in increased SenV RNA levels after 16 h of infection compared to nontreated cells (Fig. 4E). Moreover, transfection of increasing amounts of HDAC6-specific siRNA but not HDAC3-specific siRNA in Huh7 cells significantly enhanced permissiveness for HCV (JFH-1) infection as compared to NT control Huh7 cells (Fig. 4F), with the level of enhancement approximating that observed in cells with 14-3-3ε knockdown that abrogates the RIG-I translocon and concomitant RIG-I signaling (Liu et al., 2012). Notably, the reduction of IFIT2 mRNA expression we observed in our studies (see Fig. 3F), corresponds with the loss of immune signaling and enhanced permissiveness to virus infection imposed by loss of HDAC6 expression level or inhibition of its acetyl-transferase activity (Figs. 3F and 4E). We further evaluated WNV TX-02 infection of WT or HDAC KO mice. During early WNV infection, RIG-I serves a dominant role for the acute induction of innate immunity and protection against WNV infection in vivo; however, MDA5 has a nonredundant role in contributing to retain active immunity and protection against WNV infection at later times following RIG-I signaling (Errett et al., 2013, Suthar et al., 2010). Thus, we focused our assessment of the clinical scores and weight loss in mice infected with WNV TX-02 during the first week of infection. Clinical symptoms were monitored daily during the course of infection to observe the disease occurrence and neurovirulence. Both WT and HDAC6 KO mice were susceptible to infection, with HDAC6 KO mice showing increased and more rapid onset of disease during early/acute infection, consistent with the kinetics of RIG-I-dependent signaling of innate immunity in acute WNV infection (Errett et al., 2013). When compared with the WT controls, HDAC6 KO mice displayed earlier symptoms 2 to 3 days post-infection (Fig. 4G) that associated with early weight loss (Fig. 4G). These results demonstrate the positive role of HDAC6 in RIG-I-dependent activation of innate immunity to protect against RNA virus infection.
4. Discussion
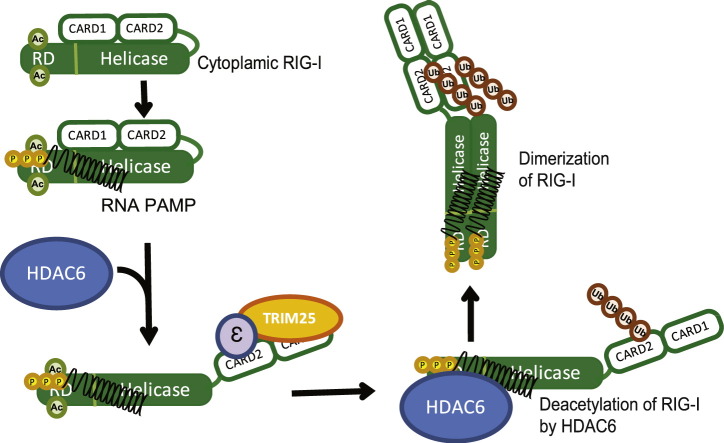
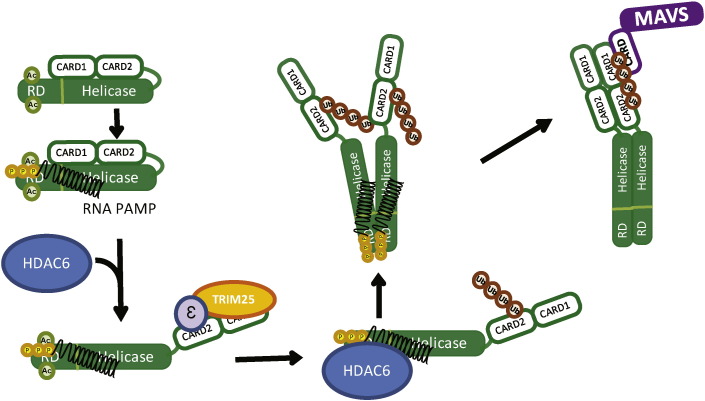
RIG-I is a major sensor of RNA virus infection. Among the RLRs, MDA5 has also been shown to act as an essential PRR for recognition of specific viruses such as picornaviruses, and to act cooperatively with RIG-I in recognition of flaviviruses and reoviruses (Loo et al., 2008). In contrast, LGP2, the third member of the RLR family, might serve as a co-factor or negative regulator of RLR signaling (Saito et al., 2007, Komuro and Horvath, 2006, Satoh et al., 2010). In terms of RIG-I, multiple steps are required for its activation, including RNA-binding dependent conformational change, polyubiquitination of the N-terminus CARD and the C-terminus RD, phosphorylation, formation of the RIG-I/14-3-3ε/TRIM25 translocon, ATPase hydrolysis, and oligomerization (Liu et al., 2012, Saito et al., 2007, Cui et al., 2008, Oshiumi et al., 2013, Gack et al., 2007, Maharaj et al., 2012, Wies et al., 2013). Homotypic complex formation of signaling molecules is a feature of innate immune signaling activation wherein complex formation is subject to regulation by post-translational protein modifications (Takashima et al., 2015, Gay et al., 2014, Blander, 2014). Our results show that RIG-I specific deacetylation at residues K858 and K909 by HDAC6 is required for its oligomerization that is essential for activation in response to virus infection and PAMP RNA transfection, and that HDAC6 directly regulates the IRF3-dependent IFN-β gene expression by reversing RIG-I acetylation. We found that the K858Q and K909Q acetyl-mimetic mutants of RIG-I showed moderate RNA binding activities but reduced homo-oligomerization and ATPase activities. That K858-909Q mutant RIG-I cannot signal downstream gene induction while failing to mediate virus-induced oligomer formation suggests that oligomeric RIG-I is the active unit that engages MAVS for innate immune activation. Our results suggest a model in which in resting cells, RIG-I is acetylated within the RD at K858 and K909 to regulate intramolecular interactions between the RD-Heli2 domains and hold RIG-I in a signaling-off conformation for preventing PAMP RNA ligand-independent homo-oligomerization and signaling activation. Binding to PAMP ligand RNA promotes a conformation change (Saito et al., 2007) that permits HDAC6 interaction for deacetylation of the RD (Fig. 5). This process then releases RIG-I autorepression and allows RIG-I to undergo ATP hydrolysis and form signaling-active homo-oligomers within the RIG-I translocon that facilitates MAM targeting and interaction with MAVS for innate immune signaling (Liu et al., 2012, Bhatia et al., 2011). During viral infection, RIG-I expression levels are increased in response to IRF3 activation and IFN signaling, such that newly-synthesized RIG-I may have a lower threshold for activation due to having lower acetylation levels. Thus, during virus infection the deacetylation of RIG-I by HDAC6 represents a critical step in the process of innate immune activation to relieve autorepression.
Fig. 5.
Model of HDAC6-depende t RIG-I activation. Details are described in the text.
Lysine acetylation, which neutralizes the positive charge of the amino acid, has been shown to be involved in regulation of protein interactions, DNA binding, RNA bindings, and many other various protein functions (Choudhary et al., 2009, Shakespear et al., 2011, Schwartz et al., 2010). Eighteen HDAC genes have been identified and are grouped into four classes based on their sequence homology; among these HDACs, HDAC6 and HDAC10 (class IIb HDACs) are distributed primarily in the cytoplasm, and other classes of HDACs are distributed in the nucleus (class I and IV), in the mitochondria (class III), or shuttling in between the nucleus and cytoplasm (class IIa) (reviewed in (Shakespear et al., 2011)). Besides the important functions of HDACs in histone modification and regulation of gene expression, emerging roles of HDACs in regulating inflammation and innate immune response are now being revealed (Klampfer et al., 2004, Nusinzon and Horvath, 2006, Vlasakova et al., 2007). For example, HDAC1, HDAC2, HDAC3, and HDAC8 repress the transcription factor NF-κB and MTA-1 (reviewed in (Shakespear et al., 2011)). HDAC6, however, was reported to promote IFN production in an IRF3-dependent manner (Nusinzon and Horvath, 2006), in which we now assign RIG-I as an HDAC6 target that facilitates IRF3 activation and IFN production. In our study, IRF3 nuclear translocation during SenV infection was blocked by treatment of cells with the HDAC6 specific inhibitors Tucastatin A and Tubacin, consistent with recent reports (Chattopadhyay et al., 2013). However, these HDAC6-associated effects on IRF3 were noted to be indirect, and our data now indicate that the regulatory events by HDAC6 occur upstream of IRF3 activation at the level of RIG-I signaling activation. HDAC6-inhibition enhanced the susceptibility of cells to both HCV and SenV infections. In fact, some viruses have developed strategies to control host immune response by modulating the activities of HDACs to disregulate the transactivator functions of IRFs, such as human papillomavirus E7 viral protein and respiratory syncytial virus-inducing cellular BCL-3 (Jamaluddin et al., 2005, Park et al., 2000). HDAC6 was also reported to be targeted and cleaved by influenza A virus-induced Caspase-3 (Husain and Harrod, 2009) and to regulate HIV-1 infection by reversing α-tubulin acetylation and therefore preventing HIV-1 envelope-dependent infection (Valenzuela-Fernandez et al., 2005). In addition, a recent report showed that overexpression of HDAC6 enhances resistance to adenovirus and H5N1 Influenza virus infections in murine embryonic stem cells, suggesting an important role of HDAC6 in anti-viral activities in both RIG-I dependent and independent actions (Wang et al., 2015). Of note is that while this current report was in revision, Choi, et al. reported their study showing that HDAC6 serves to deacetylate lysine 909 of RIG-I to facilitate innate immune induction in response to RNA virus infection (Choi et al., 2016). Our results validate and extend these observations to define both K858 and K909 as acetyl-lysine residues targeted by HDAC6 to regulate RIG-I function.
HDAC6 contains two tandem catalytic domains, an ubiquitin-binding domain (BUZ), and a dynein-binding domain (reviewed in (Boyault et al., 2007)). Besides α-tubulin, which is the major substrate of HDAC6, β-catenin, peroxiredoxin, Hsp90, and cortactin have been described as the substrates of HDAC6 (Boyault et al., 2007). In consideration of the fact that the post-translation modification of ubiquitination regulates many proteins in the RIG-I signaling pathway, HDAC6 may interact with these proteins through polyubiquitin chains that could serve to coordinate lysine acetylation and ubiquitination. Our observations show that virus infection induces the HDAC6/RIG-I interaction, and we speculate that this process could be regulated by various RIG-I modifications. Our data also indicates that the acetylation of RIG-I in resting, non-infected cells likely restricts RIG-I from random self-oligomerization to prevent IFN induction. So far we have not identified which acetyltransferases may be responsible for RIG-I acetylation. It is believed that Type B HATs, e.g. HAT1 and MEC-17, are cytoplasmic actyltransferases and are responsible for acetylating nascent proteins. These HATs can recognize unacetylated newly synthesized proteins such that they could be candidate RIG-I acetylases.
In summary, we propose that RIG-I deacetylation by HDAC6 serves to promote its “signaling-on” conformation initiated by RNA ligand-binding to RIG-I, resulting in K63-linked polyubiquitination, deacetylation, and oligomerization of RIG-I leading to activation (Fig. 5). Also, we propose that the unmodified RIG-I lysine residues, K858 and K909, are important in governing RIG-I activation and signaling function, consistent with both biochemical and structural models of RIG-I activation (Saito et al., 2007, Jiang et al., 2011). Upon RNA ligand binding, RIG-I deacetylation by HDAC6, K63 ubiquitination, and 14-3-3ε chaperone interactions offer multiple checkpoints for regulation of the innate immune response. However, deacetylation of RIG-I at the RD and translocon formation at the CARDs are two independent regulatory actions. Deacetylation of RIG-I is critical for its self-oligomerization via RD and for later CARD-dependent interactions with MAVS to signal downstream. As our studies implicate a role for HDAC6 at each of these steps, virus and host processes that affect HDAC6 function are likely to control RIG-I signaling pathway (Husain and Harrod, 2009).
The following are the supplementary data related to this article.
Funding Sources
Supported by NIH grants AI118916, AI083019, AI098943, AI100625, AI104002, by MOST grant 102-2320-B-002-044-MY2, and by NHRI grant NHRI-EX104-10417SC.
Conflict of Interest Statement
The authors whose names are listed immediately below certify that they have no affiliations with or involvement in any organization or entity with any financial interest, or non-financial interest in the subject matter or materials discussed in this manuscript.
Author Contributions
H.M.L. conducted molecular studies of RIG-I protein-interaction and signaling analyses. F.J., Y.M.L., T.H., S.H. and J.M. performed experiments. H.M.L and M.G. directed the research. H.M.L. and M.G. wrote the manuscript.
Acknowledgements
We thank our colleagues for reagents, and members of the Gale laboratory and Liu laboratory for discussion.
Contributor Information
Helene Minyi Liu, Email: mliu@ntu.edu.tw.
Michael Gale, Jr., Email: mgale@uw.edu.
References
- Bantscheff M., Hopf C., Savitski M.M., Dittmann A., Grandi P., Michon A.M., Schlegl J., Abraham Y., Becher I., Bergamini G., Boesche M., Delling M., Dumpelfeld B., Eberhard D., Huthmacher C., Mathieson T., Poeckel D., Reader V., Strunk K., Sweetman G., Kruse U., Neubauer G., Ramsden N.G., Drewes G. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat. Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- Bhatia R., Shaffer T.H., Hossain J., Fisher A.O., Horner L.M., Rodriguez M.E., Penfil S., Theroux M.C. Surfactant administration prior to one lung ventilation: physiological and inflammatory correlates in a piglet model. Pediatr. Pulmonol. 2011;46:1069–1078. doi: 10.1002/ppul.21485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander J.M. A long-awaited merger of the pathways mediating host defence and programmed cell death. Nat. Rev. Immunol. 2014;14:601–618. doi: 10.1038/nri3720. [DOI] [PubMed] [Google Scholar]
- Boyault C., Zhang Y., Fritah S., Caron C., Gilquin B., Kwon S.H., Garrido C., Yao T.P., Vourc'h C., Matthias P., Khochbin S. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21:2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.K., Gack M.U. RIG-I-like receptor regulation in virus infection and immunity. Curr. Opin. Virol. 2015;12:7–14. doi: 10.1016/j.coviro.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S., Fensterl V., Zhang Y., Veleeparambil M., Wetzel J.L., Sen G.C. Inhibition of viral pathogenesis and promotion of the septic shock response to bacterial infection by IRF-3 are regulated by the acetylation and phosphorylation of its coactivators. MBio. 2013;4 doi: 10.1128/mBio.00636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Lienlaf M., Wang H.W., Perez-Villarroel P., Lee C., WOAN K., Rock-Klotz J., Sahakian E., Woods D., Pinilla-Ibarz J., Kalin J., Tao J., Hancock W., Kozikowski A., Seto E., Villagra A., Sotomayor E.M. A novel role for histone deacetylase 6 in the regulation of the tolerogenic STAT3/IL-10 pathway in APCs. J. Immunol. 2014;193:2850–2862. doi: 10.4049/jimmunol.1302778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.J., Lee H.C., Kim J.H., Park S.Y., Kim T.H., Lee W.K., Jang D.J., Yoon J.E., Choi Y.I., Kim S., Ma J., Kim C.J., Yao T.P., Jung J.U., Lee J.Y., Lee J.S. HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG-I. EMBO J. 2016;35:429–442. doi: 10.15252/embj.201592586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Civril F., Bennett M., Moldt M., Deimling T., Witte G., Schiesser S., Carell T., Hopfner K.P. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12:1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S., Eisenacher K., Kirchhofer A., Brzozka K., Lammens A., Lammens K., Fujita T., Conzelmann K.K., Krug A., Hopfner K.P. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Daffis S., Samuel M.A., Keller B.C., Gale M.,.J.R., Diamond M.S. Cell-specific IRF-3 responses protect against West Nile virus infection by interferon-dependent and -independent mechanisms. PLoS Pathog. 2007;3:e106. doi: 10.1371/journal.ppat.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E., Boulant S., Zhang Y., Lee A.S., Odendall C., Shum B., Hacohen N., Chen Z.J., Whelan S.P., Fransen M., Nibert M.L., Superti-Furga G., Kagan J.C. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errett J.S., Suthar M.S., Mcmillan A., Diamond M.S., Gale M.,.J.R. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J. Virol. 2013;87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterl V., Sen G.C. The ISG56/IFIT1 gene family. J. Interf. Cytokine Res. 2011;31:71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., Jung J.U. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gay N.J., Symmons M.F., Gangloff M., Bryant C.E. Assembly and localization of Toll-like receptor signalling complexes. Nat. Rev. Immunol. 2014;14:546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- Guo D., Chen T., Ye D., Xu J., Jiang H., Chen K., Wang H., Liu H. Cell-permeable iminocoumarine-based fluorescent dyes for mitochondria. Org. Lett. 2011;13:2884–2887. doi: 10.1021/ol200908r. [DOI] [PubMed] [Google Scholar]
- Haggarty S.J., Koeller K.M., Wong J.C., Grozinger C.M., Schreiber S.L. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner S.M., Liu H.M., Park H.S., Briley J., Gale M.,.J.R. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain M., Harrod K.S. Influenza A virus-induced caspase-3 cleaves the histone deacetylase 6 in infected epithelial cells. FEBS Lett. 2009;583:2517–2520. doi: 10.1016/j.febslet.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Jamaluddin M., Choudhary S., Wang S., Casola A., Huda R., Garofalo R.P., Ray S., Brasier A.R. Respiratory syncytial virus-inducible BCL-3 expression antagonizes the STAT/IRF and NF-kappaB signaling pathways by inducing histone deacetylase 1 recruitment to the interleukin-8 promoter. J. Virol. 2005;79:15302–15313. doi: 10.1128/JVI.79.24.15302-15313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Ramanathan A., Miller M.T., Tang G.Q., Gale M., Patel S.S., Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell A., Stoddard M., Li H., Marcotrigiano J., Shaw G.M., Gale M.,.J.R. Pathogen-associated molecular pattern recognition of hepatitis C virus transmitted/founder variants by RIG-I is dependent on U-core length. J. Virol. 2015;89:11056–11068. doi: 10.1128/JVI.01964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller B.C., Fredericksen B.L., Samuel M.A., Mock R.E., Mason P.W., Diamond M.S., Gale M.,.J.R. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J. Virol. 2006;80:9424–9434. doi: 10.1128/JVI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klampfer L., Huang J., Swaby L.A., Augenlicht L. Requirement of histone deacetylase activity for signaling by STAT1. J. Biol. Chem. 2004;279:30358–30368. doi: 10.1074/jbc.M401359200. [DOI] [PubMed] [Google Scholar]
- Komuro A., Horvath C.M. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 2006;80:12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E., Lunardi T., Mccarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Liu H.M., Gale M. Hepatitis C virus evasion from RIG-I-dependent hepatic innate immunity. Gastroenterol. Res. Pract. 2010;2010:548390. doi: 10.1155/2010/548390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.M., Loo Y.M., Horner S.M., Zornetzer G.A., Katze M.G., Gale M.,.J.R. The mitochondrial targeting chaperone 14-3-3epsilon regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe. 2012;11:528–537. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.-M., Gale M.,.J.R. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M.A., Garcia-Sastre A., Katze M.G., Gale M.,.J.R. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Ranjith-Kumar C.T., Hao L., Kao C.C., Li P. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′ triphosphate. Nucleic Acids Res. 2011;39:1565–1575. doi: 10.1093/nar/gkq974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharaj N.P., Wies E., Stoll A., Gack M.U. Conventional protein kinase C-alpha (PKC-alpha) and PKC-beta negatively regulate RIG-I antiviral signal transduction. J. Virol. 2012;86:1358–1371. doi: 10.1128/JVI.06543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumi A. Histone acetyltransferases as regulators of nonhistone proteins: the role of interferon regulatory factor acetylation on gene transcription. J. Biomed. Biotechnol. 2011;2011:640610. doi: 10.1155/2011/640610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinzon I., Horvath C.M. Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation. Mol. Cell. Biol. 2006;26:3106–3113. doi: 10.1128/MCB.26.8.3106-3113.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H., Miyashita M., Matsumoto M., Seya T. A distinct role of Riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.S., Kim E.J., Kwon H.J., Hwang E.S., Namkoong S.E., Um S.J. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J. Biol. Chem. 2000;275:6764–6769. doi: 10.1074/jbc.275.10.6764. [DOI] [PubMed] [Google Scholar]
- Saito T., Hirai R., Loo Y.M., Owen D., Johnson C.L., Sinha S.C., Akira S., Fujita T., Gale M.,.J.R. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U. S. A. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Owen D.M., Jiang F., Marcotrigiano J., Gale M.,.J.R. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T., Kato H., Kumagai Y., Yoneyama M., Sato S., Matsushita K., Tsujimura T., Fujita T., Akira S., Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell G., Loo Y.M., Marcotrigiano J., Gale M.,.J.R. Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C.E., Kunwar P.S., Greve D.N., Moran L.R., Viner J.C., Covino J.M., Kagan J., Stewart S.E., Snidman N.C., Vangel M.G., Wallace S.R. Structural differences in adult orbital and ventromedial prefrontal cortex predicted by infant temperament at 4 months of age. Arch. Gen. Psychiatry. 2010;67:78–84. doi: 10.1001/archgenpsychiatry.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakespear M.R., Halili M.A., Irvine K.M., Fairlie D.P., Sweet M.J. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 2011;32:335–343. doi: 10.1016/j.it.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Shigemoto T., Kageyama M., Hirai R., Zheng J., Yoneyama M., Fujita T. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J. Biol. Chem. 2009;284:13348–13354. doi: 10.1074/jbc.M809449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H.S., Choi S., Khattar P., Choi N., Lee S.C. Histone deacetylase inhibitors suppress the expression of inflammatory and innate immune response genes in human microglia and astrocytes. J. NeuroImmune Pharmacol. 2010;5:521–532. doi: 10.1007/s11481-010-9192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R.,.J.R., Loo Y.M., Foy E., Li K., Yoneyama M., Fujita T., Lemon S.M., Gale M.,.J.R. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar M.S., Ma D.Y., Thomas S., Lund J.M., Zhang N., Daffis S., Rudensky A.Y., Bevan M.J., Clark E.A., Kaja M.K., Diamond M.S., Gale M.,.J.R. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima K., Oshiumi H., Takaki H., Matsumoto M., Seya T. RIOK3-mediated phosphorylation of MDA5 interferes with its assembly and attenuates the innate immune response. Cell Rep. 2015;11:192–200. doi: 10.1016/j.celrep.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Valenzuela-Fernandez A., Alvarez S., Gordon-Alonso M., Barrero M., URSA A., Cabrero J.R., Fernandez G., Naranjo-Suarez S., Yanez-Mo M., Serrador J.M., Munoz-Fernandez M.A., Sanchez-Madrid F. Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection. Mol. Biol. Cell. 2005;16:5445–5454. doi: 10.1091/mbc.E05-04-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasakova J., Novakova Z., Rossmeislova L., Kahle M., Hozak P., Hodny Z. Histone deacetylase inhibitors suppress IFNalpha-induced up-regulation of promyelocytic leukemia protein. Blood. 2007;109:1373–1380. doi: 10.1182/blood-2006-02-003418. [DOI] [PubMed] [Google Scholar]
- Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T., Hartmann G., Patel D.J. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Meng Q., Huo L., Yang M., Wang L., Chen X., Wang J., Li Z., Ye X., Liu N., Li Q., Dai Z., Ouyang H., Li N., Zhou J., Chen L., Liu L. Overexpression of Hdac6 enhances resistance to virus infection in embryonic stem cells and in mice. Protein Cell. 2015;6:152–156. doi: 10.1007/s13238-014-0120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wies E., Wang M.K., Maharaj N.P., Chen K., Zhou S., Finberg R.W., Gack M.U. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Guan K.L. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J. Cell Biol. 2012;198:155–164. doi: 10.1083/jcb.201202056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Coyne C.B., Sarkar S.N. PKC alpha regulates Sendai virus-mediated interferon induction through HDAC6 and beta-catenin. EMBO J. 2011;30:4838–4849. doi: 10.1038/emboj.2011.351. [DOI] [PMC free article] [PubMed] [Google Scholar]