Abstract
The general transcription factor TFIID provides a regulatory platform for transcription initiation. Here we present the crystal structure (1.97 Å) and NMR analysis of yeast TAF1 N-terminal domains TAND1 and TAND2 when bound to yeast TBP, together with mutational data. The yTAF1-TAND1, which in itself acts as a transcriptional activator, binds into the DNA-binding TBP concave surface by presenting similar anchor residues to TBP as E. coli Mot1 but from a distinct structural scaffold. Furthermore, we show how yTAF1-TAND2 employs an aromatic and acidic anchoring pattern to bind a conserved yTBP surface groove traversing the basic helix region, and we find highly similar TBP-binding motifs also presented by the structurally distinct TFIIA, Mot1 and Brf1 proteins. Our identification of these anchoring patterns, which can be easily disrupted or enhanced, provides compelling insight into the competitive multiprotein TBP interplay critical to transcriptional regulation.
Initiation of eukaryotic gene transcription at a core promoter requires the assembly of a preinitiation complex (PIC). These complexes are biologically dynamic assemblies, and their composition can be altered during development and thereby drive cell-specific programs of transcription1,2. All PICs include TATA-binding protein (TBP), either of RNA polymerase I, II or III, as well as polymerase and promoter specific co-activators, which include the core factor complex of RNA polymerase I, the SAGA or TFIID assemblies of RNA polymerase II, or the TFIIIB complex of RNA polymerase III3–6. The megadalton-sized TFIID is a multiprotein assembly and the predominant regulator of protein expression in eukaryotes, comprising TBP and thirteen evolutionary conserved TBP-associated factors (TAFs)4,5. TFIID is assembled in a stepwise manner, where a symmetric core-TFIID complex recruits additional TAFs to form the complete and asymmetric holo-TFIID that nucleates the PIC7.
TBP is the only protein required for transcription by all polymerases, and mutations in critical TBP interaction sites suggest that a substantial amount of global gene regulation in yeast and in higher eukaryotes occur via direct binding to TBP8,9. Recent work has structurally described TBP anchoring to PIC by interactions with general transcription factors Rnr7 (Pol-I), TFIIB (Pol-II) or the Brf1 core domain (Pol-III)3, enhanced by TFIIA (pol-II) or the Brf1 c-terminal extension (pol-III)3, and competitively regulated by NC2 and Mot1 (named BTAF1 in human)10. Direct TBP contacts with a large number of transcriptional activators have long been recognized as a key feature of TBP functionality5,11,12. According to the ‘activation by recruitment’ model, transcriptional activators use separate domains for DNA-binding and recruitment of the transcriptional machinery13. However, while the DNA anchoring in this model is well established12, recruiting activation domain complexes have only been characterized at low resolution14–17. Furthermore, although TBP has been shown to be a major target for transcriptional activators5,8,9,12, no TBP complex with a transcriptional activating domain has yet been structurally characterized.
TAF1, with homologues in all eukaryotes, is the largest and functionally most diverse TBP-associated factor and is considered the anchor point for TBP in TFIID4,5. Interactions between TAF1 and TBP are required for activated transcription in both yeast and mammalian cells9. The so-called ‘hand-off’ hypothesis suggest that TAFs in TFIID help release TBP from autoinhibited or unproductive complexes8,18,19 and then competitively release TBP to transcriptional activators that bring the complex to the promoter18. The yeast TAF1 (yTAF1) N-terminal domain 1 (TAND1; residues 10–37) independently acts as a transcriptional activation domain18,20, which is functional also when attached to several other TAFs in TFIID21. In contrast, the yeast TAND2 region (residues 46–71) alone has an inhibitory effect on transcription18,20 and competes with TFIIA in binding to TBP22,23. The dTAF1-TAND1 region alone binds to the concave, DNA-binding yTBP surface using TATA-box mimicry24, but is in itself a very poor transcriptional activator18. While initial NMR data suggest that yTAF1-TAND1 and -TAND2 bind to the concave and convex surface of TBP respectively20,25, due to the unstable nature of the yTAF1-yTBP complex, its detailed structural features have hitherto not been revealed.
To gain specific insight into TBP binding by TAF1, and thereby by transcriptional activators and repressors, we have determined the structure of a biologically active fusion protein comprising the yTBP core domain and residues 8–71 of yTAF125. By joint use of structural and biological techniques, our work describes the first high-resolution structure of a TAF1 protein bound to TBP containing both transcriptionally activating and repressing regions. The current structure and mutational analysis reveals detailed and specific molecular patterns of interactions to TBP, which by their structural diversity and competitive versatility provide an extended basis for our understanding of transcriptional regulation.
RESULTS
Crystal structure of the yTBP–yTAF1-TAND1-TAND2 complex
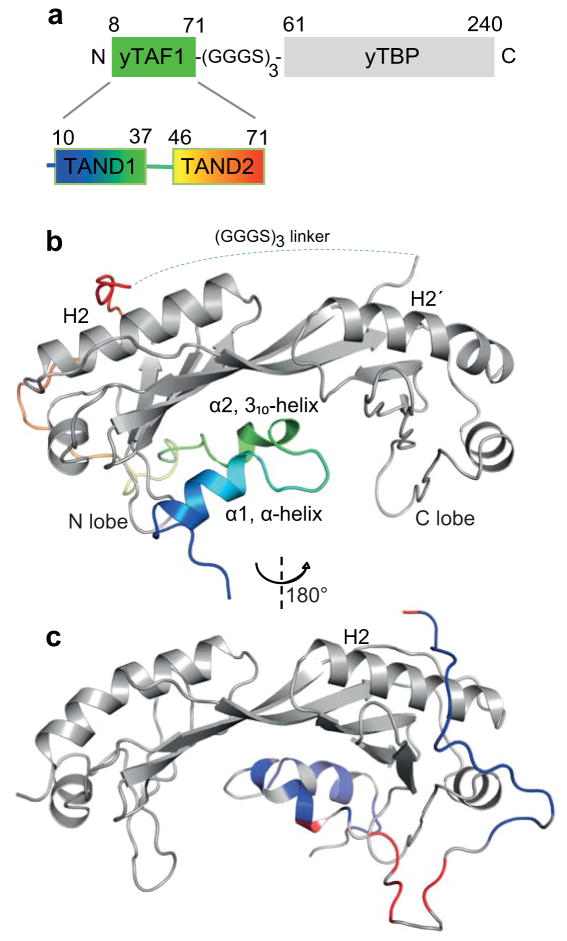
The structure of a biologically fully functional yTBP-yTAF1 fusion protein25, comprising the yTBP core domain and the yTAF1-TAND1 and -TAND2 regions (Fig. 1a), was determined by X-ray crystallography at 1.97 Å resolution (Fig. 1b, Table 1), and its dynamical properties were analysed by Nuclear Magnetic Resonance (NMR) (Fig. 1c, Supplementary Fig 1). The fusion (GGGS)3 linker between yTAF1 and TBP is disordered as judged by random-coil chemical shifts and complete lack of electron density. While yTAND1 bound TBP in the DNA-binding groove, yTAND2 bound the convex outer surface of the N-terminal lobe (Fig. 1b,c). NMR relaxation experiments showed that the yTAND1 and yTAND2 regions are stably anchored towards yTBP while the yTAND1-TAND2 linker region formed a highly dynamic, loop-like structure extending from the yTBP core domain (Fig. 1c). While yTAF1 regions that bound to TBP showed similar NMR relaxation decay rates as TBP, suggesting similar dynamic properties as the overall complex, the TAND1-TAND2 linker region showed deviating decay rates suggesting increased flexibility (Supplementary Fig. 1). Our NMR relaxation data further supported a stable compact structure of the 1:1 complex in solution with no detectable millisecond exchange (Supplementary Fig. 1). Electron density suggested three tentative Ca2+ ions support TBP-TAF1 crystal contacts in this region, but no such bound ions were observed in solution (Supplementary Fig. 2). The yTBP structure showed a markedly higher similarity to DNA-bound yTBP (RMSD 0.50) than to dimeric yTBP in the absence of DNA (RMSD 0.93–1.46) while each TBP lobe was virtually identical to corresponding lobes in DNA-bound or –free yTBP. Binding of yTAF1 to yTBP thus appeared to stabilize the relative orientations of the N- and C-terminal lobes towards the TATA-bound state (Supplementary Fig. 3).
Figure 1. Structure and dynamics of yTBP–yTAF1 binding.
(a) Schematic representation of yTBP-yTAF1 fusion protein. The yTBP-yTAF1 fusion protein comprises the yTBP core domain and the yTAF1-TAND1 and -TAND2 regions. A (GGGS)3 fusion linker connects the two proteins. (b) Cartoon representation of yTBP (grey) -yTAF1 (N-blue to C-red). The approximate location of the disordered fusion linker connecting yTAF1 and yTBP is indicated (dashed line). Key yTBP helices 2 (H2) and 2′ (H2′) as well as secondary structure elements of yTAF1 are annotated. (c ) same as (b) but rotated 180° in y (indicated) and −55° in x and colored by NMR relaxation rates. Residues in yTAF with mean R1 decay rates within ±2σ of the mean value for yTBP in blue, > +4σ in red; R1ρ and NOE data show the same pattern (Supplementary Fig. 1).
Table 1.
Data collection and refinement statistics
| yTBP-yTAF1a | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 32.89, 74.25, 99.67 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 60–1.97 (2.07–1.97)b |
| Rmerge | 0.083 (0.581) |
| I / σI | 19.3 (3.2) |
| Completeness (%) | 99.5 (98.3) |
| Redundancy | 7.1 (6.9) |
| Refinement | |
| Resolution (Å) | 60–1.97 |
| No. reflections | 17873 |
| Rwork / Rfree | 0.164/0.223 |
| No. atoms | |
| Protein | 1936 |
| Ligand/ion | 40 |
| Water | 213 |
| B-factors | |
| Protein | 26.7 |
| Ligand/ion | 49.6 |
| Water | 39.5 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.020 |
One crystal was used for data collection and refinement.
Values in parentheses are for highest-resolution shell.
TAND1 binds TBP using buried, TATA-box-mimicking residues
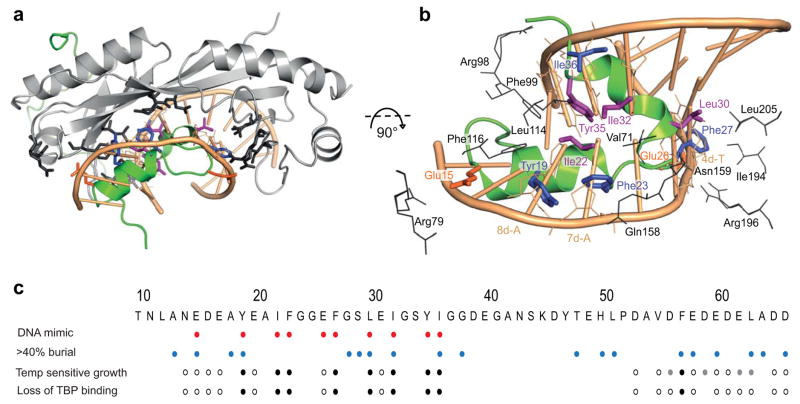
The yTAF1-TAND1 region bound the hydrophobic concave surface of yTBP by means of two helices α1 (residues 16–23) and α2 (residues 30–35; partly a 310 helix), thus occupying the same structural space as the TATA-box in the yTBP-DNA structure (Fig. 2a). A set of yTAF1-TAND1 residues structurally mimicked DNA bases, riboses and phosphates on both strands of the widened minor groove in yTBP-bound TATA (Fig. 2b). Consequently, yTBP contacts to yTAF1-TAND1 also mimicked those to TATA-box DNA26; specifically, residues Phe99, Leu114 and Phe116 of the yTBP N-terminal lobe interacted with Tyr19, Ile22, Ile32, Tyr35 and Ile36 of yTAF1-TAND1, whereas yTBP-Val71 and TBP-Gln158 from the ‘roof’ of the TBP concave surface interacted with TAND1-Phe23; all of these yTAF residues mimicked base or ribose moieties of TATA-box DNA (Fig. 2b). Earlier mutation studies support both TBP residues Val71 and Leu114 as key residues in yTAF1 binding8,9. Similarly, interactions mimicking TATA phosphate or base oxygen contacts included yTBP-Arg79 and yTAF1-Glu15, yTBP-Arg98 and yTAF1-Gly38(CO), yTBP-Asn159 and yTAF1-Ser29(OH), and finally yTBP-Arg196 and yTAF1-Glu26, which together with hydrophobic interactions involving yTBP-Ile194, -Leu205 and yTAF1-Phe27 connected yTAF1 to the c-terminal yTBP stirrup (Fig. 2b, Supplementary Table 1). The degree of burial of yTAF1 residues on yTBP binding correlates well with both DNA-mimicking and biological activity, and residues in yTAF1-TAND1 where alanine mutations impair Gal4-dependent transcription by 70% or more and lead to both temperature sensitive growth and loss of TBP binding18 all directly participated in the TBP-binding interface (Fig. 2c). The phosphate-mimicking properties of the buried TAF1-Glu15 may be dynamically substituted by neighboring Asp16 and Glu17 in the E15A mutant, reducing the biological effect of this mutation. Thus, the structure of yTAF1-TAND1 in complex with yTBP was in complete agreement with biological data.
Figure 2. TATA-box mimicry of yTAF1-TAND1.
(a) Superposition of yTBP (grey) -yTAF1 (green) onto the yTBP-DNA complex (PDB 1YTB; only the TATA-box is shown). Interacting residues are shown in sticks and colored as below. (b) Close-up view of the TAND1 and TBP interaction, with overlayed TATA-box as in (a). Interacting residues in yTBP (grey) and residues in yTAF1-TAND1 mimicking TATA-box riboses (blue), bases (purple) and phosphates (orange) parts are labelled. (c) DNA-mimicking residues in yTBP-TAF1-TAND (red) and TAF1 side chains >40% buried in the TBP complex (blue) are indicated together with previous results from single alanine mutation screening showing temperature sensitive growth and/or lost TBP binding (black: severe effects, grey: milder effects, empty: no effect)18,29.
TAND2 binds a TBP surface groove critical for PIC binding
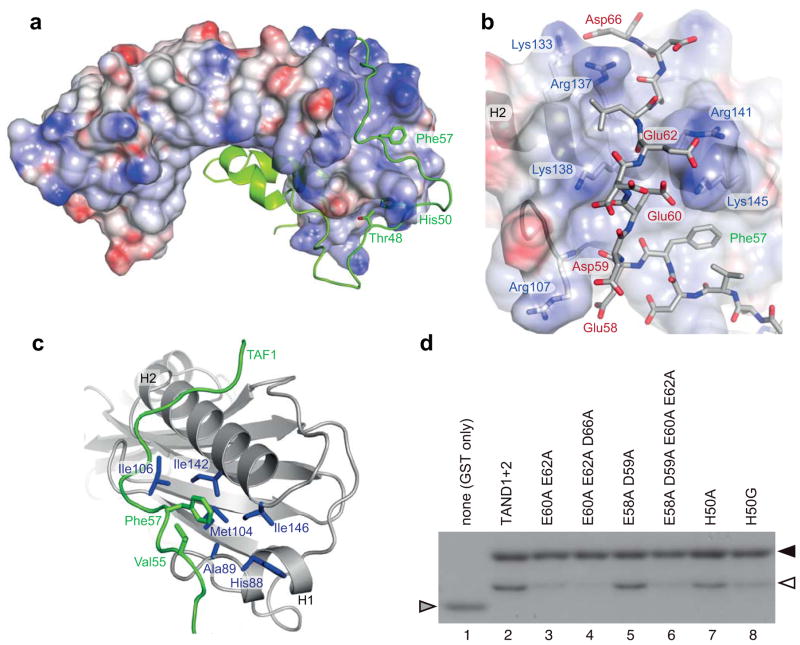
The highly negatively charged yTAF1-TAND2 region (Fig. 1), which is required for efficient yTAND1 binding to yTBP18, bound to the yTBP convex surface of the N-terminal lobe, where it meandered through a winding groove traversing helix 2 of yTBP (Fig. 3a,b). The yTBP groove is lined by positively charged residues including Lys133, Arg137, Lys138, Arg141 and Lys145, which were involved in electrostatic interactions with yTAF1-TAND2 residues Glu60, Glu62, and Asp66 (Fig. 3b and Supplementary Table 1). Among these, residues Lys133, Lys138 and Lys145 on yTBP helix 2 have been shown to be crucial for interactions within the PICs of polymerase I, II and III27,28. A network of electrostatic interactions linking yTBP and yTAF1-TAND2 was observed between the sidechains of Glu60-Lys145-Glu62-Arg141 (Fig. 3b). Adjacent to this charge-interaction network, we found a near-complete burial of yTAF1-TAND2-Phe57 in a groove depression in yTBP, lined with aliphatic and aromatic yTBP residues and supported by hydrophobic interactions with yTAF-Val55 (Fig. 3c). The backbone of Phe57 was further anchored by a well-defined hydrogen bond to yTBP Asn91 (Supplementary Table 1). Our structural observation of yTAF1-Phe57 as a TBP-anchoring residue is supported by severe growth defects of F57A in yTAF1 of magnitudes similar to a ΔTAND1 deletion29. No regular secondary structure was formed by yTAF1-TAND2 in its extended interaction with yTBP, but the complex was well formed with low B-factors.
Figure 3. Electrostatic and hydrophobic anchoring of yTAF1-TAND2 to TBP.
(a) Electrostatic surface representation of yTBP, with the bound yTAF1-TAND2 in green and with anchoring TAND2 residues annotated. (b) Detailed view of the surface groove interaction connecting yTAF1-TAND2 (sticks) and yTBP (cartoon). Participating side chains in the charge-charge interaction network are shown in sticks and labelled, as is the Phe57 anchor residue. Hydrogen bonds are listed in Supplementary Table 1. (c) The yTBP hydrophobic surface pocket, lined by residues (blue) in helix 1 (His88 and Ala89), strand 2 (Met104 and Ile106), and helix 2 (Ile142 and Ile146), binds the anchoring Phe57 residue supported by Val55 in yTAF1-TAND2 (green). (d) GST-pulldown assays. GST-TAND1-TAND2 fusion proteins carrying TAND2 mutations as indicated (lanes 3–8, lane 2 is the wild type) or GST alone (lane 1) were incubated with equimolar TBP. Triangles indicate GST-TAND1-TAND2 (black), TBP (white) and GST (grey) positions.
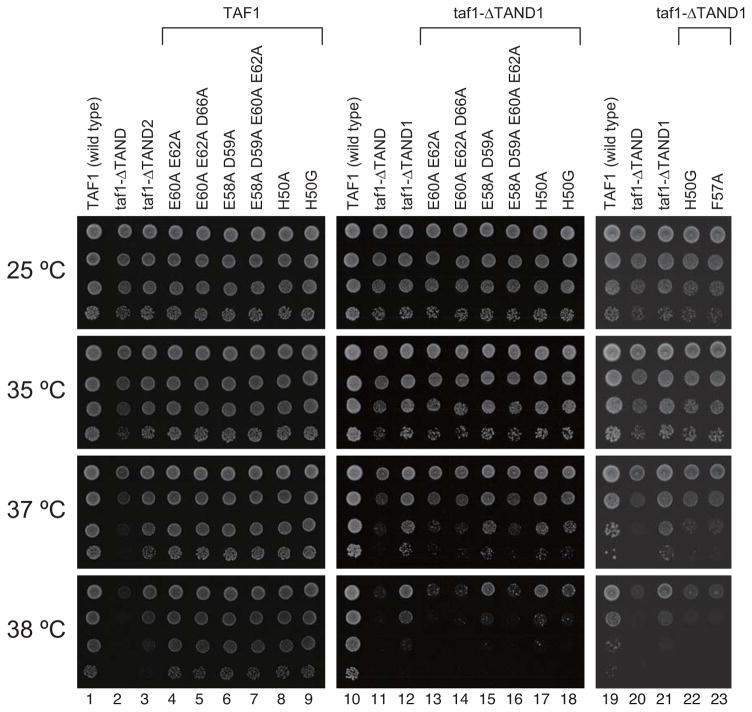
To investigate the role of the interleaved charge-interaction networks in yTAF1 interactions to the convex yTBP surface, we designed a set of TAND2 mutations. While previous experiments show that E60A or E62A alone have little effect on TBP binding29, the double mutation E60A+E62A, which would efficiently disrupt the Glu60-Lys145-Glu62-Arg141 network (Figure 3b) indeed drastically reduced both yTBP binding (Fig. 3d) and yeast growth rates in yTAF1-ΔTAND1 (Fig. 4), suggesting a principal role of this charge-interaction network in TAND2-TBP anchoring. In contrast, the double mutant E58A+D59A, designed based on its structural interaction with yTBP-Arg107 (Fig. 3b), did not release yTBP binding (Fig. 3d) and had little effect on yeast growth (Fig. 4). None of the multiple charge mutants significantly affected yeast growth in intact yTAF1, even when combined, suggesting that the charge-interaction networks mainly consolidate yTAF1-TAND2 anchoring (Figure 4). Furthermore, His50 was nearly completely buried in the complex (Fig. 2c) and structurally anchored the TAND1-TAND2 linker to yTBP (Fig. 3a). H50A did not significantly affect growth, but a H50G mutation, removing all hydrophobicity, released yTBP binding (Fig. 3d) and affected yeast growth in a yTAF1-ΔTAND1 strain by similar magnitude as a F57A mutation (Fig. 4). Taken together, our experiments suggest three major TAND2 binding determinants: 1) hydrophobic anchoring of Phe57 into the groove on the yTBP convex surface, 2) a charge-interaction network linking negative charges Glu60 and Glu62 in yTAND2 with positive surface charges Lys145 and Arg141 on yTBP, and 3) burial of His50 in the yTAND1-TAND2 linker region. The yTAF-TAND2 motif is highly conserved in hTAF1 and dTAF129, which we thus expect to anchor into the TBP convex surface groove similar to yTAF-TAND2.
Figure 4. Multiple acidic residues in TAND2 affect yeast growth.
Growth phenotype of yeast strains carrying TAND mutations as indicated. The strains were serially diluted (10-fold), spotted onto YPD medium, and grown at the indicated temperatures for 3 days. Since both TAND1 and TAND2 regions affect TBP binding29, effects of site-specific TAND2 mutations on growth were assayed both in the presence (lanes 4–9) and absence (lanes 13–18, 22–23) of TAND1. For direct comparison, a joint experiment was performed with the previously studied F57A mutant29.
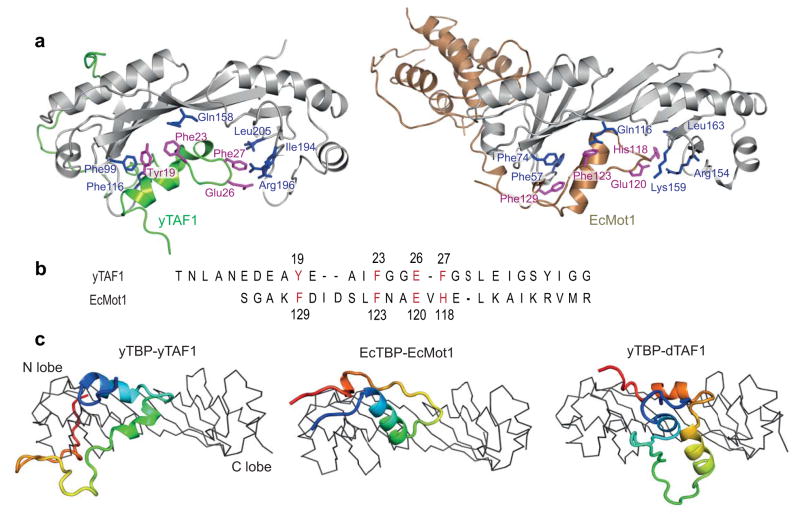
TAND1 uses same anchor points as Mot1 but in reverse
As described above, the yTAND1 region of yTAF1 appeared to disrupt TBP interactions by replacing them with DNA-mimicking residues, a strategy that has previously been suggested both for dTAF and EcMot110,24. The interactions of Mot1 and yTAF-TAND1 with TBP were particularly similar: Mot1-F123 interacts with EcTBP-Gln116 similarly as yTAND1-Phe23 to yTBP-Gln158, and EcMot1-Phe129 forms an aromatic stack with EcTBP-Phe57 and -Phe74 in the same way as TAND1-Tyr19 with yTBP-Phe99 and yTBP-Phe116 (Fig. 5a). Furthermore, a corresponding EcMot1 equivalent to the electrostatic interaction between yTAND1-Glu26 and yTBP-Arg196, connecting yTAND1 to the c-terminal yTBP stirrup, is found in EcMot1-Glu120 and EcTBP-Lys159, which is further stabilized by the hydrogen bond between EcMot1-His118 and EcTBP-Arg154 (Fig. 5a).
Figure 5. Similar TBP-anchoring residues in yTAF1-TAND1 and Mot1 despite reverse sequence tracing.
(a) Cartoon representation of TBP complexes with yTAF1 (green, current structure) and Mot1 (sand, 3OC3), highlighting interacting residues of TBP (blue) and yTAF1 or Mot1 (magenta). Only part of the Mot1 structure is shown for clarity. (b) Structure-based sequence alignment between the TBP interacting residues of TAF1 and Mot1. TBP anchoring residues are annotated and the Mot1 sequence is reversed as prompted by the reversed structural sequence tracing shown in a. (c) Ribbon style representation of TBP and cartoon yTAF1, Mot1 and dTAF with rainbow coloring (N-terminus:blue, C-terminus:red). The TBP lobes are labeled and the orientation is 90° rotated compared to (a).
Interestingly, a closer look revealed that the peptide chain orientation of yTAF1 is reversed compared to Mot1 and dTAF, although both the relative orientation of their helices and the mode of TBP interactions on a per-residue level are highly similar between the two proteins. Remarkably, the yTAF1-TAND region could be aligned to the reverse sequence of Mot1 with TBP contacting residues in conserved positions (Fig. 5b). We also found that transcriptional activator domains in EBNA2, VP16 and Gal4, which are functionally similar to yTAF-TAND1 in transcriptional activation and also bind the TBP concave surface18, could be aligned to yTAF1-TAND1-TAND2 with conserved contact residues to TBP (Supplementary Fig. 4). In the dTAF1-TBP structure, dTAF-TAND1 covers an extended surface compared to yTAF1 and Mot1 (Fig. 5c), which is consistent with its higher affinity to TBP29. However, although several of the yTAF1 and Mot1 contacts to TBP are also found in dTAF24, the different threading of dTAF-TAND1 into the TBP DNA-binding groove (Fig. 5c) precludes dTAF sequence alignment to yTAF1-TAND1 or Mot1, since its DNA-mimicking patches are nonlinearly permutated in sequence compared to yTAF1 and Mot1. The longer hTAF1 TAND1 motif (residues 1–100) showed sequence similarities both to yTAF1-TAND1 and dTAF1, suggesting the presence of two TBP-binding motifs N-terminal to hTAF1-TAND2: one N-terminal motif similar to dTAF1-TAND1, and a second one resembling yTAF1-TAND1 which is directly connected to hTAF1-TAND2 (Supplementary Fig. 4).
TAND2 presents a conserved regulatory TBP-binding motif
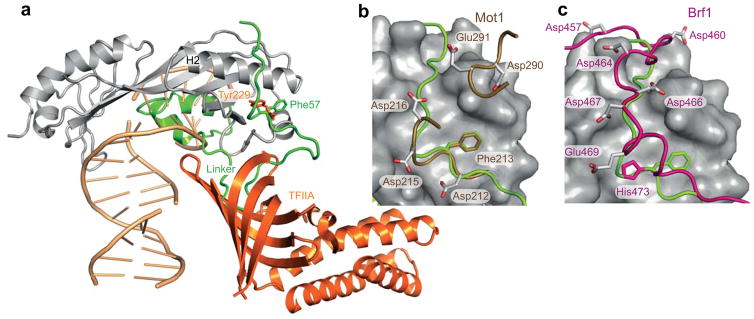
The structure of yTBP-yTAF1 together with previous TBP-TFIIA, TBP-Brf1 and TBP-Mot1 structures gives new insight into the specificity of convex surface interactions by TBP-binding proteins. TFIIA binding to TBP competes with yTAF1-TBP binding in yeast transcriptional initiation23,30, but the specific molecular determinants for this have hitherto been unknown. In our structure, we found that yTAF1 anchors into the same TBP convex surface groove as the acidic loop (228DYLI231) of TFIIA, and furthermore wraps around the TBP N-terminal stirrup region in a way that renders simultaneous binding of TFIIA impossible (Fig. 6a). A closer analysis suggested direct molecular competition for the aromatic binding pocket between Phe57 of yTAF1 and Tyr229 of TFIIA (Fig. 6a). Superimposing the backbone of the TBP entities in the TBP-EcMot1 and the yTBP-yTAF1 complexes resulted in structural overlap of Phe213 and Phe57 of EcMot1 and yTAF-TAND2, respectively, which both inserted into the same hydrophobic cleft in TBP (Fig. 6b). The crystal structure of ternary Brf1-TBP-DNA complex (pdb entry: 1NGM),31 reveals a similar association of Brf1 in the same surface groove. Brf1 His473 replaces the Phe57 (yTAF1-TAND2) key residue in the corresponding TBP hydrophobic cleft but with a 180° reversal of the sidechain orientation in the hydrophobic cleft, possibly due to the main chain running in reverse through the groove (Fig. 6c).
Figure 6. Conserved surface groove and anchoring residues in competitive TBP binding.
(a) Superposition of TBP-TAF1 on to the TBP-TFIIA (orange) –DNA (wheat) ternary complex (from PDB 1NH2). Aromatic TBP-anchoring residues of TAF1 and TFIIA on the convex TBP surface are shown as sticks and labeled. The linker region between TAND1 and TAND2 of TAF1 protruding into the space occupied by the β-barrel TFIIA structure is highlighted. (b) Mot1 (sand, PDB 3OC3) and (c) Brf1 (magenta, PDB 1NGM) complexes with TBP superimposed onto yTAF1 (green) – yTBP (surface), highlighting in stick representation the common anchoring aromatic residue in these transcription activators and repressors as well as hydrogen binding sidechains connecting to the basic region of TBP. All superpositions were made by structural alignment of the TBP backbone in the respective complexes onto yTBP in the current structure. Only yTBP from the current structure is shown in a–c.
Our structure and mutational analysis showed that efficient binding of yTAND2 (55VDFEDEDEDELADD66) to the yTBP convex surface groove in addition to aromatic anchoring (bold) also requires charge-charge interactions between acidic residues on yTAND2 (underligned) and conserved lysines and arginines on TBP (Fig. 3, 4; Supplementary Table 1). The TAND2 core region is well conserved in yTAF1, dTAF1 and hTAF1, suggesting similar TBP binding29. Interestingly, we found that both EcMot1 and Brf1 show similar stretches of acidic residues which interact with the same basic TBP surface as does yTAF1-TAND2. While tracing the same groove but in reverse compared to TAND2, Brf131 displays acidic residues N-terminal to the aromatic histidine anchor (457DDPDNLEDVDDEELNAHLLNEE478) interacting similarly as TAND2 with the TBP basic patch (Fig. 6c). In EcMot1, acidic residues C-terminal to the buried phenylalanine (211NDFVDD216) are complemented by an acidic loop from the EcMot1 heat-repeat region (288SPDEDI293), which jointly match the TBP basic patch (Fig. 6b). Finally, although the TFIIA structure does not show electron density beyond the aromatic anchor provided by (228DYLI231), mutations of TBP lysines on helix 2 were early shown to disrupt TBP-TFIIA interactions27,32 suggesting electrostatic contributions to binding. Interestingly, a 30-fold increase of TFIIA-TBP binding is observed on phosphorylation of TFIIA at Ser220, Ser225 and Ser23233 surrounding the aromatic anchor residue, and a S220A S225A S232A multiple mutant protein showed greatly reduced transcriptional activity34. We noted that although structurally disordered in the crystal35, these residues envelop the 228-DYLI motif that competes with TAF1 binding to the TBP convex surface groove (Fig. 6). Extrapolating from our yTAF1-yTBP structure, phosphorylation of these TFIIA serines could provide the vital negative charge required for interaction with the conserved lysines on TBP (Fig. 3, 4), leading to enhanced TBP binding and transcription. Similar structural mechanisms may well govern PTEN-mediated regulation of Brf1, where phosphorylation controls the association between TBP and Brf136, and phosphorylation adjacent to a critical tryptophane residue in the glucocortoid receptor transactivation domain AF1 dramatically increases TBP binding37.
DISCUSSION
The current high-resolution structure of the yTBP–yTAF1-TAND1-TAND2 complex shows how TBP is anchored to the TFIID complex, and provides structural details on how an intrinsically disordered transcriptional regulatory yTAF1 domain20 interacts with TBP. By favoring the presence of the bound state in a fusion protein, where increased association rates have shifted the equilibrium towards the complex state25, we could identify and characterize critical anchor points in the TBP-TAF1 interaction by structural and mutational analysis. Induced folding is considered a major driving force for interaction between a disordered and a folded protein38. For yTAF1 binding to TBP, strong coupling of folding and binding is manifested by near-complete disruption of both binding and biological function by removal of any single hydrophobic anchoring sidechain in yTAF1-TAND1-TAND2 (Fig. 2, 3, 4). The charge-charge anchoring interactions identified on the convex TBP surface are less sensitive to single mutations and seem to contribute to binding in a more additive way with lower levels of structural rigidity (Fig. 3, 4). We find that the anchoring interactions identified in yTAF1 are present also in other structurally diverse TBP-binding general transcription factors (Fig. 5, 6), and may well be widely present in TBP-binding transcriptional activation domains. In particular, the detailed structural and mutational characterization of the binding of the TAND2 acidic and aromatic peptide regions to the TBP convex surface (Fig. 3, 4, 6) provides a novel molecular framework for addresing the as yet unresolved question how apparently casually assembled charged peptides with key aromatic residues can recruit TBP and thereby activate transcription11,12.
At first glance, one might consider some features of the current structure to reflect suboptimal yTAF-yTBP interactions, such as the structural and sequence diversity of TBP interactors using similar anchor points, and the sensitivity of complex formation and biological activity to mutations at these sites. However, we suggest instead that these properties may all be required for biological function, since they enable rapid dynamic competition between positively and negatively acting factors binding to common TBP interaction sites. Such competition has been suggested to be a cornerstone for biologically adequate responsiveness in the regulation of transcriptional output8. Since TBP is jointly required by PolI, -II and –III complexes, a simultaneously selective and dynamic TBP recruitment mechanism is essential to direct alternate usage of PolI, II and III as biologically required. The structural and sequence divergence in proteins competitively binding to identical TBP interaction surfaces as shown here (Fig. 2, 5, 6, 7) provides differential handles and thereby a biologically versatile means of differentially directing recruitment of the jointly utilized TBP into various multiprotein complexes targetting transcription. Targetting of anchoring positions by proteins competing for the same binding site can readily be exploited in-vivo to direct the timing of biologically appropriate interactions. Such anchoring posesses a biological advantage, since multiple TBP binding equilibria can then easily be shifted simply by competition at single sites in the interaction surface, by altered expression levels of competing proteins, or by posttranslational modifications affecting the affinity of one of the competitors. Efficient and regulated competition at anchoring positions on TBP provides for rapid alteration of cell fate in response to biological needs by directing TBP recruitment to various transcription complexes and promotor sites.
While the current structure of the stabilized yTAF1-yTBP fusion protein presents a snapshot of the bound state, we know that dynamics in regulatory TBP binding is biologically crucial8. In fact, transcriptional activity appears inversely correlated with the formation of stable TBP complexes18. The yTAF1-TAND1 and the transcriptional activation domains from EBNA2, VP16 and Gal4, which all form less stable complexes with TBP, are all still able to efficiently activate transcription in a Gal4 assay18. In contrast, dTAF1-TAND1, which binds the same TBP surface as yTAF1-TAND1 but with nanomolar affinity, is unable to activate transcription29. Thus, the formation of a stable TBP complex is not consistent with high transcriptional activity; indeed a complete structural description of an efficient TBP transcriptional activator complex may require an ensemble of states 8,14. In fact, the frequent presence of multiple transcriptionally activating regions with low degree of sequence homology in the same transcriptional regulator12 may promote intrinsic, multivalent competition. In agreement with the ‘handoff’ model13,17,23, such rapid dynamic shielding of critical TBP-binding surfaces might be required to avoid the formation of autoinhibited or unproductive TBP complexes while retaining access for downstream general transcription factors. We hypothesize that competitive conformational exchange at anchoring positions may provide the clue for how TBP, despite its rigid fold compared to other multirecognition proteins39,40 is able to bind such a wide variety of dynamic interactors in a timely and productive manner.
Taken together, our data on TBP binding extends the view of transcriptional competition of TBP interactors to a common target. In particular, our structure identifies and highlights how TBP-anchoring residues are similarily presented to TBP by varied TBP-interacting folds, which allows for TBP interactions to be easily disrupted or enhanced by competition or by posttranscriptional modification. In biophysical terms, our data agrees with transcriptional activation as a highly dynamic process where ensembles of multiprotein equilibria are shifted as a response to biological signals, rather than to a hierarchic initiation of a single-way, sequential cascade of events. Increased or decreased binding of transcriptional regulators may shift the TBP multiprotein equilibria and thereby promote TBP relocation, in agreement with transcription sites being rapidly assembling and disassembling entities. For transactivating domains to interfere in the regulation of such a dynamic entity as the PIC41, flexibility may be a prerequisite42. Increased structural and biophysical knowledge in this area is critical to understanding how transcription factors and transcriptional activators collaborate in regulating the transcription of individual genes as well as entire gene programs.
ONLINE METHODS
Protein preparation
The fusion protein encoding yTAF1-TAND1-TAND2 (residues 8–71) linked to the core domain of yTBP (residues 61–270) with a (GGGS) 3 linker as well as GST-TAF1-TAND and TBP proteins were expressed and prepared as previously described25. For crystallization, the yeast fusion yTBP-yTAF1 protein was prepared to a final concentration of 1.5 mM in 100mM NaCl, 0.5mM TCEP, 10% (v/v) Glycerol and 20mM HEPES, pH 7.5.
Crystallization, Data Collection and Structure Determination
Crystals were obtained at 4°C by sitting-drop vapor diffusion against a well solution containing 0.2 M cesium chloride, 0.1 M MES pH 6.5, 26% PEG350MME. A native diffraction dataset were collected at the temperature of 100 K from a single crystal diffracting to 1.97 Å at beamline ID14-1 (λ = 0.9334 Å) in ESRF, Grenoble. The initial structure was determined by molecular replacement using the Molrep program integrated in the CCP4i package43. The initial Fo-Fc map revealed the electron density for the yTAND12 domain. No electron density was observed for the (GGGS)3 fusion linker connecting yTAF1-71 and yTBP-61 or for residues yTBP-61, yTAF1-67-71. The distance between yTBP-61 and yTAF-66 in the crystal structure is 38Å, thus the disordered fusion linker including flanking disordered residues, which would cover 65 Å in an extended state, comfortably covers this distance. The uncleaved N-terminal hexahistidine tag is also not observed but may have contributed to stabilizing crystallization, since no crystals were obtained for the hexahistidine-free fusion protein construct. The peptide was modeled and the entire model was manually adjusted using program Coot44. The models were subjected to several rounds of refinement and validation using autoBUSTER45 and MolProbity46 respectively to obtain the final model. The final model had 98.7% of the residues in the Ramachandran favored region with no Ramachandran outliers. The overall MolProbity score of the final model was of 0.93, and the structure belongs to the best scoring structures at comparable resolution (100% percentile). Side chain surface accessibility was calculated from the coordinates using VADAR47. All protein structure figures were generated using PyMOL (www.pymol.org).
NMR experiments and data analysis
All NMR experiments were performed using a Varian INOVA spectrometer, operating at a proton Larmor frequency of 600 MHz, at 25°C. Data was processed and analysed as described48. Assignments for yTBP-yTAF149 (BMRB entry 6702) were confirmed and slightly extended by HNCO, HNCA, HN(CO)CA, HN(CA)CB and HN(COCA)CB experiments on a deuterated sample. The 15N-R1 and 15N-R1ρ relaxation experiments for yTBP-yTAF1 were recorded with a protein concentration of 320 μM using 15N,13C,2H-labeled protein. For the R1 relaxation experiments, 18 data points, including four duplicates, were recorded, with relaxation delays between 10–1500 ms. The spin lock field strength used for measurements of R1ρ was 1730 Hz and the number of recorded data points 19 with relaxation delays between 5–50 ms including five duplicates. The 15N{1H}-NOE measurements were performed by recording experiments including or not including a 5 s period of 120° 1H saturation pulses. The total recovery delay was 12 s in both cases. For the evaluation of relaxation data, the program PINT50 was used where peaks were integrated by line shape fitting. R1 and R1ρ rate constants were obtained by fits to exponential functions. The jack-knife approach was used for estimation of errors in the decay rates51. R2 was calculated using the following equation: R1ρ = R1 cos2θ + R2 sin2θ, where θ is the tilt-angle, defined as arctan(B1/ Ω). B1 is the spin lock field strength and Ω the resonance offset from the radio frequency carrier52. For the calculation of the rotational correlation time (τc), the software Tensor2 was used53. The correlation time was calculated from the ratio R2/R1 using an axially symmetric rotational diffusion tensor assuming that the yTBP-yTAF1 protein is arranged as a prolate ellipsoid. Residues with NOE<0.65 were excluded from the analysis.
Yeast strains and plasmid construction
TAF1 deletion strains, YTK11411 and YTK12029, were described previously54. New strains used in this work are summarized in Supplementary Table 2) and were constructed as follows: YTK12803, YTK13412, YTK13413, YTK13414, YTK13415, YTK13416, YTK13417 and YTK13444 were generated from YTK11411 by replacing the URA3-marked plasmid (pYN1/TAF1) with HIS3-marked plasmids; pM7121/taf1-ΔTAND, pM7280/taf1-E60A E62A, pM7281/taf1-E60A E62A D66A, pM7282/taf1-E58A D59A, pM7283/taf1- E58A D59A E60A E62A, pM7284/taf1-H50A, pM7285/taf1-H50G and pM7298/taf1-ΔTAND2, respectively, using a plasmid shuffle technique. Similarly, YTK13428, YTK13429, YTK13430, YTK13431, YTK13432, YTK13433, YTK13434, YTK13435, YTK13436 and YTK13536 were generated from YTK11411 by replacing pYN1/TAF1 with LEU2-marked plasmids; pM7118/TAF155, pM7119/taf1-ΔTAND, pM7286/taf1-ΔTAND1, pM7287/taf1-ΔTAND1 E60A E62A, pM7288/taf1-ΔTAND1 E60A E62A D66A, pM7289/taf1-ΔTAND1 E58A D59A, pM7290/taf1-ΔTAND1 E58A D59A E60A E62A, pM7291/taf1-ΔTAND1 H50A, pM7292/taf1-ΔTAND1 H50G and pM7320/taf1-ΔTAND1 F57A, respectively.
pM4770/TAF154, which contains the entire TAF1 sequence including its native promoter and terminator (5.3 kb in total), was subjected to site-specific mutagenesis56 to create pM7121, pM7279, pM7280, pM7281, pM7282, pM7283, pM7284, pM7285 and pM7319. The oligonucleotides used in this study are listed in Supplementary Table 3. The 5.1 kb NotI-SalI fragment from pM7121/taf1-ΔTAND was ligated into the NotI/SalI sites of pRS31557 to generate pM7119. pM7286/taf1-ΔTAND1 was created by replacing the 3.4 kb NotI-XbaI fragment of pM7118/TAF155 with the 3.3 kb NotI-XbaI fragment of pM7279/taf1-ΔTAND1. The 2.2 kb BssHII-XbaI fragments including the mutated TAND2 region were amplified by PCR from pM7280, pM7281, pM7282, pM7283, pM7284, pM7285 and pM7319 using the primer pair TK43-TK125, and then ligated into the BssHII/XbaI sites of pM7286 to generate pM7287, pM7288, pM7289, pM7290, pM7291, pM7292 and pM7320, respectively. The 2.2 kb BamHI-XbaI fragment of pM977/taf1-ΔTAND258 was ligated into the BamHI/XbaI sites of pRS313 to generate pM7298.
To prepare GST-tagged TAND proteins harboring TAND2 mutations, the 198 bp BamHI-EcoRI fragments were amplified from pM7280, pM7281, pM7282, pM7283, pM7284 and pM7285 using the primer pair TK344-T869, and then ligated into the BamHI/EcoRI sites of pGEX2T (GE Healthcare), generating pM7305, pM7306, pM7307, pM7308, pM7309 and pM7310, respectively. The plasmids expressing wild type GST-TAND (pM1431) or wild type TBP (pM1578) were generated as described previously58.
GST pull-down assay
Purified TBP (20 pmol) was incubated for 1 h with bacterial lysate expressing GST-TAF1-TAND or GST (20 pmol) in 150 μl of 0.2 M KCl,20 mM Tris-HCl at pH 7.9, 12.5 mM MgCl2, 0.2 mM EDTA, 10 % (v/v) glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride at 4 °C. After incubation with 10 μl of glutathione-Sepharose 4B beads (GE Healthcare) for another 1 hr, the beads were washed with 3×500 μl buffer as above, boiled in SDS sample buffer, and eluates were separated on SDS-PAGE.
Supplementary Material
Acknowledgments
This work was supported by grants from the Swedish Research Council (621-2011-6028, 621-2012-5250 M.S., 621-2012-5136 P.L.), VINNOVA (P32045-1 M.S.), the Swedish Cancer Foundation (11 0681 M.S.), the Swedish Child Cancer Foundation (PROJ09/092 M.S.), Forum Scientium Award (C.A.), Canadian Institutes for Health Research grant MT-13611 (M.I.), the Grant-in-Aid for Scientific Research 23370077 from Japan Society for the Promotion of Science (T.K.), and equipment grants to LiU from the Knut and Alice Wallenberg foundation. M. I. holds a Canada Research Chair. We thank H. Th. M. Timmers, L. Penn, C. H. Arrowsmith and P. de Graaf for critical discussion, and acknowledge the Swedish NMR Centre (www.nmr.gu.se), the Protein Science Facility (psf.ki.se) and beamline ID14-1 at the European Synchrotron Radiation Facility (www.esrf.eu).
Footnotes
ACCESSION CODE
The atomic coordinates are deposited in the RCSB Protein Data Bank with the PDB code 4B0A.
AUTHOR CONTRIBUTIONS
M. I. and M. S. conceived and designed the study. Experiments and data evaluation were designed and performed by M. A., M. I. S., and M. M. (crystallography), Y. O. and T. K. (yeast mutations) and M. A., C. A., S. H., P. L. and M. S. (NMR). M. A., C. A., S. H., M. M. and M. S. wrote the paper. All authors discussed the interpretations and implications of the results and edited the manuscript at all stages.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Goodrich JA, Tjian R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet. 2010;11:549–58. doi: 10.1038/nrg2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D’Alessio JA, Ng R, Willenbring H, Tjian R. Core promoter recognition complex changes accompany liver development. Proc Natl Acad Sci U S A. 2011;108:3906–11. doi: 10.1073/pnas.1100640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45:439–46. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 4.Papai G, Weil PA, Schultz P. New insights into the function of transcription factor TFIID from recent structural studies. Curr Opin Genet Dev. 2011;21:219–24. doi: 10.1016/j.gde.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–78. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 6.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieniossek C, et al. The architecture of human general transcription factor TFIID core complex. Nature. 2013;493:699–702. doi: 10.1038/nature11791. [DOI] [PubMed] [Google Scholar]
- 8.Chitikila C, Huisinga KL, Irvin JD, Basehoar AD, Pugh BF. Interplay of TBP inhibitors in global transcriptional control. Mol Cell. 2002;10:871–82. doi: 10.1016/s1097-2765(02)00683-4. [DOI] [PubMed] [Google Scholar]
- 9.Martel LS, Brown HJ, Berk AJ. Evidence that TAF-TATA box-binding protein interactions are required for activated transcription in mammalian cells. Mol Cell Biol. 2002;22:2788–98. doi: 10.1128/MCB.22.8.2788-2798.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wollmann P, et al. Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature. 2011;475:403–7. doi: 10.1038/nature10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sigler PB. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988;333:210–2. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- 12.Hahn S, Young ET. Transcriptional Regulation in Saccharomyces cerevisiae: Transcription Factor Regulation and Function, Mechanisms of Initiation, and Roles of Activators and Coactivators. Genetics. 2011;189:705–36. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ptashne M, Gann AA. Activators and targets. Nature. 1990;346:329–31. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- 14.Brzovic PS, et al. The acidic transcription activator gcn4 binds the mediator subunit gal11/med15 using a simple protein interface forming a fuzzy complex. Mol Cell. 2011;44:942–53. doi: 10.1016/j.molcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu WL, et al. Structures of three distinct activator-TFIID complexes. Genes Dev. 2009;23:1510–21. doi: 10.1101/gad.1790709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baek HJ, Kang YK, Roeder RG. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem. 2006;281:15172–81. doi: 10.1074/jbc.M601983200. [DOI] [PubMed] [Google Scholar]
- 17.Elmlund H, et al. Cryo-EM reveals promoter DNA binding and conformational flexibility of the general transcription factor TFIID. Structure. 2009;17:1442–52. doi: 10.1016/j.str.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Kotani T, et al. A role of transcriptional activators as antirepressors for the autoinhibitory activity of TATA box binding of transcription factor IID. Proc Natl Acad Sci U S A. 2000;97:7178–83. doi: 10.1073/pnas.120074297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muldrow TA, Campbell AM, Weil PA, Auble DT. MOT1 can activate basal transcription in vitro by regulating the distribution of TATA binding protein between promoter and nonpromoter sites. Mol Cell Biol. 1999;19:2835–45. doi: 10.1128/mcb.19.4.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mal TK, et al. Structural and functional characterization on the interaction of yeast TFIID subunit TAF1 with TATA-binding protein. J Mol Biol. 2004;339:681–93. doi: 10.1016/j.jmb.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 21.Takahata S, Kasahara K, Kawaichi M, Kokubo T. Autonomous function of the amino-terminal inhibitory domain of TAF1 in transcriptional regulation. Mol Cell Biol. 2004;24:3089–99. doi: 10.1128/MCB.24.8.3089-3099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagby S, et al. TFIIA-TAF regulatory interplay: NMR evidence for overlapping binding sites on TBP. FEBS Lett. 2000;468:149–54. doi: 10.1016/s0014-5793(00)01213-8. [DOI] [PubMed] [Google Scholar]
- 23.Kokubo T, Swanson MJ, Nishikawa JI, Hinnebusch AG, Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol Cell Biol. 1998;18:1003–12. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu D, et al. Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell. 1998;94:573–83. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 25.Mal TK, et al. Functional silencing of TATA-binding protein (TBP) by a covalent linkage of the N-terminal domain of TBP-associated factor 1. J Biol Chem. 2007;282:22228–38. doi: 10.1074/jbc.M702988200. [DOI] [PubMed] [Google Scholar]
- 26.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–7. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 27.Buratowski S, Zhou H. Transcription factor IID mutants defective for interaction with transcription factor IIA. Science. 1992;255:1130–2. doi: 10.1126/science.1546314. [DOI] [PubMed] [Google Scholar]
- 28.Kim TK, Roeder RG. Involvement of the basic repeat domain of TATA-binding protein (TBP) in transcription by RNA polymerases I, II, and III. J Biol Chem. 1994;269:4891–4. [PubMed] [Google Scholar]
- 29.Kotani T, et al. Identification of highly conserved amino-terminal segments of dTAFII230 and yTAFII145 that are functionally interchangeable for inhibiting TBP-DNA interactions in vitro and in promoting yeast cell growth in vivo. J Biol Chem. 1998;273:32254–64. doi: 10.1074/jbc.273.48.32254. [DOI] [PubMed] [Google Scholar]
- 30.Ozer J, Mitsouras K, Zerby D, Carey M, Lieberman PM. Transcription factor IIA derepresses TATA-binding protein (TBP)-associated factor inhibition of TBP-DNA binding. J Biol Chem. 1998;273:14293–300. doi: 10.1074/jbc.273.23.14293. [DOI] [PubMed] [Google Scholar]
- 31.Juo ZS, Kassavetis GA, Wang J, Geiduschek EP, Sigler PB. Crystal structure of a transcription factor IIIB core interface ternary complex. Nature. 2003;422:534–9. doi: 10.1038/nature01534. [DOI] [PubMed] [Google Scholar]
- 32.Lee DK, DeJong J, Hashimoto S, Horikoshi M, Roeder RG. TFIIA induces conformational changes in TFIID via interactions with the basic repeat. Mol Cell Biol. 1992;12:5189–96. doi: 10.1128/mcb.12.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solow SP, Lezina L, Lieberman PM. Phosphorylation of TFIIA stimulates TATA binding protein-TATA interaction and contributes to maximal transcription and viability in yeast. Mol Cell Biol. 1999;19:2846–52. doi: 10.1128/mcb.19.4.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solow S, Salunek M, Ryan R, Lieberman PM. Taf(II) 250 phosphorylates human transcription factor IIA on serine residues important for TBP binding and transcription activity. J Biol Chem. 2001;276:15886–92. doi: 10.1074/jbc.M009385200. [DOI] [PubMed] [Google Scholar]
- 35.Bleichenbacher M, Tan S, Richmond TJ. Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes. J Mol Biol. 2003;332:783–93. doi: 10.1016/s0022-2836(03)00887-8. [DOI] [PubMed] [Google Scholar]
- 36.Woiwode A, et al. PTEN represses RNA polymerase III-dependent transcription by targeting the TFIIIB complex. Mol Cell Biol. 2008;28:4204–14. doi: 10.1128/MCB.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garza AM, Khan SH, Kumar R. Site-specific phosphorylation induces functionally active conformation in the intrinsically disordered N-terminal activation function (AF1) domain of the glucocorticoid receptor. Mol Cell Biol. 2010;30:220–30. doi: 10.1128/MCB.00552-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–8. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoeflich KP, Ikura M. Calmodulin in action: diversity in target recognition and activation mechanisms. Cell. 2002;108:739–42. doi: 10.1016/s0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 40.Lee CW, Martinez-Yamout MA, Dyson HJ, Wright PE. Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein. Biochemistry. 2010;49:9964–71. doi: 10.1021/bi1012996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Werven FJ, van Teeffelen HA, Holstege FC, Timmers HT. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat Struct Mol Biol. 2009;16:1043–8. doi: 10.1038/nsmb.1674. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, et al. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–88. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–21. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 46.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willard L, et al. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003;31:3316–9. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andresen C, et al. Transient structure and dynamics in the disordered c-Myc transactivation domain affect Bin1 binding. Nucleic Acids Res. 2012;40:6353–66. doi: 10.1093/nar/gks263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mal TK, et al. Resonance assignments of 30 kDa complexes of TFIID subunit TAF1 with TATA-binding protein. J Biomol NMR. 2005;33:76. doi: 10.1007/s10858-005-1929-3. [DOI] [PubMed] [Google Scholar]
- 50.Ahlner A, Carlsson M, Jonsson BH, Lundström P. PINT – a software for integration of peak volumes and extraction of relaxation rates. J Biomol NMR. 2013 doi: 10.1007/s10858-013-9737-7. in press. [DOI] [PubMed] [Google Scholar]
- 51.Mosteller F, Tukey JW. Data analysis and regression : a second course in statistics. Addison-Wesley; Reading, Mass: 1977. [Google Scholar]
- 52.Palmer AG, 3rd, Massi F. Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem Rev. 2006;106:1700–19. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 53.Dosset P, Hus JC, Blackledge M, Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J Biomol NMR. 2000;16:23–8. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 54.Ohyama Y, Kasahara K, Kokubo T. Saccharomyces cerevisiae Ssd1p promotes CLN2 expression by binding to the 5′-untranslated region of CLN2 mRNA. Genes Cells. 15:1169–88. doi: 10.1111/j.1365-2443.2010.01452.x. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi H, Kasahara K, Kokubo T. Saccharomyces cerevisiae Med9 comprises two functionally distinct domains that play different roles in transcriptional regulation. Genes Cells. 2009;14:53–67. doi: 10.1111/j.1365-2443.2008.01250.x. [DOI] [PubMed] [Google Scholar]
- 56.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–82. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 57.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahata S, et al. Identification of a novel TATA element-binding protein binding region at the N terminus of the Saccharomyces cerevisiae TAF1 protein. J Biol Chem. 2003;278:45888–902. doi: 10.1074/jbc.M306886200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.