Abstract
Three dimensional hydrogels are a promising vehicle for delivery of adult human bone-marrow derived mesenchymal stem cells (hMSCs) for cartilage tissue engineering. One of the challenges with using this cell type is the default pathway is terminal differentiation, a hypertrophic phenotype and precursor to endochondral ossification. We hypothesized that a synthetic hydrogel consisting of extracellular matrix (ECM) analogs derived from cartilage when combined with dynamic loading provides physiochemical cues for achieving a stable chondrogenic phenotype. Hydrogels were formed from crosslinked poly(ethylyene glycol) as the base chemistry and to which (meth)acrylate functionalized ECM analogs of RGD (cell adhesion peptide) and chondroitin sulfate (ChS, a negatively charged glycosaminoglycan) were introduced. Bone-marrow derived hMSCs from three donors were encapsulated in the hydrogels and cultured under free swelling conditions or under dynamic com pressive loading with 2.5 ng/ml TGF-β3. hMSC differentiation was assessed by quantitative PCR and immunohistochemistry. Nine hydrogel formulations were initially screened containing 0, 0.1 or 1mM RGD and 0, 1 or 2wt% ChS. After 21 days, the 1% ChS and 0.1 mM RGD hydrogel had the highest collagen II gene expression, but this was accompanied by high collagen X gene expression. At the protein level, collagen II was detected in all formulations with ECM analogs, but minimally detectable in the hydrogel without ECM analogs. Collagen X protein was present in all formulations. The 0.1 mM RGD and 1% ChS formulation was selected and subjected to five loading regimes: no loading, 5% strain 0.3Hz (1.5%/s), 10% strain 0.3 Hz (3%/s), 5% strain 1 Hz (5%/s), and 10% strain 1Hz (10%/s). After 21 days, ~70-90% of cells stained positive for collagen II protein regardless of the culture condition. On the contrary, only ~20-30% of cells stained positive for collagen X protein under 3 and 5%/s loading conditions, which was accompanied by minimal staining for RunX2. The other culture conditions had more cells staining positive for collagen X (40-60%) and was accompanied by positive staining for RunX2. In summary, a cartilage-like biomimetic hydrogel supports chondrogenesis of hMSCs, but dynamic loading only under select strain rates is able to inhibit hypertrophy.
1. Introduction
Bone marrow-derived mesenchymal stem cells (MSCs) are a promising autologous cell source for cartilage tissue engineering, offering several advantages over cartilage cells (i.e., chondrocytes). For example, MSCs can be expanded to achieve a large number of cells, which are needed for implantation, but without losing their multipotency and ability to differentiate down the chondrogenic lineage1. MSCs offer a one-step surgical procedure instead of two-steps, which is required in autologous chondrocyte implantation. Although promising, the ability of MSCs to regenerate hyaline cartilage in vivo has been limited2,3. This finding is in part attributed to the fact that the default pathway in chondrogenically differentiating MSCs is terminal differentiation. This differentiation fate leads to a hypertrophic phenotype that is a precursor to endochondral ossification, where cartilage becomes hypertrophic, mineralizes, and eventually is replaced with new bone4-6. With the ability to deliver MSCs within a 3D scaffolding material, such as a hydrogel, the opportunity arises to design 3D environments that support a stable chondrogenic phenotype. However, the cues that lead to a stable phenotype remain to be elucidated.
The interaction of MSCs with their environment is known to play an important role in differentiation7,8. This raises the question as to whether the unique environment of cartilage is important for MSC chondrogenesis. In cartilage, chondrocytes are embedded within a dense extracellular matrix (ECM) comprised primarily of collagen type II and highly negatively charged aggrecan macromolecules. During physical activity, cartilage experiences large compressive strains that are transmitted through the tissue and converted into local physiochemical cues. These cues include deformation of cells, integrins acting as mechanoreceptors, and dynamic flow of ions and water that alters the local osmolarity and leads to fluid-induced shear stress9,10. These effects, among others, have been shown to significantly affect ECM synthesis by chondrocytes11.12 and cartilage homeostasis; and therefore may also play a role in chondrogenesis of MSCs. Several studies have explored the individual effects of different cartilage-related ECM analogs and of dynamic loading on chondrogenesis of MSCs when encapsulated in hydrogels, but the results have been mixed.
The cell adhesion moiety RGD, which is found in many ECM proteins including fibronectin and collagens, has been investigated in MSC chondrogenesis. Fibronectin is involved in the early stages of chondrogenic differentiation, but is eventually down-regulated with maturation13. The tethering of a cell adhesion peptide like RGD into a hydrogel provides a mechanism by which cells may interact with the hydrogel and this interaction has been shown to improve MSC viability within synthetic hydrogels2. RGD, however, has also been shown to inhibit MSC chondrogenesis14, but at concentrations that are sufficiently high to affect the phenotype of fully differentiated chondrocytes15. On the contrary, incorporating RGD either through degradable linkers16 or at low concentrations17 has been shown to support MSC chondrogenesis. These findings suggest that RGD supports MSC chondrogenesis, but depends on the contextual presentation of RGD.
Another ECM analog that has been investigated in MSC chondrogenesis is chondroitin sulfate (ChS), the main sulfated glycosaminoglycan found in aggrecan. Chondroitin sulfate can readily be functionalized enabling its incorporation into a hydrogel via covalent crosslinks18. Similar to native cartilage, ChS introduces a high density of fixed negative charges into a hydrogel, which elevates the local osmolarity19. Studies with fully differentiated chondrocytes have shown that osmolarities within the range of native cartilage support ECM synthesis, while lower or higher osmolarities, inhibit ECM synthesis20,21. Similarly, high osmolarities have been shown to inhibit chondrogenesis of MSCs22-24. Nonetheless, the incorporation of ChS into a synthetic hydrogel supports chondrogenesis by expression of cartilage-related genes and proteins in human MSCs25,26 and has been shown to downregulate hypertrophic genes in goat MSCs7. However, concentration effects of ChS in a synthetic hydrogel have been shown to influence ECM synthesis of fully differentiated chondrocytes27 and therefore may also impact MSC chondrogenesis, but this effect has yet to be studied.
Dynamic mechanical compression applied to MSCs embedded in hydrogels has resulted in highly variable responses, enhancing chondrogenesis in some studies28,29, while inhibiting chondrogenesis in other studies30,31. A number of factors may contribute to the variable responses in MSCs including hydrogel formulation, concentration of the ECM analog, and loading parameters. For fully differentiated chondrocytes, the combination of ECM analogs and dynamic compression has a synergistic and positive effect on ECM synthesis, when compared to either cue alone, but the improved response is dependent on the concentration of the ECM analog15,19,27. Therefore, additional studies are needed to better understand the impact of the physicochemical environment that combines ECM analogs with dynamic compression on MSC chondrogenesis.
As cells in native cartilage experience multiple cues at any given time, a synthetic hydrogel that captures multiple physicochemical cues may be provide a more biomimetic environment for MSC chondrogenesis. Thus in this work, we hypothesized that a biomimetic hydrogel containing RGD and chondroitin sulfate in combination with dynamic mechanical compression enhances chondrogenesis of human MSCs (hMSCs), but in a manner that depends on the magnitude of the cues. We employed a synthetic hydrogel made from crosslinked poly(ethylene glycol), which serves as the base hydrogel to which RGD and chondroitin sulfate are systemically incorporated. In this first part of this study, RGD and chondroitin sulfate were incorporated into a PEG hydrogel over a concentration range that was previously determined to be suitable for fully differentiated chondrocytes27 and then screened for hMSC chondrogenesis and hypertrophy in the absence of loading. In the second part of this study, we selected one combination of the ECM analogs and investigated several different dynamic loading regimes that fall within the physiological range for their impact on hMSC chondrogenesis and hypertrophy. Overall, findings from this study support the hypothesis that the introduction of ECM analogs in the form of RGD and chondroitin sulfate, improves hMSC chondrogenesis, but that dynamic loading under certain loading regimes is necessary to preserve a stable chondrogenic phenotype and inhibit hypertrophy.
2. Materials and Methods
2.1 Macromer Synthesis
Poly(ethylene glycol) dimethacrylate (PEGDM) was synthesized via microwave methacrylation32 by reacting poly(ethylene glycol) (4600 g mol−1) (Sigma) with methacrylic anhydride in the presence of hydroquinone (Sigma). The PEGDM product was recovered by dissolution in methylene chloride followed by precipitation in ethyl ether, filtration and drying. The degree of methacrylation was determined to be 94% by 1HNMR (Varian VYR-500) by comparing the area under the peak for the vinyl groups (δ=5.5-6.2 ppm) to the area under the peak for the methyl groups in the PEG backbone (δ=3.3-3.9 ppm). Acryloyl-PEG-RGD was synthesized by reacting YRGDS (Genscript) in excess in a 1.1:1 molar ratio with acryloyl-PEG-N-hydroxysuccinimide (3400 g mol−1) (Laysan Bio, Inc.) in 50 mM sodium bicarbonate. The degree of acrylation was 94% and the degree of conjugation of the peptide was 71% determined by 1HNMR. Chondroitin sulfate methacrylate (ChSMA) was synthesized by dissolving chondroitin sulfate A (Sigma) at 25% (w/v) in deionized water and reacted with excess methacrylic anhydride (1:8 molar ratio of chondroitin sulfate to methacrylic anhydride)18,27. The product was sterile filtered and recovered by lyophilization. On average, 16% of the disaccharide repeat units on each chondroitin sulfate polymer were substituted with a methacrylate as determined via 1HNMR (Bruker Ascend 400) by comparing the area of under the peak for the vinyl resonances (δ=5.5-6.2 ppm) to the area under the peak for the acetyl groups chondroitin sulfate sugar backbone (δ=1.7-2.0 ppm).
2.2 Acellular Hydrogel Formulation and Characterization
Hydrogels were fabricated via photopolymerization of macromers at concentrations of 8, 9 or 10wt% PEGDM, 0, 0.1 or 1 mM Acryloyl-PEG-RGD, and 0, 1, or 2wt% ChSMA with 0.5wt% photoinitiator Igracure 2959 (I2959) (BASF) in phosphate buffered saline (PBS, pH 7.4) with 352 nm light at 5 mW cm−2 for 10 minutes. Hydrogels were allowed to swell to equilibrium for 24 hours in PBS at 37°C prior to their characterization.
The tangent modulus under compression was determined using cylindrical hydrogels (5mm in height × 5mm in diameter) using a mechanical tester (MTS Synergie 100, 10N). The samples were strained at a constant strain rate of 0.5 mm min−1 using nonporous platens to 15% strain and the resulting modulus was determined for the linear region of the stress-strain curve. A sample size of 10 was used.
The volumetric equilibrium swelling ratio (Q) was estimated from the experimentally determined mass equilibrium swelling ratio, which was determined by measuring swollen mass after 24 hours of free swelling in PBS relative to the dry polymer mass, and assuming a density of the polymer (1.07 g ml−1) and density of the solvent density (1 g ml−1). A sample size of 10 was used.
The incorporation of RGD into the hydrogels was evaluated by qualitative and semi-quantitative fluorescence intensity using a fluorescently labelled acryloyl-PEG-RGD. In brief, cadaverine 488 was reacted with the acryloyl-PEG-RGD in the presence of 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) in 0.1 M sodium bicarbonate at a 1:1 molar ratio for 24 hours. The product was purified by dialysis and recovered by lyophilization. PEG hydrogels with fluorescently labeled RGD were created by initially making a 100 mM stock solution of RGD consisting of 1 wt% acryloyl-PEG-RGD-fluorophore and the other 99 wt% acryloyl-PEG-RGD. This stock solution was diluted to the desired concentration of 1mM and 0.1 mM RGD, and the hydrogels were polymerized using a redox initiating system of ammonium persulfate (0.025 M APS) and tetramethylethylenediamine (0.0125 M TEMED) for 10 minutes. The hydrogel were placed in PBS for 24 hours after polymerization to remove unincorporated RGD. Images were acquired using the Molecular Imager VersaDoc system and intensity measurements were used to semi-quantify the amount of fluorescently labeled RGD incorporated into the hydrogels.
The incorporation of chondroitin sulfate into the PEG hydrogels was confirmed qualitatively using toluidine blue stain and quantitatively using the DMMB dimethylmethylene blue (DMMB) assay. Hydrogels were stained with a solution of toluidine blue, which stains for negatively charged glycosaminoglycans, for 24 hours in PBS (0.1% toluidine blue, 7% ethanol, 0.1% NaCl in PBS, pH<2.5) and imaged using light microscopy (Zeiss Pascal, Olympus DP70 100x magnification). DMMB assay was used to quantify the amount of ChSMA incorporated into the hydrogels by measuring the soluble fraction of ChSMA, i.e. the fraction not covalently attached into the hydrogel network, which had diffused out into the PBS bath after 24 hours of free swelling.
2.3 hMSC Donors and Expansion
Bone marrow derived adult hMSCs (passage 1) were purchased from Texas A&M Cell Distribution Center. Three donors were investigated for the free swelling studies: (1) Donor 1 is a 24 year old female, (2) Donor 2 is a 29 year old female, (3) Donor 3 is a 24 year old female. Three different donors were investigated for the loading studies: (4) Donor 4 is a 24 year old female, (5) Donor 5 is a 22 year old male, and (6) Donor 6 is a 24 year old female. The hMSCs were thawed and expanded in stem cell culture media (20% fetal bovine serum (FBS, Atlanta Biologicals), 50 U ml−1 penicillin, 50 mg ml−1 streptomycin, 10 ng ml−1 bFGF and 20 mg ml−1 gentamicin in low glucose Dulbecco's modified Eagle media (DMEM, Invitrogen)). The hMSCs were seeded at 3000 cells cm−2 and grown to 80% confluency in T225 flasks up to passage 2. The cells were grown under standard cell conditions of 37°C with 5% CO2.
2.4 hMSC Encapsulation and Culture in Hydrogels
hMSCs were encapsulated in hydrogels at a cell concentration of 10×106 cells ml−1 of filter-sterilized (0.22 μm filter) macromer solution by photopolymerization as described above. The cell-laden constructs (5mm in diameter and 2mm in height) were cultured in chondrogenic differentiation medium composed of 1% ITS+ Premix, 100 nM dexamethasone, 2.5 ng ml−1 TGF-β3, 50 mg ml−1 l-ascorbic acid 2-phosphate, 50 U ml−1 penicillin, 50 mg ml−1 streptomycin, and 20 mg ml−1 gentamicin in high glucose Dulbecco's modified Eagle media. The hMSC-laden hydrogels were cultured under standard cell conditions of 37°C with 5% CO2 under static conditions for 7 or 14 days.
2.5 Mechanical Loading and Cell Viability
A custom-built bioreactor system33,34 was utilized to apply intermittent dynamic compressive strains to the hydrogels. The hydrogels were cultured under free swelling conditions for the first week and then subjected to an intermittent dynamic loading regime for up to an additional two weeks. Dynamic compression was applied in unconfined compression in a sinusoidal waveform for 1 hr followed by 23 hrs of rest. Four regimes were investigated with strains and frequencies of: 5% peak-to-peak strain (2.5% amplitude strain) 1Hz, 5% peak-to-peak strain (2.5% amplitude strain) 0.3 Hz, 10% peak-to-peak strain (5% amplitude strain) 1Hz, and 10% peak-to-peak strain (5% amplitude strain) 0.3 Hz. Hydrogels were cultured individually in a 24-well plate with 2 ml per well of chondrogenic differentiation media, which was changed every other day for the duration of the study. Free swelling controls were removed at 7, 14, and 21 days and loaded hydrogels were removed at 14 and 21 days. Cell viability was assessed after 21 days of culture in PBS with 4 nM ethidium homodimer and 4 nM calcein-AM at 37°C for 20 minutes. Hydrogels were imaged at 100x magnification with a water objective by confocal microscopy.
2.6 Quantitative PCR (qPCR)
RNA was extracted from constructs (n=3 technical replicates per donor) using MicroElute Total RNA Kit per manufacturer instruction (Omega). RNA was transcribed to cDNA with a High Capacity Reverse Transcription Kit (Applied Biosystems). Quantitative real-time polymerase chain reaction (qPCR) was performed with Fast SYBR Green Master Mix (Applied Biosystems) on a 7500 Fast Real-Time PCR Machine (Applied Biosystems). All genes of interest are relative to the housekeeping gene, L30. Relative expression levels were calculated from Ct values. The genes of interest were collagen I (found in undifferentiated hMSCs and in fibrocartilage), collagen II (marker for chondrogenesis), and collagen X (marker for hypertrophy) (Invitrogen) (Table S1). PCR efficiency was determined for each gene. Briefly, threshold Ct values were determined from serial dilutions of cDNA and plotted. The slope between Ct values of 15-25 was used to determine efficiency, resulting in the following efficiencies, 94% for L30, 98% for collagen I, 102% for collagen II, and 87% for collagen X. Normalized gene expression was determined using the actual efficiencies following methods described by Pfaffl35.
2.7 Immunohistochemistry (IHC)
Hydrogels were fixed overnight in 4% paraformaldehyde at 4°C and transferred to a 15% sucrose solution at 4°C for storage (n=4 technical replicates per donor). Constructs went through a series of dehydration steps and were embedded in paraffin. Sections (10 μm) were stained for the presence of aggrecan, collagen II, collagen X, and Runx2 by immunohistochemistry. For anti-collagen I, sections were pretreated with 1 mg ml−1 pepsin. For anti-collagen II sections were pre-treated with 2000 U ml−1 hyaluronidase. For anti-collagen X sections were pre-treated with 1 mg ml−1 protease followed by 1 mg ml−1 pepsin, followed by 0.25% trypsin in 1mM EDTA. For anti-collagen I sections were pre-treated with pepsin (1 mg ml−1) followed by antigen retrieval. For anti-aggrecan sections were pre-treated with antigen retrieval, followed by chondroitinase ABC (100 mU ml −1) and keratinase I (1U ml−1), and followed by hyaluronidase (2000 U ml−1). Anti-RunX2 sections required no pretreatment. After permeabilization and blocking, samples were treated overnight at 4°C with primary antibody: 1:50 anti-collagen II (Abcam, ab3092), 1:5 anti-aggrecan (USBio, A1059-53F), 1:50 anti-collagen X (Abcam, ab3092), 1:50 anti-collagen I (Abcam, ab24710), and 1:50 anti-Runx2 (Abcam, ab23981) in blocking solution. Constructs were treated for 2 hours with goat anti-mouse IgG or goat anti-rabbit IgG labeled Alexa Fluor 488 (1:200) and Alexa Fluor 546 (1:200) and counterstained with DAPI.
2.8 Enzyme Activity Analysis
A SensoLyte Plus 520 MMP-13 Assay Kit (Anaspec) was used to determine the concentration of active matrix metalloproteinase (MMP) in the media of the free swelling and loading conditions. Culture media from donor 5 and 6 was collected flash-frozen in liquid nitrogen from the loading and the free swelling cultures every other day from day 7 to day 21. The media was stored at −80°C until the assay was conducted. The media from multiple time points were pooled together as such: day 11 (media collected from days 7-11), day 17 (media from days 13-17) day 21 (media from days 17-21). The assay was performed as described in the kit protocol containing recombinant human pro-MMP-13 as the standard and a fluorescent MMP-13 substrate, but without an activation step to measure only the endogenous active form of MMP-13 in the media. The fluorescent signal was measured (Ex/Em=490nm/520nm) using a FLUOstar Optima plate reader (BMG Labtech).
2.9 Statistical Analysis
Data are presented as mean with standard deviation. Mean and standard deviation for acellular hydrogel characterization (ChS incorporation, RGD relative fluorescence, compressive modulus, and swelling ratio) are from technical replicates and were analyzed using a one-way ANOVA with a post hoc Tukey's test, α=0.05. For all qPCR gene expression and immunohistochemistry data, the mean represents average of three biological replicates, where each donor is an average of the technical replicates (n=3 for gene expression, n=4 for IHC). Data were analyzed by a one-way ANOVA with a post hoc Tukey's test, α=0.05, to determine significant difference between conditions at specific time points.
3. Results
3.1 Effects of ECM Analogs Introduced in PEG-based Hydrogels
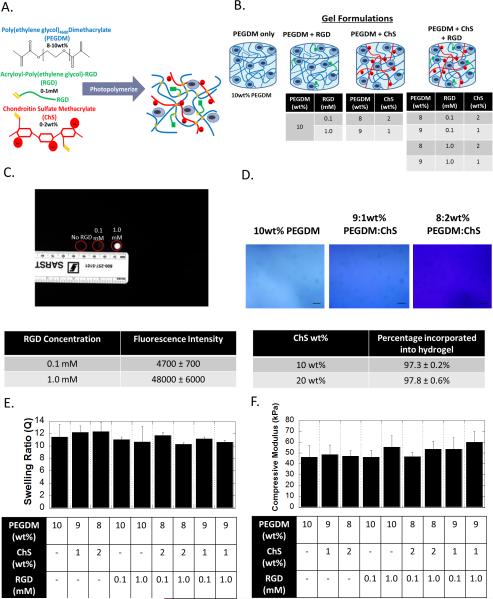
PEG hydrogels were fabricated by photopolymerization of (meth)acrylate functionalized monomers of PEGDM, monoacrylate-PEG-RGD, and ChSMA to form a crosslinked network composed of PEG and ChS with covalently tethered RGD (Fig. 1A). Hydrogels were formed with compositional variations in RGD (0, 0.1 and 1 mM) and ChS (0, 1 and 2 wt%) (Fig. 1B). After hydrogel formation, the hydrogels were allowed to free swell to remove any unreacted ECM analog. For RGD, fluorescently labeled RGD was used to confirm its incorporation into the hydrogel (Fig. 1C). Photographs of the hydrogels show much greater fluorescence in the 1 mM RGD hydrogel formulation compared to the 0.1 mM RGD hydrogel formulation with no observable fluorescence in the PEG hydrogel with no RGD. The fluorescence intensity was semi-quantified and determined to be an order of magnitude greater in the 1 mM RGD hydrogel formulation when compared to the 0.1 mM RGD hydrogel. The incorporation of ChSMA was qualitatively confirmed using toluidine blue, which stains for sulfated glycosaminoglycans, and showed increased staining with increasing concentration of ChSMA (Fig. 1D). The total amount of ChS incorporated into the hydrogel was confirmed to be ~97% indicating that nearly all of the ChSMA monomer was incorporated into the hydrogel (Fig. 1D). The macroscopic properties, specifically compressive modulus (Fig. 1E) and the equilibrium volumetric swelling ratio (Fig. 1F), were also measured for the different hydrogel formulations. The compressive modulus was ~50 kPa and the volumetric swelling ratio was ~11. Both properties were statistically similar for all hydrogel formulations.
figure 1.
formation and characterization of the biomimetic hydrogels formed from crosslinked poly(ethylene glycol) as the base chemistry and to which (meth)acrylate-functionalized rgd and chondroitin sulfate (chs) are incorporated. a) a schematic showing the monomers and the resulting network structure formed after photopolymerization. b) a schematic of the different hydrogel formulations investigated in this study. c) the incorporation of rgd was confirmed using fluorescently labeled rgd. fluorescent images and the corresponding semi-quantification of fluorescence intensity (n=3) show increased fluorescence with the increasing rgd from 0 to 0.1 to 1 mm. d) the incorporation of chs as confirmed by toluidine blue staining, which stains negatively charged molecules blue. brightfield microscopy images of toluidine blue stained hydrogels with 0, 1, or 2 wt% chs show increased blue staining. scale bar= 100μm. note that peg-only hydrogels exhibit some background staining. the amount of chs incorporated into the hydrogel was also quantitatively assessed (n=3). e) the compressive modulus was measured for the different hydrogel formulations and show no significant differences (n=10, p=0.73). f) the volumetric swelling ratio was measured for the different hydrogel formulations and show no significant differences (n=10, p=0.47). all data are presented as mean with standard deviation parenthetically or as error bars.
3.2 Effect of ECM Analogs on hMSC Chondrogenic Differentiation in PEG Hydrogels
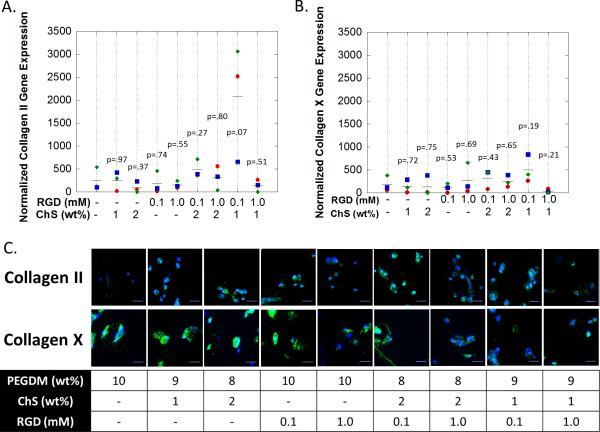
A total of nine hydrogel formulations were investigated to screen for RGD (0, 0.1 and 1 mM) and ChS (0, 1 and 2wt%) concentrations that enhance hMSC chondrogenesis in the presence of low TGF-β3 concentration (2.5 ng/ml). hMSCs from three donors were investigated. After 14 days of culture in chondrogenic differentiation media, cell-laden hydrogels were assessed by qPCR and IHC for collagen II as a marker for chondrogenesis, collagen X as a marker for hypertrophy (Fig. 2) and collagen I, which is expressed by MSCs and is a marker of fibrocartilage (Fig. S1).
figure 2.
the effects of ecm analogs on hmsc chondrogenesis under free swelling culture conditions after 14 days of culture. gene expression (by qpcr) for a) collagen ii and b) collagen x. the symbols (diamond (donor 1), square (donor 2), circle (donor 3)) represent the mean for each donor (n=4 technical replicates). the line (▬) represents the mean for all donors (n=3) and p values are relative to peg-only hydrogel. normalized expression is defined as relative expression at day 14 normalized to relative expression at 24 hours after encapsulation. c) immunohistochemistry staining for collagen ii (green) (top row) and for collagen x (green) (bottom row). cell nuclei (blue) were counterstained with dapi. scale bar = 20μm.
Gene expression data represent the change in expression that occurred between initial encapsulation (i.e., 24 hours after encapsulation) and day 14. Collagen II mRNA levels were elevated (i.e., greater than one) across all hydrogel formulations by day 14. The formulation consisting of 0.1 mM RGD and 1 wt% ChS had the highest (~10x) (p=0.07) collagen II gene expression values over the PEG-only hydrogel. In addition, this formulation had higher (p=0.05-0.08) Collagen II expression when compared to the other formulations with ECM analogs; although to a lesser extent (p=0.4) when compared to the formulation with 0.1 mM RGD and 2 wt% ChS. For collagen X, the formulation consisting of 0.1 mM RGD and 1 wt% ChS, which had the highest expression of collagen II, had higher (~4x) (p=0.14) expression of collagen X expression when compared to the PEG-only hydrogel. The mean expression was also higher when compared to the other formulations with ECM analogs, but the increased expression was less significant (p=0.10-0.45). Collagen I gene expression was reduced (i.e., less than one) across all hydrogel formulations by day 14 with no differences among the hydrogel formulations. At the protein level, there was very little if any collagen II detected in the PEG-only hydrogel. However, collagen II was present in all of the formulations with ECM analogs. Collagen X protein was present in all of the hydrogel formulations. Collagen I protein was not detectable in any of the hydrogel formulations.
3.3 hMSC Chondrogenesis under Dynamic Compressive Loading
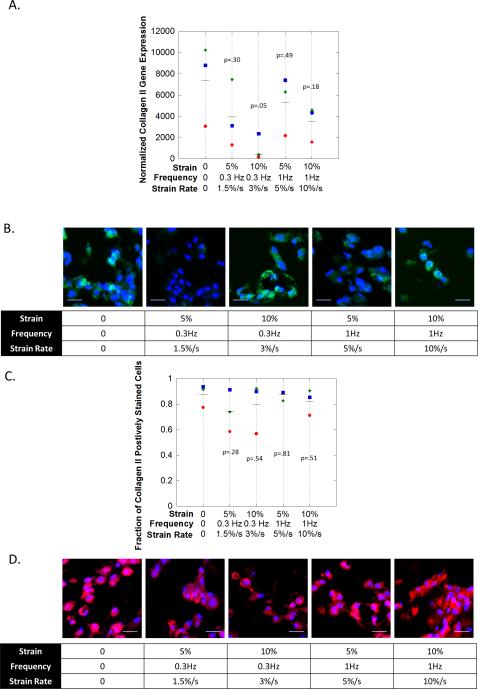
Hydrogels were subjected to unconfined dynamic compression to investigate the effect of dynamic compressive loading on hMSC chondrogenesis. One hydrogel formulation (0.1 mM RGD: 1% ChS) was selected from the screening study and subjected to dynamic compressive strains for 1 hour per day at one of five different loading regimes with strain rates included in parentheses: (1) no loading, (2) 5% strain 0.3 Hz (1.5%/s), (3) 10% strain 0.3 Hz (3%/s), (4) 5% strain 1Hz (5%/s), and (5) 10% strain 1 Hz (10%/s). The hydrogels were initially cultured in a free swelling environment for one week, and then placed in bioreactors and subjected to dynamic compression for two weeks. Here gene expression was normalized to expression levels of the pre-encapsulated hMSCs for each donor respectively to assess the overall differentiation of the hMSCs for each of the different culture conditions.
hMSC chondrogenesis was assessed after 14 days for collagen II (Fig. S2A,C) and more extensively after 21 days of culture for viability (Fig. S3), collagen II and aggrecan (Fig. 3) and for collagen I (Fig S4). At 14 days all conditions had elevated (p=0.07-0.19) collagen II gene expression levels when compared to pre-encapsulated hMSCs. However, there were no differences (p=0.57) as a function of the culture condition. Collagen II protein was present in all conditions. At 21 days, cells were mostly viable with some dead cells present in the hydrogel (Fig. S3). There were no observable differences in viability under loading. Gene expression for collagen II (Fig. 3A) was variable across all loading conditions with no significant differences, but remained several orders of magnitude higher than prior to encapsulation. In all loading conditions, collagen II protein (Fig. 3B-C) was detected with an average of ~70-90% of the cells staining positive. Although qualitatively, there appeared to be less staining for collagen II protein in the loading condition with a strain rate of 1.5%/s. Collagen I gene expression was approximately 2-3 orders of magnitude lower when compared to collagen II gene expression with no observable trends as a function of loading condition. Collagen I protein was not detectable in any of the hydrogels as a function of loading. The IHC analysis was extended to aggrecan (Fig. 3D), which was highly expressed at the protein level with no observable differences among the culture conditions. These results suggest that collagen II and aggrecan expressions are not substantially affected by dynamic compression across the loading regimes investigated.
figure 3.
the effects of dynamic compressive loading on hmsc chondrogenesis in a biomimetic peg-based hydrogel containing 0.1 mm rgd and 1 wt% chs after 21 days of culture. a) gene expression (by qpcr) for collagen ii. normalized expression is defined as relative expression at day 21 normalized to relative expression for pre-encapsulated hmscs. b) immunohistochemistry staining for collagen ii (green). cell nuclei (blue) were counterstained with dapi. scale bar = 20μm. c) semi-quantitative analysis of the fraction of cells staining positive for collagen ii. d) immunohistochemistry staining for aggrecan (red). cell nuclei (blue) were counterstained with dapi. scale bar = 20μm. d). in b and c, the symbols (diamond (donor 1), square (donor 2), circle (donor 3)) represent the mean for each donor (n=4 technical replicates). the line (▬) represents the mean for all donors (n=3) and p values are relative to peg-only hydrogel.
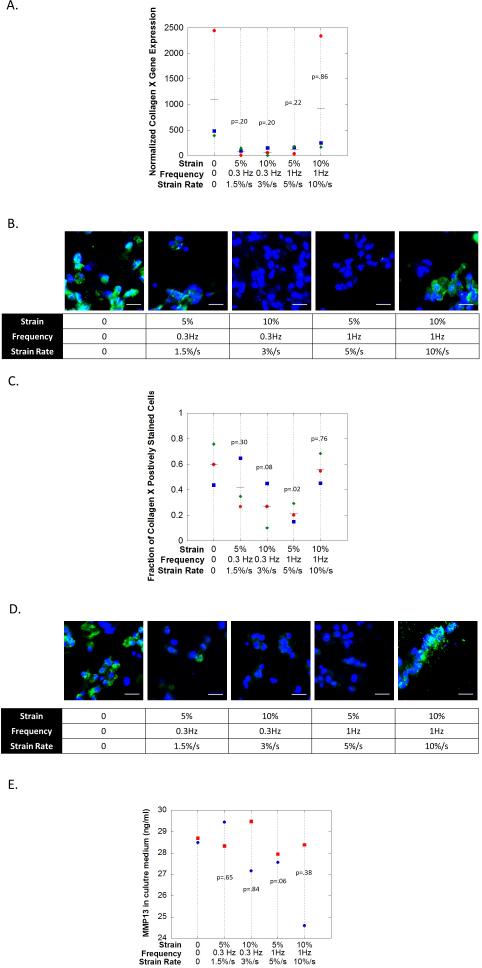
3.4 hMSC Hypertrophy under Dynamic Compressive Loading
hMSC hypertrophy was assessed after 14 days by collagen X (Fig S2B,C) and more extensively after 21 days for collagen X, RunX2, and MMP13 (Fig. 4) under the same experimental conditions as described in the previous section. At 14 days all conditions had elevated (p=0.06-0.21) collagen X gene expression levels when compared to pre-encapsulated hMSCs. However, there were no large differences (p=0.39) as a function of the culture condition. Collagen X protein was not detected in two of the loading conditions with strain rates of 3%/s and 5%/s, but was detected in the free swelling condition and loading conditions at the extremes with strain rates of 1.5%/s and 10%/s. At 21 days, collagen X gene expression was reduced (p=0.20-0.22) in the loading conditions with strain rates of 1.5-5%/s when compared to the free swelling (Fig. 4A). At the highest stain rate (10%/s) collagen X expression was similar to the free swelling condition. At the protein level, collagen X (Fig. 4B) was detected in the free swelling condition with ~60% of the cells expressing the protein. In the loading conditions at the extremes with strain rates of 1.5%/s and 10%/s, collagen X protein was also detected with 40-55% of the cells expressing the protein. On the contrary in the loading conditions with strain rates of 3 and 5 %/s, fewer cells were expressing collagen X with only ~30% and 20%, respectively. Staining for RunX2 at the protein level (Fig. 4C) followed that of collagen X with prominent staining in the free swelling condition and in the extreme loading conditions with strain rates of 1.5 %/s and 10 %/s. However minimal staining was detected in the loading conditions with strain rates of 3 and 5 %/s. MMP-13 activity was also assessed in the culture medium for two of the three donors and data for day 21 is shown (Fig. 4D). Under the 5%/s strain rate, the levels of MMP-13 activity were lower (p=0.06) when compared to the free swelling condition, while all other loading conditions were more similar (p=0.38-0.84) to the free swelling conditions. No differences were observed at the early time points of day 11 and day 17 (data not shown).
figure 4.
the effects of dynamic compressive loading on hmsc hypertrophy in a biomimetic peg-based hydrogel containing 0.1 mm rgd and 1 wt% chs after 21 days of culture. a) gene expression (by qpcr) for collagen x. normalized expression is defined as relative expression at day 21 normalized to relative expression for pre-encapsulated hmscs. b) immunohistochemistry staining for collagen x (green). cell nuclei (blue) were counterstained with dapi. scale bar = 20μm. c) semi-quantitative analysis of the fraction of cells staining positive for collagen x. d) immunohistochemisty staining for runx2 (green). cell nuclei (blue) were counterstained with dapi. scale bar = 20μm. e) the concentration of active mmp13 in culture media accumulated from days 17-21. in a, c and e, the symbols (diamond (donor 4), square (donor 5), and circle (donor 6)) represent the mean of each donor (n=4 technical repeats). the line (▬) represents the average of all donors (n=3).
4. Discussion
Human mesenchymal stem cells are a promising cell source for cartilage tissue engineering given their ability to differentiate down the chondrogenic lineage. A successful tissue engineering approach using hMSCs requires that the scaffold (e.g., hydrogel) promote a stable chondrogenic phenotype with cartilage-specific ECM synthesis. The challenge is that the default pathway for hMSCs differentiating down the chondrogenic lineage is terminal differentiation, a precursor to endochondral ossification36-41. However, the environment within healthy cartilage contains a host of physiochemical cues that maintain a stable chondrocyte phenotype throughout one's life. Herein, we show that recapitulating some of the physiochemical cues from cartilage within a synthetic hydrogel enhances hMSC chondrogenesis and simultaneously maintains a stable chondrogenic phenotype by preventing terminal differentiation. A 3D hydrogel environment with carefully tuned biochemical and biomechanical cues offers a promising strategy to achieve hMSC-mediated cartilage regeneration.
In this study, we first screened two ECM analogs, a cell adhesion peptide (RGD) and a sulfated glycosaminoglycan (ChS), for their ability to promote hMSC chondrogenesis in a synthetic PEG hydrogel, but in the absence of mechanical loading. Our results show that the presence of ECM analogs, regardless of type (RGD or ChS) or concentration, enhanced hMSC chondrogenesis. This observation was evident by positive staining for the protein, collagen II, a late marker for chondrogenesis. Collagen II was present in as early as two weeks of culture for all hydrogel formulations that contained ECM analogs, but was not detectable in the PEG-only hydrogels. Collagen II mRNA levels in the PEG-only hydrogel, however, were similar to many of the other formulations with ECM analogs. This result suggests that hMSCs are indeed undergoing chondrogenesis in all of the hydrogel formulations, but that the process may be slower in the absence of ECM analogs. This observation is supported by our previous work26, which showed that when higher TGF-β3 concentrations were employed (i.e., 10 ng/ml instead of 2.5 ng/ml used in the current study), collagen II protein was detected in PEG-only hydrogels within two weeks of culture. We have also previously shown that when TGF-β3 was absent from the culture medium, ECM analogs of ChS or RGD were not sufficient to induce expression of collagen II protein in hMSCs over this same time frame26. Since chondrogenesis is known to require TGF-β signaling1,42,43, our findings suggest that the enhanced chondrogenesis, as observed by positive staining for collagen II protein, may be a result of a synergistic effect between the ECM analogs and TGF-β signalling.
In the case of RGD, it is well known that a cross-talk exists between growth factor receptors and integrins to control downstream signaling44,45. In particular, TGF-β signalling has been shown to act synergistically with intracellular signalling pathways that depend on cell adhesion (e.g., Rho/Rock signalling)46. Therefore, when hMSCs engage with the RGD tethers present in the PEG hydrogel, it is possible that integrin-mediated signalling may act synergistically with TGF-β3 to enhance chondrogenesis. Furthermore, TGF-β binding to its receptor can lead to endogenous secretion of TGF-β, thus amplifying the signalling cascade47. Interestingly, the latent form of TGF-β that is secreted by cells, can be activated by integrins that recognize RGD48-50. We and others have reported that when cells are encapsulated in a PEG hydrogel with RGD, they up-regulate integrins that bind to RGD25,51. While the exact mechanism remains to be elucidated, we hypothesize that the presence of RGD in the PEG hydrogel may enhance chondrogenesis through a cross-talk in the signalling pathways downstream of TGF-β signalling and/or by integrin-mediated activation of endogenously secreted TGF-β. Although further studies are needed to test this hypothesis.
In the case of chondroitin sulfate, its sulfate groups may help to sequester soluble TGF-β3 and enhance its bioactivity. It is believed that the interaction between GAGs and growth factors prolongs their activity by increasing the half-life when bound to the matrix51. Studies have shown that TGF-β3 can bind to sulfated GAGs, reducing its release and improving it's effectiveness in cartilage tissue engineering53. In addition, the negatively charged chondroitin sulfate also attracts positive ions from the culture media leading to increased osmolarity within the hydrogel19, which is similar to that of cartilage19,54,55. It has been suggested that hyper-, but physiologically relevant, osmolarity can influence post-transcriptional regulation of Sox9 and therefore could lead to increased protein synthesis of collagen II56. Furthermore, hyperosmolarity has been shown to enhance TGF-β activity in other cell types and was responsible for increased collagen synthesis57. While the exact mechanism remains to be elucidated, we hypothesize that the presence of chondroitin sulfate enhances TGF-β3 activity either through contextual presentation of the growth factor (i.e., binding of the growth factor) or through a hypertonic environment that acts synergistically with TGF-β signalling. Although further studies are needed to test this hypothesis.
When the two ECM analogs were combined, the formulation containing 1wt% ChS and 0.1 mM RGD led to the highest collagen II mRNA levels across all three donors concomitant with collagen II protein expression in nearly all the cells. This observation suggests that while all formulations with ECM analogs induced hMSC chondrogenesis, there appears to be a synergistic effect between the ECM signals that depends on their magnitudes. Indeed, several studies have shown that there is an optimal osmolarity that enhances matrix synthesis by chondrocytes where supraphysical levels can be inhibitory to chondrocytes19,20 and to chondrogenically differentiating MSCs22. High concentrations of RGD have been shown to support osteogenesis of hMSCs in 3D cultures through traction dependent processes58 and thus low RGD concentrations may favor chondrogenesis17. The combination of RGD and ChS at the lower concentrations led to markedly improved chondrogenesis (as observed by the high mRNA levels for collagen II). We hypothesize that this effect occurs as a result of enhanced TGF-β signalling by the presence of both ECM analogs, where TGF-β induces Sox9-dependent transcriptional activity42 that ultimately controls collagen II expression42. However, additional experiments are needed to better understand the observed synergistic effect between the RGD and ChS on chondrogenesis.
The hypertrophic protein, collagen X, which is a hallmark of terminal differentiation and the default pathway for chondrogenically differentiating MSCs, was present in all formulations regardless of the presence of ECM analogs. This result is consistent with our previous reports26 and reports by others, which use TGF-β to induce MSC chondrogenesis 4-6,37, 59. These findings show that despite the presence of ECM analogs that create a cartilage-like biomimetic hydrogel environment, the enhanced chondrogenesis was accompanied by a hypertrophic phenotype. However, these initial studies were performed in the absence of any mechanical stimulation.
It is well known that mechanical forces are critical to cartilage homeostasis60.61 and have been shown to regulate chondrogenesis of MSCs28,29. We selected one formulation from the free swelling screening study, which showed robust chondrogenesis (i.e., 0.1 mM RGD and 1wt% ChS), and investigated the effects of dynamic compressive loading on MSC chondrogenesis and terminal differentiation. Here, a more in-depth analysis was performed. Interestingly, collagen II at the gene and protein levels did not appear to be affected by loading. This result is in contrast to our previous work, which showed inhibition of collagen II protein synthesis under dynamic compressive loading in PEG hydrogels with RGD31 and in PEG hydrogels with ChS26. In our previous work, however, loading was applied at higher strains (i.e., 15%), where others have shown that relatively high fluid flow velocities can inhibit collagen II synthesis by chondrogenically differentiating hMSCs61. Results from the current study, thus, suggest that lower strains (≤10%) and/or the presence of multiple ECM cues may be critical to maintaining chondrogenesis when hMSCs are encapsulated in PEG-based hydrogels and subjected to loading.
Most notably, dynamic mechanical loading affected terminal differentiation of the hMSCs and in a manner that depended on the loading rate. Interestingly, strain rates in the range of 3%/s (0.3 Hz 10%) to 5%/s (1 Hz 5%) inhibited collagen X protein expression across all three donors. There were notable differences between gene and protein expression levels for collagen X, which may in part be due to differences between transcriptional and translational control as shown by others63. The protein RunX2, a transcription factor involved in chondrocyte terminal differentiation and important to endochondral ossification64-66, was also inhibited under similar strain rates as collagen X. On the contrary, RunX2 and collagen X proteins were detected in the hydrogels cultured in the absence of loading as well as cultured under the two extreme loading conditions, at 1.5%/s and 10%/s strain rates. Runx2 protein appeared to be co-localized with the nucleus in some cells, but also located in the cytoplasm. This observation is consistent with prior reports that show Runx2 shuttling between the nucleus and cytoplasm67.68. These results suggest that the hypertrophic phenotype in chondrogenically differentiating hMSCs may be highly mechanosensitive, which has been suggested by others69,70. Our findings are consistent with others, which reported inhibition of hypertrophy, evident by reduced collagen X gene expression, by dynamic loading for hMSCs in a hydrogel71. However, results from this study suggest that this effect may be strain rate dependent. Since collagen X is critical to endochondral ossification, mechanical loading may play a role in controlling this process. Indeed, studies have pointed to the mechanical environment as regulating the type of cartilage tissue that is formed during development and as well during homeostasis72. Other studies have pointed to a role for non-collagenous extracellular matrix proteins (e.g., maitrilin, which is involved in matrix assembly and detected early in chondrogenically differentiating MSCs59) in regulating chondrocyte hypertrophy72 and which have been shown to be highly sensitive to dynamic loading73. Furthermore, multiple physiochemical cues are generated in the hydrogel as a result of the combined presence of dynamic loading and ECM analogs. These cues include: (a) cell deformation and fluid flow induced by dynamic loading alone, (b) integrins acting as mechanoreceptors to dynamic loading, and (c) dynamic changes in osmolarity that result from the movement of mobile cations in and out of the hydrogel during dynamic loading9. In all cases, the magnitude of the cue will depend on the loading rate. While the exact mechanism remains to be elucidated, dynamic loading will dramatically change the environment surrounding the hMSCs. We hypothesize that a combination of cues are likely contributing to the regulation of the hypertrophic phenotype, while the chondrogenic phenotype appears to be less sensitive to the loading environment at least when TGFβ3 is supplemented to the media. Additional studies, however, are needed to identify the pathways that lead to the observed load-induced inhibition of hMSC hypertrophy.
MMP-13 is also known to be upregulated in hypertrophic chondrocytes along with collagen X and RunX275. In this study, MMP-13 activity was detected in the culture medium, and appeared to be reduced under the strain rate 5%/s that also resulted in minimal collagen X and RunX2 protein levels. MMP-13 is involved in the resorption of cartilage and the remodelling of new bone76, and therefore its up-regulation in activity occurs later in this process. Indeed, others have reported that when hMSCs are cultured in pellets and in a hypertrophic-specific culture medium, MMP-13 mRNA was not detected until day 2141. Here, MMP-13 was detectable, but differences were not apparent until late in our study. It is important to note that we measured active MMP-13 and therefore it is possible that MMP-13 protein may have been affected by other loading conditions, but that this was accompanied by the secretion of molecules, such as TIMPs, that regulate its activity.
There are several limitations of this study, which are worth noting. First, the number of human donors was limited (n=3), thus reducing the statistical power of the study. The large variability observed here, however, is consistent with other reports77-80. As a result, our findings, particularly with respect to the gene expression data, are limited. However, we did observe notable differences in protein expression among different culture conditions that were consistent across all donors. While we did not quantify the total amount of protein deposited, the obvious presence or absence of a particular protein provided key insight into the effect of our culture condition on hMSC chondrogenesis. Second, our studies are limited to three weeks and therefore long-term studies are needed to determine whether the combined loading environment and biomimetic hydrogel can continue to maintain the chondrogenic phenotype while preventing terminal differentiation. Furthermore, terminal differentiation was evaluated in the presence of TGFβ3 by collagen type X, an early marker of hypertrophy. Additional studies are necessary to evaluate whether dynamic loading at the regimes identified here are sufficient to inhibit late stage hypertrophy in a more relevant hypertrophic medium81. Third, our studies were limited to non-degradable hydrogels. The use of a stable hydrogel was critical to ensure that the physiochemical cues, resulting from the hydrogel, were largely maintained throughout the culture period. However, non-degradable crosslinks prevent diffusion of large ECM molecules and thus limit their deposition to the pericellular space82. While a pericellular matrix can shield cells from mechanical strain, the relatively short culture times and limited matrix deposition by the chondrogenically differentiated MSCs are not expected to affect strain transfer to the encapsulated cells. Previously we have shown that a relatively thick PCM is necessary to inhibit strain transfer to cells. In addition, non-degradable hydrogels have also been shown to limit cell proliferation due to the restricted space; although cell proliferation was not measured in this study.
Conclusion
We describe a cartilage-biomimetic hydrogel that is composed of ECM analogs of a cell adhesion peptide based on RGD and a sulfated glycosaminoglycan based on ChS, which were incorporated, into a PEG hydrogel. This biomimetic hydrogel enhanced chondrogenesis of hMSCs over a range of ECM analog concentrations, but in all formulations and donors a hypertrophic phenotype was evident by the presence of collagen X. However when dynamic mechanical loading was introduced, chondrogenic differentiation did not appear to be affected, but hypertrophy was inhibited across all donors under moderate strain rates (i.e., 3%/s (10% 0.3 Hz) and 5%/s (5% 1 Hz). Our findings suggest that the combination of ECM analogs and dynamic mechanical loading contribute to the regulation of hypertrophy and help to promote a stable chondrogenic phenotype in MSCs.
Supplementary Material
Acknowledgements
Primary financial support for this work was provided by a NSF CAREER Award (#0847390). Additional support was provided by NIH (#1R01AR065441). The authors also acknowledge a NSF GRFP and a Department of Education Graduate Assistantships in Areas of National Need to EAA.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 2.Wakitani S, Mitsuoka T, Nakamura N, et al. Cell Transplant. 2004;13:595–600. doi: 10.3727/000000004783983747. [DOI] [PubMed] [Google Scholar]
- 3.Barry F, Murphy M. Nat Rev Rheumatol. 2013;9:584–594. doi: 10.1038/nrrheum.2013.109. [DOI] [PubMed] [Google Scholar]
- 4.Yang L, Tsang KY, Tang HC, Chan D, Cheah KSE. PNAS. 2014;111:12097–12102. doi: 10.1073/pnas.1302703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Kraan PM, van den Berg WB. Osteoarthritis and Cartilage. 2012;20:223–232. doi: 10.1016/j.joca.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Pacifici M, Golden EB, Oshima O, Shapiro IM, Leboy PS, Adams SL. Annals of the New York Academy of Sciences. 1990;599:45–57. doi: 10.1111/j.1749-6632.1990.tb42363.x. [DOI] [PubMed] [Google Scholar]
- 7.Varghese S, Hwang NS, Canver AC, Theprungsirikul P, Lin DW, Elisseeff J. Matrix Biology. 2008;27:12–21. doi: 10.1016/j.matbio.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Conor CJ, Case N, Guilak F. Stem Cell Research and Therapy. 2013;4:61. doi: 10.1186/scrt211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grodzinsky AJ, Levenston ME, Jin M, Frank EH. Annual Review of Biomedical Engineering. 2000;2:691–713. doi: 10.1146/annurev.bioeng.2.1.691. [DOI] [PubMed] [Google Scholar]
- 11.Waldman SD, Spiteri CG, Grynpas MD, Pilliar RM, Kandel RA. Tissue Engineering. 2004;10:1323–1331. doi: 10.1089/ten.2004.10.1633. [DOI] [PubMed] [Google Scholar]
- 12.Kinsiday JD, Jin M, DiMicco MA, Kurz B, Grodzinsky AJ. Journal of Biomechanics. 2004;37:595–604. doi: 10.1016/j.jbiomech.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 13.DeLise AM, Fischer L, Tuan RS. Osteoarthritis Cartilage. 2000;8:309–334. doi: 10.1053/joca.1999.0306. [DOI] [PubMed] [Google Scholar]
- 14.Connelly JT, Garcia AJ, Levenston ME. Biomaterials. 2007;28:1071. doi: 10.1016/j.biomaterials.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Villanueva I, Weigel CA, Bryant SJ. Acta Biomaterialia. 2009;5:2832–2846. doi: 10.1016/j.actbio.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salinas CN, Anseth KS. Biomaterials. 2008;29:2370–2377. doi: 10.1016/j.biomaterials.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu SQ, Tian QA, Wang L, Hedrick JL, P Hui JH, Yang YY, et al. Macromol Rapid Commun. 2010;31:1148–1154. doi: 10.1002/marc.200900818. [DOI] [PubMed] [Google Scholar]
- 18.Bryant SJ, Davise-Arehart KA, Luo N, Shoemaker RK, Arthur JA, Anseth KS. Macromolecules. 2004;37:6726–6733. [Google Scholar]
- 19.Farnsworth NL, Mead BE, Antunez LR, Palmer AE, Bryant SJ. Matrix Biology. 2014;40:17–26. doi: 10.1016/j.matbio.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Urban JPG, Hall AC, Gehl KA. J. Cell Physiology. 1993;154:262–270. doi: 10.1002/jcp.1041540208. [DOI] [PubMed] [Google Scholar]
- 21.Urban JPG, Rheumatology J. 1994;33:901–908. doi: 10.1093/rheumatology/33.10.901. [DOI] [PubMed] [Google Scholar]
- 22.Wuertz K, Godburn K, Neidlinger-Wilke C, et al. Spine. 2008;33:1843–1849. doi: 10.1097/BRS.0b013e31817b8f53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao YQ, Liange CZ, Li H, et al. Cell Biology International. 2013;37:826–834. doi: 10.1002/cbin.10110. [DOI] [PubMed] [Google Scholar]
- 24.Liang C, Li H, Tao Y, et al. Journal of Translational Medicine. 2012:10. doi: 10.1186/1479-5876-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salinas CN, Anseth KS. Journal of Tissue Engineering Regenerative Medicine. 2008;2:296–304. doi: 10.1002/term.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinmetz NJ, Bryant SJ. Biotechnology and Bioengineering. 2012;109:2671–2682. doi: 10.1002/bit.24519. [DOI] [PubMed] [Google Scholar]
- 27.Villanueva I, Gladem SK, Kessler J, Bryant SJ. Matrix Biology. 2010;29:51–62. doi: 10.1016/j.matbio.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angele P, Shuman D, Angele M, Kinner B, Englert C, Hente R. Biorheology. 2004;41:335. [PubMed] [Google Scholar]
- 29.Pelaez D, Huang CYC, Cheung HS. Stem Cells Dev. 2008;18:93. doi: 10.1089/scd.2008.0030. [DOI] [PubMed] [Google Scholar]
- 30.Thorpe SD, Buckley CT, Vinardell T, O'Brien FJ, Campbell VA, Kelly DJ. Biochem Biophys Res Commun. 2008;377:458. doi: 10.1016/j.bbrc.2008.09.154. [DOI] [PubMed] [Google Scholar]
- 31.Steinmetz NJ, Bryant SJ. Acta Biomatierialia. 2011;7:3829–3840. doi: 10.1016/j.actbio.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 32.Lin-Gibson S, Bencherif S, Cooper JA, Wetzel SJ, Antonucci JM, Vogel BM, Horkay F, Washburn NR. Biomacromolecules. 2004;5:1280–1287. doi: 10.1021/bm0498777. [DOI] [PubMed] [Google Scholar]
- 33.Nicodemus GD, Bryant SJ. J Biomech. 2008;41:1528–1536. doi: 10.1016/j.jbiomech.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villanueva I, Hauschultz DS, Mejic D, et al. Osteoarthritis and Cartilage. 2008;16:909–918. doi: 10.1016/j.joca.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfaffl MW. Nucleic Acids Research. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoo JU, Barthel TS, Nishimura K, Solchaga L, Caplan AI, Goldberg VM. J Bone Joint Surg Am. 1998;80:1745–1757. doi: 10.2106/00004623-199812000-00004. [DOI] [PubMed] [Google Scholar]
- 37.Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. Exp Cell Res. 1998;238:265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 38.Miwale F, Girard-Lauriault PL, Wang HT, Lerouge S, Antoniou J, Wertheimer MR. Tissue Engineering. 2006;12:2639–2647. doi: 10.1089/ten.2006.12.2639. [DOI] [PubMed] [Google Scholar]
- 39.Miwale F, Stachura D, Roughley P, Antoniou J. Journal of Orthopedic Research. 2006;24:1792–1798. doi: 10.1002/jor.20200. [DOI] [PubMed] [Google Scholar]
- 40.Pettari K, Winter A, Steck E, Goetzke K, Hennig T, Ochs BG. Arthritis and Rheumatology. 2006;54:3254–3266. doi: 10.1002/art.22136. [DOI] [PubMed] [Google Scholar]
- 41.Mueller MB, Tuan RS. Arthritis and Rheumatology. 2008;58:1377–1388. doi: 10.1002/art.23370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furumatsu T, Tsuda M, Taniguchi N, et al. J. Biol Chem. 2005;280:8343–8350. doi: 10.1074/jbc.M413913200. [DOI] [PubMed] [Google Scholar]
- 43.Lefebvre V, Li P, de Crombrugghe B. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alam N, Goel HL, Zarif MJ, et al. J. Cell Physiol. 2007;213:649–653. doi: 10.1002/jcp.21278. [DOI] [PubMed] [Google Scholar]
- 45.Kim S-H, Turnbull J, Guimond S. J. Endorinology. 2011;209:139–151. doi: 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
- 46.Allen JL, Cooke ME, Alliston T. Mol Biol Cell. 2012;23:3731–3742. doi: 10.1091/mbc.E12-03-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joyce ME, Roberts AB, Spron MB, Bolander ME. Journal of Cell Biology. 1990;110:2195–2207. doi: 10.1083/jcb.110.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munger JS, Sheppard D. Cold Spring Harbor Perspecives in Biology. 2011:3. doi: 10.1101/cshperspect.a005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wipff PJ, Rifkin DB, Meister JJ, et al. Journal of Cell Biology. 2007;179:1311–1323. doi: 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horiguchi M, Ota M, Rifkin DB. Journal of Biochemistry (Tokyo) 2012;152:321–329. doi: 10.1093/jb/mvs089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villanueva I. Chemical Egnineering, Vol PhD. University of Colorado; Boulder, CO: 2009. p. 294. [Google Scholar]
- 52.Hubbell JA. Curr. Opin. Biotechnol. 2003;14:551–558. doi: 10.1016/j.copbio.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 53.Park JS, Woo DG, Yang HN, et al. J Biomed Mater Res A. 2009;91:408–415. doi: 10.1002/jbm.a.32271. [DOI] [PubMed] [Google Scholar]
- 54.Yellowley CE, Hancox JC, Donahue HJ, Cell Biochem J. 2002;86:290–301. doi: 10.1002/jcb.10217. [DOI] [PubMed] [Google Scholar]
- 55.Le D, Hofbauer MA, Towle CA. Arch Biochem Biophys. 2006;445:1–8. doi: 10.1016/j.abb.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 56.Tew SR, Peffers MJ, McKay TR, et al. American Journal of Physiology-Cell Physiology. 2009;297:C898–C906. doi: 10.1152/ajpcell.00571.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugirua T, Yamauchi A, Kitamura H, et al. Kidney Int. 1998;53:1654–1660. doi: 10.1046/j.1523-1755.1998.00903.x. [DOI] [PubMed] [Google Scholar]
- 58.Huebsch N, Arany PR, Mao AS, et al. Nature Materials. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu J, Wang W, Ludeman M, et al. Tissue Eng Par A. 2008;14:667–680. doi: 10.1089/tea.2007.0272. [DOI] [PubMed] [Google Scholar]
- 60.Mauck RL, Nicoll SB, Seyhan SL, Ateshian GA, Hung CT. Tissue Egnineering. 2003;9:597. doi: 10.1089/107632703768247304. [DOI] [PubMed] [Google Scholar]
- 61.Sarraf CE, Otto WR, Eastwood M. Cell Proliferat. 2011:44–99. doi: 10.1111/j.1365-2184.2010.00740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kock LM, Malda J, Dhert WJA, et al. J Biomech. 2014;47:2122–2129. doi: 10.1016/j.jbiomech.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 63.Lomas C, Tang XD, Chanalaris A, et al. Eur Cells Mater. 2011;22:178–189. doi: 10.22203/ecm.v022a14. [DOI] [PubMed] [Google Scholar]
- 64.Tang GH, Rabie ABM. J Dent Res. 2005;84:166–171. doi: 10.1177/154405910508400211. [DOI] [PubMed] [Google Scholar]
- 65.Mackie EJ, Ahmed YA, Tatarczuch L, Chen K-S, Mirams M. International Journal of Biochemistry and Cell Biology. 2008;40:46–62. doi: 10.1016/j.biocel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 66.Kawaguchi H. Molecules and Cells. 2008;25:1–6. [PubMed] [Google Scholar]
- 67.Fujita T, Izumo N, Fukuyama R, Meguro T, Nakamuta H, Kohno T, Koida M. Biochemical and Biophysical Research Communications. 2001;280:348–352. doi: 10.1006/bbrc.2000.4108. [DOI] [PubMed] [Google Scholar]
- 68.Pockwinse SM, Rajgopal A, Young DW, Mujeeb KA, Nicerson J, Javed A, Redick S, Lian JB, van Wijnen AJ, Stein JL, Stein GS, Doxsey SJ. Journal of Cellular Physiology. 2006;206:354–362. doi: 10.1002/jcp.20469. [DOI] [PubMed] [Google Scholar]
- 69.Nowland NC, Prendergast PJ, Murphy P. PLoS Comput Biol. 2008:4. doi: 10.1371/journal.pcbi.1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong M, Siegrist M, Goodwin K. Bone. 2003;33:685–693. doi: 10.1016/s8756-3282(03)00242-4. [DOI] [PubMed] [Google Scholar]
- 71.Bian L, Zhai DY, Zhang EC, Mauck RL, Burdick JA. Tissue Engineering: Part A. 2012;18:715–724. doi: 10.1089/ten.tea.2011.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Solem RC, Eames BF, Tokita M, et al. Dev Biol. 2011;356:28–39. doi: 10.1016/j.ydbio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang X, Trehan SK, Guan Y, et al. J. Biol Chem. 2014;289:34768–34779. doi: 10.1074/jbc.M114.583104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kanbe K, Yang X, Wei L, et al. J Bone Miner Res. 2007;22:318–328. doi: 10.1359/jbmr.061104. [DOI] [PubMed] [Google Scholar]
- 75.Hellingman CA, Koevoet W, van Osch G. J Tissue Eng Regen Med. 2012;6:e1–e11. doi: 10.1002/term.502. [DOI] [PubMed] [Google Scholar]
- 76.Behonick DJ, Xing Z, Lieu S, et al. PLoS ONE. 2007:2. doi: 10.1371/journal.pone.0001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Phinney DG, Kopen G, Righter W, et al. J Cell Biohcem. 1999;75:424–436. [PubMed] [Google Scholar]
- 78.Solchaga LA, Johnstone B, Yoo JU. Cell Transplant. 1999;8:511–519. doi: 10.1177/096368979900800506. [DOI] [PubMed] [Google Scholar]
- 79.Huang JI, Kazi J, Durbhakula MM, Hering TM, Yoo JU, Jonstone B. Journal of Orthopaedic Research. 2005;23:1383–1389. doi: 10.1016/j.orthres.2005.03.008.1100230621. [DOI] [PubMed] [Google Scholar]
- 80.Schätti O, Grad S, Goldhahn J, Salzmann G, Li Z, Alini M, Stoddart MJ. Eur Cell Mater. 2011;11:214–215. doi: 10.22203/ecm.v022a17. [DOI] [PubMed] [Google Scholar]
- 81.Mueller MB, Tuan RS. Arthritis Rheum. 2008;58:1377–1388. doi: 10.1002/art.23370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nicodemus GD, Skaalure SC, Bryant SJ. Acta Biomaterialia. 2011;7:492–504. doi: 10.1016/j.actbio.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.