SUMMARY
PLK4 is the major kinase driving centriole duplication. Duplication occurs only once per cell cycle, forming one new (or daughter) centriole that is tightly engaged to the preexisting (or mother) centriole. Centriole engagement is known to block the reduplication of mother centrioles, but the molecular identity responsible for the block remains unclear. Here, we show that the centriolar cartwheel, the geometric scaffold for centriole assembly, forms the identity of daughter centrioles essential for the block, ceasing further duplication of the mother centriole to which it is engaged. To ensure a steady block, we found that the cartwheel requires constant maintenance by PLK4 through phosphorylation of the same substrate that drives centriole assembly, revealing a parsimonious control in which “assembly” and “block for new assembly” are linked through the same catalytic reaction to achieve homeostasis. Our results support a recently deduced model that the cartwheel-bound PLK4 directly suppresses centriole reduplication.
Graphical Abstract
INTRODUCTION
Centriole biogenesis in vertebrate cycling cells follows a stereotypical program (Fırat-Karalar and Stearns, 2014; Fu et al., 2015). Cells begin G1 phase with two preexisting centrioles, both of which are capable of recruiting the pericentriolar material (PCM) and thereby functioning as the centrosome or microtubule-organizing center (MTOC). In S phase, the two MTOC-competent centrioles duplicate, each generating one daughter centriole that is incapable of recruiting the PCM (MTOC non-competent) and thus must be tightly attached, or engaged, to its mother for proper segregation in mitosis (Wang et al., 2011). Duplication starts with the formation of the cartwheel, a 9-fold symmetric scaffold forming the base of the daughter centriole. The newborn, MTOC-non-competent daughter centriole is itself unable to duplicate and can further suppress the duplication potential of the mother centriole to which it is tightly engaged (Loncarek et al., 2008; Tsou and Stearns, 2006; Tsou et al., 2009), strictly limiting centriole duplication to once per cell cycle. At the end of cell cycle, daughter centrioles are converted to centrosomes (MTOC competent) (Wang et al., 2011), disengaged from their mothers (Kuriyama and Borisy, 1981), and removed of the cartwheel (Vorobjev and Chentsov, 1980; Vorobjev and Chentsov YuS, 1982), all of which depend on polo-like kinase 1 (Plk1) (Wang et al., 2011), transforming to mother-like centrioles in the following G1 phase. Centriole-to-centrosome conversion and centriole disengagement, respectively, license daughter and mother centrioles for duplication (Fu et al., 2016; Novak et al., 2014; Wang et al., 2011), but the underlying reason for cartwheel removal is not fully clear.
The newborn (MTOC non-competent) centrioles are “sterile,” as they lack the PCM and PCM-associated components such as CEP152 essential for duplication (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Fu et al., 2016; Hatch et al., 2010; Novak et al., 2014; Wang et al., 2011). In contrast, it is not understood how mother (MTOC-competent) centrioles also cease to duplicate when they are engaged to newborn centrioles. It has been shown that cells have no arithmetic ability to count the centriole number (Wong and Stearns, 2003) but can somehow differentiate the property intrinsic to duplicated centrioles from that of unduplicated centrioles, allowing “duplication” and “block for re-duplication” to occur simultaneously in the same cytoplasm (Wong and Stearns, 2003). It is possible that the physical property of the engagement, which holds two centrioles closely in short distances (Shukla et al., 2015), blocks reduplication. Alternatively, a specific chemical property intrinsic to newborn centrioles may serve as the “mark” of duplicated centrioles and locally suppress further duplication of the engaged mother. The molecular identity of such a mark, however, is unknown.
Interestingly, the cartwheel that forms and stabilizes newborn centrioles throughout S/G2 phase (Izquierdo et al., 2014) is removed during mitosis before these centrioles support duplication in the following S phase. The pattern of cartwheel assembly or disassembly correlates nicely with the formation or relief of the duplication block during the cell cycle, raising a testable model that perhaps the cartwheel is the “mark” of the newborn centriole constituting the block. It has been reported that the centriolar protein SAS-6 and its binding partner STIL form the cartwheel (Cottee et al., 2015; Kitagawa et al., 2011; Qiao et al., 2012; van Breugel et al., 2011) in a process catalyzed by the polo-like kinase PLK4 (Arquint et al., 2015; Dzhindzhev et al., 2014; Kratz et al., 2015; Moyer et al., 2015; Ohta et al., 2014) and that loss of the cartwheel during mitosis depends on Plk1 (Wang et al., 2011) and Cdk1 (Arquint and Nigg, 2014). Here, we aim to disrupt cartwheel dynamics through manipulations of PLK4 and STIL, with a goal to determine whether the cartwheel is involved in the block for centriole reduplication.
RESULTS
Persistent PLK4 Activity Stabilizes the Cartwheel and Newborn Centrioles
To address cartwheel dynamics and maintenance after centriole duplication has occurred, a number of retinal pigment epithelial (RPE1) cell lines in which PLK4 can be conditionally removed or inactivated were made (see Table S1 and Experimental Procedures for details). We generated PLK4flox/neoflox cells in which the endogenous, wild-type PLK4 can be conditionally deleted by expression of the Cre recombinase, a derived cell line in which the endogenous Plk4 is deleted, and an analog-sensitive mutant (PLK4as) that can be chemically inactivated by bulky ATP analog is expressed under the tetracycline inducible promoter (PLK4−/−; tet-PLK4as; see Figure S1 and Supplemental Experimental Procedures for details). To ensure proper expression of PLK4as under the endogenous promoter, we also obtained a PLK4as knockin cell line (PLK4asKI; unpublished data), similar to the DLD-1 cell line recently reported (Moyer et al., 2015). Moreover, using these cells, we constructed other derived cell lines in which the endogenous p53 and STIL were inactivated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 gene targeting, generating acentriolar cell lines in which centrioles made of different forms of STIL can be subsequently reconstituted (Table S1) (Izquierdo et al., 2014; Wang et al., 2015). For clarity, the brief name of each cell line is used in the main text, and detailed genetic information is listed in Table S1.
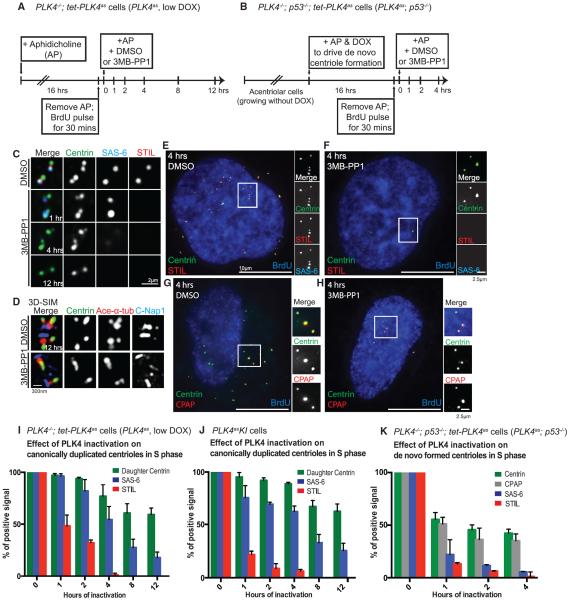
We first examined if cartwheel maintenance requires constant PLK4 activity in PLK4as cells (PLK4−/−; tet-PLK4as) arrested at S phase, when the cartwheel is present in every newborn daughter centriole (Figure 1A). Unlike control cells always carrying two pairs of duplicated centrioles in which the newly formed daughter was labeled with STIL or SAS-6 (Figure 1C, top row), inactivation of PLK4 resulted in serial losses of the cartwheel components from daughter centrioles (Figures 1C and 1I). Specifically, 1 hr after PLK4 inactivation, ~50% of daughter centrioles lost STIL (Figure 1C, second row); by 4 hr, a similar loss of SAS-6 was observed (Figure 1C, third row), consistent with the previous report (Moyer et al., 2015). Moreover, after 12 hr, daughter centrioles started to disappear (Figure 1C, bottom row; Figures 1D and 1I), leaving behind two single mother centrioles each of which was labeled with centrin, C-Nap1, and acetylated alpha tubulin, as revealed by superresolution microscopy (3D-SIM) (Figure 1D). This is consistent with our previous report that the cartwheel is required to stabilize newborn centrioles (Izquierdo et al., 2014). Rapid losses of STIL and SAS-6 from the cartwheel upon PLK4 inactivation were also observed with PLK4as knockin RPE1 cells (PLK4asKI) (Figure 1J). Note that in the absence of the cartwheel, daughter centrioles remained engaged to the mother centriole before they eventually lost stability (Figure 1C, second and third row). The requirement of PLK4 in stabilizing the cartwheel was also seen for de novo centrioles (Figures 1E, 1H, and 1K), which were reconstituted in PLK4as; p53−/− cells (Figure 1B; Table S1). Interestingly, the loss of SAS-6 and centrin from de novo centrioles upon PLK4 inactivation occurred much faster than that from canonically duplicated centrioles (Figures 1I–1K). After 4 hr of PLK4 inactivation, the number of de novo centrioles, as judged by centrin and CPAP staining, dropped from 59 per cell on average before PLK4 inactivation to 11 per cell. These results together suggest that the cartwheel is a dynamic structure requiring constant maintenance by PLK4 and that cartwheel maintenance is essential for the long-term stability of newborn centrioles.
Figure 1. Persistent PLK4 Activity Is Required to Stabilize the Cartwheel and Newborn Centrioles in Either Canonical Duplication or De Novo Synthesis.
(A and B) Schematic of the experimental design to inactivate PLK4 in late S phase cells containing either (A) canonically duplicated or (B) de novo centrioles, using the PLK4as or PLK4as; p53−/− cell line, respectively, as indicated. Doxycycline (DOX) was added as indicated to induce PLK4 expression. De novo centrioles were induced to form in acentriolar PLK4as; p53−/− cells in S phase by adding doxycycline.
(C) The serial loss of cartwheel components from canonically duplicated newborn centrioles at indicated time points after PLK4 inactivation was characterized using the indicated antibodies. Top: centrioles from DMSO control treatment. Bottom: three representative images of PLK4 inactivation by 3MB-PP1 at each time point.
(D) Superresolution 3D-SIM image of acetylated tubulin, centrin, and C-Nap1 in S phase centrioles after 12 hr of control treatment (DMSO, top) or PLK4 inactivation (bottom).
(E–H) De novo formed centrioles in S phase were treated with DMSO (E and G) or inhibited of PLK4 (F and H) and analyzed by immunofluorescence microscopy using the indicated antibodies. Upon PLK4 inactivation, cartwheel components STIL and SAS-6 (F), but not the core centriolar component CPAP (H), were quickly lost. Note also that the number of de novo centrioles (centrin/CPAP foci) was dramatically reduced after 3MB-PP1 treatments (F and H), faster than that for canonical centrioles (see I–K).
(I) Quantification of (C). Error bars represent SD; n > 50, N = 3.
(J) Similar cartwheel loss was also seen and quantified in PLK4asKI cells. Error bars represent SD; n > 50, N = 3
(K) Quantification of (E)–(H). Error bars represent SD; n > 25, N = 3.
Phosphomimetic STIL Supports Centriole Assembly Only When PLK4 Is Present
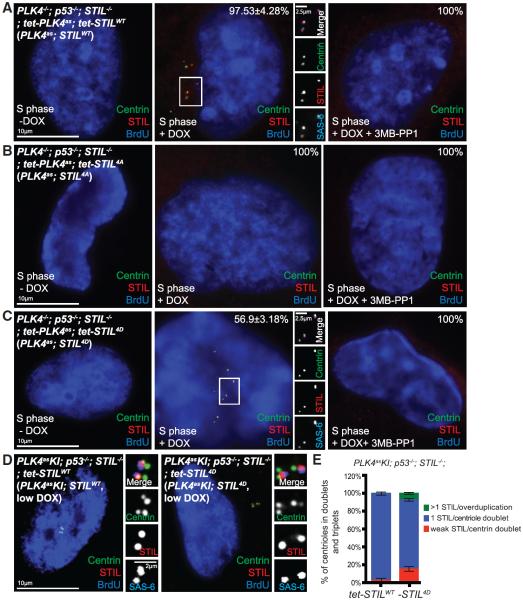
Recent studies showed that phosphorylation of STIL by PLK4 drives cartwheel assembly (Dzhindzhev et al., 2014; Kratz et al., 2015; Moyer et al., 2015; Ohta et al., 2014). While the four critical residues phosphorylated by PLK4 (S1061, S1111, S1116, and T1119) are conserved between human and Drosophila (Dzhindzhev et al., 2014), it is unclear if phosphomimetic STIL can bypass the requirement of PLK4 for centriole assembly. To address it, epitope-tagged full-length STIL carrying wild-type (STILWT), phosphonull (STIL4A) or phosphomimetic (STIL4D) residues were made and expressed in acentriolar, PLK4as; STIL−/− cells (Table S1) in the presence or absence of 3MB-PP1. As expected, in the presence of PLK4 activity (no 3MB-PP1), expression of STILWT or STIL4D, but not STIL4A, successfully rescued de novo centriole formation, as revealed by centrin, STIL, and SAS-6 staining (Figures 2A–2C, left and center). Conversely, when PLK4 was inactivated, none of the constructs, including STIL4D, could drive de novo centriole formation (Figures 2A–2C, right). These results together indicate that while the STIL4D mutant is functional, it cannot bypass the requirement of PLK4 for centriole assembly, suggesting that PLK4 may need to phosphorylate additional sites on STIL or work on other substrates.
Figure 2. Phosphomimetic STIL Supports Centriole Assembly but Cannot Bypass the Requirement of PLK4.
(A–C) Acentriolar PLK4as; STIL−/− cells stably carrying exogenous, tetracycline-inducible STIL constructs were induced to express STILWT (A), STIL4A (B), or STIL4D (C) in the presence or absence of 3MB-PP1, as indicated, and examined for centriole assembly using SAS-6, STIL, and centrin antibodies. Note that STILWT (A) and STIL4D (C) could drive centriole assembly only when PLK4 was active. STIL4A is not functional in either case (B). Error bars represents SD; n > 25, N = 3.
(D and E) The efficiency of centriole duplication during S phase (BrdU) was examined in stable PLK4asKI; STILWT or 4D cell lines in which centriole biogenesis is supported by exogenously expressed STILWT or STIL4D, using indicated antibodies (D). Note that centrioles duplicate normally in most STILWT and STIL4D cells under low DOX, forming centriole doublets each labeled with 1 STIL dot (1 STIL/Doublet). A minor centriole over-duplication is noted in ~7.5% of STIL4D cells (E, green), forming centriole triplets or rosettes each labeled with more than one STIL dots (> 1 STIL/overduplication). Error bars represent SD; n > 50, N = 3.
Phosphomimetic STIL Can Stabilize the Cartwheel in the Absence of PLK4 Activity
Next, we asked whether STIL4D is sufficient to stabilize the cartwheel in the absence of PLK4. To test this, we used the CRISPR/Cas9 method to inactivate STIL and TP53 genes in the PLK4as knockin cell line (PLK4asKI), generating PLK4asKI; p53−/−; STIL−/− cells in which centrioles made of different forms of STIL can be subsequently reconstituted under the endogenous level of PLK4 (Table S1). Tetracycline-inducible constructs of STILWT or STIL4D were reintroduced back to PLK4asKI; p53−/−; STIL−/− cells. Upon induction, centrioles derived from STILWT or STIL4D were formed first through de novo assembly, followed by canonical duplication (data not shown). PLK4asKI; STILWT or PLK4asKI; STIL4D cell lines stably carrying relatively normal numbers of canonical centrioles were obtained through long-term culture in the presence of low doxycycline (Figures 2D and 2E; Table S1). We thus examined the effect of PLK4 inactivation in these two cell lines arrested in S phase.
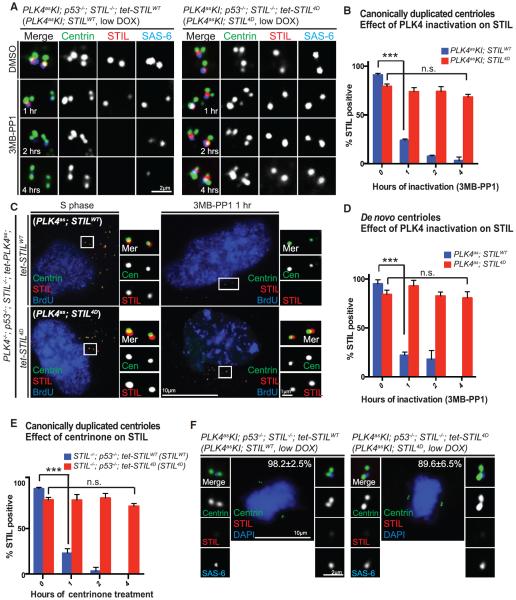
As expected, STILWT-derived centrioles quickly lost cartwheel integrity upon PLK4 inactivation, as indicated by loss of STIL or SAS-6 (Figures 3A and 3B). In contrast, surprisingly, the cartwheel made of STIL4D could stably exist in newborn centrioles in the absence of PLK4 activity, retaining STIL, SAS-6, and other centriolar components (Figures 3A and 3B; data not shown). In addition to canonical centrioles, a similar PLK4-independent stabilization of the cartwheel by STIL4D is also seen in de novo centrioles reconstituted in PLK4as; STILWT or 4D cells (Table S1; Figures 3C and 3D). Moreover, to confirm that our observation is not due to a nonspecific effect of the PLK4 analog-sensitive mutation, we inactivated PLK4 with the recently reported small molecule inhibitor centrinone (Wong et al., 2015) in cell lines expressing wild-type PLK4 but carrying centrioles made of exogenous STIL4D or STILWT as the control (see STILWT or STIL4D cell lines in Table S1). Consistently, centrinone treatment caused serial losses of the cartwheel components from STILWT centrioles, but not from centrioles made of STIL4D (Figure 3E). Interestingly, while the STIL4D-derived cartwheel is stable in the absence of PLK4, it is still efficiently removed from daughter centrioles during mitosis (Figure 3F), indicating that other mechanisms are involved in normal cartwheel removal (Arquint and Nigg, 2014). Together, our data indicate that phosphomimetic STIL can bypass the requirement of PLK4 for cartwheel maintenance, but not for centriole assembly, suggesting that perhaps PLK4 has additional substrates involved in other steps of centriole biogenesis.
Figure 3. Cartwheels Composed of Phosphomimetic STIL Are Stably Maintained at the Daughter Centrioles Independent of PLK4 Activity.
(A) Centrioles in PLK4asKI; STILWT or 4D cell lines, as indicated, were stained for STIL and SAS-6 before and after the inhibition of the endogenous PLK4 with 3MB-PP1.
(B) Quantification of STIL loss seen in (A). Error bars represent SD; n > 25, N = 3. ***p < 0.001; n.s., non-significant (two-tailed t test).
(C) De novo centrioles made of STILWT (top) or STIL4D (bottom) were induced in PLK4as; STILWT or 4D cells and analyzed with the indicated antibodies before (left) and after (right) PLK4 inhibition for 1 hr.
(D) Quantification of STIL loss seen in (C). Centrioles made of the exogenous STILWT or STIL4D were inhibited of PLK4 for indicated amounts of time and analyzed for STIL retention. Error bars represent SD; n > 25, N = 3. ***p < 0.001; n.s., non-significant (two-tailed t test).
(E) Quantification of STIL loss induced by centrinone. Wild-type PLK4 was inhibited with centrinone in STILWT or STIL4D cell lines for indicated amounts of time. Error bars represent SD; n > 25, N = 3. ***p < 0.001; n.s., non-significant (two-tailed t test).
(F) Loss of the cartwheel from STIL4D newborn centrioles occurs normally in mitosis. Stable PLK4asKI; STILWT or PLK4asKI; STIL4D cells in metaphase were examined for cartwheel loss with indicated antibodies. Note that by metaphase, both STILWT and STIL4D were removed from most centrioles.
Loss of the Cartwheel from Daughter Centrioles Induces Reduplication of the Mother Centriole
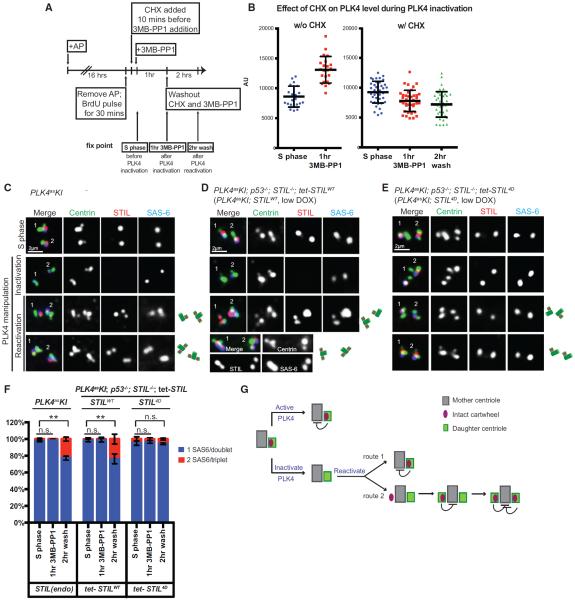
Inactivation of PLK4 induces rapid loss of the cartwheel components from daughter centrioles (Figure 1J). To test if constant maintenance of an intact cartwheel is required for the block of reduplication, PLK4 activity was resumed after cartwheel loss. A caveat of this procedure is that inactivation of PLK4 also results in gradual accumulation of PLK4 at centrosomes (Cunha-Ferreira et al., 2013; Guderian et al., 2010; Klebba et al., 2013), leading to centriole over-duplication upon PLK4 reactivation. To inactivate PLK4 without causing its accumulation, cells were treated with cycloheximide (CHX) to block protein synthesis, shortly before and during PLK4 inhibition (Figure 4A). As expected, in the absence of new translation, PLK4 levels at the centrosome after PLK4 inhibition stayed the same (Figure 4B), unlike cells untreated with CHX. Note that PLK4 and protein synthesis were inhibited for 1 hr only, followed by PLK4 reactivation (Figure 4A), as long-term inhibition of protein synthesis caused non-specific adverse effects on cells (data not shown).
Figure 4. Loss of the Cartwheel from the Daughter Centriole Induces Reduplication of the Engaged Mother Centriole.
(A) Experimental schematic inactivating PLK4 in S phase-arrested cells without causing PLK4 accumulation at the centrosome. Following S phase arrest and BrdU labeling, cycloheximide was added to the cells to block protein synthesis 10 min before PLK4 inactivation. PLK4 was then inactivated with 3MB-PP1 for 1 hr, followed by restoring of PLK4 activity and protein translation for 2 hr. At each fixation time point, PLK4 intensity at centrosomes was quantified using immunofluorescence.
(B) Scatterplot quantification of centrosomal PLK4 intensity at each fix point. Error bars represent SD; n > 20.
(C–E) Representative images of each fix point in PLK4asKI (C, endogenous STIL), PLK4asKI; STILWT (D, exogenous STILWT), and PLK4asKI; STIL4D cells (E, exogenous STIL4D). Top row: fix point 1 (S phase). Second row: fix point 2 (after 1 hr of PLK4 inhibition). Third and fourth row: fix point 3 (after 2 hr of washout; images from two independent cells are shown). Centriole reduplication was indicated by two colocalizing STIL/SAS-6 foci per centin triplet.
(F) Quantification of normally duplicated (blue) and reduplicated (red) centrioles in (C)–(E). Error bars represent SD; n > 50, N = 3. Centriole overproduction was detected in a small population of PLK4asKI; STIL4D cells (exogenously expressed STIL4D) regardless of the treatment (see Figure 2E). **p < 0.01; n.s., non-significant (two-tailed t test).
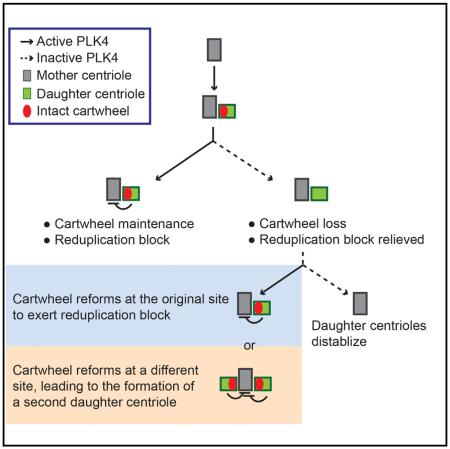
(G) Stochastic nature of centriole reduplication. In normal S phase (top), the cartwheel is actively maintained at the daughter centriole by PLK4 and exerts a block for reduplication. When PLK4 is inactivated (bottom) the cartwheel fails to remain and the mother centriole is relieved from the reduplication block. Once PLK4 activity is resumed, an intact cartwheel can be rebuilt in the preexisting daughter centriole and exert an immediate duplication block (route 1). Alternatively, the cartwheel can randomly form at a new site around the mother, resulting in the formation of an additional daughter centriole and establishing a block for reduplication (route 2). However, the recruitment of the cartwheel component back to the preexisting daughter centriole is unaffected, leading to stabilization of the two daughter centrioles.
Intriguingly, 2 hr after restoring PLK4 activity and protein synthesis, all preexisting daughter centrioles, which were still engaged to the mother centriole (Figure 4C, second row), regained intact cartwheels (100%), as revealed by STIL and SAS-6 staining. Moreover, we consistently saw that ~20% of mother centrioles reduplicated, carrying two engaged daughter centrioles, one preexisting and one newly formed, both equipped with the cartwheel containing STIL and SAS-6 (Figures 4C, third and fourth row, and 4F, left). To further test if the reduplication of mother centrioles depends on loss of STIL from the daughter, we repeated the same assay in cells where daughter centrioles were made of the exogenous STIL4D or STILWT as the control (Figures 4D and 4E). Strikingly, whereas a significant fraction of mother centrioles engaged to STILWT daughter centrioles reduplicated after a transient PLK4 inactivation and reactivation (Figure 4D; Figure 4F, center), very few mother centrioles engaged to STIL4D daughter centrioles reduplicated (Figure 4E; Figure 4F, right). Importantly, the stochastic nature of the reduplication (i.e., not all mother centrioles losing the associated cartwheel could reduplicate) is consistent with the principle of the duplication control (see Figure 4G and Discussion). Collectively, our results suggest that loss of intact cartwheel, or STIL, from daughter centrioles can induce stochastic reduplication of the engaged mother centriole in the same S phase, leading to overproduction of centrioles. Thus, the intact cartwheel carried by the daughter centriole constitutes a critical part of the duplication block, which is specifically tethered to mother centrioles through the engagement to locally suppress reduplication.
DISCUSSION
Phosphorylation of STIL by PLK4 drives cartwheel assembly for centriole duplication (Dzhindzhev et al., 2014; Kratz et al., 2015; Moyer et al., 2015; Ohta et al., 2014), which occurs only once per cell cycle. Our results here showed that cartwheel maintenance at the newborn daughter centriole is essential for the duplication block, requiring persistent PLK4 activity. Inactivation of PLK4 after duplication causes rapid loss of cartwheel components from daughter centrioles. Resuming PLK4 activity, intriguingly, induces cartwheel assembly not only back to the preexisting daughter centriole but also, stochastically, at a new site around the mother, where the second daughter forms. Conversely, the cartwheel made of phosphomimetic STIL can exist stably, continuously exerting duplication block independent of PLK4. Thus, PLK4 not only promotes centriole duplication but also preserves the “mark” of duplicated centrioles, the cartwheel, to prevent reduplication, revealing a counterintuitive control in which “assembly” and “block for new assembly” are both driven by the same machinery to ensure homeostasis.
We reason that the reduplication process induced by loss of cartwheel integrity occurs stochastically through one of the following two routes (Figure 4G): First, the intact cartwheel is rebuilt in the preexisting daughter centriole, where it exerts a block for reduplication, leading to stabilization of only one daughter centriole. In the second route, the cartwheel is randomly formed at a new site around the mother and quickly establishes a block for reduplication, but it has no effect on the recruitment of cartwheel components back to the preexisting daughter, leading to stabilization of two daughter centrioles. It is likely that the first route is preferred, as the preexisting daughter still carries some cartwheel components, such as PLK4 or SAS-6, which may provide high-affinity binding sites for efficient reassembly. The stochastic nature of the process is consistent with the basic principle of the “once-only” control for centriole duplication and, more importantly, is supported by our observations that only a fraction of mother centrioles losing the cartwheel from the associated daughter could reduplicate to form the second daughter.
How does the cartwheel locally block the duplication potential of the mother centrioles to which it is engaged? A recent model proposes that STIL-bound PLK4 at the cartwheel may specifically target the PCM-bound (or CEP152-bound) PLK4 around the mother centriole for degradation and thereby suppress centriole reduplication (Arquint et al., 2015; Ohta et al., 2014). This model was deduced from observations that PLK4 can induce self-destruction through trans-autophosphorylation (Cunha-Ferreira et al., 2013; Guderian et al., 2010; Holland et al., 2010; Klebba et al., 2013), that the kinase activity of PLK4 is greatly enhanced by binding to STIL (Moyer et al., 2015), and that the binding of PLK4 to STIL prevents PLK4 from self-destruction (Arquint et al., 2015; Ohta et al., 2014). Consistent with this model, overexpression of PLK4 or STIL should and is indeed known to alleviate the duplication block, allowing multiple daughters to be born by the same mother. However, in this case, the model would also predict that when more daughter centrioles are born, more cartwheel-associated inhibitory activities would arise to counteract PLK4 at the mother centriole, which in turn would require even higher levels of PLK4 to drive the formation of more daughter centrioles, explaining why a higher level of PLK4, rather than a longer period of time in S phase, is the key to amplify daughter centrioles. Our studies here provide us an angle at which the model can be tested, and the results are largely consistent. More efforts are required to engineer specific reagents and tools to fill in the remaining gap.
EXPERIMENTAL PROCEDURES
Details of experimental procedures regarding cell culture, drug treatment, antibodies, microscopy, immunoblots, and statistical analysis are provided in Supplemental Experimental Procedures.
Transgenic Cell Lines and Plasmid Constructs
To generate PLK4 conditional knockout cell lines, exon 3 and 4 of PLK4 were targeted by adeno-associated virus (AAV)-mediated homologous recombination as described previously (Berdougo et al., 2009; Tsou et al., 2009). A PLK4as knockin cell line (PLK4asKI) in which centrosomes biogenesis are supported by PLK4as expressed under the endogenous promoter was obtained from the Andrew J. Holland lab (unpublished data), similar to the PLK4as DLD-1 cell line the Andrew J. Holland lab recently reported (Moyer et al., 2015). CRISPR/Cas9-mediated gene targeting was used to inactivate p53, STIL, or both in various cell lines (see Table S1), as described previously (Izquierdo et al., 2014). For further details, see Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Persistent PLK4 is required to stabilize the cartwheel and newborn centrioles
Phosphomimetic STIL requires active PLK4 to support centriole duplication
Phosphomimetic STIL can bypass the requirement of PLK4 for cartwheel maintenance
Loss of intact cartwheel induces stochastic reduplication of mother centrioles
ACKNOWLEDGMENTS
We thank A. Shiau and K. Oegema at Ludwig Institute for Cancer Research for sharing the PLK4 inhibitor centrinone. We also thank A. Holland at Johns Hopkins University for providing PLK4 antibody and the RPE1 PLK4asKI cell line. We are grateful to A. North (Rockefeller University) for the use of the OMX superresolution microscope, supported by the award S10RR031855 from the National Center for Research Resources. P.V.J. was supported by NIH grant R01GM094972. This work was supported by the National Institute of Health grant GM088253, American Cancer Society grant RSG-14-153-01-CCG, and a Geoffrey Beene Cancer Research Center grant (to M.-F.B.T.).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes Supplemental Experimental Procedures, one figure, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.06.069.
AUTHOR CONTRIBUTIONS M.K. and M.-F.B.T. designed experiments. M.K. performed most of the experiments and analyzed the data. B.P.O., R.K.S., and R.C.H. helped perform the experiments. P.V.J. designed the strategy to generate PLK4 conditional knockout allele. M.K., B.P.O., and M.-F.B.T. wrote the manuscript.
REFERENCES
- Arquint C, Nigg EA. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr. Biol. 2014;24:351–360. doi: 10.1016/j.cub.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Arquint C, Gabryjonczyk AM, Imseng S, Böhm R, Sauer E, Hiller S, Nigg EA, Maier T. STIL binding to Polo-box 3 of PLK4 regulates centriole duplication. eLife. 2015;4:e07888. doi: 10.7554/eLife.07888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdougo E, Terret ME, Jallepalli PV. Functional dissection of mitotic regulators through gene targeting in human somatic cells. Methods Mol. Biol. 2009;545:21–37. doi: 10.1007/978-1-60327-993-2_2. [DOI] [PubMed] [Google Scholar]
- Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 2010;191:731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee MA, Muschalik N, Johnson S, Leveson J, Raff JW, Lea SM. The homo-oligomerisation of both Sas-6 and Ana2 is required for efficient centriole assembly in flies. eLife. 2015;4:e07236. doi: 10.7554/eLife.07236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I, Bento I, Pimenta-Marques A, Jana SC, Lince-Faria M, Duarte P, Borrego-Pinto J, Gilberto S, Amado T, Brito D, et al. Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr. Biol. 2013;23:2245–2254. doi: 10.1016/j.cub.2013.09.037. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, Riparbelli M, Rodrigues-Martins A, Bettencourt-Dias M, Callaini G, Glover DM. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467:714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev NS, Tzolovsky G, Lipinszki Z, Schneider S, Lattao R, Fu J, Debski J, Dadlez M, Glover DM. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 2014;24:2526–2532. doi: 10.1016/j.cub.2014.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fırat-Karalar EN, Stearns T. The centriole duplication cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130460. doi: 10.1098/rstb.2013.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Hagan IM, Glover DM. The centrosome and its duplication cycle. Cold Spring Harb. Perspect. Biol. 2015;7:a015800. doi: 10.1101/cshperspect.a015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Lipinszki Z, Rangone H, Min M, Mykura C, Chao-Chu J, Schneider S, Dzhindzhev NS, Gottardo M, Riparbelli MG, et al. Conserved molecular interactions in centriole-to-centrosome conversion. Nat. Cell Biol. 2016;18:87–99. doi: 10.1038/ncb3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guderian G, Westendorf J, Uldschmid A, Nigg EA. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J. Cell Sci. 2010;123:2163–2169. doi: 10.1242/jcs.068502. [DOI] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 2010;188:191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo D, Wang WJ, Uryu K, Tsou MF. Stabilization of cartwheel-less centrioles for duplication requires CEP295-mediated centriole-to-centrosome conversion. Cell Rep. 2014;8:957–965. doi: 10.1016/j.celrep.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Flückiger I, Gönczy P, Steinmetz MO. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba JE, Buster DW, Nguyen AL, Swatkoski S, Gucek M, Rusan NM, Rogers GC. Polo-like kinase 4 autodestructs by generating its Slimb-binding phosphodegron. Curr. Biol. 2013;23:2255–2261. doi: 10.1016/j.cub.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz AS, Bärenz F, Richter KT, Hoffmann I. Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol. Open. 2015;4:370–377. doi: 10.1242/bio.201411023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Borisy GG. Microtubule-nucleating activity of centrosomes in Chinese hamster ovary cells is independent of the centriole cycle but coupled to the mitotic cycle. J. Cell Biol. 1981;91:822–826. doi: 10.1083/jcb.91.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat. Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer TC, Clutario KM, Lambrus BG, Daggubati V, Holland AJ. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 2015;209:863–878. doi: 10.1083/jcb.201502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak ZA, Conduit PT, Wainman A, Raff JW. Asterless licenses daughter centrioles to duplicate for the first time in Drosophila embryos. Curr. Biol. 2014;24:1276–1282. doi: 10.1016/j.cub.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ashikawa T, Nozaki Y, Kozuka-Hata H, Goto H, Inagaki M, Oyama M, Kitagawa D. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 2014;5:5267. doi: 10.1038/ncomms6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao R, Cabral G, Lettman MM, Dammermann A, Dong G. SAS-6 coiled-coil structure and interaction with SAS-5 suggest a regulatory mechanism in C. elegans centriole assembly. EMBO J. 2012;31:4334–4347. doi: 10.1038/emboj.2012.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Kong D, Sharma M, Magidson V, Loncarek J. Plk1 relieves centriole block to reduplication by promoting daughter centriole maturation. Nat. Commun. 2015;6:8077. doi: 10.1038/ncomms9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell. 2009;17:344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov YS. The ultrastructure of centriole in mammalian tissue culture cells. Cell Biol. Int. Rep. 1980;4:1037–1044. doi: 10.1016/0309-1651(80)90177-0. [DOI] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov YuS. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, Soni RK, Uryu K, Tsou MF. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 2011;193:727–739. doi: 10.1083/jcb.201101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, Acehan D, Kao CH, Jane WN, Uryu K, Tsou MF. De novo centriole formation in human cells is error-prone and does not require SAS-6 self-assembly. eLife. 2015;4:10586. doi: 10.7554/eLife.10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat. Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
- Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, Seo CP, Hsia JE, Kim SK, Mitchell JW, et al. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 2015;348:1155–1160. doi: 10.1126/science.aaa5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.