Abstract
Dynamic Combinatorial Chemistry (DCC) has proven to be a reliable method for identifying hit compounds for target nucleic acid (DNA and RNA) sequences. Typically, these hit compounds are subjected to a lengthy process of optimization via traditional medicinal chemistry. Here, we examine the potential of DCC to also generate and test variations on a hit compound as a method for probing the binding site of an RNA-targeted compound. Specifically, we demonstrate that addition of linker dithiols to a disulfide library containing a known binder to the HIV-1 frameshift-stimulatory RNA (a critical regulator of the HIV life cycle) can yield a mixture of new bridged structures incorporating the dithiol, depending on dithiol structure. Equilibration of this library with the HIV FSS RNA resulted in selection of the original disulfide in preference to bridged structures, suggesting incorporation of the bridge is not compatible with this particular binding site. Application of this strategy to other RNA targets should allow for rapidly profiling the affinity of modified compounds.
Keywords: RNA, Dynamic Combinatorial Chemistry, peptides, HIV
1. Introduction
Nature employs a process of mutation and selection to evolve chemical structures with specific properties. Dynamic Combinatorial Chemistry (DCC) was originally introduced as a method to mimic evolution in the identification of small molecules with novel properties by allowing binding-induced selection (or, in other cases, self-selection) to act on an equilibrating system of structures.1, 2 This field has grown to encompass a broad range of studies known as “dynamic covalent chemistry”,3 “systems chemistry”,4 or “constitutional dynamic chemistry”.5 Systems employing dynamic exchange of covalent bonds have enabled the discovery of polymers with novel materials properties,6, 7, 8 new molecular architectures,9 highly selective host-guest systems, and new ligands for binding macromolecules.
We have been engaged, for several years, in the use of dynamic molecular systems (resin-bound dynamic combinatorial libraries, or RBDCLs) for identifying sequence-selective RNA-binding compounds from interconverting mixtures of compounds inspired by known nucleic acid-binding natural products. Combined with follow-on medicinal chemistry efforts, this work has led to structures able to bind CUG repeat RNA, displace the CUG-binding splicing factor MBNL1, and thereby enhance MBNL1-dependent splicing in vivo.10, 11 A similar process resulted in compounds able to bind an RNA critical to the life cycle of HIV, yielding a new class of antiviral agents.12, 13, 14
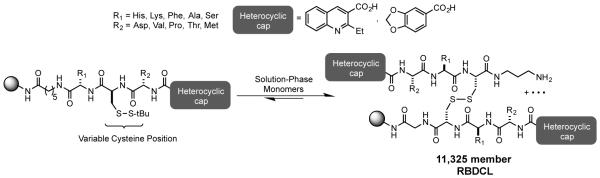
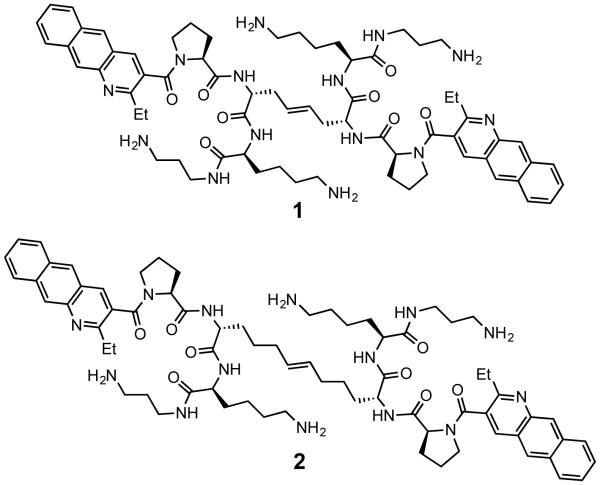
Our original RNA-targeted RBDCL design,15 loosely inspired by the structure of echinomycin and other nucleic acid binding, bisintercalating peptide antibiotics, employs cysteine-mediated disulfide exchange as its “mutation” mechanism (Figure 1). Once hit compounds have been identified from the library, we have focused on the synthesis of structures in which the disulfide is replaced by a hydrocarbon bioisostere (olefin or alkyl chain) in order to produce compounds stable to the reducing environment within a cell. While successful, this strategy requires somewhat laborious synthesis, limiting the throughput of new compounds that can be made, specifically testing changes to the geometry of the linker on RNA-binding ability. The importance of varying this parameter was highlighted in our work on CUG RNA binders, in which we observed that extending the length of the carbon linker (from 4 carbons as in compound 1, to 8 carbons as in compound 2) dramatically enhanced both the affinity and selectivity of the compound for its intended target.
Figure 1.
RNA-targeted Resin-Bound Dynamic Combinatorial Library (RBDCL)
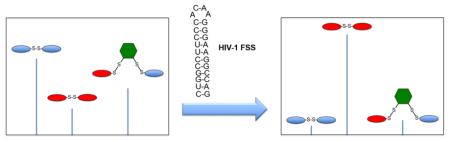
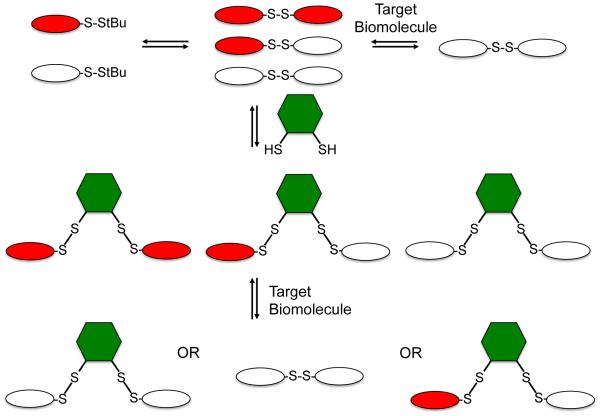
This led us to consider the question: is it possible to incorporate variation of linker identity and geometry into a dynamic combinatorial library (DCL)? Seen in view of systems chemistry, a broader question is: given a dynamic system with a known outcome (in this case, a molecule able to bind a specific RNA sequence), what happens when a new “evolutionary opportunity” is provided? We set out to answer that question in the context of RNA recognition by introducing a new, geometrically distinct set of exchangers. Conceptually, this is shown in Figure 2. By introducing a dithiol into an existing DCL, a new library is created in which structures incorporating the linker coexist (absent self-selection against such structures) with the original disulfides. Introduction of a target receptor can now yield one of three outcomes: the original molecule plus the new linker, a new molecule incorporating the linker, or, if none of the molecules incorporating the linker binds with higher affinity than the originally selected compound, the original molecule will be amplified. A variety of dithiols have been studied extensively in the context of self-selecting DCLs,16, 17, 18 thus suggesting that one could expand the geometric opportunities available to our RNA-targeted DCL by incorporating one or more dithiol linkers.
Figure 2.
Linker DCC
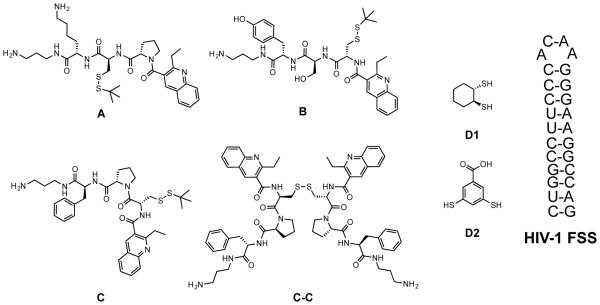
To test this concept, we sought to determine whether adding dithiols to a simplified subset of our RNA-targeted DCL would generate new, bridged structures, and subsequently to determine their fate when the new bridge-incorporated DCL was exposed to a known RNA target (i.e., if the bridged structure would be amplified, indicating a preference for the new geometry, or if the population would shift in favor of a dimer selected absent the dithiol bridge). To that end, we examined simple libraries consisting of three monomers from our initial RBDCL (A, B, and C, Figure 3), and one of two dithiol linkers. Library member C was chosen as a disulfide dimer formed from it (C-C) was strongly selected by an RNA stemloop known as the HIV-1 frameshift-stimulatory stemloop, or FSS (Figure 3). This sequence is part of a long mRNA that encodes two polyproteins: Gag, which contains the structural proteins used to make the HIV viral capsid, and Pol, which contains several critical viral enzymes including HIV-protease. Pol is only produced as a fusion with Gag (Gag-Pol), and its production is controlled by a tightly regulated −1 ribosomal frameshift event.19 Thus, during translation of the gag-pol mRNA, the ribosome reaches a stop codon after producing Gag, and terminates. This occurs with a frequency of 90 – 95%. However, 5 – 10% of the time, the ribosome pauses when it encounters the FSS RNA, allowing it to shift backwards 1 nucleotide and then bypass the gag stop codon to allow production of Gag-Pol. Previous researchers had demonstrated that altering the ratio of Gag:Gag-Pol in either direction resulted in the production of virus with significantly reduced infectivity;20 our work confirmed that compounds targeting the FSS RNA could alter Gag:Gag-Pol ratios and inhibit the infectivity of pseudotyped HIV.13 Monomers A and B were incorporated to provide a modicum of structural diversity, while maintaining aspects of the structure of C. For example, A retains the proline residue while changing the position of the cysteine and substituting lysine for phenylalanine; B maintains the position of the cysteine, but replaces the proline with a serine residue. A subtle additional modification in B is the substitution of tyrosine for phenylalanine. With regard to the choice of linker dithiols, dithiol linker D2 has been studied by Sanders and Otto,21 while D1 has, to our knowledge, not previously featured in a dynamic library or systems chemistry experiment.
Figure 3.
Library structures and target RNA
2. Materials and Methods
2.1. Reagents
Peptide synthesis reagents were obtained from P3 BioSystems (Louisville, KY), and Wang resin (0.8 mmol/g, 100-200 mesh) was obtained from Advanced ChemTech (Louisville, KY). Other reagents were purchased from Sigma-Aldrich (St. Louis, MO), Alfa Aesar (Ward Hill, MA), TCI America (Portland, OR), and Thermo Fisher Scientific (Waltham, MA). All reagents were used without additional purification unless specifically stated. All water used in the production of buffer solutions and in HPLC was deionized and then glass-distilled (abbreviated here as ddH2O). Frameshift stimulatory signal (FSS) RNA was obtained from Integrated DNA Technologies (Coralville, IA). The RNA was purified by the supplier through RNase free HPLC. The sequence is as follows: 5’-rGrGrC rCrUrU rCrCrC rArCrA rArGrG rGrArA rGrGrC rC −3’.
2.2. Synthesis of Monomers
Compounds A, B, and C were synthesized on Wang resin using standard Fmoc solid phase peptide synthesis (SPPS), by analogy to our previous work. Each compound was cleaved from the resin and simultaneously deprotected using 95% trifluoroacetic acid (TFA) with 5% triethylsilane. The identity of each compound was verified by MALDI-TOF mass spectrometry. The compounds were determined by analytical HPLC to be >85% pure, and were used in the libraries as such in early experiments (DCLs 1-3). The concentrations of these compounds in stock solutions were determined by UV-Vis spectroscopy, based on the known extinction coefficient of the quinoline moiety. Monomers A, B, and C were later purified using preparative reverse phase HPLC with a C18 column (Waters, XBridge Prep C18 5 m OBD 19×250mm). A gradient method was utilized from 3 to 95% acetonitrile/0.1% TFA in ddH2O/0.1% TFA. After preparative HPLC, compounds were confirmed to be >95% pure by analytical HPLC. The concentration of these compounds in their stock solutions was determined by UV-Vis spectroscopy as above. These stock solutions were utilized in DCL 4 and in control experiments.
2.3. Synthesis of Dithiols
Dithiol D1 (trans-1,2-cyclohexanedithiol) was synthesized by literature methods reported by Whitesides22. Methyl 3,5-dihydroxybenzoate was synthesized as described by Bojja23 and colleagues. Dithiol D2 (3,5-dimercaptobenzoic acid) was synthesized by the method of Otto and Sanders,21 beginning with the methyl 3,5-dihydroxybenzoate described above.
2.4. General Library Setup
All library experiments were conducted in polypropylene microcentrifuge tubes. DCLs 1, 2, and 3 were produced by dissolving compounds A, B, and C in HBS buffer (20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 0.01% Tween-20, pH 7.4), then mercaptopropanol, a particular dithiol (D1 or D2, where indicated), and FSS RNA were added as described in each experiment. DCL 4 was produced in a similar manner to other libraries, but all components were dissolved in PBS buffer (10 mM phosphate, 150 mM NaCl, 5 mM MgCl2, pH 7.4). Mercaptopropanol was present in indicated experiments at 1 mM or varied as indicated; dithiol concentration was varied as discussed below. Frameshift stimulatory signal RNA is present at 20 μM in some experiments as noted.
2.5. Library Analysis
MALDI-TOF mass spectrometry was performed using a Bruker AutoFlex III instrument. For each experiment, 1 μL of the library solution was combined with 1 μL of matrix solution (50 mM α-cyano-4-hydroxycinnamic acid in 1:1 ddH2O/acetonitrile 0.1% TFA) and thoroughly mixed by pipetting before being spotted onto a Bruker MSP 96 ground steel target. Analytical HPLC was performed on a Shimadzu LC-2010A equipped with a reverse phase Shimadzu Shim-pack CLC-ODS M column (C18, 5 μm).
2.6. Ratiomeric Analysis of MALDI-TOF Spectra
The following process was performed manually or using an Excel macro to analyze MALDI-TOF spectra of the DCLs. Each species observed by mass spectrometry was assumed to have a charge of +1 for this analysis. Peaks from the spectra were compared with a mass table of predicted structures (Monomers M+, reduced monomers M-SH+, dimers M-M+, bridged structures M-D-M+, and associated sodium peaks). A ratio was calculated for the intensity of each M-M+ peak relative to the intensity of every other M-M+ peak present. The same was done to compare the sodium adduct peaks, M-MNa+, to each other. The peaks of monomers bearing a free thiol, M-SH+, were also compared to the M-M+ peaks, and again the sodium peaks were separately compared. The peaks resulting from bridged structures, M-D-M+ and M-D-MNa+, were compared to M-M+ and M-MNa+ peaks respectively. These ratios were then compared between spectra. If the comparison of two species was increased in a DCL containing a template, by more than 3 fold, the numerator species is said to be amplified by the template.
3. Results and Discussion
3.1. Library Equilibration
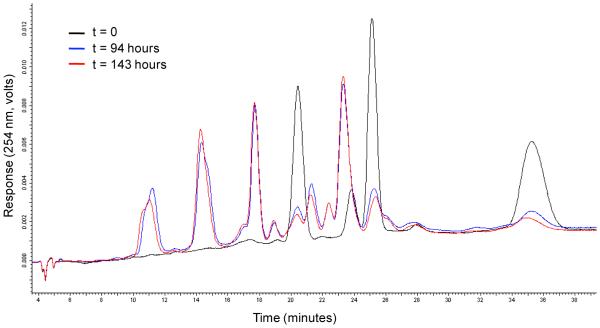
We began our study by examining the equilibration time necessary for DCLs incorporating dithiol linkers. In a representative experiment, monomers A, B, and C (40 μM) were equilibrated with dithiol D1 (2.5 mM) and mercaptopropanol (1 mM; included to facilitate exchange by analogy to the work of Balasubramanian24 and others) in buffer (20 mM HEPES, 5 mM MgCl2, 150 mM NaCl, 0.01% Tween 20, pH 7.4) in a microcentrifuge tube at room temperature. Aliquots of the solution were removed at intervals, acidified to halt disulfide exchange, and analyzed by HPLC. As shown in Figure 4, disulfide exchange enabled conversion of monomers to lower-polarity molecules (dimers and presumably higher-order structures). This process had essentially reached equilibrium at 94 hours (approximately 4 days), consistent with our prior results. Changing conditions (monomer, dithiol, and mercaptopropanol concentration) did not significantly alter the observed equilibration time; thus, this was employed as the minimum equilibration time for all further experiments.
Figure 4.
Library equilibration is complete after 94 hours. HPLC of a DCL consisting of A, B, C, and D1, with mercaptopropanol as an exchange catalyst.
MALDI-MS was used in all subsequent experiments both to provide evidence for the existence of particular structures in a library, and to indicate via ratiomeric analysis which structures were favored as a function of changes in concentration, linker dithiol identity, and the presence or absence of RNA. Control experiments were carried out to verify that observed changes in mass spectra did not simply result from solution constituent – mediated changes in our ability to measure specific molecular ions.
3.2. Optimization of Library Component Concentrations
Initial experiments with linker dithiol 2 (D2) produced little observable bridged products. In order to enhance the production of bridged structures, libraries containing only monomers and a range of concentrations of a single dithiol were assembled. All of the vials used to optimize the dithiol concentrations contained 20 μM of each monomer A, B, and C. Dithiol 2, D2, was tested at concentrations of 50, 100, 250 and 500 μM. After a minimum 6 days of equilibration, samples of each library were analyzed by MALDI-MS. Vials containing the highest intensity of bridged structures relative to dimers were considered to have the optimum concentration for the specific dithiol. The optimum concentration of D2 was found to be 250 μM.
Experiments were also performed to determine the effect of varying concentrations of mercaptopropanol on the libraries, in the presence of a dithiol. A constant concentration of 4 mM D2 was utilized while the concentration of mercapropropanol was varied from 1 to 4 mM. By increasing the concentration of mercaptopropanol, more monomer species (M-SH, M-mercaptopropanol) are formed with a reduction in the presence of dimeric species. Therefore, the concentration of mercaptopropanol was limited to 1 mM in subsequent experiments. As discussed below, complete elimination of mercaptopropanol from the library allowed for some bridged species to persist in the presence of RNA.
3.3. Library selections in the presence of RNA
Previous efforts within our group have shown that a dynamic, exchanging system containing monomers such as A, B, and C undergo selection in the presence of RNA.12 This has led to amplification of dimers with an affinity for the templating RNA. Libraries containing bridged structures contain greater geometric diversity, and we set out to determine what species of these libraries would be amplified by the presence of a biologically relevant RNA sequence.
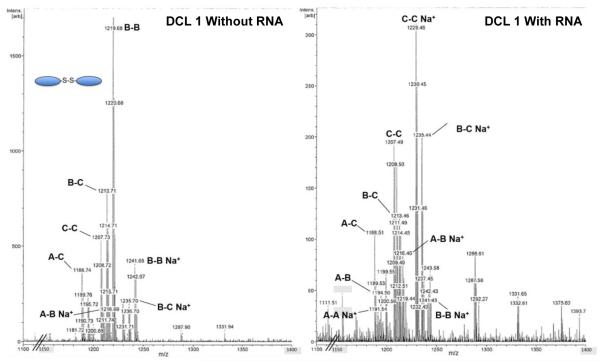
As discussed above, dimer C-C was selected from an 11,325-member RBDCL by the FSS RNA stemloop. To confirm that this also occurred with the simple library in solution prior to introducing bridging dithiols, we assembled a library (DCL 1) consisting of 1 mM mercaptopropanol, and 40 μM of each monomer A-C. A second identical library was set up, but also contained 20 μM FSS RNA. After 5 days, the library had reached equilibrium and was analyzed by MALDI-MS. These spectra revealed a significant enhancement of molecular ion and Na+ peaks corresponding to C-C relative to other species (Figure 5 and Table 1). Heterodimers incorporating monomer C were also enhanced. This confirmed that selection of C-C by RNA occurred in solution as well as on solid phase, as previously observed. To verify that the presence of RNA did not interfere with observation of molecular and sodium ion peaks, we also carried out two DCL experiments consisting of just 20 μM monomer C, 1 mM mercaptopropanol, and either 20 or 35 μM FSS RNA. We observed no statistically significant differences in the ion peak intensities from mass spectra obtained following equilibration, suggesting that the RNA did not interfere with our ability to observe library species.
Figure 5.
MS spectra from DCL1 equilibrated with (right) and without (left) FSS RNA.
Table 1.
Ratiomeric analysis of DCL 1.
| Species | Amplification ratio |
|---|---|
| C-C:B-B+Na+ | 15.5 |
| A-C:B-B+Na+ | 11.4 |
| B-C:B-B+Na+ | 10.5 |
| A-B:B-B+Na+ | 5.19 |
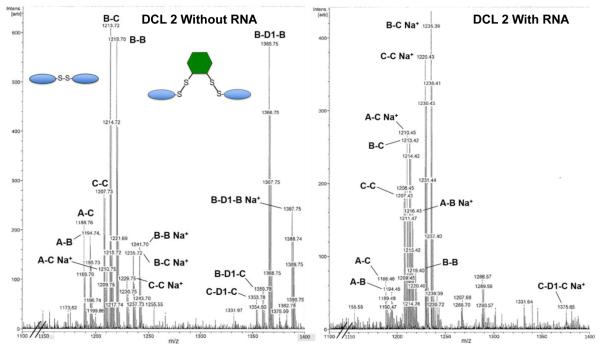
After successfully replicating previous results in a solution-phase DCL, we began to test the effect of incorporating dithiols into library selections with RNA. DCL 2 was designed to be similar to DCL 1, but also included 2.5 mM D1. Analysis of an equilibrated library aliquot lacking RNA by MALDI-MS (Figure 6, left) revealed the presence of three of the six possible bridged structures (X-D1-Y), as well as five of the expected six homo- and heterodimers (X-Y). Monomers derivatized with mercaptopropanol or D1 were also observed. The A-A homodimer was not observed, presumably due to self-selection for other compounds in the library. Mass spectra obtained from this library equilibrated with FSS RNA (Figure 6, right) suggest that introduction of the RNA drives the library towards amplification of the originally identified compound C-C. An unanticipated result was that even bridged structures incorporating other monomers (i.e. B-D1-B, etc.) were dramatically disfavored in the presence of RNA. We attribute this to incorporation of monomers from these bridged structures into C-X .
Figure 6.
Mass spectral analysis of DCL 2. Libraries equilibrated with (right) and without (left) FSS RNA reveal selection for the C-C dimer, and against bridged structures.
As discussed above, mercaptopropanol was incorporated into the DCLs as an exchange catalyst. Given the large excess of ditihiol D1, it seemed likely that mercaptopropanol was not actually needed. Therefore, we tested this by constructing DCL 3 to contain 2.5 mM D1 and 40 μM of each monomer A, B, and C, with no mercaptopropanol present in the solution. After equilibration, analysis of the mass spectrometry results from this library again showed that dimer C-C had the highest amount of amplification with the RNA template. Absent mercaptopropanol, some bridged dimers persist (A-D1-C and C-D1-C) but are not amplified to the extent that C-C is (C-C:C-D1-C amplification ratio is 4.1:1).
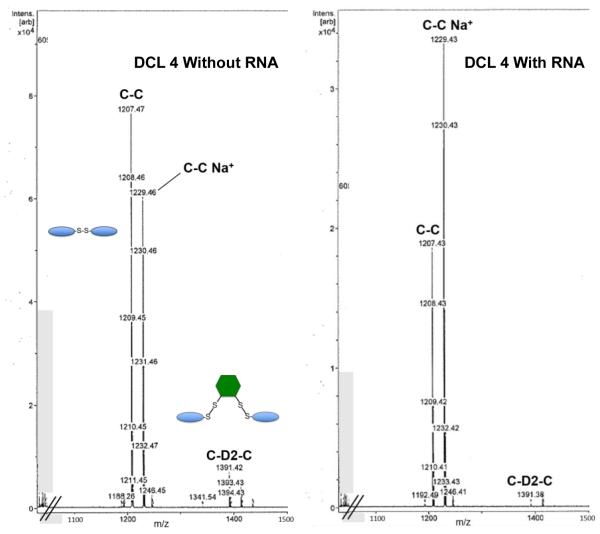
Next, we tested whether altering the linker dithiol would change the outcome of the experiment. DCL 4 consisted of 250 μM D2 and 70 μM monomer C only, in order to simplify analysis and explicitly test formation of structures incorporating only this monomer. Based on the results for DCL3, mercaptopropanol was not used. Once again the library was equilibrated with and without added HIV FSS RNA. Mass spectral analysis of the equilibrated libraries showed that although C-D2-C was formed in the mixture absent RNA, dimer C-C was significantly amplified relative to the bridged structure C-D2-C in the presence of HIV FSS RNA (Figure 7).
Figure 7.
Results for DCL4 with (right) and without (left) HIV FSS RNA. A small amount of C-D2-C remains in the presence of RNA, but is diminished relative to C-C.
4. Conclusions
We have demonstrated that the introduction of dithiols D1 and D2 to a simple disulfide-based, RNA-targeted DCL allows the production of bridged structures incorporating the new linker. When allowed to equilibrate in the presence of HIV FSS RNA, the equilibrium strongly shifts in favor of a previously selected dimer ligand C-C, to the detriment of bridged structures present in the mixture at the outset of the experiment. In addition to recapitulating the result of the original resin-bound library selection in the solution phase, these experiments confirm that dynamic library selections may be used to examine the geometric constraints of ligand binding to an RNA target. As such, related efforts may enable structural hypotheses to be generated and tested more quickly and simply than is possible with traditional medicinal chemistry. Given the simplicity of DCL experiments, a variety of linkers or other modifying groups may be incorporated into hit structures and tested rapidly, simply by examination of mass spectra following library equilibration. While conversion of the resulting structures to non-reducible analogs is still a necessity prior to biological testing in the context of disulfides, we have shown that this is a relatively straightforward exercise once an optimum ligand is identified.
Acknowledgement
This research was supported by the National Institutes of Health through research grant GM100788.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Corbett PT, Leclaire J, Vial L, West KR, Wietor J-L, Sanders JKM, Otto S. Chem. Rev. 2006;106:3652. doi: 10.1021/cr020452p. [DOI] [PubMed] [Google Scholar]
- 2.Miller BL. Dynamic Combinatorial Chemistry. John Wiley & Sons; New York: 2009. [Google Scholar]
- 3.Black SP, Sanders JKM, Stefankiewicz AR. Chem. Soc. Rev. 2014;43:1861. doi: 10.1039/c3cs60326a. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Nowak P, Otto S. J. Am. Chem. Soc. 2013;135:9222. doi: 10.1021/ja402586c. [DOI] [PubMed] [Google Scholar]
- 5.Lehn JM. Chem. Soc. Rev. 2007;36:151. doi: 10.1039/b616752g. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez-Sanchez A, Fulton DA, Pomposo JA. Chem. Commun. 2014;50:1871. doi: 10.1039/c3cc48733d. [DOI] [PubMed] [Google Scholar]
- 7.Moulin E, Cormos G, Giuseppone N. Chem. Soc. Rev. 2012;41:1031. doi: 10.1039/c1cs15185a. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Carnall JMA, Stuart MCA, Otto S. Angew. Chem. Int. Ed. 2011;50:8384. doi: 10.1002/anie.201103297. [DOI] [PubMed] [Google Scholar]
- 9.Wilson A, Gasparini G, Matile S. Chem. Soc. Rev. 2014;43:1948. doi: 10.1039/c3cs60342c. [DOI] [PubMed] [Google Scholar]
- 10.Gareiss PC, Sobczak K, McNaughton BR, Palde PB, Thornton CA, Miller BL. J. Am. Chem. Soc. 2008;130:16254. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ofori LO, Hoskins J, Nakamori M, Thornton CA, Miller BL. Nucl. Acids Res. 2012;40:6380. doi: 10.1093/nar/gks298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNaughton BR, Gareiss PC, Miller BL. J. Am. Chem. Soc. 2007;129:11306. doi: 10.1021/ja072114h. [DOI] [PubMed] [Google Scholar]
- 13.Ofori LO, Hilimire TA, Bennett RP, Brown NW, Smith HC, Miller BL. J. Med. Chem. 2014;57:723. doi: 10.1021/jm401438g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilimire TA, Bennett RP, Stewart RA, Garcia-Miranda P, Blume A, Becker J, Sherer N, Helms ED, Butcher SE, Smith HC, Miller BL. ACS Chem. Biol. 2016;11:88. doi: 10.1021/acschembio.5b00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNaughton BR, Miller BL. Org. Lett. 2006;8:1803. doi: 10.1021/ol060330+. [DOI] [PubMed] [Google Scholar]
- 16.Solá J, Lafuente M, Atcher J, Alfonso I. Chem. Commun. 2014;50:4564. doi: 10.1039/c4cc00245h. [DOI] [PubMed] [Google Scholar]
- 17.Hamieh S, Saggiomo V, Nowak P, Mattia E, Ludlow RF, Otto S. Angew. Chem. Int. Ed. 2013;52:12368. doi: 10.1002/anie.201305744. [DOI] [PubMed] [Google Scholar]
- 18.Postma TM, Galloway WRJD, Cougnon FBL, Pantoş GD, Stokes JE, Spring DR. Synlett. 2013;24:765. [Google Scholar]
- 19.Brakier-Gingras L, Charbonneau J, Miller BL, Rahman A-U. Frontiers in Clinical Drug Research: HIV. Vol. 1. Bentham Science Publishers Ltd.; Sharjah, U.A.E.: 2015. pp. 67–82. [Google Scholar]
- 20.Dulude D, Berchiche YA, Gendron K, Brakier-Gingras L, Heveker N. Vaccine. 2006;345:127. doi: 10.1016/j.virol.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 21.Corbett PT, Sanders JKM, Otto S. Chem. Eur. J. 2008;14:2153. doi: 10.1002/chem.200701413. [DOI] [PubMed] [Google Scholar]
- 22.Houk J, Whitesides GM. J. Am. Chem. Soc. 1987;109:6825. [Google Scholar]
- 23.Srinivas BTV, Maadhur AR, Bojja S. Tetrahedron. 2014;70:8161. [Google Scholar]
- 24.Whitney AM, Ladame S, Balasubramanian S. Angew. Chem. Int. Ed. 2004;43:1143. doi: 10.1002/anie.200353069. [DOI] [PMC free article] [PubMed] [Google Scholar]