SUMMARY
Three receptors (Rpn1/S2/PSMD2, Rpn10/S5a, Rpn13/Adrm1) in the proteasome bind substrates by interacting with conjugated ubiquitin chains and/or shuttle factors (Rad23/HR23, Dsk2/PLIC/ubiquilin, Ddi1) that carry ubiquitinated substrates to proteasomes. We solved the structure of two such receptors with their preferred shuttle factor, namely hRpn13Pru:hPLIC2UBL and scRpn1 T1:scRad23UBL. We find that ubiquitin folds in Rad23 and Dsk2 are fine-tuned by residue substitutions to achieve high affinity for Rpn1 and Rpn13 respectively. A single substitution in hPLIC2 yields enhanced interactions with the Rpn13 ubiquitin contact surface and sterically blocks hRpn13 binding to its preferred ubiquitin chain type, K48 diubiquitin. Rpn1 T1 binds two ubiquitins in tandem and we find that Rad23 binds exclusively to the higher affinity Helix28/Helix30 site. Rad23 contacts at Helix28/Helix30 are optimized compared to ubiquitin by multiple conservative amino acid substitutions. Thus, shuttle factors deliver substrates to proteasomes through fine-tuned ubiquitin-like surfaces.
Keywords: Proteasome, ubiquitin, Rpn1, Rpn13, Dsk2, Rad23
eTOC Blurb
Chen et al. solve the hRpn13Pru:hPLIC2UBL and Rpn1 T1:Rad23UBL structures, which reveal how proteasome substrate receptors are targeted by substrate carrying shuttle factors. Within each shuttle factor, a common ubiquitin-like surface is fine-tuned by conservative amino acid substitutions to bind a specific receptor.
INTRODUCTION
Regulated protein degradation in eukaryotes is performed by the ubiquitin-proteasome pathway, reviewed in (Finley et al., 2016). Substrates are ubiquitinated by an enzymatic cascade and ultimately recognized by ubiquitin receptors Rpn1/S2/PSMD2 (Shi et al., 2016), Rpn10/S5a (Deveraux et al., 1994), and Rpn13/Adrm1 (Husnjak et al., 2008; Schreiner et al., 2008) of the proteasome 19S regulatory particle (RP). The RP abuts either or both ends of the proteasome 20S catalytic core particle (CP), a large cylindrical structure with a hollow interior where substrates are proteolyzed (Groll et al., 1997).
The three RP ubiquitin receptors have distinct modes of substrate recognition. A site in the Rpn1 toroid (T1, toroid 1) binds preferentially to K6 and K48 ubiquitin chains (Shi et al., 2016); hRpn10 binds ubiquitin at either of two helical ubiquitin-interacting motifs (UIMs) (Wang et al., 2005; Zhang et al., 2009b); Rpn13 binds ubiquitin through loops of a pleckstrin-like receptor for ubiquitin (Pru) domain (Husnjak et al., 2008; Schreiner et al., 2008). Each receptor is proximal to one of the three deubiquitinating enzymes (DUBs) of the proteasome RP. Rpn11 is close to Rpn10 (Lander et al., 2012; Lasker et al., 2012) and acts at the substrate end of ubiquitin chains to promote degradation (Verma et al., 2002; Yao and Cohen, 2002). Ubp6/Usp14 binds to a second toroid site in Rpn1 (T2, toroid 2) that is proximal to the T1 site (Leggett et al., 2002; Shi et al., 2016) and acts on substrates with more than one ubiquitin chains (Lee et al., 2016). Uch37/UCHL5 (Lam et al., 1997) binds to a deubiquitinase adaptor domain (DEUBAD)(Sanchez-Pulido et al., 2012) of Rpn13 (Chen and Walters, 2015; Hamazaki et al., 2006; Qiu et al., 2006; Sahtoe et al., 2015; VanderLinden et al., 2015; Yao et al., 2006). Ubp6/Usp14 acts within ubiquitin chains (Hanna et al., 2006; Lee et al., 2016) whereas Uch37 deconjugates ubiquitin chains at a distal location relative to the substrate (Lam et al., 1997); both can either promote or antagonize degradation (Hanna et al., 2006; Koulich et al., 2008; Lam et al., 1997; Lee et al., 2010; Lee et al., 2016). As ubiquitins are removed from substrates by the DUBs, a heterohexameric ring of AAA+ ATPase proteins (Rpt1-Rpt6) promotes substrate unfolding and transit into the CP (Finley et al., 2016; Gillette et al., 2008; Rabl et al., 2008; Smith et al., 2007).
Rpn13 and Uch37 are required for proper cell cycle progression (Randles et al., 2016) and are upregulated in human cancers (Chen et al., 2009; Chen et al., 2012; Fejzo et al., 2013; Fejzo et al., 2008; Jang et al., 2014; Sacco et al., 2010; Zheng et al., 2015). Moreover, cell permeable molecules that restrict tumor growth in mice xenograft models of multiple myeloma and ovarian cancer were found to conjugate to an Rpn13 cysteine (Anchoori et al., 2013) and similarly, an hRpn13-targeting peptoid inhibitor exerts selective cytotoxicity to multiple myeloma cells (Trader et al., 2015). Collectively, these studies offer promise for Rpn13 as a therapeutic target for the prevention of certain cancer types.
It is not yet clear how frequently ubiquitinated substrates are recruited to proteasomes by directly binding to Rpn1, Rpn10, or Rpn13. It is likely that ubiquitinated substrates are commonly delivered to the three RP receptors by ubiquitin-like (UBL)- ubiquitin-associated (UBA) family members (Figure 1A). The UBA domains of such shuttle factors bind ubiquitin (Bertolaet et al., 2001; Wilkinson et al., 2001) while their UBL domains bind to Rpn1, Rpn10, or Rpn13 (Elsasser et al., 2002; Hiyama et al., 1999; Husnjak et al., 2008; Mueller and Feigon, 2003; Rosenzweig et al., 2012; Shi et al., 2016; Walters et al., 2002). UBL-UBA proteins have different specificities for ubiquitin chains (Raasi et al., 2005). hHR23a (Rad23 in yeast) C-terminal UBA domain preferentially binds K48-linked chains (Raasi and Pickart, 2003) by sandwiching between neighboring ubiquitin moieties (Varadan et al., 2005) whereas hPLIC1/ubiquilin-1 (Dsk2 in yeast) UBA domain binds with significantly higher affinity to monoubiquitin and does not exhibit notable preference for K48 versus K63 ubiquitin chains (Zhang et al., 2008).
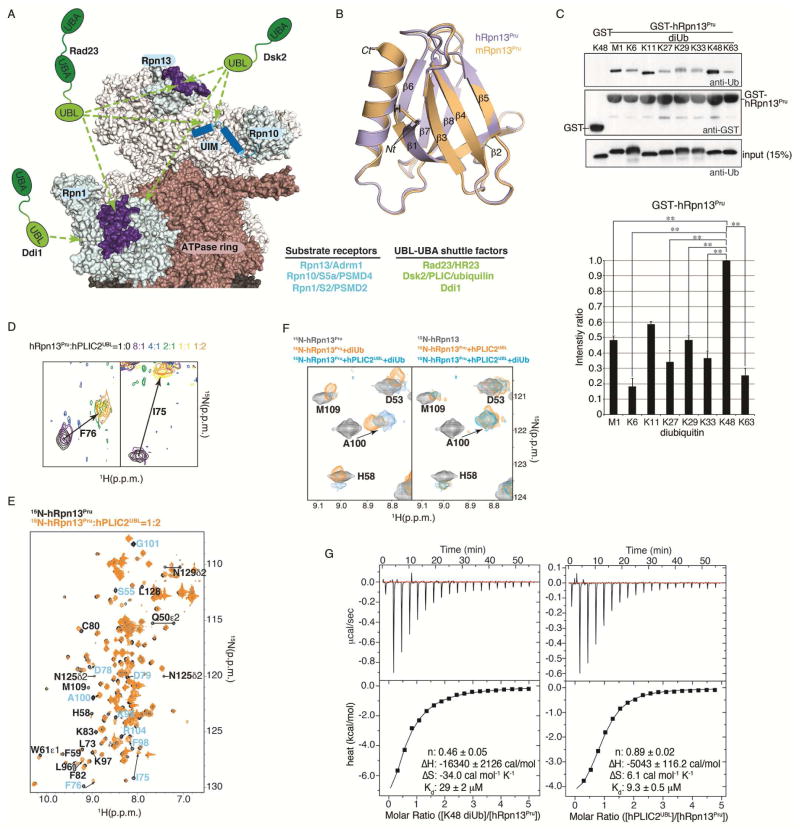
Figure 1. hRpn13Pru preferentially binds to hPLIC2UBL over K48 diubiquitin.
(A) Model of the proteasome highlighting substrate receptors Rpn1, Rpn10 and Rpn13Pru (light-blue) generated by using a cryoEM-based model (PDB 4CR2). The Rpn1 T1 and Rpn13Pru ubiquitin-binding loops are indicated in indigo whereas cartoon representations are displayed for the hRpn10 UIMs. The ATPase ring and CP are burgundy and grey respectively with the remaining RP constituents colored white. The three shuttle factors are represented as green cartoon images.
(B) Superimposed structure of hRpn13Pru (periwinkle blue) and mRpn13Pru (orange, PDB 2R2Y) with secondary structural elements labeled.
(C) GST pull-down assay with GST-hRpn13Pru and M1, K6, K11, K27, K29, K33, K48, and K63 diubiquitin, as indicated (top). Immunoblotting was done with anti-ubiquitin (top panel) or anti-GST (second panel) antibodies. GST protein was used as a negative control with K48 diubiquitin, as indicated. Direct loading for 15% of the diubiquitin input for each chain type with immunoblotting by anti-ubiquitin antibody is included (third panel). The pull-down assay was repeated three times and the diubiquitin signal intensities were separately normalized to the strongest signal by using ImageJ and plotted (bottom). **, indicates statistical significance with a p-value < 0.05 by a two-tailed, two sample Student’s t-test.
(D) hRpn13Pru I75 and F76 demonstrate intermediate-slow exchange upon binding to the hPLIC2UBL. A color code for the molar ratio of hRpn13Pru to hPLIC2UBL is indicated above the spectra.
(E) 1H, 15N HSQC spectra of 15N-labeled hRpn13Pru (black) and with 2-fold molar excess unlabeled hPLIC2UBL (orange). hRpn13Pru signals that shift away from their unligated state following the addition of hPLIC2UBL are labeled, with amino acids from the ubiquitin-binding region highlighted in blue.
(F) Selected regions from 1H, 15N HSQC spectra of 15N-labeled hRpn13Pru (black) with equimolar K48 diubiquitin (orange, left) or hPLIC2UBL (orange, right), and both hPLIC2UBL and K48 diubiquitin (blue).
(G) ITC analysis of the hRpn13Pru binding to K48 diubiquitin (left) or hPLIC2UBL (right). 1.12 mM K48 diubiquitin or 1.07 mM hPLIC2UBL was injected into a calorimeter cell containing 0.0469 mM hRpn13Pru and the data were fit to a one site binding mode with the indicated thermodynamic parameters.
Each of the UBL-UBA shuttle factors have been linked to functions that impact human health. hPLIC proteins are associated with protein quality control pathways through their connections to neurodegenerative diseases (Deng et al., 2011; Hiltunen et al., 2006; Mah et al., 2000; Stieren et al., 2011), cytoprotective activity during stress (Lu et al., 2009), aggresome formation (Heir et al., 2006), targeting of aggregated proteins to autophagosomes (N’Diaye et al., 2009; Rothenberg et al., 2010), and clearance of expanded polyglutamine proteins (Wang et al., 2006; Wang and Monteiro, 2007). Rad23 proteins are similarly associated with neuroprotection activities (Tsou et al., 2015), but are most well known for their function in nucleotide excision repair (Mueller and Smerdon, 1996; Watkins et al., 1993). hHR23b (a human ortholog of Rad23) is downregulated in highly invasive breast cancer cell lines (Linge et al., 2014), associated with risk of lung, laryngeal and bladder cancer (Abbasi et al., 2009; Chang et al., 2008; Chen et al., 2007), and substitution of A249 for valine in this protein is associated with increased esophageal cancer risk (Pan et al., 2009).
Here, we solve the structure of proteasome substrate receptor complexes with the shuttle factors that deliver ubiquitinated proteins to proteasomes, namely hRpn13Pru complexed with hPLIC2UBL and Rpn1 T1 with Rad23UBL. These structures reveal the molecular basis of proteasome receptor preferences for specific shuttle factors, and how the ubiquitin fold was optimized in these two shuttle factors for enhanced binding to ubiquitin-binding surfaces in Rpn1 and Rpn13.
RESULTS
hRpn13Pru binds preferentially to hPLIC2UBL over K48 diubiquitin, its preferred linkage type
We solved the structure of hRpn13Pru to 1.8 Å resolution by x-ray crystallography (Table 1, Figures 1B, periwinkle blue, and S1A). hRpn13Pru and mouse Rpn13Pru (mRpn13Pru) share 98% sequence identity and the overall root-mean-square-deviation (r.m.s.d.) between the Cα atoms of residues Y22 to N130 is 0.31 Å (Figure 1B); deviations for Cα atoms per residue upon superposition of these two structures are provided in Figure S1B. Like mRpn13Pru, hRpn13Pru forms a two-layer β-sandwich PH domain fold (Schreiner et al., 2008), and its N- and C-terminal 20 amino acids are disordered.
Table 1.
Data collection and refinement statistics for hRpn13Pru with values for the highest resolution shell included in parentheses.
| Diffraction data | |
| Resolution range (Å) | 50 – 1.80 (1.83 – 1.80) |
| Space group | P212121 |
| Unit cell | |
| a, b, c (Å) | 42.56, 56.48, 64.09 |
| Total reflections | 88796 |
| Unique reflections | 14838 |
| Multiplicity | 6.0 (4.9) |
| Completeness (%) | 99.06 (96.35) |
| Mean I/σI | 15.72 (1.82) |
| Rmerge (%) | 6.6 (96.0) |
| Refinement | |
| Reflections | 14831 |
| Rwork | 0.168 (0.255) |
| Rfree | 0.209 (0.282) |
| Number of atoms | 1015 |
| Macromolecules | 932 |
| Ligands | 16 |
| Water | 67 |
| r.m.s.d. | |
| Bonds length (Å) | 0.013 |
| Bonds angle (°) | 1.50 |
| Ramachandran plot | |
| Most favored (%) | 100 |
| Outliers (%) | 0 |
| Average B-factor (Å2) | 36.00 |
| Macromolecules | 35.00 |
| Solvent | 41.10 |
We next investigated the hRpn13 preference for ubiquitin chains. Ubiquitin chains are formed by conjugating the C-terminal glycine in one ubiquitin molecule to the N-terminal methionine or one of seven lysines in another ubiquitin molecule, as reviewed in (Liu and Walters, 2010). To evaluate hRpn13 ubiquitin chain preference, resin-bound GST-hRpn13Pru or GST (as a negative control) was incubated with M1, K6, K11, K27, K29, K33, K48, or K63 diubiquitin and complex formation assayed by immunoblotting with anti-ubiquitin antibody. hRpn13Pru bound preferentially to K48 diubiquitin with a statistically significant reduction for M1, K6, K27, K29, K33 and K63 chain types (Figure 1C).
We also found previously that hRpn13Pru binds to the UBL domains of hPLIC2 and hHR23a (Husnjak et al., 2008). In NMR titration experiments, hRpn13Pru was found to bind hHR23aUBL in the fast exchange regime, with gradual shifting of signals from free to bound states and a Kd value of 36 μM (Husnjak et al., 2008). By contrast, hRpn13Pru binds to hPLIC2UBL in the intermediate-slow exchange regime with bound state peaks appearing at early titration points as exemplified in Figure 1D for I75 and F76, suggesting a stronger interaction compared to hHR23aUBL. F76 is required for hRpn13 interaction with ubiquitin (Schreiner et al., 2008) and its shifting, along with that of other amino acids in the hRpn13Pru ubiquitin-binding loops (blue labels in Figures 1E and S1C), suggested an overlapping binding surface for hPLIC2UBL and ubiquitin. Therefore, we tested whether binding to hPLIC2 could interfere with hRpn13 binding to ubiquitin chains.
Unlabeled hPLIC2UBL and K48 diubiquitin were added individually or together to 15N-labeled hRpn13Pru and binding assessed by 1H, 15N HSQC experiments (Figure 1F). The mixture of all three proteins was identical to that of 15N hRpn13Pru:hPLIC2UBL (Figure 1F, right panel, blue compared to orange) and not 15N hRpn13Pru:K48 diubiquitin (Figure 1F, left panel, blue compared to orange). For example, shifting observed for hRpn13 residues D53 and A100 caused by K48 diubiquitin addition to 15N hRpn13Pru was lost when hPLIC2UBL was present (Figure 1F, left panel, blue compared to orange). Such comparisons demonstrate that hRpn13Pru binds preferentially to hPLIC2UBL over K48 diubiquitin.
To test directly the relative affinity of hRpn13Pru for hPLIC2UBL and K48 diubiquitin, we used isothermal titration calorimetry (ITC). This technique revealed hRpn13Pru to prefer hPLIC2UBL, with a 3-folder higher affinity (9.3 ± 0.5 μM, Figure 1G right panel) compared to K48 diubiquitin (29 ± 2 μM, Figure 1G left panel). The hRpn13Pru affinity for K48 diubiquitin detected by ITC is significantly reduced compared to that measured by tryptophan quenching (90 nM) (Husnjak et al., 2008), most likely because of the 50-fold higher concentration needed for the ITC measurements and because the concentration is above the Kd value (Haq et al., 1997).
The structure of hRpn13Pru:hPLIC2UBL reveals a common binding surface with ubiquitin
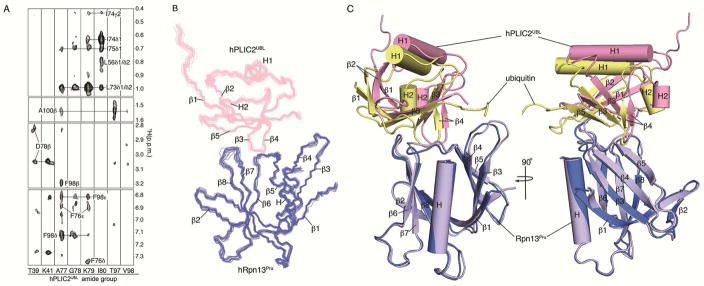
We implemented a method developed to assign intermolecular NOE interactions (Walters et al., 1997; Wang et al., 2005) to solve the hRpn13Pru:hPLIC2UBL structure in which 15N, 100% 2H-labeled hPLIC2UBL was produced and mixed with equimolar unlabeled hRpn13Pru. A standard 15N-dispersed NOE spectroscopy (NOESY) (Sklenar et al., 1993) experiment was acquired on this sample to reveal 32 unambiguous intermolecular interactions between hPLIC2UBL amide groups and side chain atoms of hRpn13Pru (Figure 2A, Table 2). The results were consistent with the titration experiments (Figures 1E, S1C–E) and indicated interaction between hPLIC2UBL β1 – β2 loop, β3, β4 and β5 and the hRpn13Pru ubiquitin-binding loops (Figure 2A). We complemented our NOE information with distance-dependent paramagnetic relaxation enhancement (PRE) effects (shown in Figure S2) to obtain long-range distance constraints between hPLIC2UBL and hRpn13Pru, as described in Experimental Procedures. PRE- and NOE-derived distance constraints, effects of the titration experiments (Table S1), and backbone ϕ and ψ torsion angle constraints (Table 2) derived by TALOS+ (Shen et al., 2009) were used to solve the hRpn13Pru:hPLIC2UBL structure by HADDOCK2.2, as described in Experimental Procedures. The structure calculations returned one cluster of 199 structures from 200 starting structures with a backbone r.m.s.d. of 0.6 ± 0.1 Å. A backbone trace of the 20 lowest energy structures are shown in Figure 2B.
Figure 2. Structure of hRpn13Pru:hPLIC2UBL reveals similarities with ubiquitin.
(A) Selected 15N planes from a 3D 15N-dispersed NOESY spectrum recorded on a sample of 0.5 mM 2H, 15N-labeled hPLIC2UBL and equimolar unlabeled hRpn13Pru. Labels inside and outside of the strips correspond to hRpn13Pru and hPLIC2UBL, respectively.
(B) Superposition of the 20 lowest energy hRpn13Pru:hPLIC2UBL structures depicted as backbone trace diagrams with hRpn13Pru in blue and hPLIC2UBL in pink. The r.m.s.d. to the average structure for this bundle is 0.45 Å.
(C) Superposition of the hRpn13Pru (periwinkle blue):hPLIC2UBL (pink) and mRpn13Pru (indigo):ubiquitin (yellow) (PDB 2Z59) complexes.
Table 2.
Structural Statistics for hRpn13Pru:hPLIC2UBL
| Distance constraints | |
| Intermolecular NOE | 32 |
| Intermolecular PRE | 64 |
| Ambiguous Interaction Restraints | 9 |
| Total dihedral angle restraints | 200 |
| ϕ (°) | 100 |
| ψ (°) | 100 |
| Structure statistics§ | |
| Ramachandran plot (%) | |
| Most-favorable region | 93.3 |
| Additionally allowed region | 6.7 |
| Generously allowed region | 0 |
| Disallowed region | 0 |
| RMSD from average structure (Å) | |
| Backbone | 0.45 ± 0.09 |
| Heavy | 0.69 ± 0.09 |
Statistics for the II° structural elements of hRpn13Pru and hPLIC2UBL from the 20 lowest energy structures.
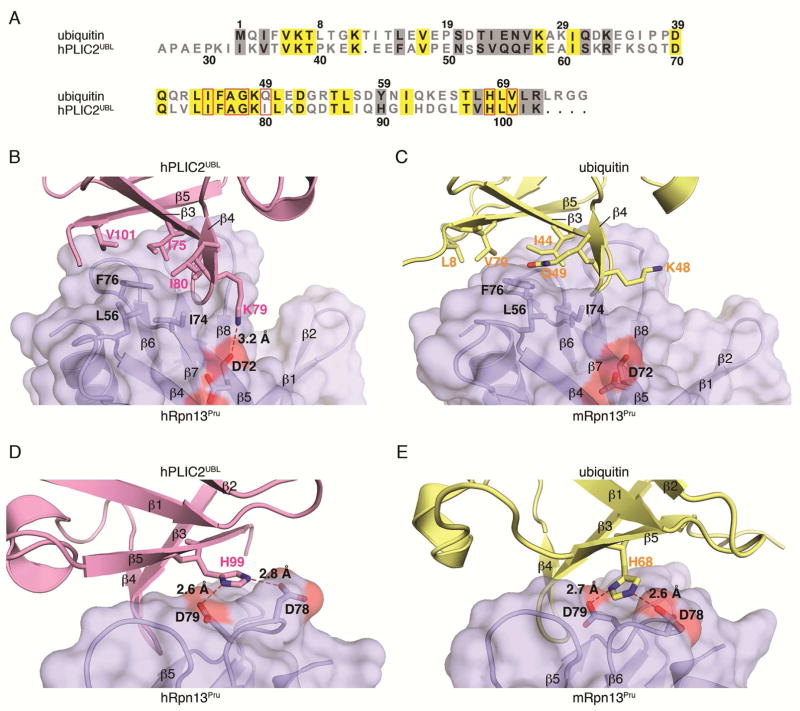
We compared our hRpn13Pru:hPLIC2UBL structure to the mRpn13Pru:ubiquitin structure to find an overall r.m.s.d. between these two protein complexes of 3.4 Å (Figure 2C). hPLIC2UBL and ubiquitin occupy approximately the same site on Rpn13Pru with similar contact surface areas, namely 1238 Å2 for hPLIC2UBL and 1256 Å2 for ubiquitin. Many formative interactions are conserved in the hRpn13Pru:hPLIC2UBL complex compared to ubiquitin, as expected by the high degree of sequence conservation between hPLIC2UBL and ubiquitin (Figure 3A). Rpn13Pru F76 located within the β5–β6 loop is strictly conserved (Husnjak et al., 2008) and interacts with an isoleucine and valine amino acid of both hPLIC2UBL (Figure 3B, I75 and V101) and ubiquitin (Figure 3C, I44 and V70). Similarly, Rpn13Pru L73 (on β5) and F98 (on β7) interact with an alanine and glycine in both hPLIC2UBL (Figure S3A, A77 and G78) and ubiquitin (Figure S3B, A46 and G47). At the core of the contact surface, ubiquitin H68 is preserved as H99 in hPLIC2UBL (Figure 3A), and Nδ1 and Nε2 from these conserved histidines form hydrogen bonds to Rpn13 D78 and D79 side chain oxygen atoms (Schreiner et al., 2008) (Figures 3D and 3E). This histidine, together with I75 and A77 of hPLIC2UBL, are required for binding to hRpn10 and the proteasome (Walters et al., 2002).
Figure 3. Dsk2 proteins mimic and enhance ubiquitin interactions with hRpn13.
(A) Sequence alignment of hPLIC2UBL and ubiquitin by ClustalW2 and Boxshade3.21. Identical and conserved residues are highlighted against a yellow and grey background, respectively. hPLIC2UBL residues that interact with Rpn13Pru and their aligned residues in ubiquitin are boxed with red rectangles.
(B–E) Expanded views of the hRpn13Pru:hPLIC2UBL (B and D) and mRpn13Pru:ubiquitin (C and E) complexes to illustrate interactions at the contact surface with key amino acids displayed and labeled. Electrostatic interactions are indicated with red dashed lines and the hRpn13Pru surface displayed in light blue.
Despite the similarities of the two contact surfaces, Rpn13Pru forms a greater number of contacts with hPLIC2UBL by replacing Q49 in ubiquitin with I80 (Figure 3A). This isoleucine establishes the core of the contact surface by engaging Rpn13Pru L56, I74, and F76 (Figure 3B). These interactions place hPLIC2UBL β4 closer to the center of Rpn13Pru β5 (Figure 2C) and in turn, enables an additional hydrogen bond between hPLIC2UBL K79 Nζ and the hRpn13Pru D72 (Figure 3B).
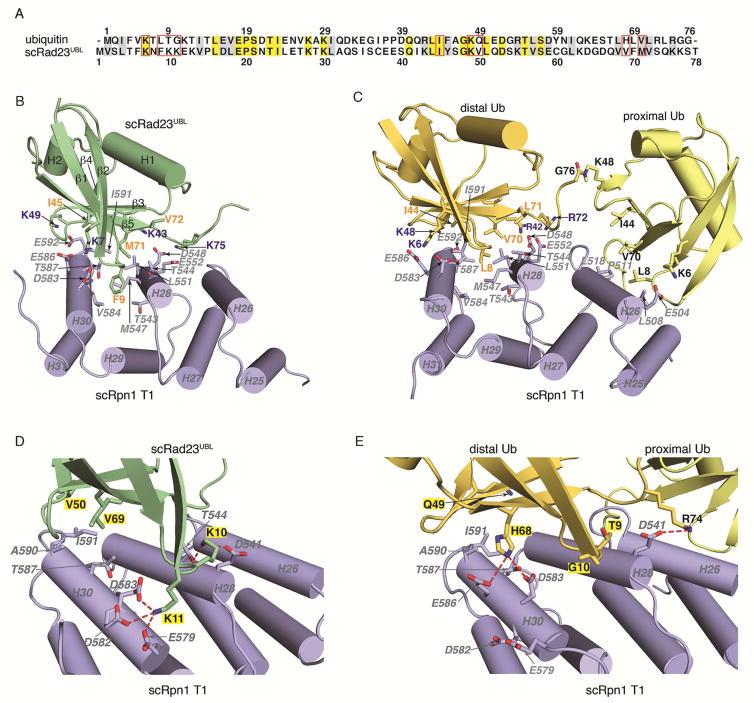
Rad23UBL binds to Helix28/Helix30 of the Rpn1 T1 site
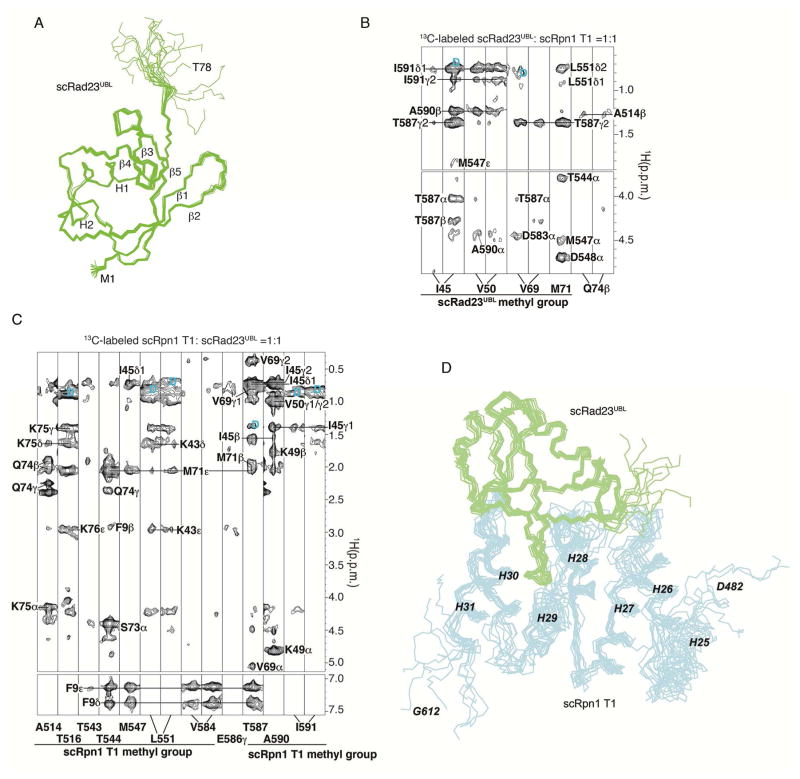
We used NMR to solve the structure of S. cerevisiae Rad23UBL (scRad23UBL) to yield twenty lowest energy structures with an r.m.s.d. of 0.21 Å for the backbone atoms of Q2-R72 (Figure 4A, Table 3). This structure is highly similar to that of ubiquitin, comprising a five-strand β-sheet, one α-helix, and one 310-helix (Figure 4A). The structures of scRad23UBL complexed with the E4 ubiquitin ligase Ufd2 (Hanzelmann et al., 2010) and of hHR23a (Walters et al., 2003) have been reported previously. Both are similar to our structure, but β1, β2 and β5 are extended in the free scRad23UBL (Figures S4A and S4B).
Figure 4. Structure of scRad23UBL and its complex with the scRpn1 T1 site.
(A) Backbone trace diagrams for the twenty lowest energy structures of scRad23UBL with the secondary structural elements superimposed. The N- and C-terminal residues and the individual secondary structural elements are labeled.
(B–C) Selected regions from 1H, 13C half-filtered NOESY experiments acquired with either 13C-labeled scRad23UBL and equimolar unlabeled scRpn1 T1 (B) or 13C-labeled scRpn1 T1 and equimolar unlabeled scRad23UBL (C) displaying intermolecular NOE interactions, as labeled. Breakthrough diagonal peaks are labeled (blue ‘D’).
(D) Superposition of the 10 lowest energy scRpn1 T1:scRad23UBL structures depicted as backbone trace diagrams with scRpn1 T1 in blue and scRad23UBL in green. The r.m.s.d. to the average structure for this bundle is 0.7 Å.
Table 3.
Structural statistics for the scRad23UBL NMR structures
| NOE-derived distance constraints | |
| Total NOE | 1314 |
| Intra-residue | 556 |
| Inter-residue | 758 |
| Sequential (|i − j| = 1) | 268 |
| Medium-range (|i − j| ≤ 4) | 197 |
| Long-range (|i − j| > 5) | 293 |
| Hydrogen bonds | 24 |
| Total dihedral angle restraints | 138 |
| ϕ (°) | 69 |
| ψ (°) | 69 |
| Structure statistics§ | |
| Ramachandran plot (%) | |
| Most-favorable region | 88.9 |
| Additionally allowed region | 11.1 |
| Generously allowed region | 0 |
| Disallowed region | 0 |
| RMSD from average structure (Å) | |
| Backbone | 0.21 ± 0.04 |
| Heavy | 0.9 ± 0.1 |
Statistics for the II° structural elements of scRad23UBL for the 20 lowest energy structures.
scRad23UBL binds to three outer helices of the scRpn1 T1 site where ubiquitin binds with a 64 nM binding affinity (Shi et al., 2016). We solved the structure of the scRpn1 T1 site complexed with scRad23UBL by NMR, as described in Experimental Procedures. Half-filtered NOESY experiments were used to record interactions between scRpn1 T1 and scRad23UBL and revealed many contacts between scRpn1 Helix28/Helix30 and scRad23UBL β1–β2 loop, β2, β3, β4 and β5 (Figures 4B and 4C), consistent with NMR titration experiments performed with 15N-labeled scRad23UBL and unlabeled scRpn1 T1 (Figures S4C–E). In total, 74 NOE-derived intermolecular constraints were used in XPLOR-NIH 2.33 to produce ten lowest energy structures with an r.m.s.d of 0.7 Å (Figure 4D and Table 4).
Table 4.
Structural statistics for scRpn1 T1:scRad23UBL
| NOE-derived distance constraints | |
| Intermolecular | 74 |
| Intramolecular | |
| scRpn1 T1 | 1237 |
| scRad23UBL | 1310 |
| Hydrogen bonds | |
| scRpn1 T1 | 59 |
| scRad23UBL | 24 |
| Total dihedral angle restraints | |
| scRpn1 T1 | 155 |
| scRad23UBL | 138 |
| Structure statistics§ | |
| Ramachandran plot (%) | |
| Most-favorable region | 82.9 |
| Additionally allowed region | 17.1 |
| Generously allowed region | 0 |
| Disallowed region | 0 |
| RMSD from average structure (Å) | |
| Backbone | 0.7 ± 0.2 |
| Heavy | 1.1 ± 0.2 |
Statistics for the II° structural elements of scRpn1 T1 for the region spanning Helix26-Helix31 and scRad23UBL for the ten lowest energy structures.
Binding to Rad23UBL mimicked that of ubiquitin involving similar amino acids, including the classic hydrophobic (Rad23UBL F9, I45, M71 and ubiquitin L8, I44, V70) and basic (Rad23UBL K7, K43, K49 and ubiquitin K6, R42, K48) interfaces (Figure 5A); however, F9 and M71 in Rad23UBL formed a greater number of favorable contacts with Rpn1 compared to ubiquitin (Figures 5B and 5C). Two ubiquitin moieties can bind to Rpn1 T1, such that one ubiquitin occupies a high affinity site at Helix28/Helix30 while the other binds a low affinity site at Helix26 (Shi et al., 2016). Rad23UBL binds exclusively to the Helix28/Helix30 site with F9 interdigitated between the two helices (Figure 5B). Although ubiquitin L8 also occupies this site (Figure 5C) (Shi et al., 2016), the F9 side chain of Rad23UBL inserts deeper into the Helix28/Helix30 pocket, enabling additional hydrophobic interactions with Rpn1 T543, M547 and V584 (Figure 5B). Similarly, the longer Rad23UBL M71 is placed closer to Rpn1 T544, M547 and T587 than ubiquitin V70 (Figures 5B and 5C). Favorable hydrophobic interactions and hydrogen bonds of the Rpn1 T1:K48 diubiquitin complex are preserved in Rpn1 T1: Rad23UBL; Rad23UBL I45 mimics ubiquitin I44 by forming similar hydrophobic contacts with Rpn1 T1 Helix30 (Figures 5B and 5C) and Rad23UBL K7, K43, K49 and K75 form hydrogen bonds with Rpn1 T1 D548, E552, D583, E586, and E592, respectively, as do ubiquitin K6, R42, K48 and R72 (Figures 5B and 5C).
Figure 5. scRad23 occupies the Helix28/Helix30 high affinity ubiquitin-binding site of Rpn1 T1.
(A) Sequence alignment of ubiquitin and scRad23UBL by ClustalW2 and Boxshade3.21. Identical and conserved residues are highlighted against a yellow and grey background, respectively. scRad23UBL residues that interact with scRpn1 T1 and their aligned residues in ubiquitin are boxed with red rectangles.
(B and C) Ribbon diagrams of the lowest energy scRpn1 T1:scRad23UBL (B) and scRpn1 T1:K48 diubiquitin (C) structures. scRpn1 (blue), scRad23UBL (green) and K48 diubiquitin (yellow) side chains at the contact surfaces are displayed and labeled with oxygen and nitrogen atoms in red and blue respectively. Displayed side chains of basic and hydrophobic residues that are conserved in scRad23UBL and ubiquitin are labeled in blue and orange, respectively, whereas scRpn1 side chains are labeled in grey.
(D and E) Expanded regions to display the contact surfaces for scRpn1 T1:scRad23UBL (D) and scRpn1 T1:K48 diubiquitin (E). Side chains of scRad23UBL K10, K11, V50, and V69 are displayed and labeled (D), as are the corresponding amino acids T9, G10, Q49, and H68 from K48 diubiquitin (E). The acidic residues in scRpn1 T1 that form hydrogen bonds with scRad23UBL or K48 diubiquitin are displayed and labeled. Oxygen and nitrogen atoms are colored red and blue, respectively.
Rad23UBL contains more positively charged residues than ubiquitin in the β1–β2 loop (K10 and K11 versus ubiquitin T9 and G10) (Figure 5A), which form five additional hydrogen bonds with acidic residues of Rpn1 T1 Helix28/Helix30 (Figures 5D and 5E). Hydrophilic residues Q49 and H68 in ubiquitin located at Rpn1 T1 are hydrophobic (V50 and V69) in Rad23UBL (Figures 5D and 5E). These two Rad23UBL residues are favored by nearby hydrophobic residues in Rpn1 T1 Helix30.
DISCUSSION
Preferences of shuttle factors for substrate receptors in the proteasome
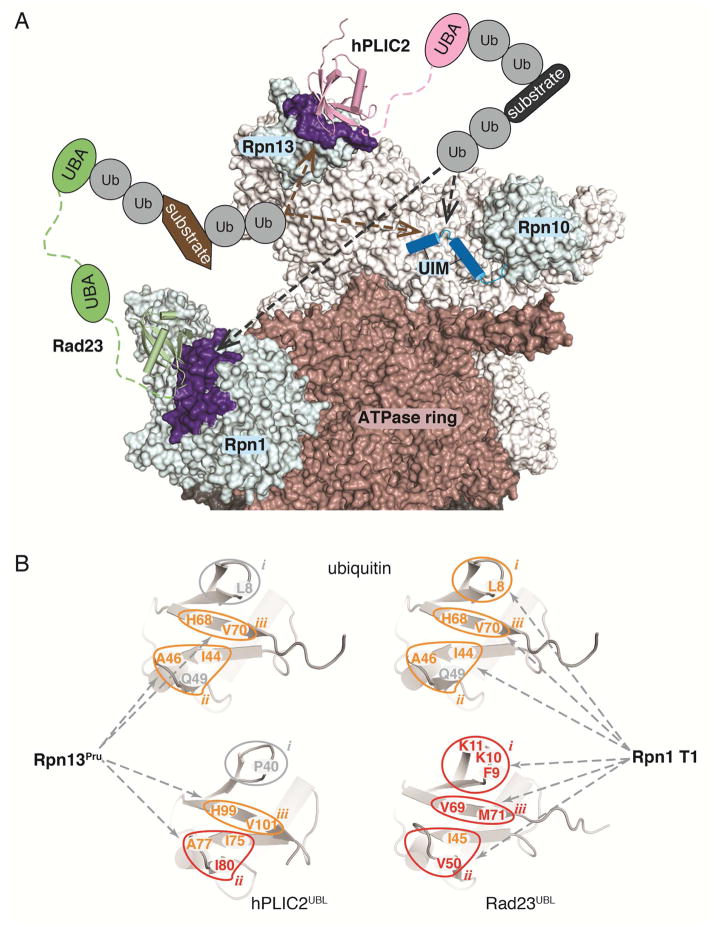
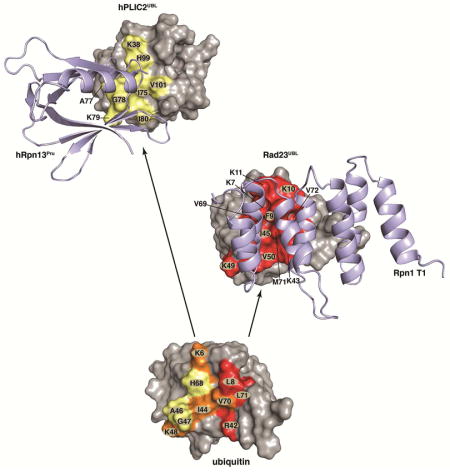
Here, we solve the structures of Rpn1 and Rpn13 bound to their preferred ubiquitin shuttle factors. We demonstrate that these shuttle factors bind to the same amino acids as does ubiquitin in proteasome substrate receptors Rpn1 and Rpn13. Consistent with our results, we previously showed that a Rpn1 mutant defective for binding to ubiquitin was unable to bind to Rad23 in S. cerevisiae (Shi et al., 2016). We find that the UBL domains in Rad23 and hPLIC2 have conservative mutations to enhance their interaction with Rpn1 and hRpn13, respectively. In the case of hPLIC2, only one amino acid change is sufficient to yield a 3-fold increase in affinity, namely Q49 to I80. The Dsk2 protein family has significantly higher sequence identity with ubiquitin than do Rad23 proteins (Figures 3A, 5A and S5). These conserved amino acids in hPLIC2 mimic ubiquitin interactions with hRpn13, including I75 (I44 in ubiquitin), H99 (H68 in ubiquitin), and V101 (V70 in ubiquitin) (Figures 2C and 3). The conserved histidine residue is at the center of the Rpn13Pru-binding surfaces, but is replaced with valine in Rad23/hHR23a/hHR23b (Figure S5), which enhances binding to the Rpn1 T1. The stronger conservation between ubiquitin and the Dsk2 proteins provides a rationale for their Rpn13 preference over Rpn1, which is a weaker ubiquitin binder (Shi et al., 2016).
One amino acid that is not preserved in Dsk2 proteins is ubiquitin L8 (Figures 3A and 3C), which interacts with P77 in Rpn13Pru. This interaction is not preserved in hPLIC2UBL where L8 is substituted with a proline (Figure 3A). This substitution aids in the formation of the β1 – β2 turn. L8 is preserved in Rad23 proteins as a hydrophobic amino acid (Figure S5) that interdigitates between Rpn1 Helix28 and Helix30, and is a phenylalanine in Rad23 from S. cerevisiae (Figure 5B). Residues adjacent to this hydrophobic amino acid in Rad23 proteins also form favorable interactions with the Rpn1 T1 including K10 and K11, which form hydrogen bonds with acidic residues (Figure 5D). In ubiquitin, K10 and K11 are T9 and G10 respectively; these amino acids cannot make as favorable interactions with the acidic amino acids in Rpn1 Helix28/Helix30 (Figure 5E). In Dsk2 proteins, K11 is replaced with an acidic residue (E42 in hPLIC2, Figures 3A and S5) and disfavored by the acidic Rpn1 Helix30.
Collectively, multiple proteasome substrate receptors and shuttle factors enhance proteasome versatility and affinity for ubiquitinated substrates
Ubiquitin binds to all three proteasome substrate receptors with distinct interactions from a common binding surface (Husnjak et al., 2008; Schreiner et al., 2008; Shi et al., 2016; Wang et al., 2005; Zhang et al., 2009a; Zhang et al., 2009b). Our structures demonstrate the UBL shuttle factors to mimic and enhance interactions with preferred substrate receptors at the expense of versatility for all receptors. Why would it be advantageous to have distinct high affinity shuttle factors for Rpn1 and Rpn13? One possibility is that these relationships enhance interactions between substrates and the proteasomes without hindering subsequent release of substrate-attached ubiquitins (Figure 6A). It is conceivable that strengthening the affinity of Rpn1 or Rpn13 for ubiquitin itself may interfere with subsequent release of ubiquitin chains following their cleavage from substrate. hHR23b and hPLIC2 co-purify with proteasomes from mammalian cells (Besche et al., 2009; Hiyama et al., 1999; Kleijnen et al., 2003; Kleijnen et al., 2000; Wang et al., 2007; Wang and Huang, 2008; Yu et al., 2016). Thus, their affinity for the proteasome receptors apparently leads to a certain level of basal occupancy of these shuttle factors at the proteasome. Such occupancy by ubiquitin chains would likely be detrimental to protein turnover. Moreover, each UBL-UBA shuttle factor contributes ubiquitin-binding site(s) to the proteasome and therefore is not expected to reduce avidity effects or overall affinity for ubiquitin chains at the proteasome.
Figure 6. Shuttle factor targeting to preferred substrate receptors of the proteasome.
(A) Model of substrate recruitment to the proteasome. Shuttle factors (pink, hPLIC2 and green, Rad23) use ubiquitin folds (UBL domains) to interact with ubiquitin-binding surfaces (indigo and Rpn10 UIM) while carrying ubiquitinated (grey) substrates (black and brown). High affinity for specific UBL domains leads to enhanced interaction between substrates and the proteasome as ubiquitin-associated (UBA) domains contribute additional binding surfaces for ubiquitin. This model used PDB 4CR2 with Rpn1, Rpn10, and Rpn13 displayed in light blue, the ATPase ring in burgundy, and CP and RP in dark grey and white, respectively. Brown and black arrows indicate available ubiquitin-binding surfaces in the proteasome for an Rpn1- and Rpn13-bound substrate, respectively.
(B) The β-strand face of the ubiquitin fold for ubiquitin (top) and shuttle factor UBL domains (bottom) highlighting three regions with circles (i, ii, iii) that define affinity for proteasome substrate receptors Rpn1 (right) and Rpn13 (left). Orange and red are used to highlight regions and amino acids that bind to the indicated shuttle factor equivalently and better respectively compared to ubiquitin, with a correspondingly weaker binding residue in ubiquitin in grey (Q49). Region i is indicated in grey on the left for its minimal contacts with Rpn13, but plays a role in the low affinity of hPLIC2 for Rpn1.
K48-linked ubiquitin chains are an established signal for substrate degradation by proteasomes and we have found that both Rpn1 and Rpn13 bind well to this chain type (Figure 1C and (Shi et al., 2016)). Nonetheless, all linkage types can support proteasome-mediated protein degradation (Kirkpatrick et al., 2006; Kulathu and Komander, 2012; Meierhofer et al., 2008; Xu et al., 2009). Consistent with this discovery, Rpn1 binds well to K6-linked chains (Shi et al., 2016), complementing the reduced binding of hRpn13 for this chain type (Figure 1C). UBL-UBA shuttle factors may further expand the versatility of proteasomes for diverse ubiquitin linkage types.
Substrates modified by two diubiquitin chains are superior substrates for proteasome compared to those modified by a single tetraubiquitin chain (Lu et al., 2015). Similarly branched chains, namely those formed with multiple ubiquitin linkage types, promote more rapid degradation by proteasomes than homogeneous chains (Meyer and Rape, 2014). Altogether, these results suggest that substrate processing by proteasomes is most efficient when more than one receptor is engaged and no doubt the ubiquitin-binding sites contributed by the shuttle factor UBA domains contribute to these effects (Figure 6A).
Tuning of the ubiquitin fold for a proteasome receptor
Our structures of the shuttle factors complexed with Rpn1 and Rpn13 in comparison to those with ubiquitin highlight three critical regions in the ubiquitin fold comprised of the β1 – β2 loop (i), β3, the β3 – β4 loop, and β4 (ii), and β5 (iii) (Figure 6B). Residues of sites ii and iii in ubiquitin are used to bind hRpn13. In hPLIC2UBL site ii is optimized for hRpn13 binding by a single glutamine (Q49 in ubiquitin) to isoleucine (I80 in hPLIC2UBL) substitution. When ubiquitin binds Rpn1, all three sites are engaged (Figure 6B), but the interactions are not ideal. In Rad23UBL sites i and iii are optimized for Rpn1 binding by including two lysines in site i and a valine in site ii (Figure 6B). Rpn1 is a weaker ubiquitin binder, requiring a greater number of amino acid substitutions in Rad23UBL compared to hPLIC2UBL to achieve a high affinity for its preferred receptor site in the proteasome.
Certain amino acid substitutions in the shuttle factor sites i, ii, and iii weaken affinity for the non-preferred substrate receptor in the proteasome, namely hPLIC2 for Rpn1 and Rad23 for Rpn13. In hPLIC2UBL, L8 from ubiquitin (F9 in Rad23UBL) of site i is substituted with a proline (P40) and an acidic amino acid (E42) included close by; these changes are not favored by the acidic amino acids of Rpn1 Helix28/Helix30. The histidine in site iii involved in hPLIC2 and ubiquitin binding to hRpn13 is substituted with a valine in Rad23UBL, which weakens its affinity for Rpn13. Thus, the shuttle factors appear to have fine-tuned affinity and preference for specific receptors in the proteasome by manipulating three key interaction sites.
EXPERIMENTAL PROCEDURES
Crystal growth
Crystallization of hRpn13Pru was achieved at 20°C by using sitting drop vapor diffusion with a low profile Intelli-Plate 96 (Art Robbins Instruments). Equal volume ratio of protein (10mg/ml) was mixed with varying reservoir solutions from 96 optimization conditions that contained 100 – 300 mM sodium acetate, 10 – 25% (w/v) PEG 4000, 1 mM DTT and 100 mM Tris-HCl buffer, pH 8.0 – 9.0. Crystals suitable for X-ray data collection appeared within 24 hours. For data collection, crystals were transferred into a cryoprotectant solution composed of the reservoir solution supplemented with 20% glycerol (v/v) and flash-frozen by liquid nitrogen.
X-ray data collection and crystal structure refinement
Diffraction data were collected on the hRpn13Pru crystal at the 24-ID-C beamline of the Advance Photon Source and processed with HKL2000 (Otwinowshi and Minor, 1997). The crystal space group is P212121 with unit cell dimensions of a=42.56 Å, b= 56.48 Å, c=64.09 Å, which is isomorphous to the Rpn13Pru crystal from murine (2R2Y) (Schreiner et al., 2008). The protein portion of mRpn13Pru was used as a starting model for refinement with Refmac (Murshudov et al., 1997). Python-based Hierarchical ENvironment for Integrated Xtallography (PHENIX) (Adams et al., 2010) and Crystallographic Object-Oriented Toolkit (COOT) (Emsley and Cowtan, 2004) were used to complete the model building and TLS (Translation/Libration/Screw) refinement was performed with the TLS Motions Server (Painter and Merritt, 2006). Hydrogen atoms were added by PHENIX before the final round of refinement. The final Rwork and Rfree are 0.168 and 0.209, respectively. The crystal data and structure refinement statistics are listed in Table 1 and the electron density map provided in Figure S1A.
hRpn13Pru:hPLIC2UBL structure calculations
hRpn13Pru:hPLIC2UBL complexes were generated by using HADDOCK2.2 (High Ambiguity Driven protein-protein DOCKing) (Dominguez et al., 2003) in combination with CNS (Brunger et al., 1998) as described in (Chen and Walters, 2012). The atomic coordinates for hPLIC2UBL were obtained from PDB entry 1J8C (Walters et al., 2002). Ambiguous Interaction Restraints (AIRs) were generated to complement our unambiguous NOE- or PRE-based distance constraints (Table 2). Briefly, residues with chemical shift perturbation values greater than one standard deviation value above the average were defined as “active” (Table S1) and their neighbors as “passive” provided they have >50% accessibility. AIRs impose that at least one atom (including hydrogen) of an active residue be within 2.0 Å of an atom from an active or passive residue of the binding partner, as described (Dominguez et al., 2003). Backbone ϕ and ψ torsion angle constraints for hRpn13Pru and hPLIC2UBL were derived by TALOS+ from the HN, Hα, Cα, Cβ, C′ and N chemical shift values (Shen et al., 2009).
For the first step of rigid-body energy minimization, 1000 structures were generated. The 200 structures with lowest energy from the rigid-body docking were subjected to semi-flexible simulated annealing in torsion angle space followed by refinement in explicit water. During semi-flexible simulated annealing, atoms at the interface were allowed to move but constrained by the AIRs, unambiguous NOE- and PRE-derived distance constraints, and backbone ϕ and ψ torsion angle constraints. After water refinement, the resulting structures were sorted and clustered using a 1.5 Å r.m.s.d. pairwise cut-off criterion. This treatment resulted in one cluster of 199 structures with an r.m.s.d. for the backbone atoms of 0.6 ± 0.1 Å to the average structure. The 20 lowest energy structures are provided as Figure 2B.
Structure determination of scRad23UBL and its complex with scRpn1 T1
As described in (Shi et al., 2016), the structure of scRad23UBL and scRpn1 T1: scRad23UBL were determined with XPLOR-NIH 2.33 (Schwieters et al., 2003) on a Linux operating system by using NOE and hydrogen bond constraints as well as backbone ϕ and ψ torsion angle constraints derived from TALOS+ (Shen et al., 2009) (Table 3 and 4). Hydrogen bonds were generated by using secondary structure assignments and NOE connectivities with defined distances from the acceptor oxygen to the donor hydrogen and nitrogen of 1.8–2.1 Å and 2.5–2.9 Å, respectively. Hydrogen bonds restraints were not included in the initial calculation, but were in the final round of structure calculations. When calculating the structures of scRad23UBL, a total of 50 linear starting structures were subjected to 50 steps of initial energy minimization to ensure full spatial sampling and appropriate coordinate geometry. The structures were next confined according to the inputted restraint data (Table 3) by subjecting them to 50,000 simulated annealing steps of 0.005 ps at 3,000 K, followed by 50,000 cooling steps of 0.005 ps. 1,000 steps of energy minimization was subsequently applied and the final structures recorded as coordinate files. All resulting structures had no distance or dihedral angle violation greater than 0.3 Å or 5°, respectively. The ten lowest energy structures were chosen for visualization and statistical analyses.
When calculating the structures of scRpn1 T1:scRad23UBL, intermolecular distance constraints determined from the 13C-half-filtered 3D NOESY experiments were used, in addition to intramolecular constraints for scRpn1 T1 and scRad23UBL that were generated from 15N/13C NOESY spectra acquired on the complexes (Table 4). All complexed structures were calculated from 50 linear starting structures and subjected to 2,000 steps of initial energy minimization to ensure full spatial sampling and appropriate coordinate geometry. The structures were next confined according to the inputted data by subjecting them to 55,000 simulated annealing steps of 0.005 ps at 3,000 K, followed by 5,000 cooling steps of 0.005 ps. 5,000 steps of energy minimization was applied to produce the final structures, which were recorded as coordinate files. The resulting structures had no distance or dihedral angle violation greater than 0.3 Å or 5°, respectively. The ten lowest energy structures were chosen for visualization and statistical analyses.
Protein sample preparation, NMR titration experiments, NOESY and Spin-labeling experiments for the hRpn13Pru:hPLIC2UBL complex, NMR experiments for scRad23UBL assignment and structure, ITC experiments and GST pull-down assays are described in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
UBL shuttle factors bind to ubiquitin-interacting residues of proteasome receptors
hRpn13:hPLIC2 structure reveals an optimized UBL for hRpn13 binding
hPLIC2 sterically blocks Rpn13 binding to K48 diubiquitin, its preferred chain type
Rpn1 T1:Rad23UBL structure reveals Rad23 binding to Helix28/Helix30 of Rpn1 T1
Acknowledgments
This research was funded by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research to K.J.W. and a grant from the NIH to H.A. (GM095558). X-ray data were collected at the Advanced Photon Source (APS) NE-CAT beamlines, which are supported by the NIGMS (P41 GM103403). APS is a US Department of Energy Office of Science User Facility operated by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
ACCESSION NUMBERS
Atomic coordinates for hRpn13Pru, hRpn13Pru:hPLIC2UBL, scRad23UBL, and scRpn1 T1: scRad23UBL are available through the PDB by accession codes 5IRS, 2NBV, 2NBU, and 2NBW, respectively.
AUTHOR CONTRIBUTIONS
X.C. performed, analyzed, and interpreted all NMR experiments, calculated NMR structures of hRpn13Pru:hPLIC2UBL, scRad23UBL, and scRpn1 T1: scRad23UBL, as well as performed GST pull-down assays; L.R. performed NMR experiments shown in Figure 1E–F and prepared 15N, 100% 2H-labeled hPLIC2UBL; K.S. and H.A. determined the crystal structure of hRpn13Pru; S.G.T performed the ITC experiments; K.J.W. conceived of and directed the project; all authors contributed to the writing.
Supplemental information includes five figures, one table, and Supplemental Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbasi R, Ramroth H, Becher H, Dietz A, Schmezer P, Popanda O. Laryngeal cancer risk associated with smoking and alcohol consumption is modified by genetic polymorphisms in ERCC5, ERCC6 and RAD23B but not by polymorphisms in five other nucleotide excision repair genes. Int J Cancer. 2009;125:1431–1439. doi: 10.1002/ijc.24442. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anchoori RK, Karanam B, Peng S, Wang JW, Jiang R, Tanno T, Orlowski RZ, Matsui W, Zhao M, Rudek MA, et al. A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer. Cancer Cell. 2013;24:791–805. doi: 10.1016/j.ccr.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- Besche HC, Haas W, Gygi SP, Goldberg AL. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009;48:2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, Quesenberry CP, Jr, Seldin MF, Kelsey KT, Kittles RA, Silva G, et al. Nucleotide excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African Americans. Int J Cancer. 2008;123:2095–2104. doi: 10.1002/ijc.23801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Kamat AM, Huang M, Grossman HB, Dinney CP, Lerner SP, Wu X, Gu J. High-order interactions among genetic polymorphisms in nucleotide excision repair pathway genes and smoking in modulating bladder cancer risk. Carcinogenesis. 2007;28:2160–2165. doi: 10.1093/carcin/bgm167. [DOI] [PubMed] [Google Scholar]
- Chen W, Hu XT, Shi QL, Zhang FB, He C. Knockdown of the novel proteasome subunit Adrm1 located on the 20q13 amplicon inhibits colorectal cancer cell migration, survival and tumorigenicity. Oncol Rep. 2009;21:531–537. [PubMed] [Google Scholar]
- Chen X, Walters KJ. Identifying and studying ubiquitin receptors by NMR. Methods Mol Biol. 2012;832:279–303. doi: 10.1007/978-1-61779-474-2_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Walters KJ. Structural plasticity allows UCH37 to be primed by RPN13 or locked down by INO80G. Mol Cell. 2015;57:767–768. doi: 10.1016/j.molcel.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Fu D, Xi J, Ji Z, Liu T, Ma Y, Zhao Y, Dong L, Wang Q, Shen X. Expression and clinical significance of UCH37 in human esophageal squamous cell carcinoma. Dig Dis Sci. 2012;57:2310–2317. doi: 10.1007/s10620-012-2181-9. [DOI] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Müller B, Feng MT, Tübing F, Dittmar GaG, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fejzo MS, Anderson L, von Euw EM, Kalous O, Avliyakulov NK, Haykinson MJ, Konecny GE, Finn RS, Slamon DJ. Amplification Target ADRM1: Role as an Oncogene and Therapeutic Target for Ovarian Cancer. Int J Mol Sci. 2013;14:3094–3109. doi: 10.3390/ijms14023094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejzo MS, Dering J, Ginther C, Anderson L, Ramos L, Walsh C, Karlan B, Slamon DJ. Comprehensive analysis of 20q13 genes in ovarian cancer identifies ADRM1 as amplification target. Genes Chromosome Cancer. 2008;47:873–883. doi: 10.1002/gcc.20592. [DOI] [PubMed] [Google Scholar]
- Finley D, Chen X, Walters KJ. Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem Sci. 2016;41:77–93. doi: 10.1016/j.tibs.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Hanzelmann P, Stingele J, Hofmann K, Schindelin H, Raasi S. The yeast E4 ubiquitin ligase Ufd2 interacts with the ubiquitin-like domains of Rad23 and Dsk2 via a novel and distinct ubiquitin-like binding domain. J Biol Chem. 2010;285:20390–20398. doi: 10.1074/jbc.M110.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq I, Ladbury JE, Chowdhry BZ, Jenkins TC, Chaires JB. Specific binding of hoechst 33258 to the d(CGCAAATTTGCG)2 duplex: calorimetric and spectroscopic studies. J Mol Biol. 1997;271:244–257. doi: 10.1006/jmbi.1997.1170. [DOI] [PubMed] [Google Scholar]
- Heir R, Ablasou C, Dumontier E, Elliott M, Fagotto-Kaufmann C, Bedford FK. The UBL domain of PLIC-1 regulates aggresome formation. EMBO Rep. 2006;7:1252–1258. doi: 10.1038/sj.embor.7400823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen M, Lu A, Thomas AV, Romano DM, Kim M, Jones PB, Xie Z, Kounnas MZ, Wagner SL, Berezovska O, et al. Ubiquilin 1 modulates amyloid precursor protein trafficking and Abeta secretion. J Biol Chem. 2006;281:32240–32253. doi: 10.1074/jbc.M603106200. [DOI] [PubMed] [Google Scholar]
- Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SH, Park JW, Kim HR, Seong JK, Kim HK. ADRM1 gene amplification is a candidate driver for metastatic gastric cancers. Clin Exp Metastasis. 2014;31:727–733. doi: 10.1007/s10585-014-9663-4. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway Na, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Kleijnen MF, Alarcon RM, Howley PM. The ubiquitin-associated domain of hPLIC-2 interacts with the proteasome. Mol Biol Cell. 2003;14:3868–3875. doi: 10.1091/mbc.E02-11-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Lu Y, Prado MA, Shi Y, Tian G, Sun S, Elsasser S, Gygi SP, King RW, Finley D. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature. 2016;532:398–401. doi: 10.1038/nature17433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Linge A, Maurya P, Friedrich K, Baretton GB, Kelly S, Henry M, Clynes M, Larkin A, Meleady P. Identification and functional validation of RAD23B as a potential protein in human breast cancer progression. J Proteome Res. 2014;13:3212–3222. doi: 10.1021/pr4012156. [DOI] [PubMed] [Google Scholar]
- Liu F, Walters KJ. Multitasking with ubiquitin through multivalent interactions. Trends Biochem Sci. 2010;35:352–360. doi: 10.1016/j.tibs.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Hiltunen M, Romano DM, Soininen H, Hyman BT, Bertram L, Tanzi RE. Effects of ubiquilin 1 on the unfolded protein response. J Mol Neurosci. 2009;38:19–30. doi: 10.1007/s12031-008-9155-6. [DOI] [PubMed] [Google Scholar]
- Lu Y, Lee B-h, King RW, Finley D, Kirschner MW. Substrate degradation by the proteasome: a single-molecule kinetic analysis. Science. 2015;348:1250834. doi: 10.1126/science.1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah AL, Perry G, Smith MA, Monteiro MJ. Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J Cell Biol. 2000;151:847–862. doi: 10.1083/jcb.151.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhofer D, Wang X, Huang L, Kaiser P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J Proteome Res. 2008;7:4566–4576. doi: 10.1021/pr800468j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JP, Smerdon MJ. Rad23 is required for transcription-coupled repair and efficient overrall repair in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2361–2368. doi: 10.1128/mcb.16.5.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller TD, Feigon J. Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J. 2003;22:4634–4645. doi: 10.1093/emboj/cdg467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- N’Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009;10:173–179. doi: 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowshi Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Pan J, Lin J, Izzo JG, Liu Y, Xing J, Huang M, Ajani JA, Wu X. Genetic susceptibility to esophageal cancer: the role of the nucleotide excision repair pathway. Carcinogenesis. 2009;30:785–792. doi: 10.1093/carcin/bgp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S, Pickart CM. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randles L, Anchoori RK, Roden RB, Walters KJ. The Proteasome Ubiquitin Receptor hRpn13 and Its Interacting Deubiquitinating Enzyme Uch37 Are Required for Proper Cell Cycle Progression. J Biol Chem. 2016;291:8773–8783. doi: 10.1074/jbc.M115.694588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J Biol Chem. 2012;287:14659–14671. doi: 10.1074/jbc.M111.316323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, Fang S, Cuervo AM, Nixon RA, Monteiro MJ. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19:3219–3232. doi: 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco JJ, Coulson JM, Clague MJ, Urbe S. Emerging roles of deubiquitinases in cancer-associated pathways. IUBMB Life. 2010;62:140–157. doi: 10.1002/iub.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahtoe DD, van Dijk WJ, El Oualid F, Ekkebus R, Ovaa H, Sixma TK. Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol Cell. 2015;57:887–900. doi: 10.1016/j.molcel.2014.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Kong L, Ponting CP. A common ancestry for BAP1 and Uch37 regulators. Bioinformatics. 2012;28:1953–1956. doi: 10.1093/bioinformatics/bts319. [DOI] [PubMed] [Google Scholar]
- Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee BH, Shi Y, Zhang N, de Poot SA, Tuebing F, et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science. 2016:351. doi: 10.1126/science.aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenar V, Piotto M, Leppik R, Saudek V. Gradient-Tailored Water Suppression for 1H-15N HSQC Experiments Optimized to Retain Full Sensitivity. J Magn Reson A. 1993;102:241–245. [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieren ES, El Ayadi A, Xiao Y, Siller E, Landsverk ML, Oberhauser AF, Barral JM, Boehning D. Ubiquilin-1 is a molecular chaperone for the amyloid precursor protein. J Biol Chem. 2011;286:35689–35698. doi: 10.1074/jbc.M111.243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trader DJ, Simanski S, Kodadek T. A Reversible and Highly Selective Inhibitor of the Proteasomal Ubiquitin Receptor Rpn13 Is Toxic To Multiple Myeloma Cells. J Am Chem Soc. 2015;137:6312–6319. doi: 10.1021/jacs.5b02069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou WL, Ouyang M, Hosking RR, Sutton JR, Blount JR, Burr AA, Todi SV. The deubiquitinase ataxin-3 requires Rad23 and DnaJ-1 for its neuroprotective role in Drosophila melanogaster. Neurobiol Dis. 2015;82:12–21. doi: 10.1016/j.nbd.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderLinden RT, Hemmis CW, Schmitt B, Ndoja A, Whitby FG, Robinson H, Cohen RE, Yao T, Hill CP. Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol Cell. 2015;57:901–911. doi: 10.1016/j.molcel.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41:1767–1777. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- Walters KJ, Lech PJ, Goh AM, Wang Q, Howley PM. DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc Natl Acad Sci U S A. 2003;100:12694–12699. doi: 10.1073/pnas.1634989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KJ, Matsuo H, Wagner G. A simple method to distinguish intermonomer nuclear Overhauser effects in homodimeric proteins with C2 symmetry. J Am Chem Soc. 1997;119:5958–5959. [Google Scholar]
- Wang H, Lim PJ, Yin C, Rieckher M, Vogel BE, Monteiro MJ. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington’s disease by ubiquilin. Hum Mol Genet. 2006;15:1025–1041. doi: 10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- Wang H, Monteiro MJ. Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem Biophys Res Commun. 2007;360:423–427. doi: 10.1016/j.bbrc.2007.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Wang X, Chen CF, Baker PR, Chen PL, Kaiser P, Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46:3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- Wang X, Huang L. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol Cell Proteomics. 2008;7:46–57. doi: 10.1074/mcp.M700261-MCP200. [DOI] [PubMed] [Google Scholar]
- Watkins JF, Sung P, Prakash L, Prakash S. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol Cell Biol. 1993;13:7757–7765. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- Yu C, Yang Y, Wang X, Guan S, Fang L, Liu F, Walters KJ, Kaiser P, Huang L. Characterization of Dynamic UbR-Proteasome Subcomplexes by In vivo Cross-linking (X) Assisted Bimolecular Tandem Affinity Purification (XBAP) and Label-free Quantitation. Mol Cell Proteomics. 2016 doi: 10.1074/mcp.M116.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Chen T, Ziv I, Rosenzweig R, Matiuhin Y, Bronner V, Glickman MH, Fushman D. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell. 2009a;36:1018–1033. doi: 10.1016/j.molcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Raasi S, Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377:162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wang Q, Ehlinger A, Randles L, Lary JW, Kang Y, Haririnia A, Storaska AJ, Cole JL, Fushman D, et al. Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol Cell. 2009b;35:280–290. doi: 10.1016/j.molcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Guo Y, Chen Y, Chen M, Lin Z, Wu Y, Chen Y. Knockdown of Adhesion-Regulating Molecule 1 Inhibits Proliferation in HL60 Cells. Acta Haematol. 2015;134:88–100. doi: 10.1159/000369916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.