SUMMARY
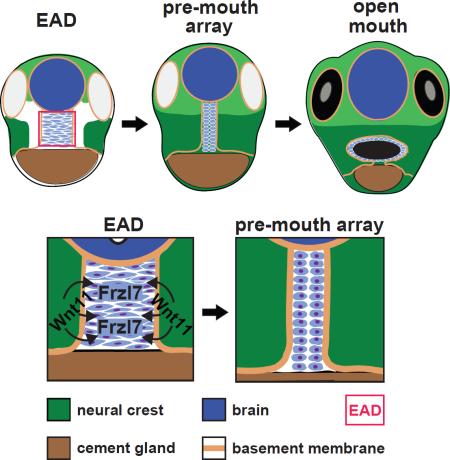
The mouth arises from the extreme anterior domain (EAD), a region where the ectoderm and endoderm are directly juxtaposed. Here we identify a ‘pre-mouth array’ in Xenopus that forms soon after the cranial neural crest has migrated to lie on either side of the EAD. Initially, EAD ectoderm comprises a wide and short epithelial mass that becomes narrow and tall with cells and nuclei changing shape, a characteristic of convergent extension. The resulting two rows of cells - the pre-mouth array - later split down the midline to surround the mouth opening. Neural crest is essential for convergent extension and likely signals to the EAD through the Wnt/PCP pathway. Fzl7 receptor is locally required in EAD ectoderm, while Wnt11 ligand is required more globally. Indeed, heterologous cells expressing Wnt11 can elicit EAD convergent extension. The study reveals a precise cellular mechanism that positions and contributes to the future mouth.
Graphical abstract
INTRODUCTION
The Extreme Anterior Domain (EAD) is the earliest facial element. It is present across the anterior midline and comprises directly juxtaposed anterior ectoderm and endoderm that develop into the mouth opening and oral cavity (Dickinson and Sive, 2006; Dickinson and Sive, 2009; Jacox and Sindelka et al. 2014). In Xenopus, the EAD is also a craniofacial organizer required for cranial neural crest (NC) ingression towards the midline (Jacox and Sindelka et al., 2014). This results in NC coming to lie on either side of the EAD prior to differentiating into facial tissues (Dickinson and Sive, 2007; Spokony, 2002).
Mouth development from the EAD occurs in multiple steps (Dickinson and Sive, 2006, 2007) that include specifying a broad anterior domain characterized by expression of pitx genes at neurula stages and disappearance of basement membrane (BM) between EAD ectoderm and endoderm at late tailbud. The stomodeal invagination then forms and ectoderm undergoes a burst of apoptosis that thins the tissue (Dickinson and Sive, 2006). Thinned ectodermal and endodermal layers intercalate to form the buccopharyngeal membrane that perforates as the mouth opens at swimming tadpole stage.
We previously noted that the EAD ectoderm forms a discrete epithelium 2-4 cells wide (Jacox and Sindelka et al., 2014). Here, we explore this observation further and show that the EAD ectoderm elongates into a ‘pre-mouth array’ that later contributes to the oral opening. Cell sheet elongation can be driven by convergent extension, a process of cell intercalation that lengthens epithelial sheets and may require the Wnt/PCP pathway (Roszko et al., 2009). We address the hypothesis that the pre-mouth array forms by convergent extension (CE) and examine the signaling role of adjacent NC in directing EAD ectoderm morphogenesis. The data reveal a precise cellular organization and identify the associated control mechanisms that position the future mouth opening.
RESULTS
EAD ectoderm becomes longer and narrower to form a ‘pre-mouth array’ that later opens into the mouth
To confirm that the Xenopus Extreme Anterior Domain (EAD) gives rise to the mouth, and to trace the origin and other derivatives of the EAD, we extended previous fate mapping analyses (Dickinson and Sive, 2006). DiI tracing from mid-neurula (stage 18) to mouth opening at swimming tadpole stage (stage 40) demonstrates that EAD ectoderm originates from the anterior neural ridge and contributes to the mouth (oral cavity), nostrils and anterior pituitary (AP) (Figure S1) consistent with data from other species (Chapman et al., 2005; Couly and Le Douarin, 1985; Eagleson et al., 1995; Osumi-Yamashita et al., 1994; Schwind, 1928).
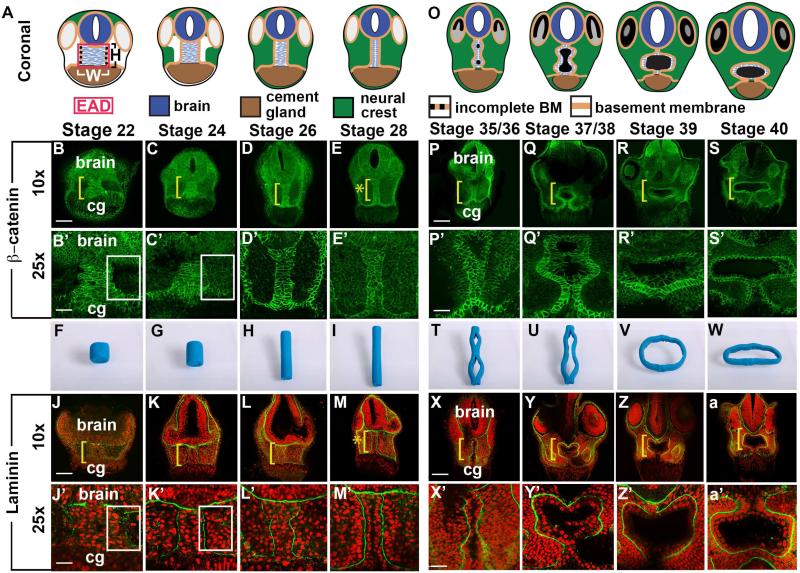
We assessed the narrowing of the EAD ectoderm previously and noticed its relationship to mouth opening (Jacox et al., 2014), through a histological time course from late neurula (stage 20) to swimming tadpole (stage 40) analyzing adherens junctions through β-catenin and basement membrane (BM) through staining for Laminin (Figure 1, S2). In coronal sections, the EAD ectoderm showed high levels of membrane-bound β-catenin (Figure 1A-E) and was bordered by BM that separated it from neural crest (NC), the brain and cement gland (Figure 1J-M). Lateral BM was patchy prior to NC ingression towards the EAD ectoderm but became continuous as NC migrated into the face at stage 24 (Figure 1J-K, 3B-E) and persisted subsequently (Figure 1L-M). Prior to NC ingression, a ‘pouch’ devoid of cells is present on either side of the EAD ectoderm (white box) (Figure 1B’-C’, J’-K’).
Figure 1.
Coronal anatomy of Xenopus face and EAD ectoderm between late neurula and swimming tadpole. (A, O) Schematic, st. 22-28 and st. 35/36-40. (B-E’, P-S’) Coronal sections with β-catenin immunolabeling (2 independent experiments, st. 22, n=10; st. 24, n=11; st. 26, n=14; st. 28 n=14). Midline region (bracket) with bright β-catenin labeling is EAD ectoderm of the pre-mouth array. Bracket: region of 10x image (B-E, P-S) enlarged in 25x view (B’-E’, P’-S’). Asterisk (E), pre-mouth array at stage 28, enlarged in E’. cg, cement gland. (F-I, T-W) Still frames from claymation of mouth opening found in Movie S2. I, pre-mouth array stage of claymation. (J-M’, X-a’); Coronal sections with Laminin (green) immunolabeling with Propidium Iodide (PI) nuclear counterstain (red) (2 independent experiments, st. 22, n=10; st. 24, n=4; st. 26, n=6; st. 28, n=7). Bracket: region of 10x image (J-M, X-a) enlarged in 25x view (J’-M’, X’-a’). Asterisk (M), pre-mouth array at stage 28, enlarged in M’. (B’, C’, J’, K’) White boxes surround lateral regions next to EAD which fill with NC cells between stages 22 (B’, J’) and 24 (C’, K’). Scale bar (10x): 170μm. Scale bar (25x): 68μm.
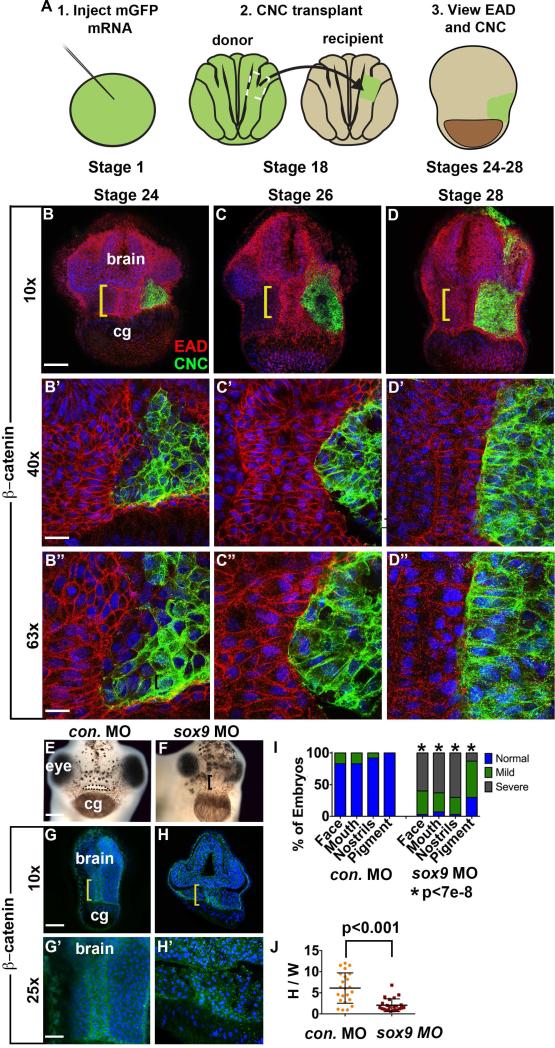
Figure 3.
EAD ectoderm undergoes convergent extension (CE) as the cranial neural crest (NC) approaches the midline and EAD CE fails to occur in sox9 LOF embryos. (A) Experimental schematic. (B-D’’) Coronal sections with mGFP-labeled NC (green) and β-catenin (red) immunolabeling from late neurula (stage 24) to late tailbud (stage 28) (2 independent experiments, st. 24, n=7; st. 26, n=7; st. 28 n=4). Midline region (bracket) with bright β-catenin labeling is EAD ectoderm. Bracket: region of 10x image (B-D) enlarged in 40x view (B’-D’) and 63x view (B’’-D’’). cg, cement gland. (E-F) Frontal view of control and sox9 LOF embryos at swimming tadpole (stage 40) assayed in 2 experiments (control MO (E) n=24; sox9 MO (F) n=30.) Dots surround open mouth. Bracket: unopened mouth. Scale bar: 200 m. (G-H’) Coronal sections assayed in 4 independent experiments (n=23) with β-catenin immunolabeling and Hoersch nuclear labeling. Midline region with bright β-catenin labeling is EAD ectoderm. Bracket: region of 10x image (G-H) enlarged in 25x view (G’-H’). cg, cement gland. (I) Graph depicting percent of embryos displaying mouth, face, nostrils and pigment formation phenotypes at st. 40 in control and sox9 LOF embryos. P values: Fisher's exact probability test. (J) Quantification of height over width of EAD (see Methods). P values: unpaired, two-tailed T test. Error bar: standard deviation. Unless otherwise specified, Scale bar (10x): 170μm. Scale bar (25x): 68μm. Scale bar (40x): 43μm. Scale bar (63x): 27μm.
At early tailbud (stage 22) the EAD ectoderm comprises a wide and short region (8-9 cells wide and 9-10 cells high), while at late tailbud stage (stage 28) it forms a tall, thin column of cells (2-3 cells wide and 20-21 cells high) (Table 1, Figure 1B-M). We call this ~2×20 cell organization the ‘pre-mouth array’ (Figure 1E, M, asterisk). BM surrounds EAD ectoderm, laterally (Figure 1J-M), dorsally and ventrally (Figure S2 and Movie S1). At hatching stage (stage 35-36), the two rows of β-catenin-positive cells separate down the middle and the oral opening is complete in the tadpole (stage 39-40) still surrounded by BM (Figure 1P-S, X-a). In contrast to BM, the apical marker ZO-1 appeared just before the cell rows separate at hatching stages (Figure S3I). Using Claymation, we animated a putative sequence of EAD ectodermal elongation and mouth opening (still frames Figure 1F-I, 1T-W, and Movie S2).
Table 1.
Average Cell Number of EAD Tissue Dimensions
| Deep Ectodermal Average | Endoderm | Outer Ectoderm | ||||
|---|---|---|---|---|---|---|
| Height | Width | Depth | Total (H*W*D) | Depth | Depth | |
| St. 22 | 9.5 (n=35, SD 2.1) | 8.2 (n=12, SD 2.3) | 6.2 (n=23, SD 1.3) | 485 | 2.0 (n=23, SD 0.8) | 1.0 (n=23, SD 0.5) |
| St. 24 | 10.9 (n=32, SD 2.1) | 6.7 (n=16, SD 2.5) | 8.0 (n=16, SD 1.7) | 579 | 2.1 (n=16, SD 1.2) | 1.7 (n=16, SD 0.7) |
| St. 26 | 15.6 (n=46, SD 3.0) | 3.8 (n=17, SD 1.4) | 8.7 (n=29, SD 2.0) | 521 | 2.3 (n=29, SD 1.3) | 1.4 (n=29, SD 0.7) |
| St. 28 | 20.3 (n=59, SD 3.4) | 2.7 (n=41, SD 0.6) | 8.2 (n=18, SD 0.8) | 446 | 1.9 (n=18, SD 1.1) | 1.4 (n=18, SD 0.5) |
These data identify a previously undescribed stage of mouth development, the ‘pre-mouth array’ that derives from organization of the EAD ectoderm and later contributes to the oral opening.
EAD ectoderm reorganization is consistent with convergent extension
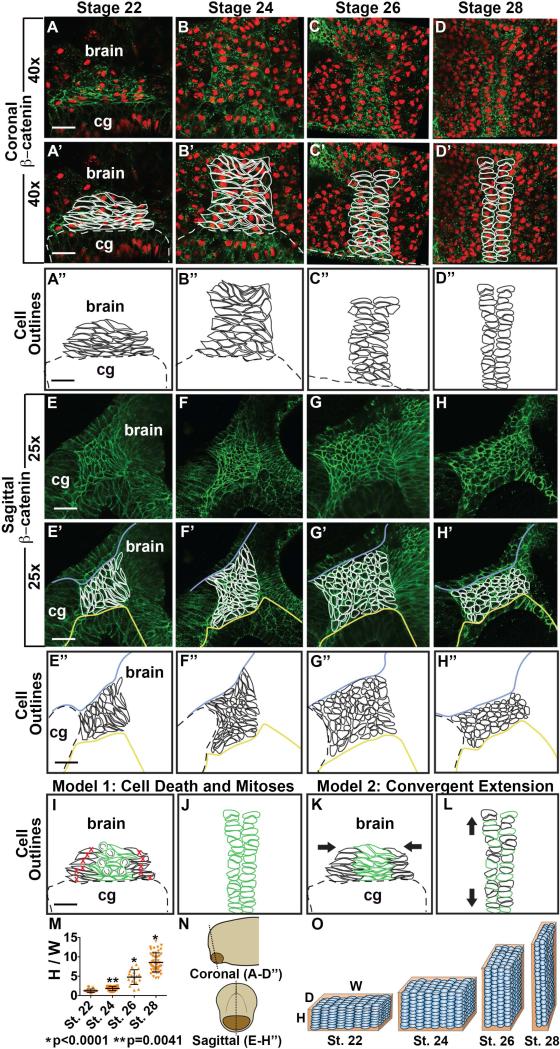
To examine EAD ectodermal reorganization at higher resolution and assess underlying mechanism, we tiled cells in the region and quantified height, width and depth. As noted at lower magnification, the EAD ectoderm begins as a wide and short region (8-9 cells wide and 9-10 cells high) at early tailbud (stage 22), while at late tailbud stage (stage 28) it forms a tall, thin column of cells (2-3 cells wide and 20-21 cells high) (Table 1, Figure 2A-D). Analysis of cell shape showed that early tailbud stage cells were laterally elongated (bipolar) with laterally elongated nuclei, whereas later tailbud stage cells were columnar with round nuclei and formed two parallel rows (Figure 2A-H, S3II). These differences are consistent with convergent extension, where cells rearrange to lengthen and narrow the EAD ectoderm while undergoing stereotypical cell and nuclear shape changes (Wallingford et al., 2002; Yin et al., 2009). The EAD ectoderm is 6-7 cells deep at stage 22 and deepens slightly to 8-9 cells at stage 28 (Table 1, Figure 2E-H). Two models that could account for observed morphogenetic changes of EAD ectoderm were considered. In model 1 (Figure 2I-J), lateral EAD ectodermal cells die, narrowing the tissue, while midline cells divide with their mitotic axes oriented to promote tissue elongation. For this model to account for changes in height and width between stages 22 and 28, 67% of EAD ectoderm cells would need to die and each midline EAD ectodermal cell would need to divide once, doubling EAD height (Table 1). In model 2 (Figure 2K-L), convergent extension transforms EAD ectoderm from a square to an elongated rectangle due to cellular intercalation and shape change with minimal cell division and death.
Figure 2.
Detailed anatomy and modeling of Xenopus EAD ectoderm between late neurula and swimming tadpole. (A-D’) Coronal sections with β-catenin (green) immunolabeling with PI nuclear counterstain (red) from stages 22-28 (2 independent experiments, st. 22, n=10; st. 24, n=11; st. 26, n=14; st. 28 n=14). (E-H’) Sagittal sections with β-catenin (green) immunolabeling from stages 22-28 (3 independent experiments, st. 22, n=12; st. 24, n=10; st. 26, n=12; st. 28, n=12). (A’-D’, E’-H’) Cell membranes traced in white. Blue line, separates deep EAD from outer ectoderm. Yellow line, separates EAD ectoderm from endoderm. (A’’-D’’, E’’-H’’) Cell outlines in black. Dotted line: top of cement gland, cg. (D-D’’) Pre-mouth array is present and indicated by cell outlines. Scale bar (25x): 68μm. Scale bars (40x): 43μm. (I-J) Model 1. I, stage 22. J, stage 28. (K-L) Model 2. K, stage 22. L, stage 28. (M) Quantification of height vs. width of EAD (see Methods) (3 independent experiments, st.22, n=12; st. 24, n=16; st. 26, n=17; st. 28, n=47). P values: unpaired, two-tailed T test comparing sequential stages. Error bar: standard deviation. (N) Diagram demonstrating coronal (A-D’) and sagittal (E-H’) sections. (O) Diagram showing the change in height (H), width (W) and depth (D) of the EAD ectoderm and its surrounding BM between stages 22 and 28. Blue ovals, EAD ectodermal cells undergoing convergent extension. Orange rectangular prism, Laminin BM surrounding EAD ectoderm.
Model 1 is not supported by the data as fewer than 1% of EAD ectodermal cells die (Dickinson and Sive, 2006, Figure S6) and there are infrequent cell divisions, on average 1-3 mitoses throughout the EAD ectoderm between stages 23 and 26, with random axis orientation (Figure S6). In contrast, observed tissue and cell changes are consistent with model 2, and we conclude that convergent extension is the likely mechanism by which EAD ectodermal elongation into the pre-mouth array occurs.
EAD ectoderm convergent extension is associated with neural crest ingress and dependent on neural crest
Previous analyses (Jacox and Sindelka et al, 2014) suggested that EAD ectoderm elongation is correlated with NC migration into the region. To examine the timing of NC association with EAD ectoderm morphogenesis, we performed detailed immunohistochemistry where mGFP-labeled NC was transplanted into unlabeled recipients, which were later immunostained for β-catenin (Figure 3A-D). By late neurula, the NC arrived at the EAD, but did not completely fill the space adjacent to it, and regions of EAD ectoderm were not touching NC. Later, NC cells closely abutted the entire EAD ectoderm which became narrower and taller by late tailbud stages. NC cells did not mingle with EAD ectoderm, but remained a distinct group with a sharp border at the EAD, suggesting this region may act as a barrier.
To characterize dynamics of NC/EAD localization, live lineage analysis was performed using a mApple-labeled EAD transplanted into a recipient that included mGFP-labeled NC (Figure S3III). Consistent with histology, the EAD transplant changed from a rectangular shape at late neurula to become narrow and tall as NC came to lie on either side of the EAD (Figure S3III). Consistent results were obtained by double in situ hybridization with the EAD marker, cpn, and a NC marker, sox9 (Figure S3III).
The close association of NC ingress and EAD elongation suggested that NC may be required for EAD ectoderm morphogenesis. NC lineages were ablated by LOF for the essential gene sox9 (Spokony et al., 2002) by injection of morpholino-modified antisense oligonucleotides (MOs). Resultant embryos did not show EAD ectoderm elongation, indicating a requirement for NC in this process (Figure 3G-H, J). A titration of sox9 MOs identified a threshold requirement for proper EAD ectoderm convergent extension (Figure S4). Older embryos did not recover midline convergent extension at stage 32 (Figure S7) and failed to form a mouth and nostrils at stage 40 (Figure 3E-F).
These data show a close association of EAD ectoderm morphogenesis and adjacent NC, and further demonstrate that NC is required for EAD convergent extension.
A localized requirement for Wnt/PCP signaling from the neural crest in EAD ectoderm convergent extension
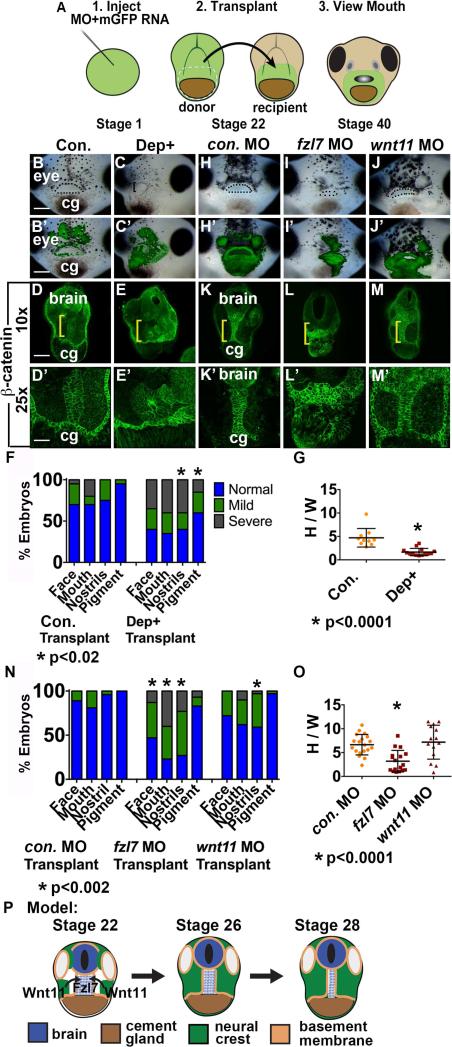
Convergent extension is dependent on Wnt/PCP signaling in other morphogenetic events (Roszko et al., 2009) (Figure 4P), and we therefore explored necessity of this pathway during EAD ectoderm elongation. We tested whether the frizzled 7 receptor (fzl7), intracellular signaling mediator dvl and Wnt/PCP ligand wnt11 were required specifically in the EAD for convergent extension. These assays were based on expression of fzl7 RNA throughout the face, wnt11 RNA in NC but not in the EAD (Figure S4), and on whole embryo LOF phenotypes (Figure S5, S6, S7). A facial transplant protocol was used (Jacox et al., 2014) where GFP-labeled EAD was removed from control, Dep+ injected embryos, or from frzl7 and wnt11 LOF embryos at early tailbud (stage 22) and transplanted to sibling controls (Figure 4A) (Jacox et al., 2014). Control transplants underwent normal EAD ectoderm elongation (Figure 4B, D, H, K, and quantified in F, G, N, O), however, when frzl7 LOF or Dep+ EAD was transplanted into control embryos, extension did not occur and small, deformed mouths and nostrils formed (Figure 4C, E-G, I, L, N, O). In contrast, transplant of wnt11 LOF EAD led to normal extension and face formation (Figure 4J, M-O), indicating that regions outside the EAD can supply Wnt11. Local frzl7 LOF did not alter NC development, as measured by sox9 expression (Figure S7). Failure of EAD ectoderm to undergo convergent extension is correlated with reduced or absent nostrils, however, it is unclear whether these events are mechanistically connected.
Figure 4.
Fzl7 is locally required in the EAD ectoderm for convergent extension. Local requirement of Dsh, fzl7, and wnt11 expression tested with an EAD transplant technique. (A) Experimental design: donor LOF tissue was transplanted to uninjected sibling recipients. (B-C’) EAD transplant outcome from control or Dep+ RNA donor tissue assayed in 3 experiments. ((B, B’) control RNA n=23; (C, C’) Dep+ RNA n=22.) (B’-C’) Overlay of (B-C) with GFP fluorescence indicating location of donor transplant in recipient. Dots surround open mouths. Bracket: unopened mouth. Frontal view. cg, cement gland. Scale bar: 200 m. (D-E’) Coronal sections of EAD transplants with control or Dep+ donor tissue assayed in 3 independent experiments ((D, D’) control RNA n=10; (E, E’) Dep+ RNA n=14) with β-catenin immunolabeling. Midline region with bright β-catenin labeling is EAD ectoderm. Bracket: region of 10x image (D-E) enlarged in 25x view (D’-E’). (F) Quantification of normal or abnormal structure development depending on background of facial tissue. P values: Fisher's exact probability test. (G) Quantification of height over width of EAD (see Methods). P value: unpaired, two-tailed T test. Error bar: standard deviation. (H-J) EAD transplant outcome from control, fzl7 or wnt11 LOF donor tissue assayed in 4 independent experiments. ((H, H’) control MO n=27; (I, I’) fzl7 MO n=30; (J, J’) wnt11 MO n=30.) (H’-J’) Overlay of (H-J) with GFP fluorescence indicating location of donor transplant in recipient. Dots surround open mouths. Bracket: unopened mouth. Frontal view. Scale bar: 200μm. (K-M’) Coronal sections of EAD transplants with control, fzl7 or wnt11 donor tissue assayed in 4 independent experiments with β-catenin immunolabeling ((K, K’) control MO n=19; (L, L’) fzl7 MO n=17; (M, M’) wnt11 MO n=14). Midline region with bright β-catenin labeling is EAD ectoderm. Bracket: region of 10x image (K-M) enlarged in 25x view (K’-M’). (N) Quantification of normal or abnormal structure development depending on LOF background of facial tissue. P values: Fisher's exact probability test. (O) Quantification of height over width of EAD midline tissue in transplants. P values: unpaired, two-tailed T test. (P) Schematic of model. NC releases Wnt11 which acts on Fzl7 receptors expressed on midline EAD cells. Unless otherwise specified, Scale bar (10x): 170μm. Scale bar (25x): 68μm.
After sox9, wnt and frzl7 LOF, Laminin expression adjacent to the EAD was disordered and spotty (Figure S6), suggesting that NC may be required for formation of BM lateral to the EAD. We further asked whether oriented cell division or death occurred during EAD ectoderm elongation. Phospho-Histone-3 (PH3) immunolabeling before or during EAD ectoderm elongation showed few dividing cells, randomly oriented (Figure S6). Minimal cell death was observed in the EAD ectoderm of control or LOF embryos (Figure S6). The very low levels of cell division and death and lack of significant change after Wnt/PCP LOF further supports the conclusion that the EAD ectoderm undergoes convergent extension.
These results indicate that a Wnt11 signal originating outside the EAD, likely in the NC, engages the Fzl7 receptor on EAD ectodermal cells to activate convergent extension via the Wnt/PCP pathway.
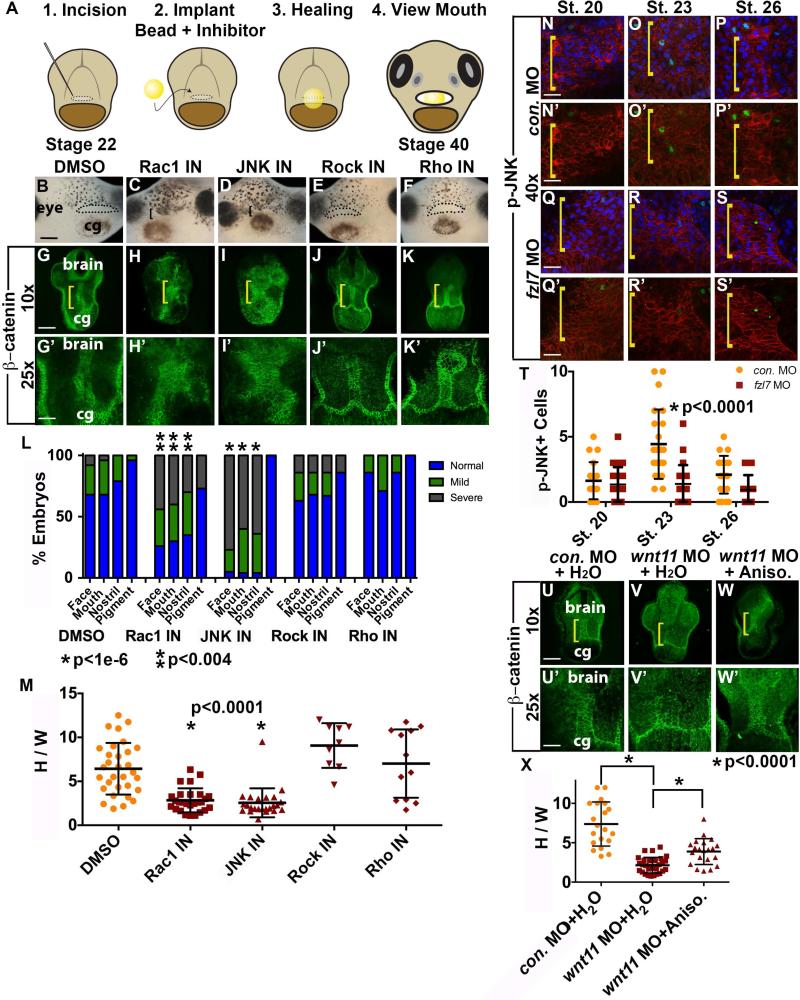
The GTPase effectors Rac and JNK are required for EAD ectoderm convergent extension
The Wnt/PCP signaling pathway bifurcates (Wallingford and Habas, 2005), and we asked which branch was involved in EAD convergent extension by assaying relevant GTPases using inhibitors and a bead-implantation assay (Figure 5A). Beads soaked in DMSO control, Rock or Rho inhibitors did not interfere with EAD ectoderm morphogenesis or face formation (Figure 5B, G, E-F, J-K, L, M), however Rac1 and JNK inhibitors were associated with reduced convergent extension and failure of mouth and nostril formation (Figure 5C-D, H-I, L, M). Implanting JNK inhibitor beads at stages 22, 24, or 28 caused marked craniofacial abnormalities at stage 40, while inhibitor application after convergent extension at stage 32 resulted in a less severe phenotype, suggesting JNK signaling is crucial specifically during tailbud stages for mouth development (Figure S8). Immunostaining for active, nuclear localized p-JNK (Yin et al., 2009) demonstrated few positive nuclei prior to EAD ectoderm elongation (Figure 5N, T), and maximal nuclear localization in EAD ectodermal cells during elongation (stage 23, Figure 5O, T). The number of p-JNK-positive nuclei peaked by stage 23 as elongation is beginning and diminished by stage 26 when extension is largely complete (Figure 5P, T). frzl7 LOF embryos demonstrated significantly lower levels of positive nuclei at stage 23 relative to control, concomitant with absence of EAD ectoderm convergent extension (Figure 5Q-T). When a bead containing the JNK-activator Anisomycin was implanted in Wnt11 LOF faces, there was an improvement in convergent extension (Figure 5U-X). These data indicate that the Rac1 and JNK branch of Wnt/PCP signaling is necessary for EAD ectoderm convergent extension.
Figure 5.
Inhibition of GTPases JNK and Rac1 is associated with a reduction in EAD ectodermal convergent extension. (A) Experimental schematic of inhibitor loaded bead implantation in the presumptive mouth, EAD region. (B-F) Frontal view of swimming tadpole (stage 40) embryos with inhibitor loaded beads implanted in presumptive mouths, assayed in 3 experiments. ((B) control DMSO n=97; (C) Rac1 n=40; (D) JNK inhibitor n=44; (E) Rock inhibitor n=75; (F) Rho inhibitor n=39.) Bracket: unopened mouth. Dots surround open mouths. cg, cement gland. Scale bar: 200μm. (G-K’) Coronal sections, stage 28, assayed in 3 independent experiments. ((G, G’) control DMSO n=31; (H, H’) Rac1 inhibitor n=27; (I, I’) JNK inhibitor n=24; (J, J’) Rock inhibitor n=9; (K, K’) Rho inhibitor n=12)) with β-catenin immunolabeling. Midline region with bright β-catenin labeling is EAD ectoderm. Bracket: region of 10x image (G-K) enlarged in 25x view (G’-K’). Scale bar (10x): 170μm. Scale bar (25x): 68μm. (L) Graph depicting percentage of embryos, displaying mouth, face, nostrils and pigment formation phenotypes at stage 40. P values: Fisher's exact probability test. (M) Quantification of height over width of EAD (see Methods). P values: unpaired, two-tailed T test. Error bar: standard deviation. (N-S’) Control and fzl7 LOF embryos at stage 20 (N, N’, n=19; Q, Q’, n=26), stage 23 (O, O’, n= 23; R, R’, n=23), and stage 26 (P, P’, n=21; S, S’, n=16) with p-JNK immunolabeling (green), mApple cell membranes (red), and Hoechst nuclear counterstain (blue in N-S) assayed in 2 experiments. Bracket: EAD. Scale bars (40x): 43μm. (T) Quantification of cells with p-JNK positive nuclei in the EAD ectoderm. Total number of EAD nuclei was equivalent between stage matched control and frzl7 LOF embryos. p<0.0006, control stage 23 compared to stages 20 and 26. P values: unpaired, two-tailed T test. (U-W’) Coronal sections, stage 28, assayed in 2 independent experiments. ((U, U’) control water n=20; (V, V’) wnt11 MO + control water n=30; (W, W’) wnt11 MO + Anisomycin JNK-activator n=21) with β-catenin immunolabeling. Midline region with bright β-catenin labeling is EAD ectoderm. Bracket: region of 10x image (U-W) enlarged in 25x view (U’-W’). Scale bar (10x): 170μm. Scale bar (25x): 68μm. (X) Quantification of height over width of EAD (see Methods). Error bar: standard deviation. P values: unpaired, two-tailed T test.
Wnt11 can substitute for neural crest and is sufficient to direct EAD ectoderm convergent extension
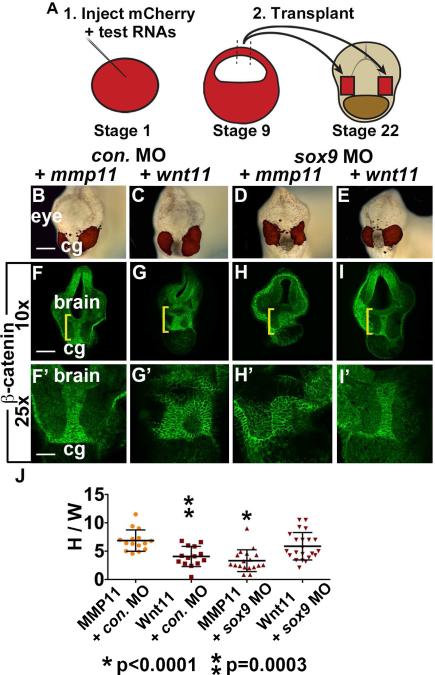
Since Wnt11 is necessary for EAD ectoderm elongation, we asked whether Wnt11 is sufficient to elicit this process by testing whether animal caps expressing ectopic Wnt11 could substitute for NC. One cell embryos were injected with RNA encoding a control secreted protein (MMP11) or Wnt11; animal caps (blastula stage ectoderm) were removed at stage 9 and implanted on either side of the EAD in control or sox9 LOF embryos at stage 22 (Figure 6A). Embryos were assayed at stage 28 when EAD ectoderm convergent extension is normally complete. While caps expressing MMP11 did not alter control embryos (Figure 6B, F) or rescue EAD shape in sox9 LOF embryos (Figure 6D, H), Wnt11-expressing caps were sufficient to restore EAD ectoderm extension when implanted into sox9 LOF embryos (Figure 6E, I, J). Wnt11-expressing caps reduced EAD ectoderm elongation in control embryos (Figure 6C, G, J), consistent with other overexpression phenotypes (De Calisto et al., 2005; Garriock et al., 2005). These data show that Wnt11 signaling is sufficient to direct EAD ectoderm convergent extension, and highlight the conclusion that the Wnt/PCP pathway directs the pre-mouth array stage of oral development.
Figure 6.
Wnt11 is sufficient for EAD ectoderm convergent extension. (A) Sufficiency of Wnt11 for midline CE was tested with an animal cap transplant technique. Experimental schematic of bilateral transplants with mApple, animal cap overexpressing Wnt11 or a control, secreted protein (inactive MMP11). (B-E) Overlay of brightfield images with mApple fluorescence indicating location of donor transplant in late tailbud recipients (stage 28). Scale bar: 200μm. (F-I’) Coronal sections of animal cap transplants with mmp11 or wnt11 overexpressing donor tissue assayed in 3 experiments with β-catenin immunolabeling ((F, F’) control MO+mmp11 n=15; (G, G’) control MO+wnt11 n=14; (H, H’) sox9 MO+mmp11 n=18; (I, I’) sox9 MO+wnt11 n=22). Midline region with bright β-catenin labeling is EAD ectoderm. Bracket: region of 10x image (F-I) enlarged in 25x view (F’-I’). (J) Quantification of height over width of EAD (see Methods). P values: unpaired, two-tailed T test. Error bar: standard deviation. Unless otherwise specified, Scale bar (10x): 170μm. Scale bar (25x): 68μm.
DISCUSSION
Mouth development is a lengthy process - three days in Xenopus or five weeks in humans, and includes many steps that position the mouth-forming region and later define where the opening will occur. This careful sequence of events ensures that when the mouth opens, it connects productively with the digestive system. In this study, we clarify one mechanism underlying mouth formation and reach four major conclusions. First, Extreme Anterior Domain (EAD) ectoderm forms an organized cell arrangement, the pre-mouth array that will later split down the midline and contribute to the oral opening. Second, pre-mouth array formation is elicited by convergent extension. Third, the signal for EAD convergent extension originates in the neural crest (NC) as it comes to lie on either side of the EAD. Fourth, Wnt/PCP signaling is both necessary and sufficient to elicit EAD convergent extension. These findings reveal an unexpected level of precision controlling vertebrate mouth development.
Organization of the presumptive mouth ectoderm into two rows of cells that will later open down the apical midline has not been observed previously. We termed this the pre-mouth array to reflect that organization of the Xenopus mouth is established long before it opens. Indeed, the pre-mouth array forms very early, prior to breakdown of the basement membrane between ectoderm and endoderm (Dickinson and Sive, 2009) and persists until stomodeum formation begins, when the apical surfaces of the pre-mouth array cells begin to separate. Localization of apical junction proteins is associated with separation (Figure S3I) and future analyses will address when the apical domain appears and whether it is necessary for mouth opening. The pre-mouth array forms from EAD ectoderm that overlies and connects with pharyngeal (gut) endoderm, suggesting that a signal from the underlying endoderm may also be involved in mouth opening.
Morphogenetic mechanisms by which the pre-mouth array forms could not be accounted for by directed proliferation or apoptosis, but EAD ectodermal cell shape and rearrangements appear similar to mediolateral intercalation resulting in convergent extension of axial mesoderm during Xenopus and zebrafish gastrulation (Keller et al., 2000; Tada and Heisenberg, 2012; Yin et al., 2009). We therefore conclude that the EAD ectoderm undergoes convergent extension. Absence of EAD ectoderm convergent extension is tightly associated with an abnormally small or absent mouth opening, suggesting it is a necessary part of mouth development.
The EAD ectoderm that undergoes convergent extension is a multilayered array, 6-8 cells deep, which is bounded by basement membrane (BM). Thus, there is BM above the EAD ectoderm (abutting the epidermal ectoderm and brain), below it (abutting the pharyngeal endoderm) and on either side (abutting the NC) (Movie S1). These BMs form a 3D ‘cage’ around the EAD ectoderm and perhaps constrain its morphogenetic possibilities. It will be interesting to examine how EAD morphogenesis is coordinated between ectodermal layers. Interestingly, prior to NC ingress, the region around the EAD ectoderm is devoid of cells, forming a pouch that is filled by NC cells as they become tightly apposed to the basal sides of the EAD (Figure 1). The BM along the lateral edges of EAD ectoderm is patchy prior to NC ingress, suggesting that NC may promote BM formation on either side of the EAD.
Since EAD ectoderm convergent extension does not occur in the absence of NC, the NC likely sends the signal for morphogenesis. The disposition of NC relative to EAD ectoderm suggests that it signals along the basal-apical axis, and not along the planar axis as suggested for convergent extension during gastrulation (Tada and Heisenberg, 2012). It will be important to examine the localization of EAD Wnt/PCP signaling components to further understand the direction of the signal.
The requirement for wnt11 and the ability of Wnt11 to substitute for NC in eliciting EAD ectoderm extension indicates that this is a pivotal signaling factor, however, other Wnt/PCP ligands may also be involved. Wnt/PCP signaling is required for proper NC migration in Xenopus. For example, Wnt/PCP signaling at contact points between NC cells halts their movement and leads to a change in their direction; this contact inhibition promotes coherent NC migration (Carmona-Fontaine et al., 2008). In another example, through a “chase-and-run” reciprocal interaction, placodal cells attract NC, but direct contact facilitates N-Cadherin junction formation, activating the PCP pathway that repulses placodal cells (Theveneau et al., 2013; Steventon et al., 2014). NC and placodes remain in close proximity and signal reciprocally to coordinate morphogenesis of sensory systems (Steventon et al., 2014). Wnt/PCP signaling functions in an additional way to modulate EAD ectoderm morphogenesis, although it is unclear whether factors used are identical in each of these cases. Many additional factors are required for mouth formation, including sonic hedgehog (Eberhart et al., 2006; Tabler et al., 2014), retinoic acid (Kennedy and Dickinson, 2012; Dickinson and Sive, 2009) and β-catenin Wnt signaling (Dickinson and Sive, 2009). However, the role of these factors in EAD ectoderm morphogenesis is not known.
Previous analyses demonstrated that the EAD is an organizer, which promotes ingression of first arch NC into the nascent face to lie on either side of the EAD (Jacox et al., 2014, Figure 3). This study describes a subsequent reciprocal signaling from NC to the EAD ectoderm to elicit convergent extension. There may be additional NC/EAD interactions. We note that NC cells and EAD ectodermal cells do not mingle and the NC does not cross the midline, implying that the EAD is a barrier to incoming NC, perhaps maintaining distinct identities in the two halves of the face.
In sum, we have identified a pre-mouth array stage in oral development that forms through local signaling interactions and readies the embryo to later make an open mouth. The precision involved is striking, and future analyses will define details of the mechanism underlying pre-mouth array formation and mouth opening.
EXPERIMENTAL PROCEDURES
Embryo Preparation
Xenopus laevis embryos were cultured using standard methods (Sive et al., 2000). Xenopus embryos were staged according to Nieuwkoop and Faber, 1994.
DiI Labeling and Transplants
All fate mapping and transplants were done in 0.5x Modified Barth's Solution (MBS). Fate mapping was performed by injecting a 5-10nl drop of 1,1-dioctadecyl-3,3,3,3-tetramethylindocarbocyanin (DiI; 2mg/ml, Molecular Probes) or DiO (2mg/ml, Molecular Probes) into the EAD or anterior neural ridge ectoderm at early or late neurula stages. Embryos were photographed and fixed at tailbud and tadpole stages for immunohistochemistry. EAD transplants were performed according to Jacox et al., 2014. NC transplants were performed according to Mancilla and Mayor, 1996. Animal cap transplants were performed on late neurula control or sox9 LOF embryos, injected with 5ng of morpholino. Tissue lateral to the EAD in LOF embryos was extirpated using a 1mm diameter capillary tube pulled to a fine point. Animal caps were removed from embryos injected with 1μg of either inactive MMP11 (control) or Wnt11 mRNA plus 1μg of mApple mRNA. Animal cap tissue was transplanted into the face of extirpated LOF embryos, and held in place with glass bridges for 1-2 hours. Transplants were cultured until late tailbud, photographed, fixed and sectioned for immunohistochemistry.
Height, depth and width measurements of the EAD were made as follows. Height: number of cells between the top of the cement gland and bottom of the brain in coronal and sagittal sections. Depth: number of cells between the top/left and bottom/right boundaries of the EAD deep ectoderm with high β-catenin labeling, in sagittal sections. Width: number of cells between the left and right borders of the bright, β-catenin positive midline or between the left and right midline Laminin basement membranes, in coronal sections.
In Situ Hybridization
cDNAs were used to transcribe in situ hybridization probes including cpn (BC059995), sox9 (AY035397), frzl7 (De Calisto et al., 2005), wnt11 (Tada and Smith, 2000), pitx1 (Schweickert et al., 2001), pitx2c (Schweickert et al., 2001), frzb1 (BC108885), and XCG (Sive, 1989). In situ hybridization was performed as described by Sive et al., 2000, without proteinase K treatment. Double-staining protocol adapted from Wiellette and Sive, 2003.
Morpholinos and RNA Rescues
Xenopus antisense morpholino-modified oligonucleotides (“morpholinos; MOs”) included start site MOs targeting fzl7 (31ng, Winklbauer et al., 2001), wnt11 (9ng, Pandur et al., 2002), and sox9 (5ng, Spokony et al., 2002) were injected at the one cell stage. Murine mRNA (500ng fzl7 mRNA, 700ng wnt11 mRNA, generated from pRK5 plasmids, gifts of Chris Garcia and Jeremy Nathans, Addgene plasmids #42259 and #42290) (Yu et al., 2012) and morpholino (fzl7 MO 31ng, wnt11 MO 9ng) were coinjected at the one-cell stage to test morpholino specificity via RNA rescue. wnt11 mRNA (De Rienzo et al., 2011), truncated Dep+ disheveled mRNA (Sokol, 1996; Tada and Smith, 2000) and noncatalytic mmp11 (gift of Malcolm Whitman) were generated from plasmids. RNA was generated in vitro using the mMESSAGE mMACHINE kit (Ambion).
Immunohistochemistry
Immunohistochemistry was performed as described (Dickinson and Sive, 2006). Caspase-3 and PH3 labeling were performed according to Kennedy and Dickinson, 2012 and Dickinson and Sive, 2009. Primary antibodies included a rabbit, polyclonal anti-Laminin antibody (Sigma L-9393) diluted 1:150, rabbit, polyclonal anti-β-catenin (Invitrogen) diluted 1:100, a mouse, monoclonal anti-brdu antibody (Becton Dickinson 347580) diluted 1:750, and a mouse, monoclonal anti-Zo-1 antibody (Invitrogen) diluted 1:100. Secondary antibodies included Alexa 488 and 647 goat anti-rabbit (Molecular Probes) and Alexa 488 goat anti-mouse (Molecular Probes) diluted 1:500 with 0.1% propidium iodide (Invitrogen) or Hoersch (Life Technologies) as a counterstain. Phalloidin (Life Technologies), an actin dye, was used in combination with DiI labeling. Sections were imaged on a Zeiss LSM 710 Laser Scanning Confocal microscope. Images were analyzed using Imaris (Bitplane) and Photoshop (Adobe).
Wnt/PCP Inhibitor and Activator Assays
Rock inhibitor (Y-27632 Calbiochem, stock: 20mM in DMSO, working: 200μM in 2% DMSO, −80°C storage), Rac1 inhibitor (NSC23766 Santa Cruz, stock: 37mM in DMSO, working: 200μM in 2% DMSO, −20°C storage), Rho inhibitor (CCG-1423 Calbiochem, stock: 22mM in DMSO, working: 200μM in 2% DMSO, −20°C storage), and JNK inhibitor (SP600125 Sigma, stock: 20mM in DMSO, working: 200μM in 2% DMSO, 4°C storage) were resuspended in DMSO, aliquoted, and stored at stock concentrations until incubation with beads. JNK activator, anisomycin (#sc-3524 Santa Cruz Biotechnology, stock 67microg/ml, working: 67ng/ml in water, 4°C storage) (Liao et al., 2006), was resuspended in water, aliquoted, and stored at 1000x stock concentrations until incubation with beads. AG 1-X2 Resin beads (Bio Rad, 140-1231, 50-100 mesh) were washed in ethanol, dried, mixed with diluted, working concentration inhibitor solution and incubated overnight at 4°C. Affi-gel blue agarose beads (50–100 mesh, Bio-Rad) loaded with anisomycin activator were prepared according to Carmona-Fontaine (2011). Late neurula embryos had a small incision cut in their facial midline where a bead was inserted into the foregut behind the EAD. Embryos were grown to late tailbud for fixation and immunohistochemistry and to swimming tadpole for live imaging.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues for discussion, reagents and critical input, especially Ryann Fame, Jasmine McCammon and Christian Cortes-Campos. We thank George Bell for help with bioinformatics, Tom diCesare for assistance with figures and Wendy Salmon for imaging support. We also thank Sergei Sokol, Masazumi Tada, Malcolm Whitman, Axel Schweickert, Jean-Pierre Saint-Jeannet, Carl-Phillipe Heisenberg, Naoto Ueno and Richard Harland for gifts of plasmids. We are grateful to the NIDCR for support (1R01 DE021109-01 to HLS and F30DE022989 to LJ) and to Harvard University for the Herschel Smith Graduate Fellowship (to LJ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
H.S. directed and supervised the study. L.J. and H.S designed the experiments. L.J. carried out all experiments except for those conducted by J.C. featured in Figure 3A-D’’’. L.J. and J.C. contributed to Figure S4A-P’’’, S4d-h. Technical assistance was provided by A.R. for in situ hybridizations and immunohistochemistry in Figure 2, S3III G-L, S4Q-X, S5I-J’, S5P-R’, S6, and S7. H.L. created the claymation movie demonstrating EAD convergent extension shown in Figure 1X-a and Movie S2. H.L. and L.J. created Movie S1 using data collected by L.J. L.J. and H.S wrote and revised the manuscript with input from all authors.
REFERENCES
- Carmona-Fontaine C. PhD Thesis. University College of London; 2011. Epub. http://carloscarmonafontaine.wikispaces.com/Thesis. [Google Scholar]
- Carmona-Fontaine C, Matthews H, Kuriyama S, Moreno M, Dunn G, Parsons M, Stern C, Mayor R. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456(7224):957–961. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S, Sawitzke A, Campbell D, Schoenwolf G. A three-dimensional atlas of pituitary gland development in the zebrafish. J Comp. Neurol. 2005;487:428–440. doi: 10.1002/cne.20568. [DOI] [PubMed] [Google Scholar]
- Couly G, Le Douarin N. Mapping of the early neural primordium in Quail-Chick chimeras: 1. Developmental relationships between placodes, facial ectoderm, and prosencephalon. Dev. Bio. 1985;110:422–439. doi: 10.1016/0012-1606(85)90101-0. [DOI] [PubMed] [Google Scholar]
- De Calisto J, Araya C, Marchant L, Riaz CF, Mayor R. Essential role of non- canonical Wnt signaling in neural crest migration. Dev. 2005;132:2587–2597. doi: 10.1242/dev.01857. [DOI] [PubMed] [Google Scholar]
- De Rienzo G, Bishop JA, Mao Y, Pan L, Ma TP, Moens CB, Tsai LH, Sive H. Disc1 regulates both β-catenin-mediated and noncanonical Wnt signaling during vertebrate embryogenesis. FASB. 2011;25(12):4184–97. doi: 10.1096/fj.11-186239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson AJ, Sive H. Development of the primary mouth in Xenopus laevis. Dev Biol. 2006;295(2):700–713. doi: 10.1016/j.ydbio.2006.03.054. [DOI] [PubMed] [Google Scholar]
- Dickinson AJ, Sive H. Positioning the extreme anterior in Xenopus: cement gland, primary mouth and anterior pituitary. Semin. Cell Dev. Biol. 2007;18(4):525–533. doi: 10.1016/j.semcdb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Dickinson AJ, Sive HL. The Wnt antagonists Frzb-1 and Crescent locally regulate basement membrane dissolution in the developing primary mouth. Dev. 2009;136(7):1071–1081. doi: 10.1242/dev.032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagleson G, Ferreiro B, Harris WA. Fate of the anterior neural ridge and the morphogenesis of the Xenopus forebrain. J. Neuro. 1995;28(2):146–158. doi: 10.1002/neu.480280203. [DOI] [PubMed] [Google Scholar]
- Eberhart J, Swartz M, Crump J, Kimmel C. Early Hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Dev. 2006;133:1069–1077. doi: 10.1242/dev.02281. [DOI] [PubMed] [Google Scholar]
- Garriock R, D'Agostino S, Pilcher K, Krieg P. Wnt11-R, a protein closely related to mammalian Wnt11, is required for heart morphogenesis in Xenopus. Dev. Biol. 2005;279:179–192. doi: 10.1016/j.ydbio.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Jacox LA, Dickinson AJ, Sive H. Face transplants in Xenopus laevis embryos. JOVE epub; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacox LA, Sindelka R, Chen J, Rothman A, Dickinson A, Sive H. The extreme anterior domain is an essential craniofacial organizer acting through kinin-kallikrein signaling. Cell reports. 2014;8(2):596–609. doi: 10.1016/j.celrep.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R, Davidson L, Edlund A, Elul T, Ezin M, Shook D, Skoglund P. Mechanisms of convergence and extension by cell intercalation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:897–922. doi: 10.1098/rstb.2000.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A, Dickinson A. Median facial clefts in Xenopus laevis: Roles of retinoic acid signaling and homeobox genes. Dev. Biol. 2012;365(1):229–240. doi: 10.1016/j.ydbio.2012.02.033. [DOI] [PubMed] [Google Scholar]
- Liao G, Tao Q, Kofron M, Chen J, Schloemer A, Davis R, Hsieh J, Wylie C, Heasman J, Kuan C. Jun Nh2-terminal kinase (JNK) prevents nuclear β-catenin accumulation and regulates axis formation in Xenopus embryos. PNAS. 2006;103(44):16313–16318. doi: 10.1073/pnas.0602557103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancilla A, Mayor R. Neural crest formation in Xenopus laevis: mechanisms of Xslug induction. Dev. Biol. 1996;177:580–589. doi: 10.1006/dbio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin): A Systematical & Chronological Survey of the Development from the Fertilized Egg till the end of Metamorphosis. Garland Publishing; New York: 1994. [Google Scholar]
- Osumi-Yamashitna N, Ninomaya Y, Do H, Eto K. The contribution of both forebrain and midbrain crest cells to the mesenchyme of the frontonasal mass of mouse embryos. Dev. Biol. 1994;164:409–419. doi: 10.1006/dbio.1994.1211. [DOI] [PubMed] [Google Scholar]
- Pandur P, Läsche M, Eisenberg LM, Kühl M. Wnt-11 activation of a non-canonical Wnt signaling pathway is required for cardiogenesis. Nature. 2002;418:636–41. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- Roszko I, Sawada A, Solnica-Krezel L. Regulation of convergence and extension movements during vertebrate gastrulation by the Wnt/PCP pathway. Sem. Cell & Dev. Biol. 2009;20:986–997. doi: 10.1016/j.semcdb.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweickert A, Steinbeisser H, Blum M. Differential gene expression of Xenopus Pitx1, Pitx2b and Pitx2c during cement gland, stomodeum and pituitary development. Mech. Dev. 2001;107(1-2):191–194. doi: 10.1016/s0925-4773(01)00461-0. [DOI] [PubMed] [Google Scholar]
- Schwind JL. The development of the hypophysis cerebri of the albino rat. Am. J. Anut. 1928;41:295–319. [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis: A laboratory manual. Cold Spring Harbor Press; Cold Spring Harbor: 2000. [Google Scholar]
- Sive HL, Hattori K, Weintraub H. Progressive determination during formation of the anterioposterior axis in Xenopus laevis. Cell. 1989;58(1):171–180. doi: 10.1016/0092-8674(89)90413-3. [DOI] [PubMed] [Google Scholar]
- Sokol SY. Analysis of Dishevelled signaling pathways during Xenopus development. Cur. Biol. 1996;6(11):1456–1467. doi: 10.1016/s0960-9822(96)00750-6. [DOI] [PubMed] [Google Scholar]
- Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink E, Saint-Jeannet J. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Dev. 2002;129:421–432. doi: 10.1242/dev.129.2.421. [DOI] [PubMed] [Google Scholar]
- Steventon B, Mayor R, Streit A. Neural crest and placode interaction during the development of the cranial sensory system. Dev. Biol. 2014;390(1):28–38. doi: 10.1016/j.ydbio.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler J, Bolger T, Wallingford J, Liu K. Hedgehog activity controls opening of the primary mouth. Dev. Biol. 2014;396(1):1–7. doi: 10.1016/j.ydbio.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, Heisenberg C-P. Convergent extension: using collective cell migration and cell intercalation to shape embryos. Dev. 2012;139:3897–3904. doi: 10.1242/dev.073007. [DOI] [PubMed] [Google Scholar]
- Tada M, Smith JC. Xwnt11 is target of Xenopus Brachyury: regulation of gastrulation movements via Dishevelled, but not through the canonical Wnt pathway. Dev. 2000;127:2227–2238. doi: 10.1242/dev.127.10.2227. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Steventon B, Scarpa E, Garcia S, Trepat X, Streit A, Mayor R. Chase-and-run between adjacent cell populations promotes directional collective migration. Nature Cell Bio. 2013;15:763–772. doi: 10.1038/ncb2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB, Fraser SE, Harland RM. Convergent extension: The molecular control of polarized cell movement during embryonic development. Dev. Cell. 2002;2(6):695–706. doi: 10.1016/s1534-5807(02)00197-1. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Habas R. The developmental biology of Disheveled: an enigmatic protein governing cell fate and cell polarity. Dev. 2005;132:4421–4436. doi: 10.1242/dev.02068. [DOI] [PubMed] [Google Scholar]
- Wiellette EL, Sive H. Vhnf1 and Fgf signals synergize to specify rhombomere identity in the zebrafish hindbrain. Dev. 2003;130(16):3821–3829. doi: 10.1242/dev.00572. [DOI] [PubMed] [Google Scholar]
- Winklbauer R, Medina A, Swain RK, Steinbeisser H. Frizzled-7 signaling controls tissue separation during Xenopus gastrulation. Nature. 2001;413:856–60. doi: 10.1038/35101621. [DOI] [PubMed] [Google Scholar]
- Yin C, Ciruna B, Solnica-Krezel L. Convergence and extension movements during vertebrate gastrulation. Cur. Topics Dev. Biol. 2009;89:163–92. doi: 10.1016/S0070-2153(09)89007-8. [DOI] [PubMed] [Google Scholar]
- Yu H, Ye X, Guo N, Nathans J. Frizzled 2 and frizzled 7 function redundantly in convergent extension and closure of the ventricular septum and palate: evidence for a network of interacting genes. Dev. 2012;139(23):4383–94. doi: 10.1242/dev.083352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.