Abstract
With the advent of the combination antiretroviral therapy era (cART), the development of AIDS has been largely limited in the United States. Unfortunately, despite the development of efficacious treatments, HIV-1 associated neurocognitive disorders (HAND) can still develop and as many HIV-1 positive individuals age, the prevalence of HAND is likely to rise because HAND manifests in the brain with very low levels of virus. However, the mechanism producing this viral disorder is still debated. Interestingly, HIV-1 infection exposes neurons to proteins including Tat, Nef, Vpr which can drastically alter mitochondrial properties. Mitochondrial dysfunction has been posited to be a cornerstone of the development of numerous neurodegenerative diseases. Therefore, we investigated mitochondria in an animal model of HAND. Using an HIV-1 transgenic rat model expressing 7 of the 9 HIV-1 viral proteins, mitochondrial functional and proteomic analysis were performed on a subset of mitochondria that are particularly sensitive to cellular changes, the neuronal synaptic mitochondria. Quantitative mass spectroscopic studies followed by statistical analysis revealed extensive proteome alteration in this model paralleling mitochondrial abnormalities identified in HIV-1 animal models and HIV-1 infected humans. Novel mitochondrial protein changes were discovered in the electron transport chain (ETC), the glycolytic pathways, mitochondrial trafficking proteins, and proteins involved in various energy pathways, and these findings correlated well with the function of the mitochondria as assessed by a mitochondrial coupling and flux assay. By targeting these proteins and proteins upstream in the same pathway, we may be able to limit the development of HAND.
Keywords: HIV-1 associated neurocognitive disorders, mitochondria, neurodegeneration, neuroAIDS
INTRODUCTION
Mitochondria are important to the proper function of neural tissue and mitochondrial dysfunction has been implicated in the pathology of many neurodegenerative disorders including Alzheimer’s disease [1], amyotrophic lateral sclerosis [2], Huntington’s disease [3], and Parkinson’s disease [4]. However the role of mitochondria in the pathogenesis of HIV-1 infection leading to HIV-1 associated neurocognitive disorders (HAND) is unclear. This question is important because HAND displays similarities to the previously mentioned neurodegenerative disorders including the delayed onset and increased prevalence with time.
Interestingly, despite efficacious HIV-1 treatments, HIV-1 related neurocognitive issues persist. Long-term HIV-1 infection often manifests as HAND [5, 6] with symptoms ranging from mild deficits to HIV-1-associated dementia. During HAND, neurons experience synaptodendritic abnormalities and damage that can lead to cell death [6–9]. One strong correlating factor of HAND is thought to be oxidative stress [10–12]. Because mitochondria are the primary source of the reactive oxygen species (ROS) responsible for oxidative stress, mitochondria abnormalities likely have a strong contribution to the pathogenesis of HAND.
Previous work has demonstrated the centrality of mitochondria to the progression of the HIV-1 pathology. Mitochondria can directly influence the infectivity of HIV-1, the course of HIV-1 infection, and the prevalence of side effects from the primary HIV-1 therapy, combination antiretroviral therapy (cART). Mitochondrial DNA single nucleotide polymorphisms (SNPs) can decrease the efficacy of infection as well as slow the resulting AIDS progression [13]. Furthermore, depletion of mitochondrial DNA has been demonstrated to block HIV-1 infection [14]. In those experiments, nuclear- and mitochondria-encoded proteins were found to be able to impact the infectivity of HIV-1 both individually and synergistically [13, 15]. The importance of mitochondria also extends to HIV-1 treatment as mitochondrial proteins can alter the side-effects of combination antiretroviral therapy (cART) [16]. These results demonstrate the centrality of mitochondria to HIV-1 infection, progression, and treatment, but less research has focused on the mitochondria in the effects of HIV-1 infection on the brain.
We hypothesized that HIV-1 viral proteins would dramatically alter the synaptic brain mitochondria in a multifaceted manner thereby altering cellular properties. To test this hypothesis, the synaptic mitochondrial proteome was quantified in the HIV-1 transgenic (HIV-1 Tg) rats. The HIV-1 Tg rat expresses 7 or the 9 HIV-1 viral proteins (not Gag or Pol) in all cells. As such, the HIV-1 Tg rat can model aspects of HAND related to the effects of expression of HIV-1 viral proteins without productive viral infection [17–19]. Importantly 4 of the 7 HIV proteins expressed in this model (Tat, Nef, Vpr and gp120) have been shown to induce damaging ROS [20–26].
MATERIALS AND METHODS
Animals
All animal experiments were conducted with HIV-1 Tg animals and litter mate controls. This model was created using a non-infectious pro-virus expressing 7 of the 9 HIV-1 viral proteins and then backcrossed to the F344 strain [27]. Animals were bred in-house with animals coming from 3 separate litters of each genotype for both experiments. For the purposes of our experiments, all controls are wild-type non-transgenic littermates. All animals were 320 days old at the time of the experiments. This time point was chosen because the HIV-1 Tg rats have generally good health for the first year of life [28]. Only male animals were used for the experiments with 3 animals being utilized for the mitochondrial functional assays (n=3) and 4 animals being used for the mitochondrial proteomics (n=4). All protocols were conducted within NIH-approved guidelines with the approval and oversight of the University of Nebraska Medical Center IACUC.
Data-dependent analysis for building a library
Details on the composition of and the methods utilized to build the SWATH-MS library have been previously published [29]. Additional samples were added to the SWATH-MS library to enrich for synaptic mitochondrial proteins. Synaptic mitochondria from two rat brains were digested and fractionated by isoelectric point with the resulting peptide fractions being included in the library as performed previously.
Isolation of synaptic mitochondria for SWATH mass spectrometry and mitochondrial functional analysis
Brains were rapidly isolated from HIV-1 Tg and litter mate control animals. The cerebellum was removed. After extraction, brains were immediately rinsed with ice-cold PBS to remove blood. The meninges were removed. Tissue was chopped and homogenized using a Dounce homogenizer. Synaptic mitochondria were isolated using a synaptosomal mitochondria preparation as previously performed [30–32]. Mitochondria for mass spectrometry were lysed in 4% sodium dodecyl sulfate (SDS), and protein concentration was quantified using a using a Pierce 660 assay with bovine serum albumin standards (Thermo Fisher Scientific, Rockford, IL). Mitochondria for functional analysis were placed in a MAS buffer and treated according to manufacturers’ protocols for the experiments.
Sample preparation for mass spectrometry
Sample preparation was conducted by using a modified version of the filter-aided sample preparation (FASP) protocol as previously described [29, 33, 34]. In short, samples were loaded onto a 20-μm filter (Pall Corporation, Ann Arbor, MI), trypsin digested (Promega, Madison, WI), and the resulting peptides were cleaned with an Oasis mixed-mode weak cation exchange cartridge (Waters, Milford, MA). Peptides were quantified with a NanoDrop (Thermo Fisher Scientific) using Scopes’ method for protein quantification [35].
Analysis of mitochondrial function
Mitochondrial function was analyzed with a Seahorse XF 24 analyzer based on the protocol of Rogers with minor alterations [36] as previously described [37]. The concentration of ADP and mitochondria were optimized for this assay. For the Seahorse experiments, 3 animals were used per group were utilized for mitochondrial coupling and mitochondrial flux assays. Each biological replicate had 4 technical replicates for the experiments. Statistical significance was determined using a repeated measures two-way ANOVA followed by Sidak’s post-hoc test with α=0.05. Graphs were generated in Prism.
For all assays mitochondria were isolated as described above and quantified via a Pierce BCA Protein Assay (Thermo Scientific). Equal amounts of mitochondria were then loaded onto Seahorse plates, centrifuged for 15 min, followed by 5 min incubation at 37°C, and loaded onto the Seahorse XF24. For the coupling assay, mitochondria were subjected to subsequent injections of ADP, oligomycin, FCCP and antimycin A while measuring the oxygen consumption rate (OCR) in the presence of succinate and rotenone. OCR is an indirect measurement of ATP production as higher oxygen consumption correlates with increased ATP production. Treatments of mitochondria with ADP, oligomycin, FCCP and antimycin A mimic various respiratory states. State 2, or basal respiration, is the amount of respiration at rest or in the presence of no treatment. State 3 respiration is the ADP-stimulated respiration in the presence of saturating substrate or during ADP treatment. State 4o is the oxygen consumption in the absence of oxidative phosphorylation as ADP consumption at complex 4 is inhibited by oligomycin binding. State 3u is the maximal uncoupled respiratory state of the mitochondria as denoted by treatment with FCCP (for review of respiratory states see [38]). Antimycin A treatment mimics the absence of electron transport by blocking complex III of the ETC.
To measure electron flux, after a baseline measurement, mitochondria are exposed to subsequent treatments of rotenone, succinate, antimycin A and ascorbic acid/tetramethylphenylenediamine (ASC/TMPD). These treatments inhibit certain electron transport subunits allowing us to measure the function of the complex I, complex II and complex IV when each complex act as the rate-limiting complex by providing complex-appropriate substrates in excess such as pyruvate and malate (basal), succinate (complex II), or ASC/TMPD (complex IV) treatment respectively after inhibition of other complexes by rotenone (complex I) or antimycin A (complex III).
SWATH-MS Data-independent analysis
SWATH-MS data-independent analysis (DIA) was conducted as previously performed [29]. In short, 5 μg of peptide from each sample was resuspended in 6 μL of 0.1% formic acid (Honeywell, Muskegon, MI). Both the HIV-1 Tg and control group had four biological replicates analyzed by SWATH DIA. Peakview (Version 1.1.0.0) was utilized to extract and integrate the fragment ion chromatograms and generated the raw expression data for each protein. No data processing (neither denoising nor smoothing) of any kind was applied to the extracted ion chromatograms. Retention time was calibrated as previously stated [29] using synthetic peptides (BiognoSYS; Zurich, Switzerland) spiked-in samples in accordance with the manufacturer’s protocol. Quantitative analysis for each protein was set at 5 peptides and 5 transitions in accordance with previously published work [29, 39] for targeted data extraction. Fragment ion chromatograms were extracted using an isolation window with of 10 min and 70 ppm accuracy for quantification purposes. Heat maps for representation of proteomic data were constructed in Multiple Experiment Viewer (www.tm4.org) [40].
Statistical analysis
CyberT was used to perform an unpaired two-condition Bayesian analysis [41, 42]. The Bayesian confidence value and the sliding window were set to 12 and 101, respectively. To assess statistical difference, the posterior probability of differential expression (PPDE) was calculated. Changes were noted as significant if the p≤0.05 and cumulative PPDE ≥ 0.95 (corresponding to α=0.05). Histograms and corresponding Pearson’s correlation coefficients were generated in GraphPad Prism (version 6.0f).
Bioinformatic analysis
To identify enrichment of proteins, data was uploaded into Panther Biological Classification system (http://www.pantherdb.org/) [43, 44]. Ingenuity Pathway Analysis (IPA) was utilized to predict upstream pathway activation (http://www.ingenuity.com/products/ipa) [45]. Data was uploaded into the software as stated previously [29]. Activation z-scores were generated based upon the proteomic data and indicate the likelihood of activation or deactivation of any particular pathway in IPA. To represent these data graphically, the IPA-determined z-scores were uploaded into Multiple Experiment Viewer to build heat maps.
RESULTS
Research involving HAND commonly is reductionist in nature, focusing on a single HIV-1 viral protein. However, the HIV-1 infection involves many proteins producing a variety of effects that may be more pronounced or dulled. As such, identification of a single cause of HIV-1-associated neurocognitive deficits is difficult. The transgenic rat model expressing 7 of the 9 proteins of the HIV-1 provirus successfully models the various effects produced by exposure to multiple proteins. Using this model, it is possible to examine the in vivo effects of HIV proteins causing functional abnormalities in mitochondria and shed light on how mitochondria elicit effects on cellular function to produce HAND.
For this experiment, the synaptic mitochondrial proteome was analyzed from HIV-1 Tg rats and littermate transgene-negative controls. Synaptic mitochondria were the focus of the proteomics because they are crucial to synaptic signaling [46–49], and synaptic signaling abnormalities are present in HIV-1-infected individuals [50–53]. In this experiment, 1003 proteins were identified and 594 proteins were annotated by MitoMiner as mitochondrial in nature (Supplemental Table 1). We identified 241 proteins with significantly altered protein expression with a large number of proteins being from the ETC, glycolysis-associated pathways, energy transfer pathways or mitochondrial trafficking pathways (Supplementary Table 1).
Our initial step in these experiments was to demonstrate our sample preparation was reproducible. To assess this property of our samples, we performed correlation analyses of the samples. Our results demonstrated in both the HIV-1 Tg animals and the litter mate controls there was a high degree of correlation between identified proteins within experimental groups (Supplementary Figure 1). Our next step in determining the significance of our results was to determine which processes were highly represented in our group of proteins. By uploading our proteins into the Panther classification system, the Gene Ontology (GO) biological processes were determined. We identified metabolic processes as being significantly enriched (Supplementary Figure 2) and decided to focus upon the ETC because this process is the key to mitochondrial function.
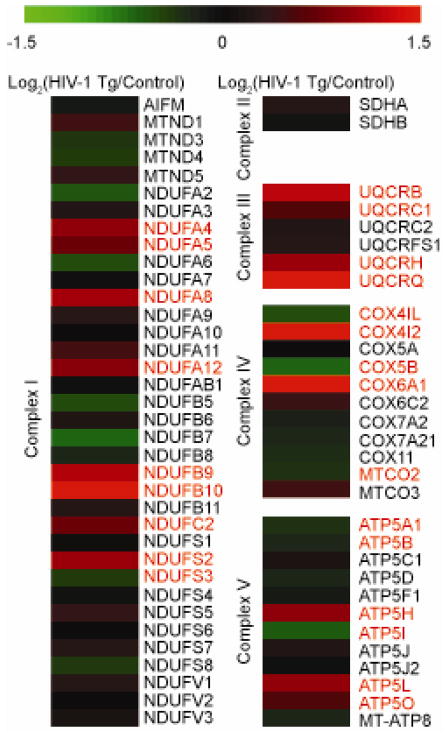
Extensive alterations were measured in the protein subunits of the ETC. Proteomic analysis demonstrated varied expression of the ETC complex subunits (Figure 1). In complex I, NDUFA4, NDUFA5, NDUFA8, NDUFA12, NDUFB9, NDUFB10, NDUFC2, and NDUFS2 were found to be significantly increased in the HIV-1 Tg rat while NDUFS3 was decreased. In complex III, UQCRB, UQCRC1, UQCRH and UQCRQ were found to be significantly increased in HIV-1 Tg animals. In complex IV, two subunits were found to be increased in HIV-1 Tg animals: COX4I2 and COX6A1, whereas three subunits of complex IV were decreased: COX4I1, COX5B and MTCO2. Finally, complex V had three significantly increased subunits in HIV-1 Tg animals: ATP5H, ATP5L and ATP5O, whereas another three were decreased: ATP5A1, ATP5B and ATP5I. These results suggest widespread mitochondrial metabolic alterations are occurring the in the HIV-1 Tg rat model.
Figure 1. Electron transport chain subunit alterations in synaptic mitochondria of HIV-1 Tg rats.
Heat map displaying the log2 change in protein expression of mitochondrial electron transport chain subunits between HIV-1 Tg and litter mate control synaptic protein as quantified via SWATH mass spectrometry. All values are displayed on a log2 scale. Names in red denote significantly different proteins (p<0.05 and cumulative PPDE>0.95) as assessed by CyberT. n=4 biological replicates.
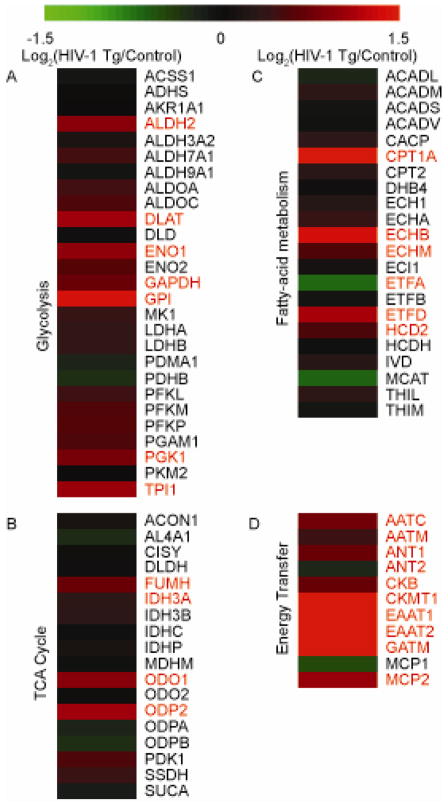
Although the ETC expression is indicative of mitochondrial alterations, the system is dependent on the influx of electrons largely generated through the glycolytic pathway, the tricarboxylic acid (TCA) cycle, and the fatty acid metabolic process. We identified a general increase of expression of glycolytic proteins in the HIV-1 Tg animals with seven proteins being significantly increased: ALDH2, DLAT, ENO1, GAPDH, GPI, PGK1, and TPI1 (Figure 2A). These results suggest a general increase in glycolysis and suggest ETC flux is likely increased in HIV-1 Tg rats. In addition, while interrogating the TCA cycle (Figure 2B) and the fatty-acid metabolic process (Figure 2C), general increases in protein expression were noted; however, the increased expression profiles were less severe than the increase in glycolysis proteins. In the TCA cycle, FUMH, IDH3A, ODO1 and ODP2 were significantly increased. IDH3A is particularly important because it is a subunit of the rate-limiting enzyme of the TCA cycle. For proteins involved in fatty acid metabolism, CPT1A, ECHB, ECHM, ETFD and HCD2 were significantly increased while ETFA was significantly decreased in the HIV-1 Tg rats. The fatty acid metabolism protein profiles suggest abnormal fatty acid metabolism is occurring.
Figure 2. Glycolytic, tricyclic carboxylic acid (TCA) cycle, and energy transfer protein expression profiles in synaptic mitochondria of HIV-1 Tg rats.
Heat map displaying the log2 change in protein expression of proteins involved in (A) glycolysis, (B) the TCA cycle, and (C) energy transfer proteins between HIV-1 Tg and litter mate control synaptic protein as quantified via SWATH mass spectrometry. All values are displayed on a log2 scale. Names in red denote significantly different proteins (p<0.05 and cumulative PPDE>0.95) as assessed by CyberT. n=4 biological replicates.
Due to the extensive alterations in the ETC, we hypothesized synaptic mitochondria in HIV-1 Tg animals may be experiencing addition energy demands throughout the cells and as a result, proteins involved in mechanisms to transport energy throughout the cell would be increased. Unsurprisingly, proteins involved in transferring high energy phosphates in through creatine transport or the malate-aspartate shuttle were largely increased in expression (Figure 2D). Of the proteins involved in creatine energy transfer, CKB, CKMT1, and GATM were significantly increased in HIV-1 Tg animals. As for the malate-aspartate shuttle proteins, AATC, AATM, and MCP2 were significantly increased in HIV-1 Tg animals. Additionally, the ADP/ATP translocases ANT1 and ANT2 displayed altered expression profiles in HIV-1 Tg animals with ANT1 being increased whereas ANT2 decreased (Figure 2D).
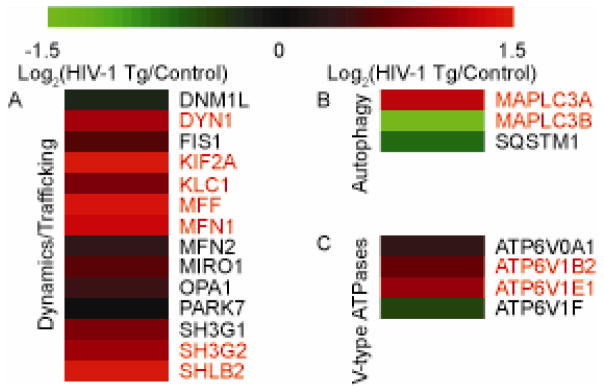
Proper trafficking of mitochondria is also important in cellular homeostasis and responses to stress. To examine whether the presence of HIV-1 proteins in the HIV-1 Tg rat could alter this property, proteins involved in mitochondrial trafficking and dynamics were interrogated (Figure 3A). Of the proteins identified, the majority displayed an increase in expression suggesting altered mitochondrial network properties. Six of these proteins were increased significantly: DNM1, KIF2A, KLC1, MFF, MFN1, SH3G2, and SHLB2. Together these proteins suggest altered mitochondrial dynamics and trafficking. These results are important because the localization of mitochondria alters many properties of the neuron such as excitability and can have profound effects on the viability of neurons.
Figure 3. Proteins expression of various processes in synaptic mitochondria of HIV-1 Tg rats.
Heat map displaying the log2 change in protein expression of mitochondrial (A) dynamics/trafficking, (B) V-type ATPases, and (C) autophagy-related proteins between HIV-1 Tg and litter mate control synaptic protein as quantified via SWATH mass spectrometry. All values are displayed on a log2 scale. Names in red denote significantly different proteins (p<0.05 and cumulative PPDE>0.95) as assessed by CyberT. n=4 biological replicates.
Although we observed considerable alteration in trafficking proteins suggesting altered dynamics and distribution, it was unclear if the trafficking changes could be related to mitochondrial autophagy, or mitophagy. To interrogate this process, we identified known autophagy regulators (Figure 3B) as well as V-type ATPases (Figure 3C) which would be present interacting with mitochondria during lysosomal fusion, the final stage of autophagy. Interestingly, within the V-type ATPases, the majority displayed increased expression with two significantly increased in HIV-1 Tg animals: ATP6V1B2 and ATP6V1E1. However, the results for the autophagy proteins were varied with SQSTM1 and MAPLC3B being decreased while MAPLC3A was increased. Together, these results largely suggest mitochondria are getting primed for autophagy as denoted by increased MAPLC3A and trafficked to lysosomes as denoted by the increases in ATP6V1B2 and ATP6V1E1.
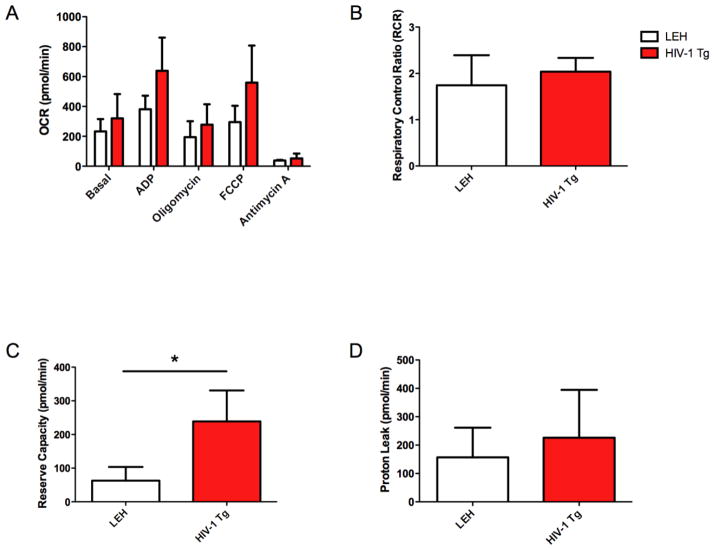
Because the proteomic analysis suggested altered energy dynamics within synaptic mitochondria, the functional properties of synaptic mitochondria were interrogated with a Seahorse Bioanalyzer. Analysis of the mitochondrial respiratory states through the mitochondrial coupling assay demonstrated that HIV-1 Tg animals had higher oxygen consumption than controls during State 2 (Basal), State 3 (ADP), State 3u (FCCP) and State 4 (Oligomycin), although the difference did not reach statistical significance (Figure 4A). No changes were observed in the respiratory control ratio (RCR) (Figure 4B), a general measure of ETC coupling, or proton leak (Figure 4D), a measure of the inner mitochondrial membrane function. However, the respiratory reserve capacity, the ability of the mitochondria to respond to a demand for increased energy production, was increased in HIV-1 Tg animals (Figure 3B). These results largely suggest the synaptic mitochondria have a greater ability to respond to insults as denoted by an increased reserve capacity. Additionally, when mitochondrial ETC complex function was assessed, significant alterations were observed in Complex 1 (Basal), Complex III (Succinate), and Complex IV (Asc/TMPD) demonstrating the ETC is functionally different in HIV-1 Tg rats (Figure 5). These results demonstrate that electron flux is greatly increased in the HIV-1 Tg animals increasing the ability of the HIV-1 Tg animals to generate ATP.
Figure 4. Mitochondrial functional coupling in the HIV-1 Tg rat brain synaptic mitochondria.
Mitochondrial oxygen consumption rate was determined using a Seahorse XF24 analyzer. For coupling (A), mitochondria were administered subsequent injections of ADP, oligomycin, FCCP, and antimycin A in the presence of succinate and rotenone. Based upon the coupling, the respiratory control ratios (RCR) (B) were calculated (RCR=OCRFCCP/OCROligomycin). Additionally, mitochondrial proton leak (C) (Proton leak = OCROligomycin-OCRAntimycin A) and the reserve capacity (D) (OCRFCCP-OCRBasal) were calculated. Listed p-values correspond to values generated by two-way repeated measures ANOVA. *: p≤0.05 on Sidak’s post-hoc comparison test. For all experiments, n=3 biological replicates.
Figure 5. Assessment of electron transport chain complex function in HIV-1 Tg rat brain synaptic mitochondria.
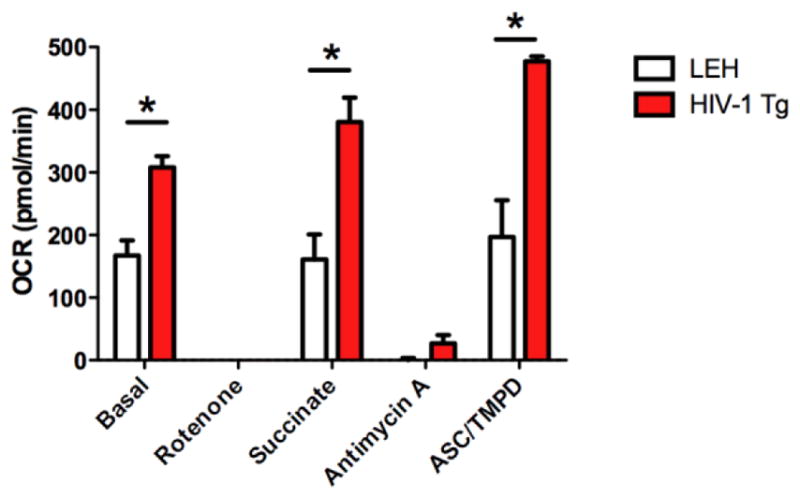
Assessment of the electron transport chain complexes of HIV-1 Tg rats as assessed by oxygen consumption rate (OCR) and measured by a Seahorse XF24 analyzer. Mitochondrial were treated to subsequent injections of rotenone, succinate, antimycin A and TMPD/asc while measuring OCR. Statistical significance was assessed by a repeated-measures two-way ANOVA followed by Sidak’s post-hoc test. *: p≤0.05 on Sidak’s post-hoc comparison test. n=3 biological replicates.
To gain further insight into mechanism regulating mitochondrial dysfunction within the context of HAND, analysis of upstream regulators that may drive the proteomic alterations was performed with IPA. Upstream analysis can reveal which network signaling pathways are contributing the most heavily to the observed phenotype or, as is our case, the phenotype in the HIV-1 Tg rat (Figure 6). In our analysis, we identified one network of pathways involved in mitochondrial biogenesis. The network pathways involved included MTOR, PPARG, PGC1A, TFAM, TFEB, and THRB. Two other network pathways impacting mitochondrial function included ADORA2A and FMR1. Together, these pathways are predicted to elicit the observed mitochondrial alterations.
Figure 6. Protein network dysregulation in the HIV-1 Tg rat brain synaptic mitochondria as predicted by Ingenuity Pathways Analysis.

Proteomic data was uploaded into Ingenuity Pathways Analysis (IPA) and activation z-scores were generated indicating the activation or deactivation of a particular pathway. n=4 biological replicates.
DISCUSSION
These experiments demonstrated significant mitochondria changes associated with the presence of HIV-1 proteins. Many alterations in the ETC were observed including, in general, increased expression of ETC complexes. These results suggest HIV-1 viral proteins directly enhance ETC protein expression, and mitochondria exposed to HIV-1 viral proteins experience a higher energy demand requiring increased ETC subunit expression. The electron flux assay further support the hypothesis that HIV-1 Tg rat mitochondria consume more oxygen. In the electron flux assay, HIV-1 Tg rat mitochondria had a higher capacity to generate ATP. These results are not wholly unexpected. Research on various illnesses has demonstrated mitochondria commonly increase oxygen production for ATP production when cellular damage occurs leading to increased ROS production and cellular dysfunction. These mechanisms are likely the mechanisms observed in these experiments.
However previous results have demonstrated HIV-1 Tat and Nef proteins decrease the amount of ATP produced [54]. There are two possible explanations for the conflicting results. First, it is likely that the observed increase in mitochondrial flux in these experiments is not directly related to ATP production. The flux assay cannot differentiate between oxygen consumed for ROS and ATP production. Therefore, it is possible the increase we observed would largely contribute to ROS generation. More support for this theory could be found in the coupling assay. The coupling assay shows the fraction of oxygen utilized for ATP production over the amount used for other cellular processes. While the fraction remains the same, the values are almost twice as high in the HIV-1 Tg rats suggesting more oxygen is consumed for ROS production. Another piece of evidence supporting the theory that HIV-1 Tg rat mitochondria are producing more ROS could be found in the reserve capacity. Reserve capacity was found to be elevated in HIV-1 Tg rats. Because the reserve capacity could be thought of as the ability of mitochondria to increase oxygen consumption, and we found that the coupling is identical between HIV-1 Tg and control animals, the potential increase in reserve capacity would largely contribute to ROS generation. Secondly, all of our experiments were conducted by providing mitochondria excess substrates for the ETC. If may be that other pathways feeding into the ETC are hindered in a way to block substrate entrance into the ETC. In the case of this study, the first hypothesis for discontinuity is the most likely as increased ROS is consistent with other data.
Despite the increased mitochondrial energy production, not all mitochondrial ETC subunits were increased. In Complex I, IV and V, certain ETC subunits were significantly decreased (Figure 1). These results suggest (1) the cell may experience subunit switching to optimize for increased ATP production and/or decrease electron flux regulation, and (2) the increased subunit directly lead to increased electron flux. Because the significance of a decrease of ETC subunit expression (Figure 1) in the presence of mitochondrial functional increases (Figure 5) is unclear, further research is necessary to elucidate the matter. However, these results are still promising in helping tease apart the individual contribution of various subunits to the flux of electrons in different cellular environments.
Interestingly, we did not find evidence of elevated proton leak (Figure 4D), an indirect measure of oxidative stress, despite increased levels of proteins that are commonly elevated in states of enhanced oxidative stress. While appearing somewhat discordant, there is a complex relationship between proton leak and ROS, with ROS inducing proton leak but proton leak leading to a decrease in ROS production. Examining the relationship is further complicated as a result of how proton leak is calculated in this experiment. In this experiment, proton leak was calculated in the absence of complex I function as all the mitochondrial coupling experiments were performed in the presence of rotenone. However, complex I and complex III both are important for contributing to proton leak and generating ROS. Therefore, the loss of complex I leak may have negated any significance despite a trend indicating increased proton leak in HIV-1 Tg rats.
In addition to the function of the ETC, it is important to look at indicators of substrates entering the ETC. To gain an indirect measure of substrates entering the ETC, we quantified the proteins involved in the glycolytic processes. Our experiments found an increase in glycolytic proteins associated with the mitochondria (Figure 2A) suggesting there is an increase in substrate availability to generate electrons. Previous work on glycolysis levels in cells exposed to HIV-1 viral proteins have produced varied results. In a variety of cells, exposure to certain HIV-1 viral proteins in vitro or HIV-1 virus in vivo results in increased glycolysis [55–57]. Conversely, isolates of blood from HIV-1 infected individual have also demonstrated decreased glycolysis [58]. However in respect to this experiment, any alteration of substrate availability could affect the ability of mitochondria to respond to various stimuli. Our data suggest HIV-1 viral proteins increase glycolysis in the brain synaptic mitochondria of the HIV-1 Tg rat model, and the increase in glycolytic proteins likely represents a shift in the metabolic processes in the synapses of neurons.
The proteomic interrogation of the TCA cycle and the fatty acid metabolic pathway agrees with the glycolysis data suggesting increased electron flux. In both of these pathways, certain proteins were significantly increased. In the TCA cycle, IDH3A, the rate-limiting enzyme of the process, was increased further increased flux through this pathway. These results largely agree with previous data suggesting the TCA cycle is abnormal in the HIV-1-infected CD4+ T cells [59, 60]. While it is unclear at this time whether the increase in TCA enzymes translates into smaller or larger metabolic pools, these data suggest there is a change in the TCA metabolite pools.
Similar to the changes observed in the TCA cycle, alterations were found in the protein expression profile of several proteins involved in fatty acid metabolism. CPT1A is important for the import of fatty acids to the mitochondria. HCD2, ECHB, and ECHM are important for oxidative steps with HCD2 and ECHB processing fatty acids of differing lengths. ETFA and ETFD are important for transferring the resultant electrons to the ETC. Alterations in any of these pathways would alter the metabolism of a cell. Our results suggest (1) fatty acid transport is abnormal suggesting storage and processing of fatty acids is altered, (2) the length of the fatty acids being metabolized is being altered, and (3) transfer of the resulting electrons to the ETC is being changed. While our results are consistent with previous results [61], our results are novel as they demonstrate that active replication is not necessary to inflict changes in the fatty acid metabolic pathway and demonstrate a profound impact of the HIV-1 viral proteins on a subset of highly active mitochondria within the brain.
Data on proteins involved in trafficking and generating high energy substrates largely suggest an altered mitochondrial energy transfer. There is an almost ubiquitous increase in expression of various proteins involved in energy transfer suggesting (1) there is increased energy demand resulting from the presence of HIV-1 proteins and (2) there is an altered energy state in these cells. Proteins involved in generating creatine phosphate and in the malate shuttle are increased specifically demonstrating enhanced shuttling of energy throughout the cell. The increased energy demand is likely a result of the damage generated by the viral proteins directly or indirectly through ROS. The altered energy state in the mitochondria is consistent with our ETC and glycolysis findings and further demonstrate the toll HIV-1 viral proteins have on synaptic mitochondria.
Mitochondrial function is regulated by many facets of the cell. One particularly underappreciated mode of mitochondrial regulation comes in the form of localization. Mitochondria in a neuron must be distributed to areas of high energy demand. In a neuron, this is particularly important as these cells have long processes, distancing the synapse from the soma. Furthermore the brain is almost wholly dependent on oxidative phosphorylation for ATP production, and has high energy demands. Intertwined with mitochondrial trafficking is mitochondrial dynamics. Mitochondrial dynamics refers to the process of remodeling the mitochondria to optimize the mitochondria for energy production, transport, or degradation. Interestingly, mitochondrial trafficking proteins were increased in the synapse. This leads to the possibility that the neuron is experiencing an altered energy state, and the mitochondria are mobilizing to better respond to these alternative demands. These characteristics may drive or be a results of certain pathologies associated with synaptic signaling in the neurons of HIV-1 infected patients.
Inspection of the proteins involved in mitochondrial dynamics revealed MFN1, a major mitochondrial fusion protein, is increased in HIV-1 Tg animals. This finding is particularly interesting because MFN1 has been demonstrated to regulate cellular responses and electrical activity of neurons by altering the metabolic response [62]. Likely, the overexpression of MFN1 would lead to extensive mitochondrial networks around the synapse which in turn could minimize damage caused by HIV-1 viral protein induced excitotoxicity. It is likely this expression pattern is a neuroprotective feature meant to minimize the damage a cell would endure.
An alternative explanation of the increased trafficking proteins is to enhance mitochondrial autophagy or mitophagy. To assess whether the increase in trafficking protein prevalence was related to mitophagy, we quantified the V-Type ATPases which would only be increased in mitochondrial proteomics if the mitochondria were interacting with lysosomes and mitophagy adaptors. We found mixed expression results in the V-type ATPases and mitophagy proteins. The low protein expression profiles of these proteins may be indicative of the model in which we are investigating synaptic proteins is inadequate to answer any questions on mitophagy. However, further work focusing on these groups of proteins in the context of HIV-1 infection is necessary.
Although the mitochondrial proteomics provide additional insight for HAND, the mechanisms regulating mitochondrial dysfunction are not directly revealed by mitochondrial proteomics. With IPA, prediction of pathways regulating mitochondrial proteomic alterations is possible. Using IPA, several pathways involved in regulating mitochondrial biogenesis such as MTOR, PPARG, PGC1A, TFAM, TFEB, and THRB were identified. Interestingly, the alterations observed in these pathways are conflicting. While PGC1A, TFAM, and TFEB are decreased suggesting decreased biogenesis, MTOR, PPARG, and THRB are increased suggesting increased biogenesis. These results may suggest dysfunction mitochondrial biogenesis is occurring in the HIV-1 Tg model and a more directed biogenesis pathways is occurring in this specific subset of mitochondria.
Undoubtedly, the presence of the HIV-1 Tg rat in which 7 of the 9 HIV-1 viral proteins are expressed raise questions about the relevance of the model. Interestingly, much of the proteomic data has been corroborated by previous publications for on the ETC [10–12] and glycolyic pathway [55–57] albeit in alternative cell types or animal models. Together these data suggest the HIV-1 viral proteins produce chronically dysfunctional mitochondria, and these mitochondrial alterations can contribute to the HAND functional pathology.
CONCLUSION
In vivo expression of HIV-1 viral proteins result a large number of mitochondrial proteomic alterations as well as mitochondrial functional changes. These results highlight the susceptibility of neuronal mitochondria to damage in HIV-1 infected individuals. We identified changes in metabolic pathways and energy transfer while identifying specific proteins which could be directly responsible for the observed alterations. Further, our work has highlighted certain pathways as targets to limit the dysfunctional mitochondria. Targeting these pathways may be useful for treatment option of HIV-1 infected individuals to alleviate HAND symptoms.
Supplementary Material
Proteomic data was analyzed through CyberT (http://cybert.ics.uci.edu/). A Bayesian analysis of the data was performed using a Bayesian coefficient of 12. Multiple testing corrections were applied. For significance, p<0.05 and cumulative posterior probability of differential expression (Cum PPDE)>0.95.
Area under curve data obtained from mass spectrometry was used to generate histograms comparing samples against one another within each experimental group. Pearson’s r coefficient was generated for each comparison.
Data from the mass spectrometry analysis were uploaded into the Panther classification system (http://www.pantherdb.org/) to determine which biological processes were most represented in our samples.
Acknowledgments
We would like to thank the Proteomics Core Facility members at the University of Nebraska Medical Center, under the direction of Dr. Pawel Ciborowski, for all their support and aid in the proteomics experiments.
Funding Sources.
This work was funded by the National Institute of Health (NIH) grants P30MH062261, R01DA027729, R01DA033150, R01DA36157, and R01MH73490.
Footnotes
Conflict of Interest.
Lance M. Villeneuve, Phillip R. Purnell, Kelly L. Stauch, Shannon E. Callen, Shilpa J. Buch, and Howard S. Fox report no conflict of interest.
References
- 1.Nunomura A, Perry G, Aliev G, Hirai K, Takeda A, Balraj EK, et al. Oxidative damage is the earliest event in Alzheimer disease. J Neuropathol Exp Neurol. 2001;60:759–67. doi: 10.1093/jnen/60.8.759. [DOI] [PubMed] [Google Scholar]
- 2.Mattiazzi M, D’Aurelio M, Gajewski CD, Martushova K, Kiaei M, Beal MF, et al. Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. The Journal of Biological Chemistry. 2002;277:29626–33. doi: 10.1074/jbc.M203065200. [DOI] [PubMed] [Google Scholar]
- 3.Seong IS, Ivanova E, Lee J, Choo YS, Fossale E, Anderson M, et al. HD CAG repeats implicates a dominant property of huntingtin in mitochondrial energy metabolism. Human Molecular Genetics. 2005;14:2871–80. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 4.Parker WD, Jr, Parks JK, Swerdlow RH. Complex I deficiency in Parkinson’s disease frontal cortex. Brain Research. 2008;1189:215–8. doi: 10.1016/j.brainres.2007.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nath A, Sacktor N. Influence of highly active antiretroviral therapy on persistence of HIV in the central nervous system. Curr Opin Neurol. 2006;19:358–61. doi: 10.1097/01.wco.0000236614.51592.ca. [DOI] [PubMed] [Google Scholar]
- 6.Kaul M, Zheng J, Okamoto S, Gendelman HE, Lipton SA. HIV-1 infection and AIDS: consequences for the central nervous system. Cell Death and Differentiation. 2005;12(Suppl 1):878–92. doi: 10.1038/sj.cdd.4401623. [DOI] [PubMed] [Google Scholar]
- 7.Saha RN, Pahan K. Tumor necrosis factor-alpha at the crossroads of neuronal life and death during HIV-associated dementia. Journal of Neurochemistry. 2003;86:1057–71. doi: 10.1046/j.1471-4159.2003.01942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattson MP, Haughey NJ, Nath A. Cell death in HIV dementia. Cell Death and Differentiation. 2005;12(Suppl 1):893–904. doi: 10.1038/sj.cdd.4401577. [DOI] [PubMed] [Google Scholar]
- 9.Ellis R, Langford D, Masliah E. HIV and antiretroviral therapy in the brain: neuronal injury and repair. Nat Rev Neurosci. 2007;8:33–44. doi: 10.1038/nrn2040. [DOI] [PubMed] [Google Scholar]
- 10.Mollace V, Nottet HS, Clayette P, Turco MC, Muscoli C, Salvemini D, et al. Oxidative stress and neuroAIDS: triggers, modulators and novel antioxidants. Trends Neurosci. 2001;24:411–6. doi: 10.1016/s0166-2236(00)01819-1. [DOI] [PubMed] [Google Scholar]
- 11.Pocernich CB, Sultana R, Mohmmad-Abdul H, Nath A, Butterfield DA. HIV-dementia, Tat-induced oxidative stress, and antioxidant therapeutic considerations. Brain Res Brain Res Rev. 2005;50:14–26. doi: 10.1016/j.brainresrev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Jana A, Pahan K. Human immunodeficiency virus type 1 gp120 induces apoptosis in human primary neurons through redox-regulated activation of neutral sphingomyelinase. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:9531–40. doi: 10.1523/JNEUROSCI.3085-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, Poole JC, Lautenberger J, Sezgin E, et al. Mitochondrial DNA haplogroups influence AIDS progression. Aids. 2008;22:2429–39. doi: 10.1097/QAD.0b013e32831940bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu G, Matsuura SE, Barrientos A, Scott WA. HIV-1 infection is blocked at an early stage in cells devoid of mitochondrial DNA. PLoS ONE. 2013;8:e78035. doi: 10.1371/journal.pone.0078035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrickson SL, Lautenberger JA, Chinn LW, Malasky M, Sezgin E, Kingsley LA, et al. Genetic variants in nuclear-encoded mitochondrial genes influence AIDS progression. PLoS ONE. 2010;5:e12862. doi: 10.1371/journal.pone.0012862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrickson SL, Kingsley LA, Ruiz-Pesini E, Poole JC, Jacobson LP, Palella FJ, et al. Mitochondrial DNA haplogroups influence lipoatrophy after highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;51:111–6. doi: 10.1097/QAI.0b013e3181a324d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vigorito M, Connaghan KP, Chang SL. The HIV-1 transgenic rat model of neuroHIV. Brain Behav Immun. 2015 doi: 10.1016/j.bbi.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roscoe RF, Jr, Mactutus CF, Booze RM. HIV-1 transgenic female rat: synaptodendritic alterations of medium spiny neurons in the nucleus accumbens. J Neuroimmune Pharmacol. 2014;9:642–53. doi: 10.1007/s11481-014-9555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran LM, Booze RM, Webb KM, Mactutus CF. Neurobehavioral alterations in HIV-1 transgenic rats: evidence for dopaminergic dysfunction. Experimental Neurology. 2013;239:139–47. doi: 10.1016/j.expneurol.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee A, Zhang X, Manda KR, Banks WA, Ercal N. HIV proteins (gp120 and Tat) and methamphetamine in oxidative stress-induced damage in the brain: potential role of the thiol antioxidant N-acetylcysteine amide. Free Radic Biol Med. 2010;48:1388–98. doi: 10.1016/j.freeradbiomed.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salmen S, Colmenares M, Peterson DL, Reyes E, Rosales JD, Berrueta L. HIV-1 Nef associates with p22-phox, a component of the NADPH oxidase protein complex. Cell Immunol. 2010;263:166–71. doi: 10.1016/j.cellimm.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Wu RF, Ma Z, Myers DP, Terada LS. HIV-1 Tat activates dual Nox pathways leading to independent activation of ERK and JNK MAP kinases. The Journal of Biological Chemistry. 2007;282:37412–9. doi: 10.1074/jbc.M704481200. [DOI] [PubMed] [Google Scholar]
- 23.Olivetta E, Mallozzi C, Ruggieri V, Pietraforte D, Federico M, Sanchez M. HIV-1 Nef induces p47(phox) phosphorylation leading to a rapid superoxide anion release from the U937 human monoblastic cell line. J Cell Biochem. 2009;106:812–22. doi: 10.1002/jcb.22041. [DOI] [PubMed] [Google Scholar]
- 24.Vilhardt F, Plastre O, Sawada M, Suzuki K, Wiznerowicz M, Kiyokawa E, et al. The HIV-1 Nef protein and phagocyte NADPH oxidase activation. The Journal of Biological Chemistry. 2002;277:42136–43. doi: 10.1074/jbc.M200862200. [DOI] [PubMed] [Google Scholar]
- 25.Deshmane SL, Mukerjee R, Fan S, Del Valle L, Michiels C, Sweet T, et al. Activation of the oxidative stress pathway by HIV-1 Vpr leads to induction of hypoxia-inducible factor 1alpha expression. The Journal of Biological Chemistry. 2009;284:11364–73. doi: 10.1074/jbc.M809266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agrawal L, Louboutin JP, Marusich E, Reyes BA, Van Bockstaele EJ, Strayer DS. Dopaminergic neurotoxicity of HIV-1 gp120: reactive oxygen species as signaling intermediates. Brain Research. 2010;1306:116–30. doi: 10.1016/j.brainres.2009.09.113. [DOI] [PubMed] [Google Scholar]
- 27.Reid W, Sadowska M, Denaro F, Rao S, Foulke J, Jr, Hayes N, et al. An HIV-1 transgenic rat that develops HIV-related pathology and immunologic dysfunction. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9271–6. doi: 10.1073/pnas.161290298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng J, Vigorito M, Liu X, Zhou D, Wu X, Chang SL. The HIV-1 transgenic rat as a model for HIV-1 infected individuals on HAART. J Neuroimmunol. 2010;218:94–101. doi: 10.1016/j.jneuroim.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 29.Villeneuve LM, Stauch KL, Fox HS. Proteomic analysis of mitochondria from embryonic and postnatal rat brains reveals response to developmental changes in energy demands. J Proteomics. 2014 doi: 10.1016/j.jprot.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stauch KL, Purnell PR, Fox HS. Quantitative proteomics of synaptic and nonsynaptic mitochondria: insights for synaptic mitochondrial vulnerability. Journal of Proteome Research. 2014;13:2620–36. doi: 10.1021/pr500295n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stauch KL, Purnell PR, Fox HS. Aging synaptic mitochondria exhibit dynamic proteomic changes while maintaining bioenergetic function. Aging (Albany NY) 2014;6:320–34. doi: 10.18632/aging.100657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stauch KL, Purnell PR, Villeneuve LM, Fox HS. Proteomic analysis and functional characterization of mouse brain mitochondria during aging reveal alterations in energy metabolism. Proteomics. 2015;15:1574–86. doi: 10.1002/pmic.201400277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wisniewski JR, Zougman A, Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. Journal of Proteome Research. 2009;8:5674–8. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- 34.Villeneuve L, Tiede LM, Morsey B, Fox HS. Quantitative proteomics reveals oxygen-dependent changes in neuronal mitochondria affecting function and sensitivity to rotenone. Journal of Proteome Research. 2013;12:4599–606. doi: 10.1021/pr400758d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scopes RK. Measurement of protein by spectrophotometry at 205 nm. Anal Biochem. 1974;59:277–82. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- 36.Rogers GW, Brand MD, Petrosyan S, Ashok D, Elorza AA, Ferrick DA, et al. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS ONE. 2011;6:e21746. doi: 10.1371/journal.pone.0021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villeneuve LM, Purnell PR, Boska MD, Fox HS. Early Expression of Parkinson’s Disease-Related Mitochondrial Abnormalities in PINK1 Knockout Rats. Mol Neurobiol. 2014 doi: 10.1007/s12035-014-8927-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chance B, Williams GR. The respiratory chain and oxidative phosphorylation. Adv Enzymol Relat Subj Biochem. 1956;17:65–134. doi: 10.1002/9780470122624.ch2. [DOI] [PubMed] [Google Scholar]
- 39.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Molecular & cellular proteomics: MCP. 2012;11:O111 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 41.Kayala MA, Baldi P. Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Res. 2012;40:W553–9. doi: 10.1093/nar/gks420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–19. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 43.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–41. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–8. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghosh A, Greenberg ME. Calcium signaling in neurons: molecular mechanisms and cellular consequences. Science. 1995;268:239–47. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 47.Zucker RS. Calcium- and activity-dependent synaptic plasticity. Curr Opin Neurobiol. 1999;9:305–13. doi: 10.1016/s0959-4388(99)80045-2. [DOI] [PubMed] [Google Scholar]
- 48.Jonas E. BCL-xL regulates synaptic plasticity. Mol Interv. 2006;6:208–22. doi: 10.1124/mi.6.4.7. [DOI] [PubMed] [Google Scholar]
- 49.Yao CK, Lin YQ, Ly CV, Ohyama T, Haueter CM, Moiseenkova-Bell VY, et al. A synaptic vesicle-associated Ca2+ channel promotes endocytosis and couples exocytosis to endocytosis. Cell. 2009;138:947–60. doi: 10.1016/j.cell.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atluri VS, Kanthikeel SP, Reddy PV, Yndart A, Nair MP. Human synaptic plasticity gene expression profile and dendritic spine density changes in HIV-infected human CNS cells: role in HIV-associated neurocognitive disorders (HAND) PLoS ONE. 2013;8:e61399. doi: 10.1371/journal.pone.0061399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong H, McCabe L, Skifter D, Monaghan DT, Gendelman HE. Activation of NR1a/NR2B receptors by monocyte-derived macrophage secretory products: implications for human immunodeficiency virus type one-associated dementia. Neurosci Lett. 2003;341:246–50. doi: 10.1016/s0304-3940(03)00194-0. [DOI] [PubMed] [Google Scholar]
- 52.Dou H, Ellison B, Bradley J, Kasiyanov A, Poluektova LY, Xiong H, et al. Neuroprotective mechanisms of lithium in murine human immunodeficiency virus-1 encephalitis. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25:8375–85. doi: 10.1523/JNEUROSCI.2164-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong H, Boyle J, Winkelbauer M, Gorantla S, Zheng J, Ghorpade A, et al. Inhibition of long-term potentiation by interleukin-8: implications for human immunodeficiency virus-1-associated dementia. Journal of Neuroscience Research. 2003;71:600–7. doi: 10.1002/jnr.10503. [DOI] [PubMed] [Google Scholar]
- 54.Tiede LM, Cook EA, Morsey B, Fox HS. Oxygen matters: tissue culture oxygen levels affect mitochondrial function and structure as well as responses to HIV viroproteins. Cell Death Dis. 2011;2:e246. doi: 10.1038/cddis.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palmer CS, Ostrowski M, Gouillou M, Tsai L, Yu D, Zhou J, et al. Increased glucose metabolic activity is associated with CD4+ T-cell activation and depletion during chronic HIV infection. Aids. 2014;28:297–309. doi: 10.1097/QAD.0000000000000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrero CA, Datta PK, Sen S, Deshmane S, Amini S, Khalili K, et al. HIV-1 Vpr modulates macrophage metabolic pathways: a SILAC-based quantitative analysis. PLoS ONE. 2013;8:e68376. doi: 10.1371/journal.pone.0068376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao W, Tan G, Zhu Z, Chen Q, Lou Z, Dong X, et al. Combined metabonomic and quantitative real-time PCR analyses reveal systems metabolic changes in Jurkat T-cells treated with HIV-1 Tat protein. Journal of Proteome Research. 2012;11:5109–23. doi: 10.1021/pr300173c. [DOI] [PubMed] [Google Scholar]
- 58.Songok EM, Luo M, Liang B, McLaren P, Kaefer N, Apidi W, et al. Microarray analysis of HIV resistant female sex workers reveal a gene expression signature pattern reminiscent of a lowered immune activation state. PLoS ONE. 2012;7:e30048. doi: 10.1371/journal.pone.0030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hollenbaugh JA, Munger J, Kim B. Metabolite profiles of human immunodeficiency virus infected CD4+ T cells and macrophages using LC-MS/MS analysis. Virology. 2011;415:153–9. doi: 10.1016/j.virol.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ringrose JH, Jeeninga RE, Berkhout B, Speijer D. Proteomic studies reveal coordinated changes in T-cell expression patterns upon infection with human immunodeficiency virus type 1. J Virol. 2008;82:4320–30. doi: 10.1128/JVI.01819-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rasheed S, Yan JS, Lau A, Chan AS. HIV replication enhances production of free fatty acids, low density lipoproteins and many key proteins involved in lipid metabolism: a proteomics study. PLoS ONE. 2008;3:e3003. doi: 10.1371/journal.pone.0003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dietrich MO, Liu ZW, Horvath TL. Mitochondrial dynamics controlled by mitofusins regulate Agrp neuronal activity and diet-induced obesity. Cell. 2013;155:188–99. doi: 10.1016/j.cell.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteomic data was analyzed through CyberT (http://cybert.ics.uci.edu/). A Bayesian analysis of the data was performed using a Bayesian coefficient of 12. Multiple testing corrections were applied. For significance, p<0.05 and cumulative posterior probability of differential expression (Cum PPDE)>0.95.
Area under curve data obtained from mass spectrometry was used to generate histograms comparing samples against one another within each experimental group. Pearson’s r coefficient was generated for each comparison.
Data from the mass spectrometry analysis were uploaded into the Panther classification system (http://www.pantherdb.org/) to determine which biological processes were most represented in our samples.