Abstract
Understanding vaginal and rectal HIV transmission and protective cellular and molecular mechanisms is critical for designing new prevention strategies, including those required for an effective vaccine. The determinants of protection against HIV infection are, however, poorly understood. Increasing evidence suggest that innate immune defenses may help protect mucosal surfaces from HIV transmission in highly exposed, uninfected subjects 1. More recent studies suggest that systemically administered type 1 interferon protects against simian immunodeficiency virus infection of macaques 2. Here we hypothesized that topically applied type 1 interferons might stimulate vaginal innate responses that could protect against HIV transmission. We therefore applied a recombinant human type 1 interferon (IFN-β) to the vagina of rhesus macaques and vaginally challenged them with pathogenic simian/human immunodeficiency virus (SHIV). Vaginal administration of IFN-β resulted in marked local changes in immune cell phenotype, increasing immune activation and HIV coreceptor expression, yet provided significant protection from SHIV acquisition as interferon response genes (IRGs) were also upregulated. These data suggest that protection from vaginal HIV acquisition may be achieved by activating innate mucosal defenses.
INTRODUCTION
Defining the innate mucosal defenses that may prevent mucosal HIV transmission is a high priority. The determinants of protection against HIV infection are, however, poorly understood. Certain alterations of host elements are known to be protective against HIV transmission. For example, a recognized 32 base pair deletion in the open reading frame of the gene encoding the HIV coreceptor CCR5 (d32) when present in the homozygous state confers nearly complete protection against HIV acquisition 3–5. Likewise topical blockade of CCR5 using modified chemokine analogues or allosteric CCR5 inhibitors can protect non-human primates from infection with simian human immunodeficiency virus (SHIV) infection 6–8. Nonetheless, most persons at apparent high risk for HIV acquisition who have remained uninfected do not have recognized polymorphisms in CCR5 or its ligands 9–11. Thus other mechanisms may exist that provide relative protection against HIV acquisition. Increasing evidence suggest that innate immune defense mechanisms may be involved in the protection of mucosal surfaces from HIV transmission in highly exposed, uninfected patients 1.
The type 1 interferons including IFN-α and IFN-β comprise a class of endogenous host elements that were first recognized for their ability to “interfere” with the replication of viruses in vitro (reviewed in 12). This class of molecules induces resistance to HIV through a number of mechanisms that include activation of several defined host factors that restrict HIV replication 13. Also type 1 interferons are capable of enhancing antiviral defenses through activation of cytolytic cells and enhancing the maturation of adaptive immune defenses 14. Type 1 interferons are expressed and secreted in response to viral infection or recognition of pathogen-associated molecular patterns (PAMPs). Following the engagement of these cytokines with the interferon receptor, the expression of a large number (>900) of interferon response genes (IRGs) with diverse functions may be induced to generate an antiviral state (reviewed in 12).
We hypothesized that type 1 interferons might be capable of mediating protection from acquisition of HIV and therefore applied a recombinant type 1 interferon (IFN-β) to the vagina of rhesus macaques, then vaginally challenged animals with pathogenic RT-SHIV. Here we show that topical administration of recombinant human beta interferon to mucosal tissues results in local changes in immune cell phenotype and activation, yet results in significant protection against RT-SHIV acquisition. This was not likely related to effects on the HIV co-receptor CCR5, as CCR5 density actually increased on mucosal and lymphoid T cells in animals that had been treated with IFN-β. These data suggest that protection from vaginal HIV infection may be achieved by activating vaginal innate defenses.
RESULTS
IFN-β treatment rapidly induces T cell activation, and increased numbers of CD4+CCR5+ T cells and macrophages in the vaginal mucosa
To assess the effects of topical IFN-β application on local immune responses, we first treated six female macaques intravaginally with either low (1.2 × 105 units/mL; n=2), intermediate (6 × 105 units/mL; n=2) or high (3 × 106 units/mL; n=2) dose recombinant human IFN-β (Rebif, Pfizer) Two controls received topical saline only. We used this type and dose of interferon as our preliminary data (not shown) indicated that IFN-β is among the most active of 12 different type 1 interferons in inhibiting HIV replication in human PBMC, and doses this high (and greater) have been well tolerated when applied intranasally in humans 15. Two days later each macaque was dosed intravaginally with 2mls of saline or IFN-β using the same concentrations as initially. Six hours later, all 8 animals were humanely euthanized. At necropsy, female reproductive tissues (FRT) were carefully examined for evidence of inflammation, but no visible lesions were detected. Vaginal tissues were processed for flow cytometry, immunohistochemistry, and RNA sequencing (RNASeq).
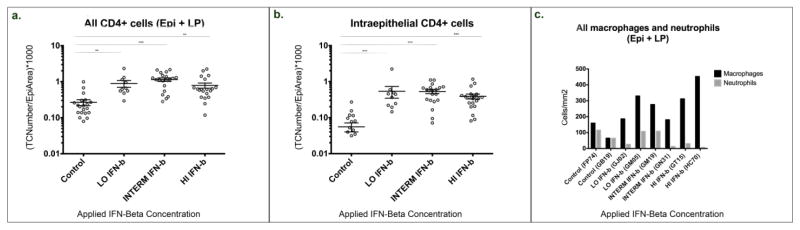
Compared to controls, all 6 IFN-β treated macaques had significantly higher numbers of CD4+ T cells and macrophages in vaginal mucosa (Fig. 1). Increased CD4+ T cell densities were detected in the epithelium, and in the lamina propria (Fig. 1a,b). Similarly, a marked, dose-dependent increase in macrophages was evident in the vaginal mucosa (Fig. 1c). By flow cytometry, vaginal mucosal CD4+ T cells of IFN-β treated animals had higher expression of HLA-DR and CCR5 (Table 1, Supplemental Figs. 1, 2) indicating these infiltrating CD4+ T cells were more activated, expressing higher levels of the HIV co-receptor CCR5, and thus potentially better targets for SHIV infection. Similarly, higher expression of HLA-DR and CCR5 was detected on CD8+ cells in vaginal tissues of IFN-β treated macaques (Table 1). Increased expression of Caspase 3 was also detected in both CD4+ and CD8+ vaginal T cells (Table 1). No significant changes were detected in expression of HLA-DR, CD38 or CCR5 on peripheral blood T cells (not shown).
Figure 1.
Absolute counts of CD4+ T cells (a, b), macrophages, and neutrophils (c) in the vagina of macaques vaginally dosed with IFN-β compared to saline treated controls. Figs a and b were generated using immunohistochemically stained frozen tissue sections of vagina and show the CD4+ T cell density in controls (n = 2), versus increasing concentrations (from left to right) as low dose (1.2 × 105 U/ml) = LO IFN-β, intermediate dose (6 × 105 U/ml) = INTERM INF-β, and high dose (3 × 106 U/ml) = HI IFN-β (n = 2) treated rhesus macaques. Sections were analyzed to count cells in the vaginal epithelium (Epi) and lamina propria (LP) combined (a), as well as the epithelium alone (b). At least ten measurements of each tissue type were measured from each macaque as indicated by individual data points. TCNumber/EpiArea refers to the number of target cells divided by the area of the epithelium analyzed. n = number of animals per condition. Each dot represents each individual image analyzed under each condition. Error bars represent standard errors of the mean (SEM). **, P≤0.01 and ***, P≤0.001, respectively. (c) Absolute numbers of macrophages (black bars) and neutrophils (white bars) were generated from immunohistochemically stained sections of formalin fixed, paraffin embedded tissue sections and are expressed as numbers of cells per mm2 tissue for each individual macaque.
Table 1.
Expression of inflammatory markers on vaginal T cells in macaques vaginally dosed with IFN-β
| T cell | Marker | IFN-β treatment | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Controls | Low dose | Interm. dose | High dose | ||||||
| GB19 | FP74 | GJ02 | GM05 | GM19 | GN31 | GT15 | HC70 | ||
| CD4+ T | CD38+ | 22.9 | 19.8 | 16.3 | 17.0 | 18.1 | 16.1 | 22.2 | 31.6 |
| HLA-DR+ | 6.2 | 2.8 | 13.1 | 15.3 | 13.6 | 15.2 | 10.8 | 12.4 | |
| CD38+/HLA-DR+ | 1.1 | 0.7 | 1.6 | 1.5 | 1.6 | 1.7 | 1.9 | 2.2 | |
| CCR5+ | 24.0 | 17.9 | 50.9 | 42.9 | 38.7 | 45.2 | 33.1 | 21.6 | |
| HLA-DR+/CCR5+ | 2.5 | 0.9 | 7.8 | 9.6 | 9.3 | 11.2 | 5.7 | 7.1 | |
| Caspase 3+ | 0.8 | 0.8 | 18.9 | 22.5 | 18.7 | 34.4 | 13.9 | 17.2 | |
| Caspase 3+/CCR5+ | 0.3 | 0.2 | 11.0 | 10.7 | 6.2 | 17.2 | 5.1 | 4.9 | |
| CD8+ T | CD38+ | 41.9 | 23.7 | 29.9 | 35.6 | 30.3 | 26.3 | 31.8 | 47.0 |
| HLA-DR+ | 5.5 | 2.5 | 14.5 | 15.5 | 13.2 | 14.9 | 11.5 | 16.6 | |
| CD38+/HLA-DR+ | 1.8 | 0.5 | 2.8 | 4.2 | 2.9 | 3.3 | 3.8 | 5.9 | |
| CCR5+ | 38.9 | 23.5 | 59.5 | 48.7 | 41.7 | 56.2 | 47.5 | 35.0 | |
| HLA-DR+/CCR5+ | 2.9 | 0.9 | 10.6 | 10.2 | 9.0 | 11.1 | 6.7 | 9.5 | |
| Caspase 3+ | 1.5 | 2.0 | 49.0 | 49.5 | 40.7 | 49.5 | 32.5 | 52.2 | |
| Caspase 3+/CCR5+ | 0.8 | 0.8 | 26.8 | 23.7 | 17.4 | 27.4 | 15.3 | 17.3 | |
Numbers indicate percentages of CD4+ or CD8+ T cell subsets co-expressing the marker(s) indicated on vaginal cells as detected by flow cytometry.
Topical IFN-β treatment protects macaques from vaginal SHIV transmission
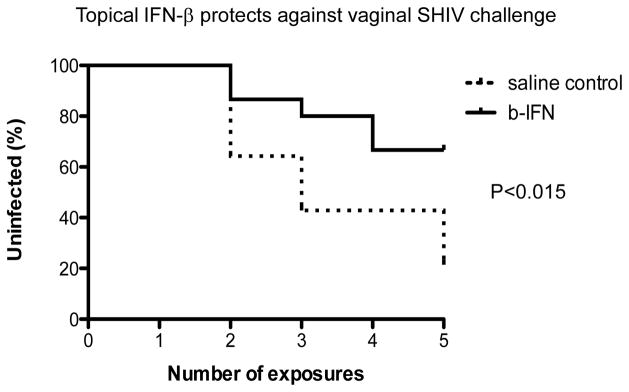
We then performed a series of topical vaginal IFN-β dosing and SHIV challenge experiments to determine whether repeated dosing could protect macaques from vaginal SHIV acquisition. Macaques were intravaginally dosed with IFN-β, or saline placebo and four hours later, all were intravaginally challenged with SHIV. One and two days after initial dosing, vaginal IFN-β or placebo dosing was repeated; this dosing and challenge regimen was repeated weekly for 5 weeks. In total, we performed this series of repeated vaginal IFN-β dosing and viral challenge studies in 15 female macaques treated with high dose 3×106 U/ml IFN-β for comparison with 14 mock treated controls. Nine of the 15 IFN-β treated macaques were completely protected from infection, whereas only 2/14 control animals escaped infection (Fig. 2). These differences were significant by Fishers Exact test (P<0.015). Note: 8/14 controls (57%) were infected by the 4th challenge, whereas only 4/15 (26%) high dose IFN-β treated animals were infected by the 4th challenge (Fig. 2). Also, the 6 IFN-β– treated animals that became infected showed a trend for lower peak viral loads compared to the controls (Fig. 3) but these results were not significant. In summary, a significant (P<0.015) level of protection from vaginal SHIV transmission was afforded to animals by repeated dosing with topical IFN-β.
Figure 2.
Kaplan Meier plots showing the infection rates of animals repeatedly intravaginally dosed with IFN-β (solid line) or mock treated controls (dashed lines) after 5 repeated dosing and challenges. After 5 challenges, 9 of 15 IFN-β treated animals remained uninfected, whereas only 2 of 14 controls remained uninfected. The difference in overall infection rates was significant by Fishers exact T test (P<0.15). Error bars indicate standard error of the means.
Figure 3.
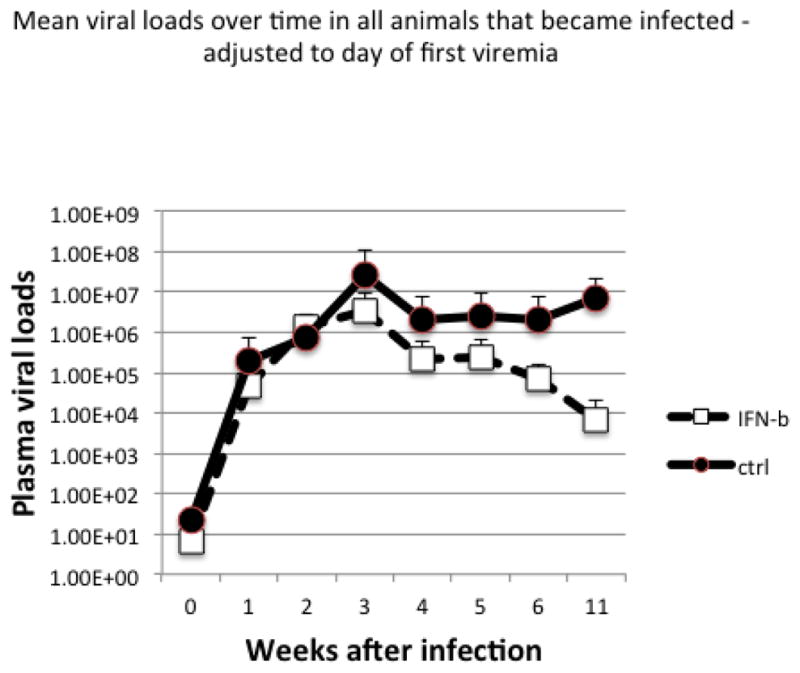
Mean plasma viral loads over time in the 6 animals that became infected despite IFN-β treatment (blue lines) versus the 12 infected placebo controls. Note animals treated with IFN-β had lower viral peaks and set points suggesting partial control of viremia.
Although topical application of IFN-β lowered the frequency of vaginal transmission, the mechanisms behind this protection are unclear. Our initial studies showed topical IFN-β resulted in an increase in absolute numbers, co-receptor expression, and activation of vaginal CD4 and CD8 T cells (Table 1). Activation was characterized more by increased HLA-DR expression than CD38 expression (Table 1, Supplemental Figs. 1, 2). Increased CCR5 expression was also observed in the initial study suggesting either that IFN-β exposure activated resident cells or enhanced the influx of activated co-receptor expressing immune cells or both. An increase in Caspase 3+, CCR5+ T cells also suggests that IFN-β exposure increased cellular activation. Conceivably, the activation of Caspase 3 by interferon resulted in the early death of cells that are plausibly good targets for HIV infection, thereby balancing or abrogating the effects of increasing target cell frequency, though some studies suggest IFN-induction of Caspase 3 may activate cells without driving cell death 16, 17.
Topical IFN-β treatment results in marked upregulation of inflammatory and myeloid activation genes
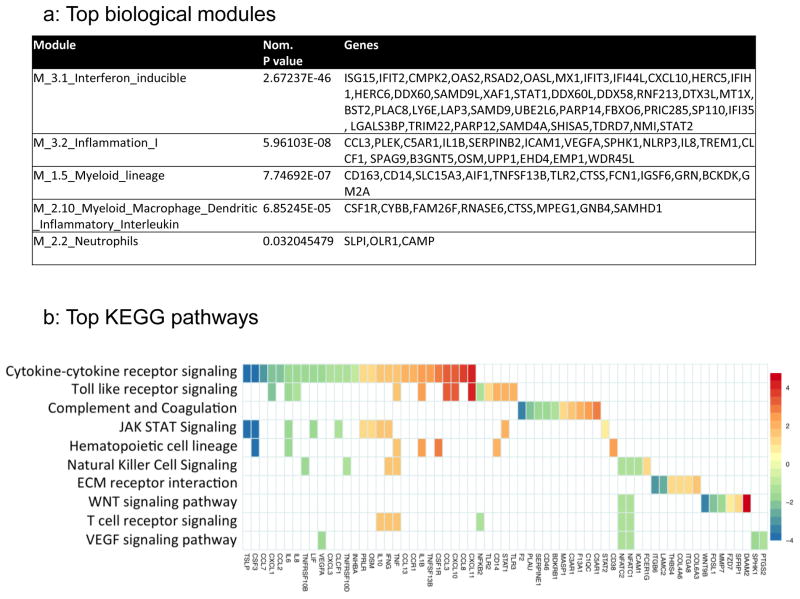
The above data suggest topical IFN-β was inducing a local protective host defenses despite increasing the pool of potentially susceptible target cells for SHIV infection. To test the hypothesis that IFN-β exposures drives expression of host IRGs that promote an overall innate antiviral state, we assessed the whole transcriptome of vaginal cell suspensions from the macaques in the pilot study using RNA-Seq. Enrichment analysis was performed using the Fisher’s exact test on significantly differential gene expression (FDR<0.05) between IFN-β-treated versus control macaque datasets, without an arbitrary cutoff in fold-change18, 19. We first applied transcriptional modules published by Chaussabel et al. 20 as broad filters to organize the enrichment results (IFN-β-treated versus controls) into top biologically relevant modules with a single multivariate score (Fig. 3A). Notably, the top 5 enriched biological modules by nominal P value via this method were interferon inducible genes (Module 3.1), inflammation I (Module 3.2), two myeloid cell lineage modules (Module 1.5 and 2.10), and neutrophils (M2.2). As expected, delineation of interferon inducible genes into type 1 or type 2 IRGs using published filters 21 revealed a higher enrichment in type 1 (P=2.19×10−57) vs type 2 (P=4.43×10−41) associated IRGs with IFN-β treatment (Fig. 4a, Supplemental Table 1).
Figure 4.
Top biological modules (a) and pathways (b) induced in vaginal cells after IFN-β treatment. (a) Marked upregulation of interferon inducible genes as well as genes associated with inflammation, and myeloid/macrophage activation were detected. (b) Checkerboard figure presents enrichment analysis of differential gene expression in FRT from IFN-β versus control macaques. The top 10 enriched Kegg pathways are plotted top to bottom on the y-axis and the gene members contributing to the enrichment are plotted along the x-axis. The scale represents log2 fold changes with red corresponding to most upregulated and blue to most downregulated genes.
We next conducted a Kegg pathway-based analysis. Kegg pathways are a collection of manually curated and drawn pathway mappings that allow a more specific assessment of potential biology following a supervised contrast than module analysis. The top 10 Kegg pathways enriched in differential gene expression between IFN-β and control treatment ranged from proinflammatory cytokine and cytokine receptor interaction (CXCL1/10/11, CCL23/7/8, IL1β, IL6, IL8, IL10, TNF, IFNγ), TLR receptor signaling and complement activation, to JAK/STAT, WNT and T cell signaling activation (Fig. 4b). The chemokines identified may promote accumulation of T cells and monocytes into the FRT as observed histologically (Fig. 1).
Lastly, since RNA-Seq was performed on unfractionated single cell suspensions, we employed an adaptation of a recent bioinformatic approach to deconvolute expression data from bulk cells into cell subset-specific subprofiles via published classifier lists 22. We discovered that gene expression changes in FRT of IFN-β-treated macaques occurred predominantly in association with a monocyte subset classifier list. Indeed, monocytes were the only significantly enriched (P=2.11 × 10−19) classifier, populated by many of the genes mentioned above (IRGs, chemokines, proinflammatory cytokines, and complement) (Supplemental Table 2). These data were consistent with the marked increase in macrophage recruitment evident by immunohistochemical analysis (Fig. 1c). Although the numbers of animals examined for immunologic changes in tissues were small, combined, these data indicate topical IFN-β was inducing a balanced local protective antiviral host defense response countering pro-inflammatory cytokine and chemokine activation, despite an increased influx of target cells for SHIV infection.
DISCUSSION
Understanding the balance between inflammatory responses that may enhance or prevent HIV transmission in local tissues is a high priority for HIV vaccine and prevention strategies. Type 1 interferons bind to a common receptor expressed on numerous cell types and activate hundreds of cellular genes including genes encoding proteins with antiviral activity. Systemic application of type 1 interferon in rhesus macaques delays SIV infection, yet sustained administration of this agent during chronic infection comes at the cost of increased immune activation, CD4 T cell losses and increased viral reservoir size 2. Numerous IRGs limit viral replication including RNaseL, Eukaryotic initiation factor 2 (EIF2a) which inhibits viral translation, APOBEC3G, Trim-5α and tetherin, factors recognized to restrict HIV replication; several that encode IFN-induced proteins with tetratricopeptide repeats (IFITs), and numerous others which play a variety of roles in mediating antiviral responses. Here we tested the hypothesis that the immune activation induced by topical interferon administration would be overcome by interferon-induced protection against SHIV acquisition. This appeared to be the case, suggesting that IFN-β is arming local mucosal defenses to protect against vaginal SHIV transmission, a model supported by demonstrating IFN-β induced upregulation of numerous IRGs including IRGs with defined antiviral activity.
Interestingly however, we also found that topical administration of IFN-β increased numbers of tissue macrophages and CD4+ T cells expressing CCR5 and HLA-DR, suggesting IFN-β promotes influx of activated cells potentially susceptible to HIV infection. As IFN-β dramatically upregulated the expression of CCL3 (promoting T cell chemotaxis via CCR5 ligation) and also CXCL10 and CXCL11 (which promote T cell chemotaxis via binding to CXCR3) we suspect these and other interferon-induced chemokines promote accumulation of target CD4 T cells into the vagina. Nonetheless, despite an increased availability of viral target cells, topical IFN-β was clearly associated with protection from SHIV challenge, likely through stimulation of innate immune defenses. In fact, the increase in myeloid (macrophage) recruitment into tissues may conceivably limit local viral replication and infection by phagocytosis and destruction of locally activated, infected T cells, but this remains speculative. To prevent interference with vaginal transmission, samples were not acquired from the animals in the challenge studies. However, the significant level of protection observed, combined with data from the pilot study indicating both an influx of macrophages and T cells (in all 6 treated animals) combined with the RNA Seq data indicating increases in monocyte activation genes, all suggest that interferon response genes and innate immune responses played a role in this protection.
This targeted, topical approach may avoid the systemic consequences of sustained systemic interferon exposure. Earlier studies exploring the role of innate defenses in protection from SIV infection attempted to induce these defenses through topical application of toll like receptor (TLR) ligands including imiquimod (a ligand for TLR 7) and CpG oligodeoxynuclotides (ligands for TLR 9). These studies not only failed to confer protection against SIV challenge but also induced a “storm” of inflammatory mucosal cytokines that were associated with increases in the magnitude of SIV replication in the infected animals 23. We considered the possibility that interferon-independent effects of TLR ligation might have obscured a potentially protective effect of type 1 interferons, and designed these experiments with this in mind. Thus we were able to examine directly the effects of interferon activity on susceptibility to SHIV infection.
The human type 1 interferon family includes several interferons that each bind the same type 1 interferon receptor but with distinguishable effects on downstream signaling 12. We chose beta interferon for these studies as in preliminary studies, this interferon was one of several type 1 interferons with the most consistent ability to block HIV replication in human lymphocytes (not shown) and because a recombinant form suitable for parenteral (systemic) administration (Rebif) was commercially available. Further, human and macaque IFN-b proteins are 95% identical and 97% similar by BLAST 24, so we felt confident in using this would be suitable for the macaque studies.
The role of type 1 interferons in HIV disease is not well understood, but sustained replication of both pathogenic HIV and SIV is associated with increased type 1 interferon production 25 and systemic administration of type 1 interferon to HIV infected persons can attenuate the magnitude of HIV replication 26. Recent studies in the rhesus SIV model indicate that blockade of type 1 interferon activity accelerates mortality in infected animals and that systemic administration of type 1 interferon may protect animals from SIV infection 2. HIV infected persons in whom HIV replication has been controlled with combination antiretroviral therapy also have a profound signature of type 1 interferon exposure and this is most prominent among those who have failed to normalize circulating CD4 T cell numbers 27. Also, in chronic SIV infection of rhesus macaques, sustained administration of type 1 interferon results in both interferon desensitization, an increase in SIV reservoirs and accelerated CD4 T cell losses 2. These works suggest that the effects of type 1 interferon are complex in HIV and SIV infections, providing both antiretroviral activity and in some settings, accelerating pathogenesis. Nonetheless, here we show that topical administration of a type 1 interferon can protect against pathogenic SHIV challenge. In these experiments, a very high dose of interferon was necessary to provide protection, likely reflecting the need for high concentrations of agents to penetrate vaginal mucosal tissues 28. Thus the costs of IFN-β as a topical strategy to prevent HIV acquisition may be too high to be clinically applicable, but developing a topical prevention strategy is not the crux of this study. A major objective of this study was to begin to delineate specific immune mechanisms and conditions that may be elicited by vaccines or other prevention strategies that result in protective host defenses. Although we did not examine sufficient numbers animals to definitively identify the mechanisms involved in this protection, our findings suggest the possibility that modulation of innate defenses, as reflected by the activity of type 1 interferons shown here, might play a role in the apparent “natural” resistance to HIV infection that has been reported in numerous high risk exposed populations 9 and that other strategies that enhance these endogenous innate defenses might prove to be of value in protection against HIV acquisition. Although we have clearly show a protective associated with topical vaginal IFN-β application, additional studies with larger numbers of animals and more frequent vaginal fluid and biopsy sampling will be needed to specifically determine the molecular mechanisms of protection elicited in the vaginal vault following IFN-β treatment.
Type 1 interferons comprise a large family of innate antiviral defense molecules that can be induced by a variety of microbial elements including viruses and their genetic sequences 29. Type 1 interferons bind to a common receptor expressed on numerous cell types and activate hundreds of cellular genes including genes encoding certain proteins with antiviral activity 30 yet at mucosal sites of challenge with the simian immunodeficiency virus, the kinetics of interferon induction by exposure to TLR ligands was too delayed to provide sufficient protection from experimental infection 31. It’s not unreasonable to propose therefore that there may be population variability in the induction, regulation and activities of such a complex defense network and to this point, a polymorphism near the gene for a type 2 interferon (interferon lambda-3, IL28B) has been linked to variability in the outcome of hepatitis C virus infection both spontaneously and with treatment using a type 1 interferon plus ribavirin 32, 33. Upregulation of the activities of cellular RNaseL and downregulation of Eukaryotic initiation factor 2 a (EIF2a) by type 1 interferons paralyze translation of both cellular and viral RNAs 29. Type 1 interferons also can activate other cellular elements such as APOBEC 3G, Trim-5a, tetherin, SAMHD1 and SLFN11 that more selectively restrict the replication of lentiviruses such as HIV 13. The importance of APOBEC3G, tetherin and SAMHD1 in antiviral defense is underscored by the existence of HIV and SIV proteins, specifically VIF, VPU and VPX respectively, that block their antiviral activities 34–37. Although human Trim-5a does not block HIV replication very effectively, certain non-human primate homologues do 38 and small changes in the human sequences result in potent restriction of HIV replication 39. In one small study, higher levels of TRIM-5a RNA were found in PBMC from high risk persons who escaped HIV infection than in PBMC from persons who were infected 40. A single nucleotide polymorphism in the APBOC3G gene (C49693T) has been associated in one study with an increased risk for HIV acquisition 41. Conceivably, other genetic variants in these restriction factors confer variable degrees of protection from infection.
In summary, these results demonstrate that repeated dosing with IFN-β results in a protective state in the vaginal environment that resists repeated vaginal challenge with a pathogenic SHIV, despite increased local accumulation of activated T cells and macrophages and heightened T cell expression of the HIV coreceptor CCR5. These results suggest that immune activation is not always deleterious and that mucosal protection against HIV acquisition may be achieved by induction of local innate host defenses. The conditions of our experiments do not allow us to identify the optimal duration of interferon exposure necessary for protection; we don’t know if shorter exposure to interferon is sufficient for a similar level of protection or if longer application of interferons might confer greater protection. In these experiments, a very high dose of interferon was necessary to provide protection, likely reflecting the need for high concentrations of agents to penetrate vaginal mucosal tissues 28. Thus the costs of IFN-β as a topical strategy to prevent HIV acquisition may be too high to be clinically practicable. Nonetheless, our findings suggest the possibility that modulation of innate defenses, as reflected by the activity of type 1 interferon shown here, might play a role in the apparent “natural” resistance to HIV infection reported in numerous high risk exposed populations 9 and that vaccine or other prevention strategies that enhance these endogenous innate defenses might prove to be of value in protection against HIV acquisition.
Despite decades of research, correlates of protection from HIV infection remain elusive, especially in mucosal tissues. Unique among all viruses, HIV is teaching us hard learned lessens, in that vaccines or topical microbicides that trigger specific types of inflammatory immune responses may in fact increase, rather than prevent HIV transmission 42, 43. Intensive work is focused on delineating the specific immune determinants that increase HIV transmission rates. However, if we could also identify specific patterns of inflammatory or immunosuppressive genes and proteins that provided protection in mucosal surfaces, perhaps these could be explored in HIV prevention trials as surrogate markers for protection, or correlated with protection in those patients who appear to repeatedly resist infection.
METHODS
Animals and experimental design
Adult, healthy female rhesus macaques free of simian immunodeficiency virus (SIV) and simian D retrovirus (SRV) were initially used to assess the immunologic effects of topically administered IFN-β on vaginal mucosal tissues in vivo (n=6). Vaginal biopsies were assessed by polychromatic flow cytometry and histopathology, and then an additional 34 female macaques (20 treated and 14 controls) were used to assess the protective effects of repeated IFN-β vaginal application followed by vaginal RT-SHIV challenge. All animals were housed at the Tulane National Primate Research Center in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International standards, and in compliance with the U.S. Department of Health and Human Services Guide for the Care and Use of Laboratory Animals. All studies were reviewed and approved by the Tulane University Institutional Animal Care and Use Committee.
Dose ranging topical IFN-β pilot study
In the first pilot experiment, 8 female macaques were intravaginally dosed with 2 mls saline containing either a low dose (1.2 × 105 units/mL; n=2), intermediate dose (6 × 105 units/mL; n=2) or high dose (3 × 106 units/mL; n=2) of recombinant human IFN-β (Rebif) and two animals were vaginally inoculated with saline only as controls. Two days later macaques were again vaginally dosed with 2mls IFN-β using the same concentrations as above for each animal. Six hours after this vaginal dosing, all 8 animals were humanely euthanized. Portions of vagina and endocervix were collected in complete RPMI medium supplemented with 5% heat inactivated fetal calf serum, l-glutamine, penicillin, streptomycin, and HEPES) buffer for immediate processing into single cell suspensions for flow cytometry, and adjacent sections were preserved in Optimum Cutting Temperature compound (OCT) and snap frozen in dry ice-cooled 2-methylbutane and processed for immunohistochemistry as described below. Parallel tissue sections were formalin fixed for paraffin embedding and sectioning for routine histopathology and immunohistochemistry.
Tissue processing and isolation of vaginal cells for flow cytometry and RNA-Seq
Single cell suspensions of vaginal tissues were prepared as previously described 44. Briefly, tissues were cut into 0.5cm pieces with paired razor blades, and incubated with 1 mM EDTA in Hanks balanced salt solution for 30 min with rapid shaking (300 RPM) in an incubated orbital shaker at 37 degrees C, followed by 2–3 sequential 30 min incubations in complete RPMI medium containing 20 U/ml collagenase (Type 1I, Sigma) again with rapid shaking at 37oC. After each incubation, tissues were further disrupted by gently pipetting 5 to 10 times with a 16-gauge feeding needle, pelleted (400g, 7 min), and supernatants discarded and medium replaced. At the end of these incubations, cell pellets were resuspended and filtered through nylon mesh and layered over a 35%/60% bilayer isotonic Percoll density cushion and centrifuged at 1000g for 30 min. The interface between the 35% and 60% Percoll layers was collected, washed, and adjusted to 107cells/ml. For flow cytometry, 100 μl aliquots (106 cells) were stained with appropriately diluted concentrations of monoclonal antibodies to CD3 (SP34), CD4 (L200), CCR5 (3A9), HLA-DR (L243)(all from BD Biosciences Pharmingen (San Diego, CA), CD38 (provided by the NIH Nonhuman Primate Reagent Resource), and LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Invitrogen, Grand Island, NY). Isotype-matched controls were included in all experiments. Samples were resuspended in BD Stabilizing Fixative (BD Biosciences) and acquired on an LSR-II flow cytometer (Becton Dickinson, San Jose, CA). Data were analyzed with Flow jo software (Tree Star, Ashland, OR).
Immunohistochemistry and quantitative image analysis
Immunohistochemistry for CD4 detection was performed on frozen tissue sections. Fresh sections of vagina tissues were embedded and snap frozen in optimum cutting temperature compound (OCT) in dry-ice cooled 2-methyl butane. Frozen sections were then prepared from OCT blocks using a cryostat, adhered to glass slides, and tissue sections were fixed in 3.7% formaldehyde in PIPES buffer and blocked with normal donkey serum prior to staining. To identify target cells, rhesus macaque tissue was stained with anti-CD4 (Biolegend, cat#317422). Images were obtained by deconvolution microscopy on a DeltaVision RT system collected on a digital camera (CoolSNAP HQ; Photometrics) using a 60x oil objective. To determine the target cell density in individual macaque sections, at least ten measurements of vaginal samples were examined per macaque. The area of the epithelium was calculated from the stratum basale to the lumen using Deltavision SoftWorX software intraepithelial target cell densities were assessed for each individual sample. To asses CD4+ cells in the lamina propria (below the stratum basale), additional area measurements were expanded to include the epithelium and ~20 microns past the basal layer. Overall, for both data sets, each image consisted of a stitched panel comprised of two 60x images across the lumen and n 60x images past the basal layer, n being dependent on the epidermal thickness of each individual sample. Per animal, 10 panel images, consisting of 3 panels across and n panels into the epithelium (n dependent of the thickness of the epithelium as this varied among animals) were taken. To calculate the density of each sample, the number of target cells in each sample was divided by the average area of the epithelium or epithelium plus ~20 microns past the stratum basale, respectively. Data are expressed as numbers of CD4+ T cells divided by the total area of the tissue analyzed in Figs 1a and 1b.
Immunohistochemistry and image analysis of macrophages and neutrophils in vaginal tissues was assessed on formalin fixed, paraffin embedded tissue sections which were de-paraffinized and hydrated. Antigen retrieval was performed in a Decloaking Chamber (Biocare Medical, Concord, CA), at 125°C and 25 psi. Endogenous peroxidase activity was blocked with buffered hydrogen peroxide (Biocare Medical) for 5 min at room temperature (RT). Unspecific binding of Abs was blocked using purified casein and other proteins (Background Sniper, Biocare Medical) for 10 min at RT. Slides were incubated with the following primary Abs for 1 h at RT: mouse monoclonal anti-human CD68 (clone KP1, Dako), mouse monoclonal anti-human neutrophil defensins (clone D21, Leica Biosystems). Anti-mouse EnVision+ Systems horseradish peroxidase was used for 45 min at RT as a secondary Ab and visualization was performed using a DAB + (diaminobenzidine) substrate chromogen (Dako). Sections were counterstained with hematoxylin, dehydrated, and mounted in mounting medium. Slides were scanned using a Zeiss MIRAX MIDI slide scanner in the brightfield mode. Cell counts were performed using HistoQuest analysis software (TissueGnostics). Nuclei were first segmented based on their hematoxylin nuclear stains. Then, intensity of colorimetric stain was determined within a defined radius of each cell nucleus and reported as cells/mm2 as shown in Fig 1c.
Viral challenge experiments
In the first series of challenge experiments, 15 female macaques were intravaginally dosed with 2ml of medium containing either the high dose (3×106 U/ml; n=6), or the low dose (1.2×105 U/ml; n=5) of IFN-β, or saline placebo (n=5) to match the dose ranging experiments above. Exactly 4 hrs later, all 15 animals were intravaginally challenged with 300 TCID50 of RT-SHIVsf162P3 45 obtained from the NIH AIDS Reagent Program. One and two days (24 and 48hrs) after initial dosing, vaginal IFN-β or placebo dosing was repeated. This dosing and challenge regimen was repeated weekly for 5 weeks. Blood was collected weekly to determine if animals were protected or infected after challenge. In these and subsequent challenge studies, no vaginal tissues were collected to ensure integrity of the FRT, and not interfere with viral challenges and infectivity. In subsequent experiments, the low dose IFN-β group was omitted, and a total of 10 additional macaques were vaginally dosed with 2 mls of 3×106 U/ml IFN-β and 9 controls were dosed with placebo, followed 4 hrs later by pathogenic challenge with RT-SHIVsf162P3. Vaginal IFN-β and placebo treatments were repeated 24 and 48 hr later, exactly as in the 2nd experiment above (no FRT samples collected). Data shown in Fig. 2 only includes the 15 animals treated with the high dose IFN-β and the 14 controls.
RNA sequencing and data analysis
RNA was isolated from the vaginal cells from two treated and two control macaques using RNeasy Micro spin columns (Qiagen) and measured for quality using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). All samples had a RIN score between 7 and 9.6 (average: 8.7). Sequencing libraries were prepared with TruSeq RNA v2 kits (Illumina, San Diego, CA) using the manufacturer’s standard protocols. Library quality was assessed by measuring insert size using a Bioanalyzer 2100 (average size: 323.6bp) and library concentration determined by qPCR (Library Quantification Kit, KR0405, Kapa Biosystems, Wilmington, MA). The libraries were run on a HiSeq 2500 with 50bp paired end reads (average reads per sample: 13.68 million). Reads were trimmed of Illumina adapter sequences using Trimmomatic v0.32 46. Trimmed paired end reads were mapped against the macaque genome (MMUL 1.0, v78) using the STAR aligner v2.3.0e 47. Transcript abundances were estimated using HTSeq v0.6.1 48. The count matrices were then tested for differentially expressed genes using edgeR v3.0.2 49 and Student’s t test. Given the small dataset, the genes with p value and false discovery rate (FDR) < 0.05 were then tested for enriched pathways using Fisher’s exact test (p < 0.05 significance level).
Supplementary Material
Acknowledgments
We thank Terri and Kelsi Rasmussen, Megan Gardner, Meagan Watkins, and Maury Duplantis and Marina Krykbaeva for technical assistance. We also thank Stephanie Richards and Andrew Smith at the VGTI Florida Collaborative Genomics Center for performing the RNA-Seq assays. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: RT Env SHIV Virus P244 derived (PHA), from Dr. James Smith. This work was supported in part by NIH grants U19 AI076981, R01 AI084793, the National Center for Research Resources, and the Office of Research Infrastructure Programs (ORIP) of the National Institutes of Health through grant no. OD011104-51, and the Center for AIDS Research at Case Western Reserve University (AI36219).
Footnotes
DISCLOSURE: The author declared no conflicts of interest.
References
- 1.Tomescu C, Abdulhaqq S, Montaner LJ. Evidence for the innate immune response as a correlate of protection in human immunodeficiency virus (HIV)-1 highly exposed seronegative subjects (HESN) Clin Exp Immunol. 2011;164(2):158–169. doi: 10.1111/j.1365-2249.2011.04379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandler NG, Bosinger SE, Estes JD, Zhu RT, Tharp GK, Boritz E, et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511(7511):601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study [comment][erratum appears in Science 1996 Nov 15;274(5290):1069] Science. 1996;273(5283):1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 4.Liu R, Paxton WA, Choe S, Ceredini D, Martin SR, Horuk R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 5.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 6.Lederman MM, Veazey RS, Offord R, Mosier DE, Dufour J, Mefford M, et al. Prevention of vaginal SHIV transmission in rhesus macaques through inhibition of CCR5. Science. 2004;306(5695):485–487. doi: 10.1126/science.1099288. [DOI] [PubMed] [Google Scholar]
- 7.Veazey RS, Ling B, Green LC, Ribka EP, Lifson JD, Piatak M, Jr, et al. Topically applied recombinant chemokine analogues fully protect macaques from vaginal simian-human immunodeficiency virus challenge. J Infect Dis. 2009;199(10):1525–1527. doi: 10.1086/598685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veazey RS, Ketas TJ, Dufour J, Moroney-Rasmussen T, Green LC, Klasse PJ, et al. Protection of rhesus macaques from vaginal infection by vaginally delivered maraviroc, an inhibitor of HIV-1 entry via the CCR5 co-receptor. J Infect Dis. 2010;202(5):739–744. doi: 10.1086/655661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lederman MM, Alter G, Daskalakis DC, Rodriguez B, Sieg SF, Hardy G, et al. Determinants of protection among HIV-exposed seronegative persons: an overview. J Infect Dis. 2010;202(Suppl 3):S333–338. doi: 10.1086/655967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salkowitz JR, Purvis SF, Meyerson H, Zimmerman P, O’Brien TR, Aledort L, et al. Characterization of high-risk HIV-1 seronegative hemophiliacs. Clin Immunol. 2001;98(2):200–211. doi: 10.1006/clim.2000.4969. [DOI] [PubMed] [Google Scholar]
- 11.Lane J, McLaren PJ, Dorrell L, Shianna KV, Stemke A, Pelak K, et al. A genome-wide association study of resistance to HIV infection in highly exposed uninfected individuals with hemophilia A. Human molecular genetics. 2013;22(9):1903–1910. doi: 10.1093/hmg/ddt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 13.van Montfoort N, Olagnier D, Hiscott J. Unmasking immune sensing of retroviruses: interplay between innate sensors and host effectors. Cytokine & growth factor reviews. 2014;25(6):657–668. doi: 10.1016/j.cytogfr.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Tough DF. Modulation of T-cell function by type I interferon. Immunol Cell Biol. 2012;90(5):492–497. doi: 10.1038/icb.2012.7. [DOI] [PubMed] [Google Scholar]
- 15.Sperber SJ, Doyle WJ, McBride TP, Sorrentino JV, Riker DK, Hayden FG. Otologic effects of interferon beta serine in experimental rhinovirus colds. Archives of otolaryngology--head & neck surgery. 1992;118(9):933–936. doi: 10.1001/archotol.1992.01880090049015. [DOI] [PubMed] [Google Scholar]
- 16.Ludwiczek O, Kaser A, Koch RO, Vogel W, Cruikshank WW, Tilg H. Activation of caspase-3 by interferon alpha causes interleukin-16 secretion but fails to modulate activation induced cell death. European cytokine network. 2001;12(3):478–486. [PubMed] [Google Scholar]
- 17.Zipp F, Beyer M, Gelderblom H, Wernet D, Zschenderlein R, Weller M. No induction of apoptosis by IFN-beta in human antigen-specific T cells. Neurology. 2000;54(2):485–487. doi: 10.1212/wnl.54.2.485. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, Park PJ. Discovering statistically significant pathways in expression profiling studies. Proc Natl Acad Sci U S A. 2005;102(38):13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29(1):150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waddell SJ, Popper SJ, Rubins KH, Griffiths MJ, Brown PO, Levin M, et al. Dissecting interferon-induced transcriptional programs in human peripheral blood cells. PLoS One. 2010;5(3):e9753. doi: 10.1371/journal.pone.0009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Abel K, Lantz K, Krieg AM, McChesney MB, Miller CJ. The Toll-like receptor 7 (TLR7) agonist, imiquimod, and the TLR9 agonist, CpG ODN, induce antiviral cytokines and chemokines but do not prevent vaginal transmission of simian immunodeficiency virus when applied intravaginally to rhesus macaques. J Virol. 2005;79(22):14355–14370. doi: 10.1128/JVI.79.22.14355-14370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witwer KW, Sisk JM, Gama L, Clements JE. MicroRNA regulation of IFN-beta protein expression: rapid and sensitive modulation of the innate immune response. J Immunol. 2010;184(5):2369–2376. doi: 10.4049/jimmunol.0902712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanderford TH, Slichter C, Rogers KA, Lawson BO, Obaede R, Else J, et al. Treatment of SIV-infected sooty mangabeys with a type-I IFN agonist results in decreased virus replication without inducing hyperimmune activation. Blood. 2012;119(24):5750–5757. doi: 10.1182/blood-2012-02-411496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asmuth DM, Murphy RL, Rosenkranz SL, Lertora JJ, Kottilil S, Cramer Y, et al. Safety, tolerability, and mechanisms of antiretroviral activity of pegylated interferon Alfa-2a in HIV-1-monoinfected participants: a phase II clinical trial. J Infect Dis. 2010;201(11):1686–1696. doi: 10.1086/652420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez S, Tanaskovic S, Helbig K, Rajasuriar R, Kramski M, Murray JM, et al. CD4+ T-cell deficiency in HIV patients responding to antiretroviral therapy is associated with increased expression of interferon-stimulated genes in CD4+ T cells. J Infect Dis. 2011;204(12):1927–1935. doi: 10.1093/infdis/jir659. [DOI] [PubMed] [Google Scholar]
- 28.Veazey RS, Klasse PJ, Schader SM, Hu Q, Ketas TJ, Lu M, et al. Protection of macaques from vaginal SHIV challenge by vaginally delivered inhibitors of virus-cell fusion. Nature. 2005;438(7064):99–102. doi: 10.1038/nature04055. [DOI] [PubMed] [Google Scholar]
- 29.Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27(3):370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95(26):15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abel K, Rocke DM, Chohan B, Fritts L, Miller CJ. Temporal and anatomic relationship between virus replication and cytokine gene expression after vaginal simian immunodeficiency virus infection. J Virol. 2005;79(19):12164–12172. doi: 10.1128/JVI.79.19.12164-12172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 33.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malim MH. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos Trans R Soc Lond B Biol Sci. 2009;364(1517):675–687. doi: 10.1098/rstb.2008.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 38.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 39.Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30. 2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79(5):3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sewram S, Singh R, Kormuth E, Werner L, Mlisana K, Karim SS, et al. Human TRIM5alpha expression levels and reduced susceptibility to HIV-1 infection. J Infect Dis. 2009;199(11):1657–1663. doi: 10.1086/598861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valcke HS, Bernard NF, Bruneau J, Alary M, Tsoukas CM, Roger M. APOBEC3G genetic variants and their association with risk of HIV infection in highly exposed Caucasians. AIDS. 2006;20(15):1984–1986. doi: 10.1097/01.aids.0000247124.35129.e1. [DOI] [PubMed] [Google Scholar]
- 42.Gray GE, Moodie Z, Metch B, Gilbert PB, Bekker LG, Churchyard G, et al. Recombinant adenovirus type 5 HIV gag/pol/nef vaccine in South Africa: unblinded, long-term follow-up of the phase 2b HVTN 503/Phambili study. Lancet Infect Dis. 2014;14(5):388–396. doi: 10.1016/S1473-3099(14)70020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramjee G, Govinden R, Morar NS, Mbewu A. South Africa’s experience of the closure of the cellulose sulphate microbicide trial. PLoS Med. 2007;4(7):e235. doi: 10.1371/journal.pmed.0040235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veazey RS, Tham IC, Mansfield KG, DeMaria M, Forand AE, Shvetz DE, et al. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4(+) T cells are rapidly eliminated in early SIV infection in vivo. J Virol. 2000;74(1):57–64. doi: 10.1128/jvi.74.1.57-64.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith JM, Dauner A, Li B, Srinivasan P, Mitchell J, Hendry M, et al. Generation of a dual RT Env SHIV that is infectious in rhesus macaques. J Med Primatol. 2010;39(4):213–223. doi: 10.1111/j.1600-0684.2010.00434.x. [DOI] [PubMed] [Google Scholar]
- 46.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.