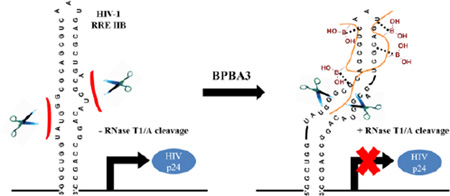
Abstract
A branched peptide containing multiple boronic acids was found to bind RRE IIB selectively and inhibit HIV-1 p24 capsid production in a dose-dependent manner. Structure-activity relationship studies revealed that branching in the peptide is crucial for the low micromolar binding towards RRE IIB, and that the peptide demonstrates selectivity towards RRE IIB in the presence of tRNA. Footprinting studies suggest a binding site on the upper stem and internal loop region of the RNA, which induces enzymatic cleavage of the internal loops of RRE IIB upon binding.
Keywords: HIV-1, RRE RNA, Branched peptides, Boronic acids, p24 inhibition
Graphical abstract
1. Introduction
RNA has gained significant attention as a potential therapeutic target because of its vital role in regulating many biological processes such as transcription, translation, splicing, catalysis, as well as transfer of genetic information within the cell. Secondary structural elements such as stems, pseudoknots, bulges, and hairpin loops afford a tertiary architecture unique to RNA. A-form RNA adopts a deeper and narrower major groove and shallower minor groove than in B-form DNA. These unique structural properties of RNA are expected to impart selective binding towards certain ligand motifs, suggesting that both the structure and function of RNA make it an ideal target in the development of therapeutics.1, 2 However, endeavors towards developing small-molecule ligands that target RNA have been met with limited success.3, 4 This predicament is exacerbated by the substantial surface area of RNAs that yield high affinity binding of cognate ligands, as well as the large conformational dynamics inherent in RNA that make it difficult to target the active conformation in efforts to design potent and selective inhibitors.5 To circumvent these challenges, several strategies have been pursued. For example, in silico studies have been employed to screen ligands against various RNA motifs, and nuclear magnetic resonance and molecular dynamics have been employed to investigate certain ligand-RNA dynamics.4, 6–8 Further, high-throughput screens using chemical libraries that contain a large number of diverse ligands are utilized to screen against various RNAs in an expedient manner.9, 10 Short interfering RNAs have also been deployed to selectively target RNA, but these are hampered by poor membrane permeability and stability.11 Several RNAs have been well-studied as therapeutic targets, including expanded nucleotide repeats r(CCUG) involved with the development of myotonic dystrophy type 2,12–15 as well as viral RNAs that contain targets such as the HIV-1 dimerization initiation site (DIS)16–19 and the HCV internal ribosome entry site (IRES).20, 21 The Rev/Rev Response Element (RRE) pathway has become a high profile drug target due to its critical role in the proliferation of HIV-1.22 RRE is a highly conserved region in the HIV-1 genome that spans approximately 351 nucleotides in the env gene.23, 24 RRE interacts with the Rev protein, which is synthesized from a fully spliced mRNA, to allow transport of singly spliced and unspliced mRNAs from the nucleus into the cytoplasm via complex formation with nuclear export factors such as Ran-GTP, eIF-5A and Crm-1.25, 26 This Rev-RRE interaction is essential to the HIV-1 life cycle as unspliced and singly-spliced mRNAs are required for translation of gag, pol, and env genes that encode structural proteins for packaging. In addition, unspliced RNA serves as the genome for new viruses.25–27 Thus, disruption of this interaction presents an avenue to inhibit viral replication. Owing to the therapeutic potential of the Rev-RRE export pathway, a variety of small molecules, such as aminoglycosides, have been designed to interrupt Rev-RRE interactions with limited clinical success.3, 28–30 Other inhibitors such as aromatic heterocycles, antisense oligonucleotides, RRE-based decoys, cyclic peptides, α-helical peptidomimetics, and others have also been identified, but also failed to yield any success in clincial applications.29, 31–46
As a general platform to develop compounds capable of selectively targeting folded structures of RNA, we utilized a branched peptide scaffold with the rationale that peptide libraries can be generated expediently on solid-phase with a wide variety of functional groups available to introduce structural diversity. Second, branching the peptide should allow for multivalent interactions with the RNA, and increase the surface area of binding towards the accessible tertiary structural elements found in the RNA. We previously disclosed the development of a first generation branched peptide library that selectively bound another HIV-1 RNA, the transactivation response element (TAR).47, 48 These peptides demonstrated little to no cytotoxicity, excellent cellular uptake, and bound TAR in the submicromolar range. Our second generation library targeted the stem-loop IIB of RRE (RRE IIB), which has been recognized as the high affinity site where Rev initially binds.27, 49–53 This library introduced two unnatural amino acid derivatives containing boronic acid, ε-(4-boronobenzoyl)-l-lysine (KBBA) and l-4-boronophenylalanine (FBPA), with the hypothesis that the empty p-orbital of boron could act as a Lewis acid and form a reversible covalent bond with the 2‘-hydroxyl group of the RNA, affording a novel mode of binding. These medium-sized branched peptide boronic acids (BPBAs) selectively bound to the tertiary structure of RRE IIB in the low micromolar regime (Table 1), encompassing a large surface area on the RNA.54, 55 Herein, we report the in vitro activity of these BPBAs via monitoring the expression of HIV-1 p24 capsid protein, as well as HIV-1 replication. One compound, BPBA3, was shown to inhibit these processes. Further characterization of this compound revealed that branching in the peptide is critical for its low micromolar binding affinity, and that binding to RRE IIB elicits a conformational change in RNA structure.
Table 1.
Sequences and dissociation constants of BPBAs
| Peptide | Sequence | Kd (µM) |
|---|---|---|
| BPBA 1 | (WKK)2*KBBAYWY | 1.4 ± 0.4 |
| BPBA 2 | (KBBAKFBPA)2*KBBAKKY | 3.3 ± 1.2 |
| BPBA 3 | (FBPAYFBPA)2*NKSY | 8.7 ± 2.3 |
| BPBA 4 | (KKKBBA)2*FBPATSY | 26.8 ± 4.4 |
| BPBA 5 | (KKBBAF)2*KKWY | 27.2 ± 6.9 |
| BPBA 6 | (WYK)2*PTWY | 28.5 ± 4.4 |
| BPBA 7 | (KKBBAK)2*KLKY | 58.4 ± 4.0 |
| BPBA 8 | (KBBAYK)2*HKKY | 86.5 ± 10 |
2. Results and discussion
2.1. Inhibition of p24 production and HIV-1 replication and toxicity of BPBA3 in cell culture
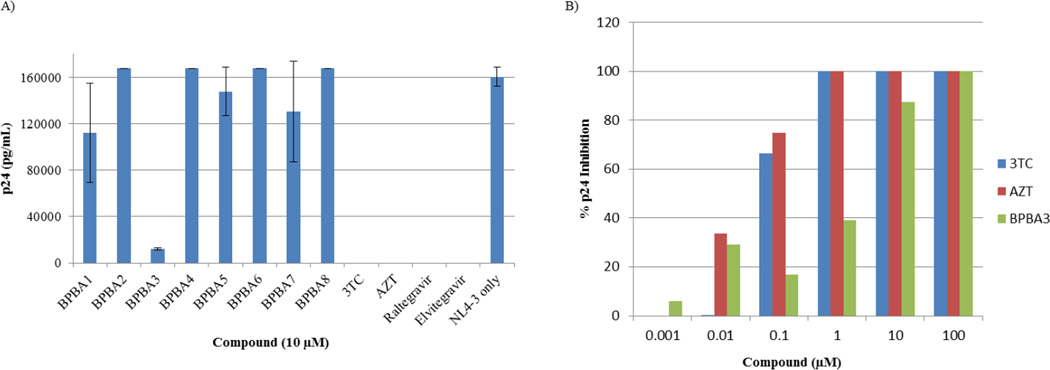
Having determined the binding affinities of BPBAs toward RRE RNA, we were interested in examining the ability of our BPBAs to inhibit HIV capsid production and HIV-1 replication in cell culture. Since the best binders had dissociation constants in the low micromolar range, we first tested our compounds at 10 µM in a p24 production and HIV particle release assay over a 24 h period using a p24 ELISA (see Supplementary Fig. SI.8).45 This initial screen revealed significant inhibition (70%) of p24 with one compound, BPBA3. An MTT assay conducted in HeLa cells over 24 h up to the concentration used in the p24 screening revealed no toxicity (see Supplementary Fig. SI.6). Encouraged by these results, a subsequent screening was conducted to determine whether inhibition of p24 could be maintained during an HIV-1 infection over a longer period of several days (Fig. 1A). As expected, nucleoside reverse transcriptase inhibitors (NRTIs), 2′,3′-dideoxy-3′-thiacytidine (3TC) and azidothymidine (AZT), and two integrase inhibitors, Elvitegravir and Raltegravir completely abolished p24 expression. Among the branched peptides tested, only BPBA3 significantly inhibited p24 production. To confirm the effect of BPBA3, a dose-dependent p24 inhibition assay was performed. As shown in Fig. 1B, increasing the concentration of BPBA3 from 1 nM to 100 µM resulted in a concomitant inhibition of p24 expression. Although 2-log less potent than AZT or 3TC, significant inhibition of p24 at 10 µM was confirmed and complete inhibition of p24 production was achieved at 100 µM. From this data, the IC50 value appears to be around 5 µM, which is in good agreement with the Kd established by dot blot (8.7 µM) and EMSA (4.6 µM) assays (see Supplementary Fig. SI.4).54 The MTT assay run in parallel with the p24 assay revealed that at higher concentrations of BPBA3, cell viability decreased; however, the inhibition of p24 production is not directly correlated to toxicity, since there is still decent cell viability (> 80 percent) at 10 µM where approximately 90 percent suppression of p24 production is observed (see Supplementary Fig. SI.7). This is exciting as this is the first peptide from our branched peptide library that demonstrated inhibition of HIV-1 p24 production and replication in cell culture.
Figure 1.
A) HIV-1 replication screening of BPBAs at 10 µM. 3TC and AZT (NRTIs), and Raltegravir and Elvitegravir (integrase inhibitors) served as positive controls. NL-4-3 only represents the U87-CD4/CXCR4 cell line infected with the HIV-1 virus (NL4-3 strain) without addition of compound/inhibitor. B) Dose-dependent inhibition of HIV-1 p24 with BPBA3. AZT and 3TC were used as reference compounds.
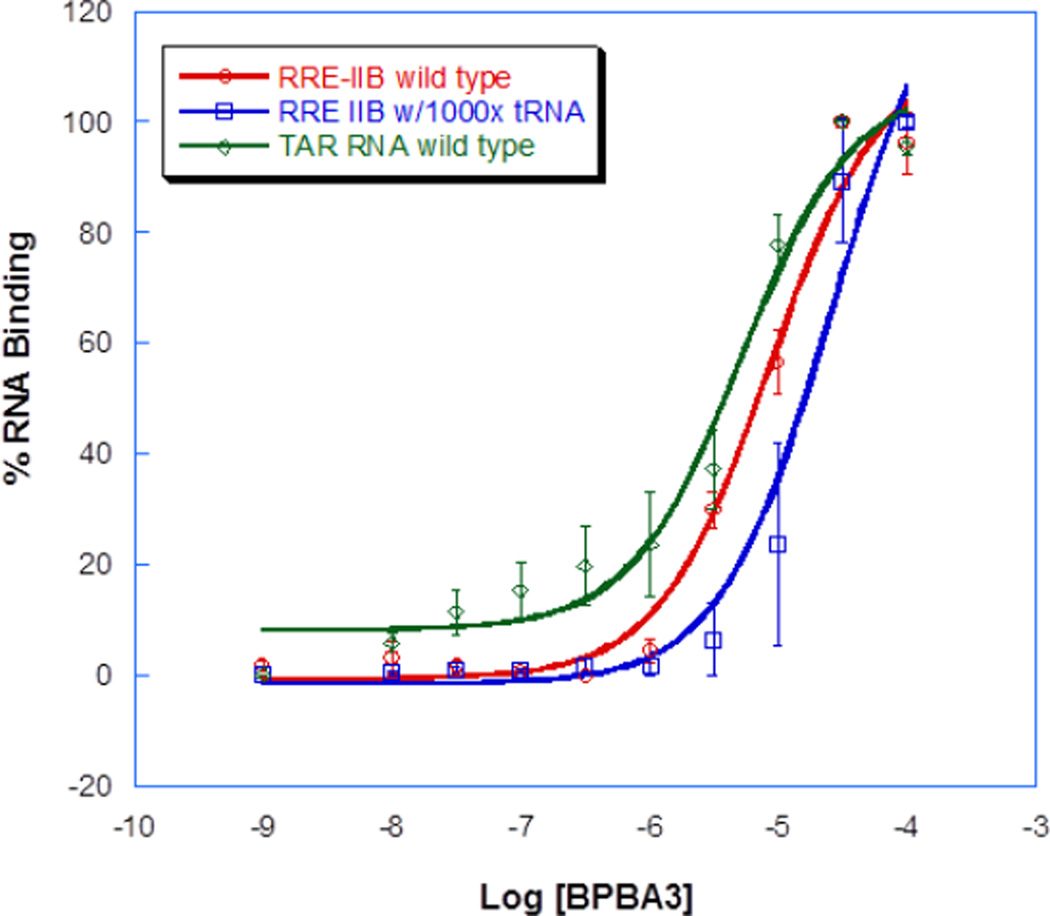
2.2. Selectivity of BPBA3 against tRNA and TAR RNA
After establishing the in vitro activity of BPBA3, we investigated its ability to selectively recognize RRE IIB, the RNA used in the high throughput screen (Fig. 2). Gratifyingly, only a 3-fold increase in Kd was observed (26.4 ± 10.2 µM) in the presence of 1000-fold excess competitor tRNA, suggesting a preference to small RNA structures. We then compared this activity with another HIV-1 conserved RNA structure, TAR (trans activating region) RNA. While the binding affinity with TAR (Kd of 5.2 ± 1.5 µM) is comparable to RRE IIB (Kd of 8.7 ± 2.2 µM), this result is not surprising since the branched peptide may interact similarly with folded stem-loop RNA structures. This binding study also highlights the potential for the binding of multiple HIV-1 RNAs by BPBA3 that could synergistically contribute towards the inhibition of HIV-1 p24 expression vide supra.
Figure 2.
Titration curves and dissociation contants of BPBA3 with wild-type RRE IIB RNA, RRE IIB RNA in the presence of 1000-fold excess competing tRNA, and TAR RNA. Each value is an average of at least three experiments.
2.3. Structure-activity of BPBA3 variants towards RRE IIB
We next probed whether the structure of BPBA3 has an effect on binding to RRE IIB. We previously explored the role of the boronic acids in BPBA3 on binding towards RRE IIB and demonstrated that removal of boronic acids in the structure completely abolished RNA binding.54 We therefore decided to perform a structure-activity relationship study to determine the effect of branching on the peptide and resulting RNA binding. The truncated and linear variants of BPBA3 are shown in Table 2. Removal of the one of the branches (LBPBA3A) led to a 2-fold loss in binding affinity, suggesting a minor binding contribution from the N-terminus branch attached to the ε-nitrogen of BPBA3. Titrations with a linear variant composed of the amino acids in BPBA3 (LBPBA3B) revealed a 3-fold loss in binding affinity towards RRE IIB. The larger decrease in binding affinity seen for LBPBA3B compared to LBPBA3A may be due to a longer peptide sequence that could alter or abolish interactions of key residues with the RNA. We suspect that the slight change in the binding affinity of these peptides is a result of an additional electrostatic attraction because of the presence of a positive charge on the ε-nitrogen of the branching lysine moiety. However, when the C-terminal branch was removed (LBPBA3C), a dramatic loss of binding affinity was observed (Kd > 100 µM). Overall, our studies suggest a critical role of the C-terminus branch of BPBA3, and amino acids on this region likely interact extensively with RRE IIB.
Table 2.
Sequences and dissociation constants of BPBA3 variants
| Peptide | Scheme | Sequence | Kd (µM) |
|---|---|---|---|
| BPBA3 | (FBPAYFBPA)2*NKSY | 8.7 ± 2.2 | |
| LBPBA3A | FBPAYFBPA*NKSY | 17.7 ± 5.0 | |
| LBPBA3B | FBPAYFBPAFBPAYFBPA*NKSY | 27.2 ± 7.1 | |
| LBPBA3C | (FBPAYFBPA)2* | >100 |
* = lysine branching unit. Each value is an average of at least three experiments.
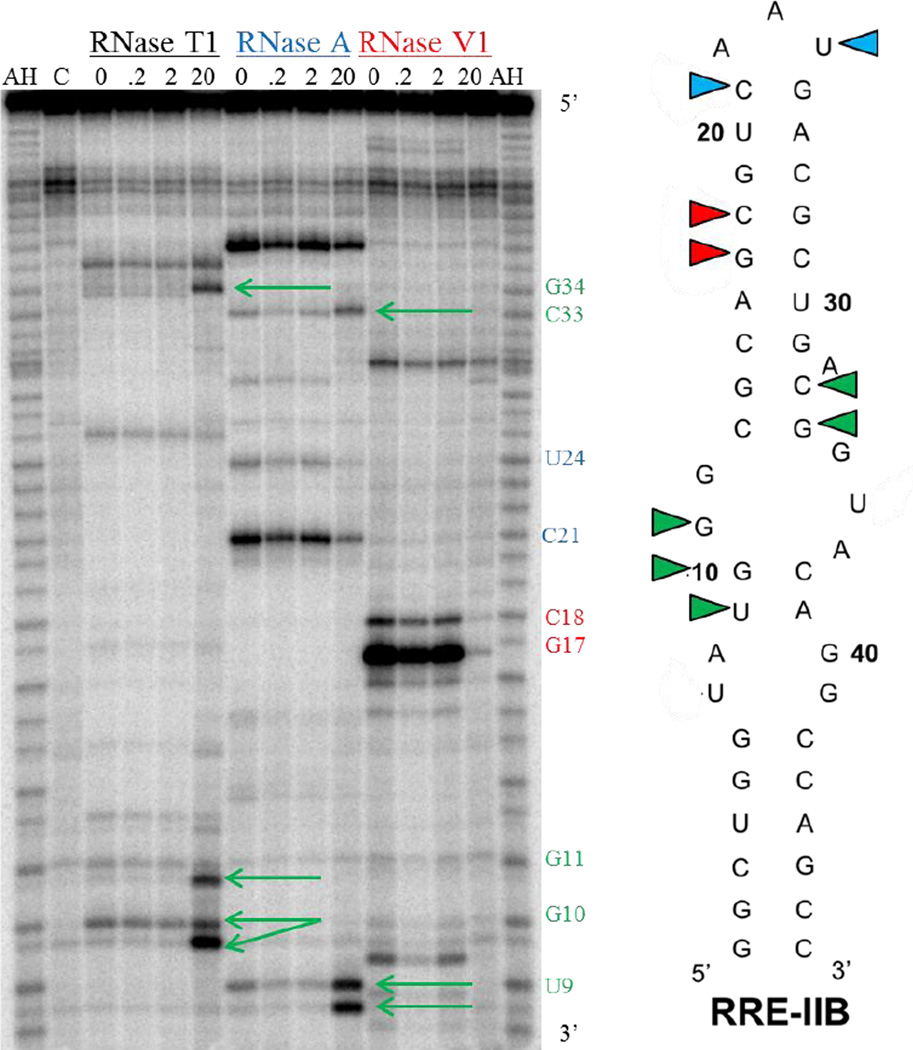
2.4. Binding site determination of BPBA3 with RRE IIB
To determine regions of specific nucleotide contacts with BPBA3, we performed ribonuclease protection assays by incubating 5’-32P-labeled RRE IIB with varying concentrations (up to 20 µM) of peptide in the presence of RNase T1, RNase A, or RNase V1 (Fig. 3). Potential binding sites were confirmed based on protection of the RNA from enzymatic cleavage. The assay revealed a concentration-dependent protection of RNA at G17 and C18 from RNase VI, which preferentially cleaves double-stranded RNA. BPBA3 also protected RRE IIB from cleavage at C21 and to a lesser extent at U24 in the presence of RNase A, which cleaves the 3’ side of unpaired pyrimidine bases. Taken together, these results indicate BPBA3 binds to the upper stem (stem B) and apical loop region of RRE IIB in a 1:1 stoichiometry (see Supplementary Figs. SI.1 and SI.5). Surprisingly, we detected additional cleavage bands when incubated with RNases T1 and A, a replicable phenomenon never observed with other peptides in this or other series. Unlike BPBA155 and other BPBAs characterized by ribonuclease protection assays (unpublished data), there was no protection from cleavage by RNases observed in the internal loops of the RNA structure. Instead, there was an increase in enzymatic cleavage observed at several bases within (U9, G10, G11, G34) and adjacent to (C33) loop B of RRE IIB when the concentration of BPBA3 was increased to 20 µM. The concentration dependency observed for the RNase activity is reasonable considering the dissociation constant for BPBA3 is in the low micromolar range. It is interesting to note that the points of contact of BPBA3 with loops A / B and the adjacent stem portion are located on opposite sides of the RNA secondary structure, indicating that the BPBA may be binding within a saddled groove and altering the tertiary conformation of the RNA. More specifically, there may be a disruption in the non-canonical G12:G35 base-pairing observed in this high binding affinity site of Rev,56, 57 that may in turn expose the neighboring nucleotides to RNase cleavage. Since the dissociation constant of the native Rev protein with RRE is in the low nanomolar range and BPBA3 has a significantly lower binding affinity in comparison, it is possible that this remodeling of tertiary structure is partially responsible for the in vitro inhibition seen with BPBA3, since this internal loop region is where the Rev protein binds.26, 58 Indeed, this may explain why other BPBAs with similar binding affinities toward RRE (BPBA1 and BPBA2) do not inhibit HIV-1 replication in cell culture. Overall, the RNA footprinting assay indicated that the binding site for BPBA3 encompasses two possible regions, stem B / apical loop region and loops A and B. However, it may be that BPBA3 is binding to other regions of the RNA that are inducing the conformation change observed; distinguishing the mechanism of increased enzymatic cleavage is beyond the scope of this assay.
Figure 3.
RNase Protection Assay autoradiograph gel of RRE IIB in the presence of increasing concentrations of BPBA3 (0–20 µM). AH is the alkaline hydrolysis ladder and C is the negative control (RRE IIB only, with no RNase or peptide). Contacts of the peptide with the RNA bases are indicated by color-coded wedges, where an increase in protection is indicated in blue (RNase A) or red (RNase VI), and a decrease in protection is indicated in green with arrows indicating bands of increased enzymatic cleavage.
3. Conclusion
In summary, a branched peptide (BPBA3) containing multiple boronic acid residues from a 3.3.4 library screening against RRE IIB RNA was found to inhibit HIV-1 p24 production and HIV replication in vitro. Biochemical characterization of this compound revealed selectivity for RRE IIB in the presence of competitor tRNA, and that the C-terminal peptide branch was critical for RNA binding. The binding interactions of BBPA3 with RRE IIB suggest a conformational change of the tertiary structure in the internal loop region that predisposes the RNA to RNase cleavage. This is a novel mode of action with branched peptide inhibitors. Further studies are currently underway to gain insight into the mechanism of action and to understand the structural elements of BPBA3 responsible for the alteration of RNA tertiary structure.
4. Experimental section
4.1. Peptide synthesis, purification, and characterization
Synthesis of the branched peptides was achieved by solid phase peptide synthesis using N-α-Fmoc protected l-amino acids (Novabiochem) (3 eq.), Pyoxim (Peptides International) (3 eq.) in DMF as coupling reagent, and DIEA (Aldrich) (6 eq.) on Rink amide MBHA resin (100–200 mesh) (Novabiochem) with 0.6 mmol/g loading. The Fmoc group was deprotected with 20% piperidine in DMF. Fmoc-Lys(Fmoc)-OH was used as a branching unit, and molar equivalencies of reagents were doubled in coupling reactions after installation of the branching unit. For linear variants LBPBA3A and LBPBA3B, Fmoc-Lys(Boc)-OH was used in place of Fmoc-Lys(Fmoc)-OH at the branching position. The solid phase synthesis was done on a vacuum manifold (Qiagen) outfitted with 3-way Luer lock stopcocks (Sigma) in either Poly-Prep columns or Econo-Pac polypropylene columns (Bio-Rad). The resin was mixed in solution by bubbling argon during all coupling and washing steps. After Fmoc deprotection of the N-terminal amino acids, the resin was bubbled in a phenylboronic acid solution (0.2 g/mL) overnight to remove the pinacol groups of boron-containing side chains. Finally, the resin was treated with 95:2.5:2.5 TFA (Trifluoroacetic acid, Acros)/H2O/TIS (Triisopropylsilane, Acros) (v/v/v) for 3 hr. The supernatant was dried under reduced pressure, and the crude peptide was triturated from cold diethyl ether. The peptides were purified using a Jupiter 4 µm Proteo 90 Å semiprep column (Phenomenex) using a solvent gradient composed of 0.1% TFA in Milli-Q water and HPLC grade acetonitrile. Peptide purity was determined using a Jupiter 4 µm Proteo 90 Å analytical column (Phenomenex), and peptide identity was confirmed by MALDI-TOF analysis. Peptide concentrations were measured in nuclease free water at 280 nm using their calculated extinction coefficients.
4.2. Preparation of 32-P labeled RNA
Wild-type RRE-IIB RNA and wild-type TAR RNA were transcribed in vitro by T7 polymerase with the Ribomax T7 Express System (Promega) using previously reported techniques.48, 59 The antisense template, sense complementary strand (5'-ATGTAATACGACTCACTATAGG-3') and RRE-IIB reverse PCR primer (5'-GGCTGGCCTGTAC-3') were purchased from Integrated DNA Technologies. The antisense template for the HIV-1 RNAs that were used are as follows: RRE IIB Wild Type RNA 5'-GGCTGGCCTGTACCGTCAGCGTCATTGACGCTGCGCCCATACCAGCCCTATAGTGAGTCGTATTACAT-3'; TAR RNA 5’-GCCCGAGAGCTCCCAGGCTCAGATCGGGCCTATAGTGAGTCGTATTACAT-3'. HIV-1 RRE-IIB Wild Type was PCR amplified using HotstarTaq DNA polymerase (Qiagen) followed by a clean-up procedure using a spin column kit (Qiagen). For the preparation of all other sequences, the antisense DNA template was annealed with the sense DNA complementary strand in reaction buffer at 95 °C for 2 min then cooled on ice for 4 min. T7 transcription proceeded at 42 °C for 1.5 hr. After transcription, DNA templates were degraded with DNase at 37 °C for 45 min and the RNA was purified by a 12% polyacrylamide gel containing 7.5 M urea. The band corresponding to the RNA of interest was excised from the gel and eluted overnight in 1× TBE buffer at 4 °C. The sample was desalted using a Sep-Pak syringe cartridge (Waters Corporation), lyophilized, and dephosphorylated with calf intestinal phosphatase (CIP) in NEBuffer 3 (New England Biolabs) according to manufacturer’s protocol. The product was recovered by a standard phenol extraction followed by ethanol precipitation. Purified RNA was stored as a pellet at −80 °C. HIV-1 RRE-IIB RNA was labeled at the 5’-end by treating 10 pmol of dephosphorylated RNA with 20 pmol of [γ-32P] ATP (111 TBq mol−1) and 20 units of T4 polynucleotide kinase in 70 mM Tris•HCl, 10 mM MgCl2, and 5 mM dithiothreitol, pH 7.6. The mixture was incubated at 37 °C for 30 min, and then at rt for 20 min. The kinase was heat-inactivated at 65 °C for 10 min. The RNA was recovered by ethanol precipitation, and the purity was examined using 12% denaturing PAGE followed by autoradiography.
4.3. Dot blot assays
Dot blot assays were performed at rt using a Whatman Minifold I 96 well Dot Blot system and Whatman 0.45 µm pore size Protran nitrocellulose membranes. To determine the binding affinities, 0.4 nM radiolabeled RNA was titrated with peptide (0.001–100 µM). First, a solution of 0.8 nM 32P-labeled RNA was refolded in 2× phosphate buffer (20 mM potassium phosphate, 200mM KCl, 1mM MgCl2, 40mM NaCl, pH 7.0) by heating at 95 °C for 3 min and then slowly cooling at rt for 20 min. Next, 25 µL of the [32P]-RNA solution was added to 25 µL of peptide in nuclease free water and incubated at rt for 4 hr. The 50 µL mixtures were filtered through the nitrocellulose membrane, which was immediately followed by two consecutive 50 µL washes with 1× phosphate buffer. Peptide binding was visualized by autoradiography using a storage phosphor screen (GE Healthcare) and a Typhoon Trio phosphorimager (GE Healthcare). Densitometry measurements were quantified using ImageQuant TL (Amersham Biosciences). Binding curves were generated using a four parameter logistic equation with Kaleidagraph (Synergy Software): y=m1+(m2−m1)/(1+10^(log(m3)−x); m1=100; m2=1; m3= 0.003 to 0.000003, where y=percentage of RNA binding, x= log[peptide], m1=percentage of RNA binding affinity at infinite concentration (nonspecific binding), m2= percentage of RNA binding affinity at zero concentration, m3=peptide concentration at 50% binding (Kd). Each experiment was performed in triplicate and error bars represent the standard deviation calculated over three replicates.
4.4. Viral replication assays
Compounds to be tested were resuspended in distilled water to a concentration of 1mM. The control drugs Zidovudine (AZT), Lamivudine (3TC), Raltegravir, and Elvitegravir were obtained from the AIDS Research and Reference Reagent Program and initially diluted in dimethyl sulfoxide (DMSO) to concentrations of 10 mM and 1 mM, respectively. All control compounds and experimental compounds were further diluted in RPMI to the appropriate working concentrations and tested against the reference HIV viral strain NL4-3, which uses the CXCR4 co-receptor. Sensitivity to inhibitors/compounds of HIV-1 replication was determined by infecting U87-CD4/CXCR4 cells with NL4-3 following a previously published procedure.60 Briefly, cells were added to 96-well flat bottom plates at a density of 1 × 104 cells/well and allowed to adhere overnight. On the next day, cells were treated with the test compounds at desired concentrations (10µM – 100µM) or controls (3TC or AZT) for 1 hour prior to infection. Cells were then exposed to virus at an MOI of 0.01 infectious units/ml (IU/ml) for 24 h, after which they were washed with phosphate-buffered saline (PBS), and fresh medium containing additional compound was added. The culture was then left to grow for 4 days and supernatant aliquots were taken at intervals from days 2 to 4. Virus production was quantified by p24 assay.61 When necessary, the fifty percent inhibitory concentration (IC50) curves were constructed for supernatant samples taken at peak virus production.
4.5 p24 Production and viral particle release assay
5BD.1 cells were re-suspended in IMDM (10% fetal calf serum, 0.2mg/ml hygromycin B, 1.5 mg/ml G418 and 0.50µg/ml gentamicin sulfate) and dispensed into 96-well plates at 20,000 cells per well in a total volume of 135µl. The cells were incubated with 10µM of each compound in triplicate, for 16 hours. After 16 hours, the medium was removed and replaced with 135µl of fresh medium containing 10µM of compound then incubated for an additional 24 hours when supernatant was collected for p24 ELISA. This assay and the 5BD.1 cells used in these experiments have been previously described.62 The 5BD.1 cells were made by calcium phosphate transfection of CMT3 COS cells with pCMVGagPol-RRE, pCMVrev and pCMVenv. The cells express pseudovirions in a Rev-dependent fashion. The pseudovirions bud into the medium and can then be measured by p24 ELISA.
4.6. Nuclease protection assays
RNA was first refolded by heating a solution of 5’-32P-labeled RRE-IIB (10 nM) and excess unlabeled RRE-IIB (200 nM) at 95 °C for 3 min and then snap cooling on ice. The refolded RNA was incubated on ice for 4 hr in a solution containing the BPBAs and buffer composed of 10 mM Tris, pH 7, 100 mM KCl, and 10 mM MgCl2. RNase (Ambion) was then added to the solution, which was further incubated on ice for 10 min (0.002 Units RNase V1), or 1 hr (1 Unit RNase T1; 20 ng RNase A). Inactivation/precipitation buffer (Ambion) was added to halt digestion, and the RNA was pelleted by centrifugation at 13,200 rpm for 15 min. Pelleted RNA was redissolved into tracking dye and run through a 12 % PAGE containing 7.5 M urea and imaged by autoradiography.
Supplementary Material
Acknowledgments
We thank K. Ray and R. Helm in the VT mass spectrometry incubator for use of their MALDI-TOF for MS analysis. We gratefully acknowledge the National Institutes of Health for their financial support under R01GM093834 and R01GM110009 and the Myles H. Thaler and Charles H. Ross Jr. Endowments at the University of Virginia.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Supplementary data (RNA structures, dot blot, EMSA, Job plot, MTT, and p24 assays) associated with this article can be found in the online version at doi:xxx
References and notes
- 1.Disney MD, Yildirim I, Childs-Disney JL. Org. Biomol. Chem. 2014;12:1029–1039. doi: 10.1039/c3ob42023j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernat V, Disney Matthew D. Neuron. 2015;87:28–46. doi: 10.1016/j.neuron.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas JR, Hergenrother PJ. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 4.Lee MM, Pushechnikov A, Disney MD. ACS Chem. Biol. 2009;4:345–355. doi: 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J, Kadakkuzha BM, Zhao L, Fan M, Qi X, Xia T. Biochemistry. 2011;50:5042–5057. doi: 10.1021/bi200495d. [DOI] [PubMed] [Google Scholar]
- 6.Seedhouse SJ, Labuda LP, Disney MD. Bioorg. Med. Chem. Lett. 2010;20:1338–1343. doi: 10.1016/j.bmcl.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, Al-Hashimi HM. Nat. Chem. Biol. 2011;7:553–559. doi: 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filikov AV, Mohan V, Vickers TA, Griffey RH, Cook PD, Abagyan RA, James TL. J. Comput.-Aided Mol. Des. 2000;14:593–610. doi: 10.1023/a:1008121029716. [DOI] [PubMed] [Google Scholar]
- 9.Galicia-Vazquez G, Lindqvist L, Wang X, Harvey I, Liu J, Pelletier J. Anal. Biochem. 2009;384:180–188. doi: 10.1016/j.ab.2008.09.037. [DOI] [PubMed] [Google Scholar]
- 10.Yen L, Magnier M, Weissleder R, Stockwell BR, Mulligan RC. RNA. 2006;12:797–806. doi: 10.1261/rna.2300406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson BL, McCray PB., Jr Nat. Rev. Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan L, Disney MD. ACS Chem. Biol. 2011;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 13.Sobczak K, Michlewski G, de Mezer M, Kierzek E, Krol J, Olejniczak M, Kierzek R, Krzyzosiak WJ. J. Biol. Chem. 2010;285:12755–12764. doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rzuczek SG, Gao Y, Tang Z-Z, Thornton CA, Kodadek T, Disney MD. ACS Chem. Biol. 2013;8:2312–2321. doi: 10.1021/cb400265y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Childs-Disney JL, Hoskins J, Rzuczek SG, Thornton CA, Disney MD. ACS Chem. Biol. 2012;7:856–862. doi: 10.1021/cb200408a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paillart J-C, Shehu-Xhilaga M, Marquet R, Mak J. Nat. Rev. Micro. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 17.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Nat. Struct. Mol. Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 19.Ennifar E, Yusupov M, Walter P, Marquet R, Ehresmann B, Ehresmann C, Dumas P. Structure. 1999;7:1439–1449. doi: 10.1016/s0969-2126(00)80033-7. [DOI] [PubMed] [Google Scholar]
- 20.Lee SJ, Hyun S, Kieft JS, Yu J. J. Am. Chem. Soc. 2009;131:2224–2230. doi: 10.1021/ja807609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyun S, Na J, Lee SJ, Park S, Yu J. ChemBioChem. 2010;11:767–770. doi: 10.1002/cbic.201000072. [DOI] [PubMed] [Google Scholar]
- 22.Rausch J, Grice S. Viruses. 2015;7:2760. [Google Scholar]
- 23.Mann DA, Mikaelian I, Zemmel RW, Green SM, Lowe AD, Kimura T, Singh M, Butler PJG, Gait MJ, Karn J. J. Mol. Biol. 1994;241:193–207. doi: 10.1006/jmbi.1994.1488. [DOI] [PubMed] [Google Scholar]
- 24.Van Ryk DI, Venkatesan S. J. Biol. Chem. 1999;274:17452–17463. doi: 10.1074/jbc.274.25.17452. [DOI] [PubMed] [Google Scholar]
- 25.Pollard VW, Malim MH. Annu. Rev. Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 26.Kjems J, Brown M, Chang DD, Sharp PA. Proc. Natl. Acad. Sci. U.S.A. 1991;88:683–687. doi: 10.1073/pnas.88.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J, Cullen BR. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 28.Hamasaki K, Ueno A. Bioorg. Med. Chem. Lett. 2001;11:591–594. doi: 10.1016/s0960-894x(01)00005-1. [DOI] [PubMed] [Google Scholar]
- 29.Zapp ML, Young DW, Kumar A, Singh R, Boykin DW, Wilson WD, Green MR. Bioorg. Med. Chem. 1997;5:1149–1155. doi: 10.1016/s0968-0896(97)00063-1. [DOI] [PubMed] [Google Scholar]
- 30.Wilson WD, Li K. Curr. Med. Chem. 2000;7:73–98. doi: 10.2174/0929867003375434. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y, Hyun S, Kim HJ, Yu J. Angew. Chem. Int. Ed. 2008;47:134–137. doi: 10.1002/anie.200703090. [DOI] [PubMed] [Google Scholar]
- 32.Xiao G, Kumar A, Li K, Rigl CT, Bajic M, Davis TM, Boykin DW, Wilson WD. Bioorg. Med. Chem. 2001;9:1097–1113. doi: 10.1016/s0968-0896(00)00344-8. [DOI] [PubMed] [Google Scholar]
- 33.Luedtke NW, Liu Q, Tor Y. Biochemistry. 2003;42:11391–11403. doi: 10.1021/bi034766y. [DOI] [PubMed] [Google Scholar]
- 34.Li K, Davis TM, Bailly C, Kumar A, Boykin DW, Wilson WD. Biochemistry. 2001;40:1150–1158. doi: 10.1021/bi002338b. [DOI] [PubMed] [Google Scholar]
- 35.Hart RA, Billaud JN, Choi SJ, Phillips TR. Virology. 2002;304:97–104. doi: 10.1006/viro.2002.1659. [DOI] [PubMed] [Google Scholar]
- 36.DeJong ES, Chang CE, Gilson MK, Marino JP. Biochemistry. 2003;42:8035–8046. doi: 10.1021/bi034252z. [DOI] [PubMed] [Google Scholar]
- 37.Prater CE, Saleh AD, Wear MP, Miller PS. Bioorg. Med. Chem. 2007;15:5386–5395. doi: 10.1016/j.bmc.2007.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Legiewicz M, Badorrek CS, Turner KB, Fabris D, Hamm TE, Rekosh D, Hammarskjold ML, Le Grice SF. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14365–14370. doi: 10.1073/pnas.0804461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee TC, Sullenger BA, Gallardo HF, Ungers GE, Gilboa E. The New biologist. 1992;4:66–74. [PubMed] [Google Scholar]
- 40.Symensma TL, Baskerville S, Yan A, Ellington AD. J. Virol. 1999;73:4341–4349. doi: 10.1128/jvi.73.5.4341-4349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giver L, Bartel DP, Zapp ML, Green MR, Ellington AD. Gene. 1993;137:19–24. doi: 10.1016/0378-1119(93)90246-y. [DOI] [PubMed] [Google Scholar]
- 42.Chaloin L, Smagulova F, Hariton-Gazal E, Briant L, Loyter A, Devaux C. J. Biomed. Sci. 2007;14:565–584. doi: 10.1007/s11373-007-9180-4. [DOI] [PubMed] [Google Scholar]
- 43.Friedler A, Friedler D, Luedtke NW, Tor Y, Loyter A, Gilon C. J. Biol. Chem. 2000;275:23783–23789. doi: 10.1074/jbc.M002200200. [DOI] [PubMed] [Google Scholar]
- 44.Mills NL, Daugherty MD, Frankel AD, Guy RK. J. Am. Chem. Soc. 2006;128:3496–3497. doi: 10.1021/ja0582051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shuck-Lee D, Chen FF, Willard R, Raman S, Ptak R, Hammarskjold M-L, Rekosh D. Antimicrob. Agents Chemother. 2008;52:3169–3179. doi: 10.1128/AAC.00274-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shuck-Lee D, Chang H, Sloan EA, Hammarskjold M-L, Rekosh D. J. Virol. 2011;85:3940–3949. doi: 10.1128/JVI.02683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bryson DI, Zhang W, Ray WK, Santos WL. Mol. BioSyst. 2009;5:1070–1073. doi: 10.1039/b904304g. [DOI] [PubMed] [Google Scholar]
- 48.Bryson DI, Zhang W, McLendon PM, Reineke TM, Santos WL. ACS Chem. Biol. 2012;7:210–217. doi: 10.1021/cb200181v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cook KS, Fisk GJ, Hauber J, Usman N, Daly TJ, Rusche JR. Nucleic Acids Res. 1991;19:1577–1583. doi: 10.1093/nar/19.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang XJ, Hope TJ, Bond BL, McDonald D, Grahl K, Parslow TG. J. Virol. 1991;65:2131–2134. doi: 10.1128/jvi.65.4.2131-2134.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Battiste JL, Mao H, Rao NS, Tan R, Muhandiram DR, Kay LE, Frankel AD, Williamson JR. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 52.Daugherty MD, Liu B, Frankel AD. Nat. Struct. Mol. Biol. 2010;17:1337–1342. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang W, Bryson DI, Crumpton JB, Wynn J, Santos WL. Chem. Commun. 2013;49:2436–2438. doi: 10.1039/c3cc00243h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang W, Bryson DI, Crumpton JB, Wynn J, Santos WL. Org. Biomol. Chem. 2013;11:6263–6271. doi: 10.1039/c3ob41053f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartel DP, Zapp ML, Green MR, Szostak JW. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 57.Ippolito JA, Steitz TA. J Mol Biol. 2000;295:711–717. doi: 10.1006/jmbi.1999.3405. [DOI] [PubMed] [Google Scholar]
- 58.Kjems J, Calnan BJ, Frankel AD, Sharp PA. EMBO J. 1992;11:1119–1129. doi: 10.1002/j.1460-2075.1992.tb05152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tebit DM, Lobritz M, Lalonde M, Immonen T, Singh K, Sarafianos S, Herchenroder O, Krausslich H-G, Arts EJ. J. Virol. 2010;84:9817–9830. doi: 10.1128/JVI.00991-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wehrly K, Chesebro B. Methods. 1997;12:288–293. doi: 10.1006/meth.1997.0481. [DOI] [PubMed] [Google Scholar]
- 62.Srinivasakumar N, Chazal N, Helga-Maria C, Prasad S, Hammarskjold M-L, Rekosh D. J. Virol. 1997;71:5841–5848. doi: 10.1128/jvi.71.8.5841-5848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.