Abstract
Inflammatory bowel disease (IBD) is associated with dysregulated macrophage responses, such that quiescent macrophages acquire a pro-inflammatory activation state and contribute to chronic intestinal inflammation. The transcriptional events governing macrophage activation and gene expression in the context of chronic inflammation such as IBD remain incompletely understood. Here, we identify Kruppel-like transcription factor-6 (KLF6) as a critical regulator of pathogenic myeloid cell activation in human and experimental IBD. We found that KLF6 was significantly upregulated in myeloid cells and intestinal tissue from IBD patients and experimental models of IBD, particularly in actively inflamed regions of the colon. Using complementary gain- and loss-of-function studies, we observed that KLF6 promotes pro-inflammatory gene expression through enhancement of NFκB signaling, while simultaneously suppressing anti-inflammatory gene expression through repression of STAT3 signaling. To study the in vivo role of myeloid KLF6, we treated myeloid-specific KLF6-knockout mice (Mac-KLF6-KO) with dextran sulfate-sodium (DSS) and found that Mac-KLF6-KO mice were protected against chemically-induced colitis; this highlights the central role of myeloid KLF6 in promoting intestinal inflammation. Collectively, our results point to a novel gene regulatory program underlying pathogenic, pro-inflammatory macrophage activation in the setting of chronic intestinal inflammation.
Introduction
The human gastrointestinal tract is home to more than one hundred trillion commensal microbes1, highlighting the need to discriminate between beneficial organisms and pathogenic, invading microbes. The innate immune system of the intestinal mucosa has therefore evolved to provide a rapid, first-line defense against harmful, infectious organisms through the recognition of conserved molecular patterns unique to invading pathogens, such as endotoxins and nucleic acids. Pattern recognition receptor (PRR)-bearing macrophages situated in the sub-epithelial lamina propria of the small and large intestine are central to this process, and in fact represent the largest pool of tissue-resident macrophages in the body2. In addition to their essential roles in maintaining host defense, scavenging dead cells and debris, and helping to maintain epithelial barrier function3, intestinal mucosal macrophages are required for shaping immune responses to the continuous barrage of antigenic challenges that breach the intestinal epithelium.
The requirement of macrophages for maintainenance of intestinal homeostasis was established by seminal work in which deletion of macrophages4, 5 or TLRs and their common signaling adapter, MyD886, resulted in severe morbidity and mortality in response to dextran sulfate sodium (DSS)-induced colitis. These studies suggest that under noninflamed conditions, intestinal macrophages recognize commensal microbes through PRR interactions and regulate mucosal immunity in response to these signals. Intestinal macrophages are also required for the development of mucosal tolerance, as macrophage-deficient mice (F4/80 knockout (KO)) fail to develop tolerance or antigen-specific regulatory T cells (Treg) following oral administration of soluble antigen7.
Innate immunity sensing mechanisms are critical for initiating and shaping the adaptive immune response in the lamina propria8, 9 and other tissues, as demonstrated by the ability of “classically” (M1) and “alternatively” (M2) activated macrophages to promote Th1 and Th2 responses, respectively. Although M1/M2 macrophage classification remains widely used, these broad categories encompass many different phenotypic and functional subsets10. Macrophages are differentiated and activated by many different stimuli, including cytokines, TLR agonists, and other exogenous stimuli. Responses to stimulation by these agents, alone or in combination, can lead to a spectrum of activation states not restricted to the classic M1/M2 classificiations. Recent efforts have therefore led to the development of a new consensus framework for the classification of activated macrophages, aimed at standardizing macrophage nomenclature10.
Under homeostatic conditions, intestinal macrophages remain “inflammation anergic”11 in response to strong signals from anti-inflammatory IL-10 and TGFβ, displaying many phenotypic and functional features of M2 macrophages. Although these cells express PRRs and display robust phagocytic activity, they fail to mount inflammatory responses to exogenous antigens12; this unique feature allows intestinal macrophages to effectively eliminate foreign invaders while simultaneously maintaining immune quiescence. Myeloid-specific deletion of M2-associated molecules such as IL-10Ra13, STAT314, and PPARγ15 leads to significant exacerbation of experimental colitis, highlighting the immunoprotective role conferred by these molecules under intestinal homeostasis.
In contrast, intestinal macrophages of inflammatory bowel disease (IBD) patients display an impairment in inflammatory anergy and an increase in pro-inflammatory, M1 activation16. This process is thought to occur due to high levels of cytokines, such as IFNγ, TNF, IL-1 and IL-6, in the gut mucosa that can divert infiltrating monocytes towards pathogenic, M1 macrophage phenotype3. These cells exhibit increased activation of NF-κB, enhanced sensitivity to TLR ligands, and elevated expression of pro-inflammatory cytokines, chemokines, MMPs and ROS/NOS17–19, leading to inappropriate inflammatory responses directed against non-harmful commensal microbes and dietary antigens. Blockade of macrophage-derived TNF is a standard therapeutic approach in the treatment of IBD20, demonstrating the pathogenic contribution of inflammatory macrophages to IBD. Experimental IBD is also dependent on pathogenic macrophages and their cytokine products, as depletion of macrophages directly21 or treatment with anti-TNF antibodies22 are both sufficient to suppress the development of spontaneous colitis in IL-10 KO mice.
Mature macrophages remain transcriptionally dynamic and can alter their activation state in response to environmental cues such as cytokines and other soluble mediators. Changes to the IBD cytokine environment, such as an upregulation of IFNγ, TNF, IL-1 and IL-6, may therefore divert macrophages away from inflammation anergy and towards a pathogenic, M1 phenotype. Given the plasticity of macrophage phenotypes and functions, it is critical to identify molecular events governing the loss of inflammatory anergy and the “switch” to pathogenic, M1 phenotypes that occurs in the setting of chronical intestinal inflammation such as IBD.
Kruppel-like transcription factors (KLFs) are a subclass of zinc-finger family transcription factors that regulate critical cellular processes including development, differentiation, proliferation, and programmed cell death23–25. Previous studies have indicated that KLF226 and KLF427 play important roles in macrophage inflammatory gene expression and function; however, it has been recently shown that KLF6 is the most highly expressed KLF family member in primary macrophages, where it simultaneously promotes M1 activation and inhibits M2 activation28. KLF6 has also been recently linked to the pathogenesis of human inflammatory diseases, including psoriasis29 and hepatic steatosis30. Nonetheless, the role of myeloid-derived KLFs, including KLF6, in the pathogenesis of IBD has never been investigated.
Our current studies seek to investigate the contribution of macrophage KLF6 to the loss of macrophage inflammatory anergy in IBD. We demonstrate a significant upregulation of KLF6 in intestinal myeloid cells from human and experimental IBD subjects, as well as a central role for KLF6 in controlling macrophage responses to pro- and anti-inflammatory stimuli associated with the development and resolution of intestinal inflammation. Thus, we propose that KLF6 may function as a molecular switch, diverting intestinal macrophages away from protective, inflammation anergic phenotypes and toward pathogenic, M1 phenotypes in response to the local IBD-associated cytokine milieu.
Results
Myeloid KLF6 is upregulated in human and experimental IBD
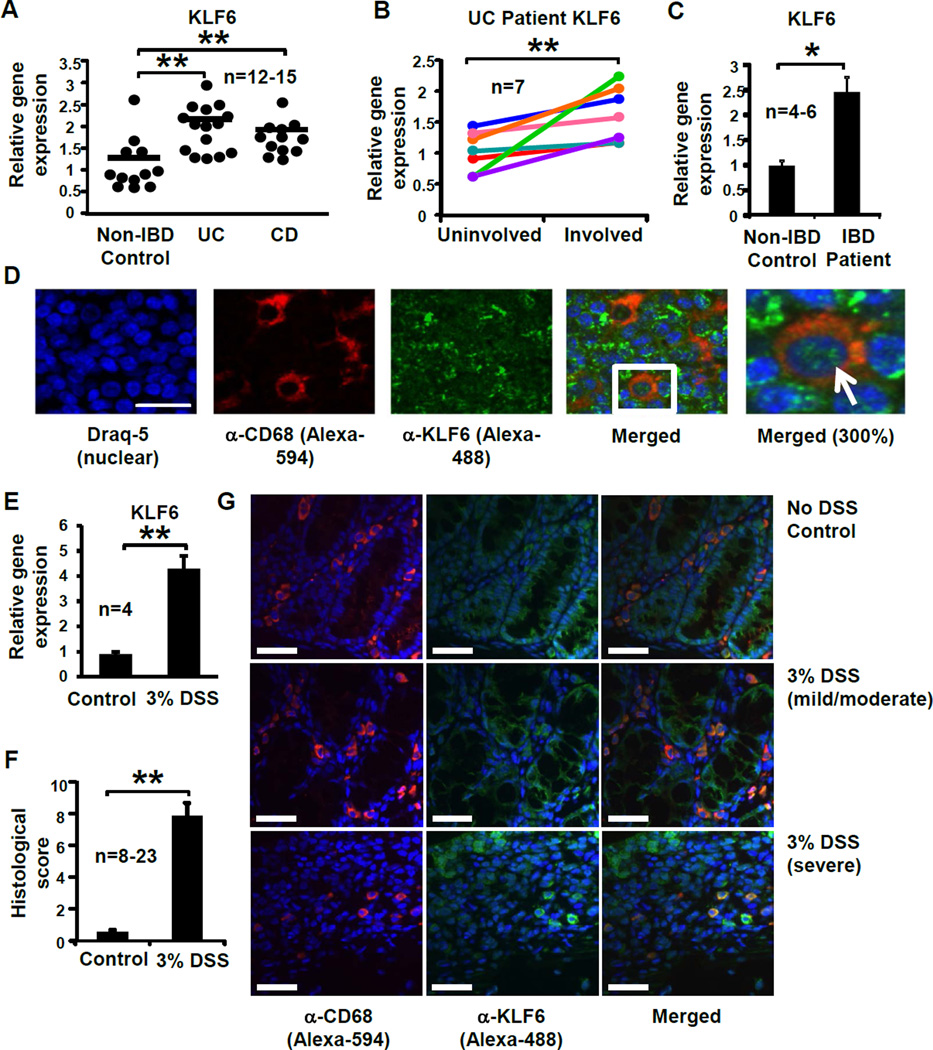
Our previous work demonstrated that KLF6 is the most highly expressed Kruppel-like transcription factor in macrophages and contributes to pro-inflammatory macrophage activation and function28. Given the role of activated macrophages in human and experimental IBD12, we sought to investigate whether myeloid cell KLF6 expression is associated with intestinal inflammation. Analysis of mRNA isolated from intestinal biopsies of ulcerative colitis (UC) and Crohn’s colitis (CD) patients, two of the primary idiopathic forms of IBD, revealed elevated KLF6 gene expression within both patient cohorts compared to intestinal tissues from non-inflamed, healthy controls (Fig. 1A). In order to determine whether higher levels of KLF6 correlate with worsened clinical disease, we obtained paired biopsy samples from uninflamed regions (“uninvolved”) and acutely inflamed colon tissue regions (“involved) from CD and UC patients. Whereas comparable levels of KLF6 mRNA were detected in involved and uninvolved samples from CD patients (Supplemental Fig. S1), significantly higher levels of KLF6 were detected in involved regions of UC patient colons, compared to uninvolved regions from the same individuals (Fig. 1B). Monocyte-derived macrophages from IBD patients also revealed a significant upregulation in KLF6 mRNA compared to macrophages from healthy controls (Fig. 1C).
Figure 1. KLF6 is upregulated in inflamed intestinal tissues and macrophages from human and experimental IBD subjects.
(A) Colon tissues from non-IBD control subjects or UC or CD patients were homogenized, cDNA prepared, and analysis of KLF6 gene expression performed by qPCR; **p≤0.001, n=12–15/group. (B) Paired biopsies from uninvolved and involved regions of UC patient colon tissue were analyzed for KLF6 gene expression by qPCR. Fold changes in involved samples were calculated relative to uninvolved (baseline) samples and normalized to expression of SHDA. Lines represent individual patients; **p≤0.03, n=7). (BC) Monocyte-derived macrophages from non-IBD control subjects or IBD patients were analyzed for KLF6 gene expression by qPCR; **p≤0.01, n=4–6/group. (D) KLF6-expressing myeloid cells were detected in human mesenteric lymph nodes by immunofluorescent confocal microscopy. Formalin-fixed, paraffin-embedded mesenteric lymph node tissues were stained with antibodies specific to CD68 (myeloid cell marker, imaged with Alexa 594 as red) and KLF6 (imaged with Alexa 488 as green). Nuclei are stained with Draq5 (imaged as blue); n=3 independent experiments. Scale bar = 100µm; white arrow indicates representative KLF6-expressing macrophage. (E) Macrophages from 3% DSS-fed mice express elevated levels of Klf6. Peripheral blood-derived macrophages from WT mice were analyzed for Klf6 gene expression by qPCR; Graphs show mean ± SEM; **p≤0.001, n=4/group. (F) Histological colon inflammation was assessed by a blinded pathologist following 5 days of 3% DSS. Score consists of active, chronic, and transmural inflammation; **p≤0.001, n=8–23 mice/group. (G) KLF6-expressing myeloid cells are observed in colon tissue from DSS-fed mice by confocal microscopy. Formalin-fixed, paraffin-embedded colon tissues from indicated mice were stained by immunofluorescent confocal microscopy using antibodies specific to CD68 and KLF6, as described above (C). Nuclei were stained with Draq5. Scale bar = 34µm. Images are representative of 3 separate experiments.
To determine whether myeloid KLF6 is predominantly nuclear or cytoplasmic, we performed immunofluorescent staining on mesenteric lymph node (MLN) tissue sections obtained from IBD patients undergoing surgical resection. Abundant KLF6 was observed throughout the MLN (green, Fig. 1D) in the cytoplasm and nuclei of CD68+ myeloid cells (red, Fig. 1D) as well as lymphocytic cells, demonstrating that numerous cell populations in the intestine express KLF6.
To determine whether these observations are recapitulated in experimental models of IBD, we assessed gene expression of Klf6 in standard models of ileitis (TNFΔARE/+)32 and colitis (DSS-fed mice)33. Monocyte-derived macrophages from DSS colitic mice (Fig. 1E) and ileal tissues from TNFΔARE/+ mice (Supplemental Fig. S2) revealed significantly elevated levels of Klf6 mRNA in diseased tissue/macrophages compared to non-diseased control tissues and cells. Histological examination of colon tissues from DSS-treated, wild-type mice revealed characteristic features of inflammation, including epithelial erosion, ulceration, immune cell infiltration, and edema, and were quantified by an established histological scoring system (34 (Fig. 1F). Immunofluorescent confocal staining of colon tissues from control (no DSS)-fed mice and DSS-fed mice revealed prominent areas of myeloid cell infiltration in healthy, as well as inflamed, colons (Fig. 1G, red). KLF6 was detected diffusely in epithelial cells in both control as well as DSS-fed mice (Fig. 1G, green), although the intensity of epithelial KLF6 staining was greater in more severe regions of DSS-fed colon tissue (lower panels G). Interestingly, CD68+ myeloid cells expressed the highest levels of KLF6 in severely inflamed, DSS-fed colon tissue, and nearly all CD68+ cells in these mice expressed robust KLF6 staining. In contrast, KLF6 expression was weak or absent in the majority of CD68+ cells in non-DSS treated control mice, as well as less inflamed areas of DSS-fed mice (upper and middle panels). Collectively, these data indicate that elevated levels of myeloid KLF6 correlate with intestinal inflammation and tissue damage in IBD patients and experimental models, particularly in inflamed colitis patients and mice.
KLF6 enhances the expression of IBD-associated pro-inflammatory genes in macrophages
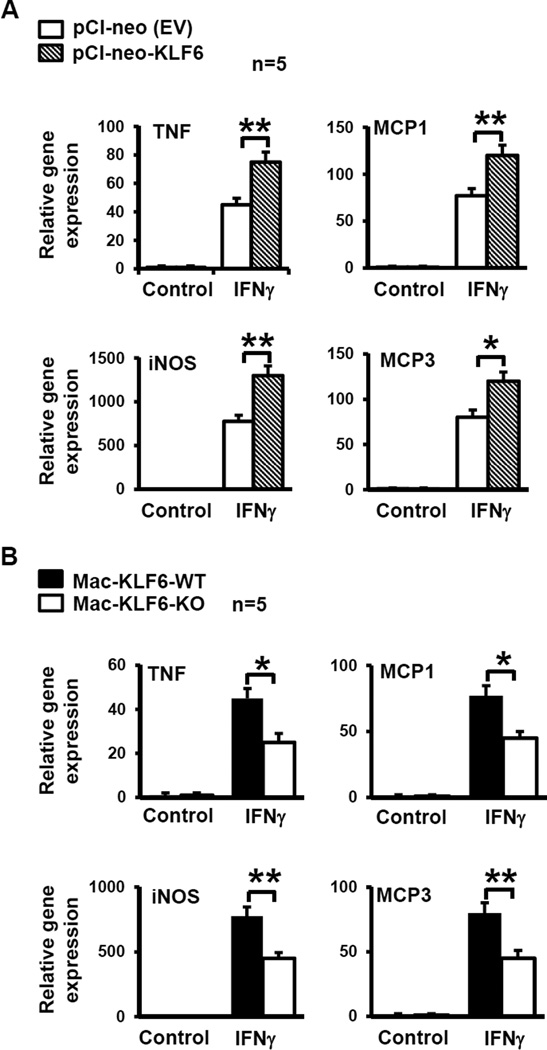
Using complementary gain- and loss-of-function approaches, we sought to test how modulation of KLF6 may influence myeloid cell responses to IFNγ, a pro-inflammatory cytokine central to the pathogenesis of IBD and inducer of M1 macrophage activation. To examine the effects of KLF6 overexpression, the RAW264.7 macrophage cell line was transiently transfected with a KLF6-expressing plasmid (pCI-neo-KLF6, Addgene) or empty-vector (EV) control, as previously described28. Transfected cells were treated with PBS (Ctrl) or IFNγ, then analyzed for Tnf, Mcp1, inos, and Mcp3 gene expression. Macrophage overexpression of KLF6 resulted in a robust and significant upregulation of all four pro-inflammatory genes in response to IFNγ, compared to EV-transfected cells (Fig. 2A).
Figure 2. KLF6 induces pro-inflammatory gene expression in macrophages.
(A) Macrophages overexpressing KLF6 show enhanced expression of pro-inflammatory genes. RAW264.7 macrophages were transfected with pCl-neo (EV plasmid control) or pCl-KLF6 to overexpress KLF6. Cells were stimulated in vitro with PBS (control) or IFNγ (100U/ml) for 6hrs, then harvested for RNA isolation. cDNA was analyzed for expression of pro-inflammatory genes Tnf, Mcp1, inos, and Mcp3 by qPCR; *p≤0.02; **p≤0.003; n=5/group. (B) Macrophages deficient in KLF6 show reduced expression of pro-inflammatory genes. Peritoneal macrophages were isolated from Mac-KLF6-WT and Mac-KLF6-KO mice following thioglycolate elicitation. Cells were stimulated in vitro with PBS (control) or IFNγ (100U/ml) for 6hrs, then harvested for RNA isolation. cDNA was analyzed for expression of pro-inflammatory genes; *p≤0.03; **p≤0.001, n=5/group. All graphs show mean ± SEM.
To evaluate the effects of KLF6 loss-of-function, we utilized myeloid-specific KLF6-null mice (Mac-KLF6-KO). These mice are generated by breeding Lyz2cre-expressing mice with KLF6 floxed mice, and display a robust reduction of KLF6 in primary macrophages28. Littermate control mice carrying only the Lyz2cre gene (Mac-KLF6-wild type (WT)) were used as controls for all experiments. Neither Mac-KLF6-WT nor Mac-KLF6-KO mice display any spontaneous, phenotypic features of IBD. Using thioglycollate-elicited peritoneal macrophages (PM) from Mac-KLF6-KO mice and littermate controls (Mac-KLF6-WT), we tested whether KLF6 expression is essential for IFNγ induced inflammatory gene expression in macrophages. In agreement with our overexpression studies (Fig. 2A), deficiency of KLF6 significantly attenuated IFNγ-induced Tnf, Mcp1, inos, and Mcp3 gene expression in macrophages (Fig. 2B). Taken together, these results implicate a role for macrophage KLF6 in IBD-associated pro-inflammatory gene expression.
KLF6 suppresses the expression of IBD-associated anti-inflammatory genes in macrophages
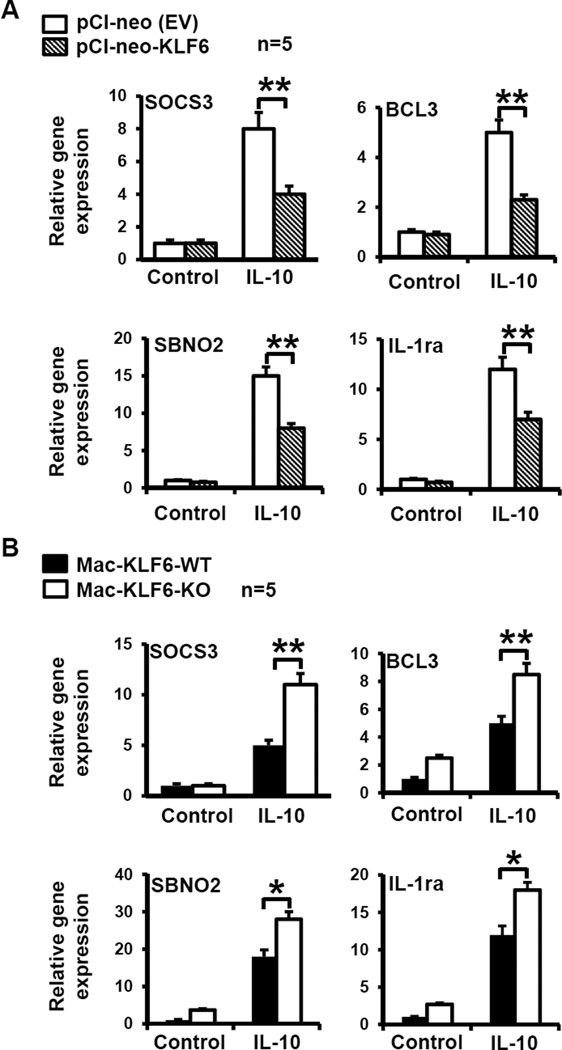
Under homeostatic conditions, high levels of IL-10 retain intestinal macrophages in an anti-inflammatory state referred to as “inflammatory anergy”11. In order to determine whether macrophage KLF6 actively inhibits IL-10-induced anti-inflammatory gene targets, we utilized gain- and loss-of function approaches. Accordingly, RAW264.7 cells overexpressing KLF6 (or EV control), as well as Mac-KLF6-KO and Mac-KLF6-WT PM, were stimulated with IL-10 and then harvested for mRNA isolation. Interestingly, we found that expression of IL-10-induced anti-inflammatory genes such as Socs3, Bcl3, Sbno2, and Il1ra was reduced in RAW264.7 macrophages overexpressing KLF6 compared to EV-transfected controls (Fig. 3A). Mac-KLF6-KO PM expressed higher levels of all four anti-inflammatory genes compared to Mac-KLF6-WT PM in response to IL-10 (Fig. 3B).
Figure 3. KLF6 inhibits anti-inflammatory gene expression in macrophages.
(A) Macrophages overexpressing KLF6 show reduced expression of anti-inflammatory genes. RAW264.7 macrophages were transfected with pCl-neo (EV plasmid control) or pCl-KLF6 to overexpress KLF6. Cells were stimulated in vitro with PBS (control) or IL-10 (10 ng/ml) for 18 hrs, then harvested for RNA isolation. cDNA was analyzed for expression of anti-inflammatory genes Sosc3, Bcl3, Sbco2, and Il1ra by qPCR; *p≤0.03; **p≤0.001, n=5/group. (B) Macrophages deficient in KLF6 show enhanced expression of anti-inflammatory genes. Peritoneal macrophages were isolated from Mac-KLF6-WT and Mac-KLF6-KO mice following thioglycolate elicitation. Cells were stimulated in vitro with PBS (control) or IL-10 (10 ng/ml) for 18hrs, then harvested for RNA isolation. cDNA was analyzed for expression of anti-inflammatory genes. Graphs show mean ± SEM; *p≤0.05, **p≤0.01, n=5/group.
Given our observations of KLF6-mediated enhancement of IFNγ-induced proinflammatory gene expression and attenuation of IL-10-induced anti-inflammatory gene expression in RAW macrophages (Fig. 2,3), we next examined whether pro- or anti-inflammatory cytokine stimulation affects macrophage KLF6 expression directly. Human monocyte-derived macrophages were stimulated with IFNγ, TNF, IL-10 or TGFβ, harvested for mRNA and protein isolation, and assayed for Klf6 expression. IFNγ and TNF stimulation of human macrophages resulted in an induction of Klf6 mRNA and protein (Supplemental Fig. S3A), whereas IL-10 and TGFβ stimulation resulted in a downregulation of Klf6 mRNA and protein (Supplemental Fig. S3B). Thus, pro- and anti-inflammatory cytokine signals can directly modulate the expression of KLF6 in human macrophages.
Deficiency of myeloid KLF6 protects against experimental colitis
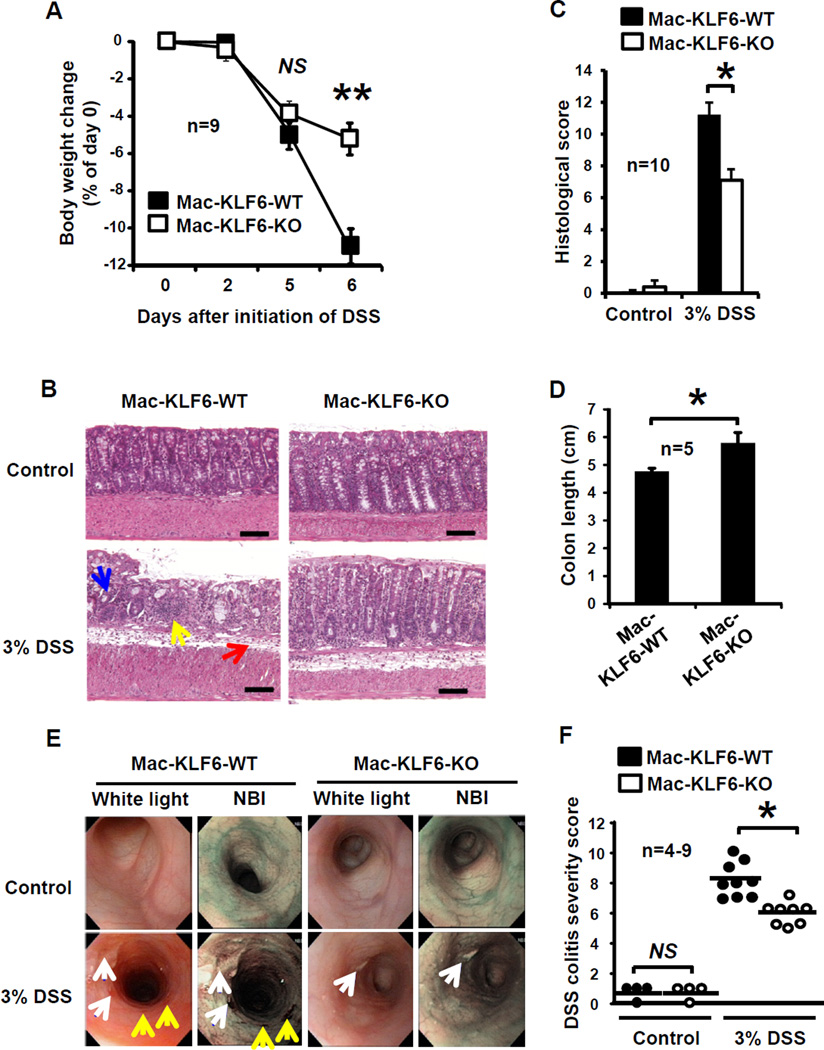
Having established that KLF6 regulates, and is regulated by, inflammatory cytokine signaling (Fig. 2–3), we next sought to test the in vivo, functional role of KLF6 in intestinal inflammation. We challenged Mac-KLF6-KO and Mac-KLF6-WT mice with 3% dextran sulfate sodium (DSS)33. When fed 3% DSS for 5 days and allowed no recovery time (acute model of colitis, with mice euthanized on day 6), Mac-KLF6-WT mice exhibited severe weight loss over time (approximately 11% from baseline, Fig. 4A). In contrast, Mac-KLF6-KO mice were partially protected from DSS-induced weight loss, averaging only a 5% loss in body weight by day 6 (Fig. 4A).
Figure 4. Myeloid deficiency of KLF6 is protective against DSS-induced colitis.
Mac-KLF6-KO and Mac-KLF6-WT mice were placed on 3% DSS for 5 days to induce colitis. (A) Body weight was measured on days 0, 2, 5, and 6, and is graphed relative to Day 0 weight; **p≤0.001, n=9 mice/group. (B) Representative H&E stained colon sections from Mac-KLF6-KO and Mac-KLF6-WT mice following 5 days of 3% DSS. Arrows indicate disruption of epithelial architecture (blue); infiltration of immune cells (yellow), and edema (red); n=10 mice/group. (C) Colon inflammation was assessed by a blinded pathologist following 5 days of 3% DSS. Score consists of active, chronic, and transmural inflammation; *p≤0.01, n=10 mice/group. (D) Total colon length was measured following 5 days of 3% DSS; *p≤0.01, n=5 mice/group. (E) Mice were subjected to colonic endoscopy following 5 days of 3% DSS. Representative images show endoscopy findings (left; arrows indicate colonic ulceration (white); mucus (blue); and erythematous lesions (yellow)) and perianal inflammation with diarrhea (right). Images are representative of 4–9 mice/group. (F) DSS colitis severity scores were assigned based on endoscopic and perianal findings. Graphs show mean ± SEM. *p≤0.03; n=8–9 mice/group. NS, not significant.
Colon tissue from water-fed Mac-KLF6-WT and Mac-KLF6-KO mice appeared healthy and exhibited no phenotypic features of colitis (Fig. 4B upper panels). In contrast, histological examination of colon tissue from Mac-KLF6-WT and Mac-KLF6-KO mice following DSS treatment revealed marked signs of inflammation in Mac-KLF6-WT compared to Mac-KLF6-KO (Fig. 4B lower panels). In agreement with histological observations, Mac-KLF6-WT mice exhibited significantly higher colitis scores than Mac-KLF6-KO mice (Fig. 4C). We also evaluated colon length in Mac-KLF6-WT and Mac-KLF6-KO mice following DSS challenge, since colonic shortening is a well-established feature of acute colitis, and found that colons from Mac-KLF6-WT mice were significantly shortened compared to Mac-KLF6-KO colons following DSS administration (Fig. 4D).
In order to examine colons in situ, we took advantage of a novel flexible colonoscopy procedure recently developed in our lab35. This technique utilizes both narrow-band imaging (NBI) as well as white light imaging, allowing for enhanced detection of erythematous regions and blood vessels (NBI) in addition to visualization of mucus and ulcerated regions. The procedure was well-tolerated and revealed significant colonic inflammation in Mac-KLF6-WT mice (Fig. 4E), including ulcerated tissue (white arrows) and erythematous regions (yellow arrows). In contrast, colon imaging of Mac-KLF6-KO mice revealed only minimal ulceration and erythema (Fig. 4E). Features of colitis observed by endoscopy were converted to decimal scores encompassing four parameters: perianal findings; wall transparency; intestinal bleeding; and focal lesions (edematous areas/ erosions/ ulcers). This quantitative endoscopic evaluation (Fig. 4F) revealed significantly worse disease in Mac-KLF6-WT mice than in Mac-KLF6-KO mice, further implicating a pathogenic contribution for KLF6 to intestinal inflammation. Mac-KLF6-KO and Mac-KLF6-WT mice fed water, in the absence of DSS, showed no detectable signs of colitis by endoscopy (Fig. 4E. upper panels).
Myeloid KLF6 modulates inflammatory gene expression in colon tissue
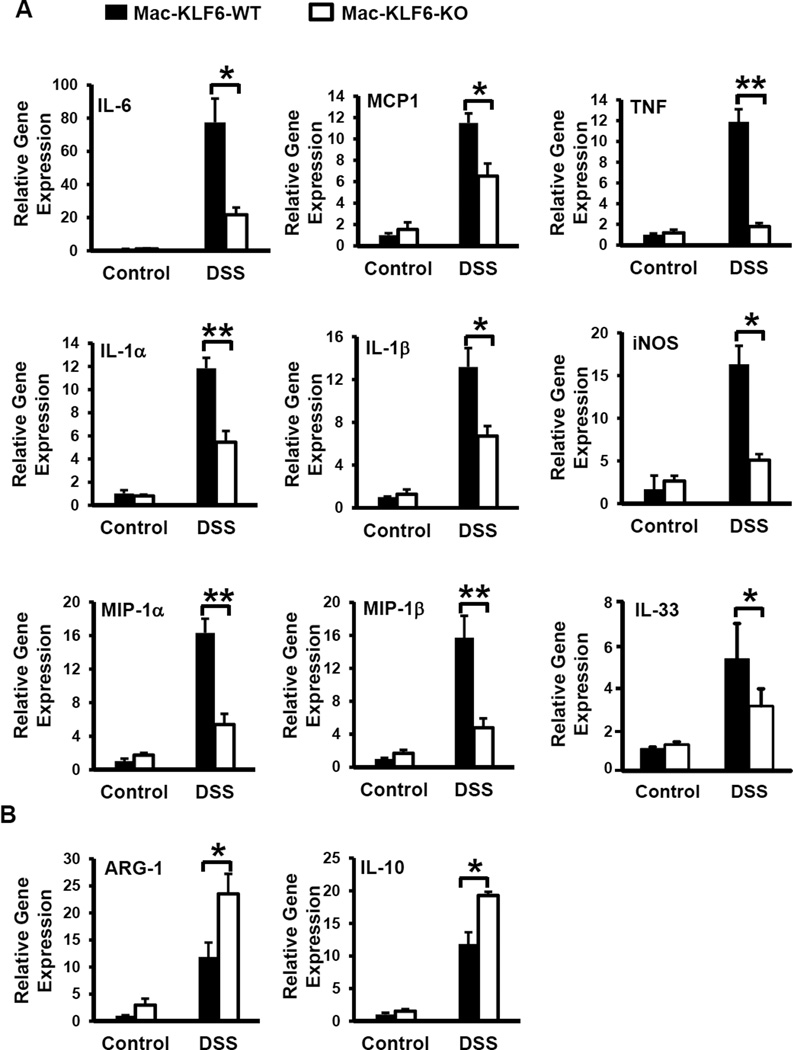
To determine effects of myeloid KLF6 deficiency on gene expression in the inflamed intestine, colon tissues were harvested from Mac-KLF6-KO and Mac-KLF6-WT mice following acute (5 day) DSS challenge. Tissues were analyzed for expression of pro- and anti-inflammatory genes known to be altered in acute colitis. Following 5 days of DSS feeding, colon tissues from both Mac-KLF6-WT and Mac-KLF6-KO mice expressed significantly elevated mRNA specific for the pro-inflammatory genes, Il1a, Il1b, Il6, Tnf, inos, Mip1a, Mip1b, Il33 and Mcp1, compared to non-DSS-fed control mice (Fig. 5A). However, DSS-fed Mac-KLF6-WT mice expressed significantly heightened expression of these genes compared to Mac-KLF6-KO mice (Fig. 5A), in agreement with exacerbated disease in Mac-KLF6-WT mice (Fig. 4). We also analyzed the expression of anti-inflammatory genes Arg1 and Il10, known to be protective in experimental and human intestinal inflammation36. As expected, given their enhanced morphological and histological colitis phenotypes, Mac-KLF6-WT mice displayed diminished expression of Arg1 and Il10 compared to Mac-KLF6-KO mice following acute DSS challenge (Fig. 5B). Taken together, our results indicate that deficiency of myeloid KLF6 both diminishes DSS-induced pro-inflammatory gene expression as well as enhances anti-inflammatory gene expression in colon tissues.
Figure 5. Myeloid KLF6 modulates inflammatory gene expression in colon tissue.
Colon tissue from Mac-KLF6-WT and Mac-KLF6-KO mice was collected following 5 days of 3% DSS challenge (or water-fed control) and analyzed for gene expression. (A) cDNA was analyzed for expression of pro-inflammatory genes Il1a, Il1b, Il6, Tnf, Mcp1, inos, Mip1a, Mip1b, and Il33. Gene expression was normalized to the housekeeping gene 36b4 and is expressed as fold change of the control (water-fed) group; *p≤0.01, **p≤0.001, n=4–5/group. (B) cDNA was analyzed for expression of anti-inflammatory genes Arg1 and Il10, then normalized to 36b4 and expressed as fold change of the control group. Graphs show mean ± SEM; *p≤0.02; **p≤0.003, n=4–5/group.
KLF6 regulates macrophage inflammatory gene expression by modulating STAT3 and NFκB signaling pathways
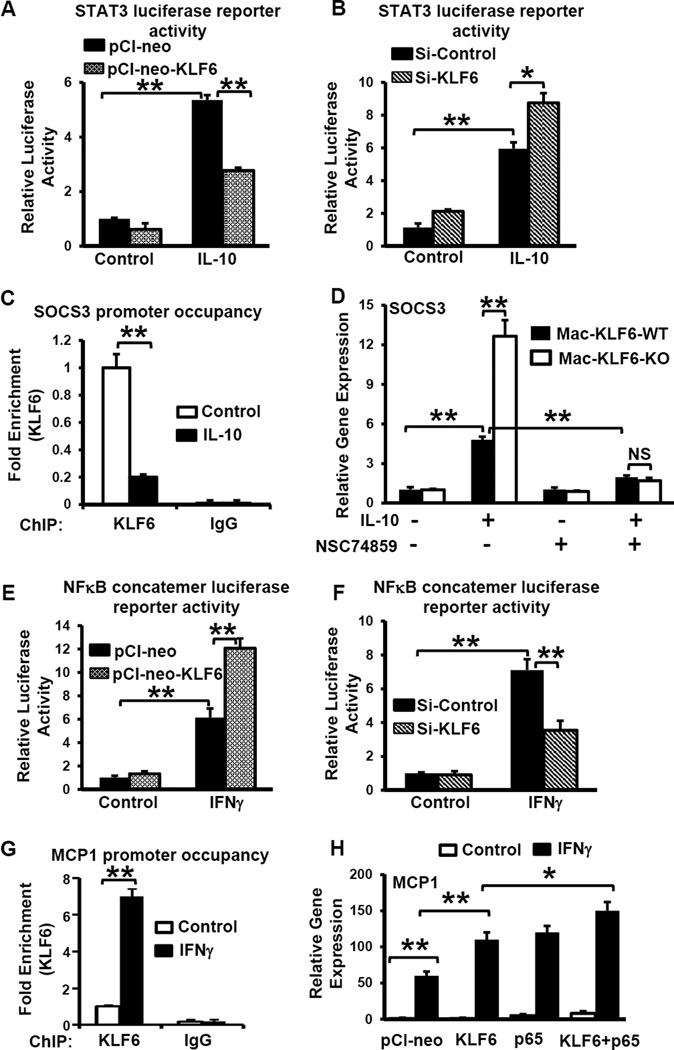
Our in vivo gene expression analysis indicated that myeloid deficiency of KLF6 enhanced the expression of Il10 in colon tissues (Fig. 5B). Many IL-10-induced anti-inflammatory genes are transcriptionally regulated by STAT337. To test whether KLF6 overexpression and/or deficiency regulates IL-10-induced STAT3 transcriptional activity, we performed gain- and loss-of-function assays. Accordingly, RAW264.7 macrophages were co-transfected with STAT3 luciferase reporter plasmid (pTL-STAT3-Luc) in the presence of pCI-neo-KLF6 or siKLF6, stimulated with IL-10, and luciferase activity was measured. Overexpression of KLF6 attenuated (Fig. 6A), whereas deficiency enhanced (Fig. 6B), IL-10-induced, STAT3-dependent luciferase activity in RAW264.7 macrophages.
Figure 6. KLF6 interacts with pro-inflammatory and anti-inflammatory transcriptional machinery to regulate gene expression in response to cytokine signaling.
(A) RAW264.7 macrophages were transfected to overexpress KLF6 (pCl-neo-KLF6) or control plasmid (pCl-neo) together with a STAT3 luciferase reporter plasmid. Cells were treated with IL-10 or PBS (control), then assessed for the relative luciferase activity, which is expressed as a ratio of control-transfected cells. (B) RAW264.7 macrophages were transfected with siRNA specific to KLF6 or control (scrambled) RNA, together with a STAT3 luciferase reporter plasmid. Cells were treated with IL-10 or PBS (Ctrl) and then assessed for luciferase activity. (C) Wild-type PMs were treated with IL-10 or PBS (Ctrl) and ChIP assays were performed on the SOCS3 promoter (−313 to −319) following immunoprecipitation with anti-KLF6 or IgG (Ctrl). (D) PMs from indicated mice were treated with IL-10 and/or the STAT3 inhibitor NSC74859 as indicated. Gene expression of SOCS3 was determined by qPCR and is expressed as fold-change over PBS-treated, wild-type PMs. (E) RAW264.7 macrophages were transfected to overexpress KLF6 (pCl-neo-KLF6) or control plasmid (pCl-neo) together with an NFκB luciferase reporter plasmid. Cells were treated with IFNγ or PBS (control), then assessed for the relative luciferase activity. (F) RAW264.7 macrophages were transfected with siRNA specific to KLF6 or control (scrambled) RNA, together with an NFκB luciferase reporter plasmid. Cells were treated with IFNγ or PBS (Control) and then assessed for luciferase activity. (G) PMs were isolated from WT mice, treated with IFNγ or PBS (Ctrl), and ChIP assays were performed on the MCP-1 promoter (−1363 to −1378) following immunoprecipitation with anti-KLF6 or IgG (Ctrl. (H) RAW264.7 macrophages were transfected to overexpress the indicated plasmids, and then stimulated with IFNγ or PBS (Control). Relative gene expression of MCP-1 was determined by qPCR and is expressed relative to PBS-treated, mock-transfected cells. Graphs show mean ± SEM; *p≤0.05; **p≤0.005, n=3.
Our in vitro and ex vivo analyses indicate that KLF6 attenuates the expression of IL-10-induced, STAT3-regulated anti-inflammatory factors, such as SOCS3, in RAW264.7 macrophages (Fig.3 A&B). We therefore evaluated whether KLF6 directly occupies the Socs3 gene promoter to repress its transcriptional activation. Results from ChIP assays using WT PMs indicate that KLF6 occupies the Socs3 promoter (−313 to −319) under unstimulated conditions and that IL-10 exposure significantly diminishes KLF6 occupancy (Fig. 6C). These findings indicate that KLF6 directly binds to STAT3 transcriptional targets, such as Socs3, to repress their expression. Deficiency of KLF6 results in a loss of this negative regulation, thereby enhancing expression of Socs3 following IL-10 exposure (Fig. 6C).
Next, we investigated whether the upregulation of IL-10-induced anti-inflammatory genes in KLF6-deficient myeloid cells (Fig. 3B) is STAT3-dependent. Mac-KLF6-WT and Mac-KLF6-KO PMs were stimulated with IL-10 in either the presence or absence of the STAT3-specific inhibitor, NSC74859. As shown in Fig. 6D, IL-10 exposure significantly elevated the STAT3 target gene Socs3 in Mac-KLF6-WT PMs. KLF6 deficiency further elevated IL-10-induced Socs3 expression in myeloid cells. Interestingly, treatment with NSC74859 significantly attenuated IL-10-induced Socs3 expression and eliminated the enhanced expression of Socs3 observed in KLF6-deficient myeloid cells. Taken together, our results indicate that KLF6 directly represses IL-10-induced anti-inflammatory gene expression by negatively regulating STAT3 transcriptional activity in myeloid cells.
Our in vitro and ex vivo analyses following IFNγ exposure indicated that KLF6 enhances pro-inflammatory gene expression in RAW264.7 macrophages (Fig. 2A&B). Therefore, RAW264.7 macrophages were co-transfected with an NFκB luciferase reporter plasmid in the presence of pCI-neo-KLF6 or siKLF6, stimulated with IFNγ, and luciferase activity was measured. Our results indicate that KLF6 overexpression significantly enhances (Fig. 6E), while deficiency attenuates (Fig. 6F), IFNγ–induced, NFκB-mediated luciferase activity in macrophages. Next, we evaluated whether KLF6 regulates the gene expression of classical NFκB pro-inflammatory targets, such as Mcp1, by directly binding to its promoter region. Our ChIP results indicate that KLF6 occupies the Mcp1 promoter (−1363 to −1378) of WT PMs under unstimulated conditions and that IFNγ exposure results in enrichment of KLF6 by at least six-fold compared to control-treated cells (Fig. 6G). Therefore, we hypothesized that KLF6 co-operates with NFκB to regulate IFNγ-induced Mcp1 expression in RAW264.7 macrophages. Our results indicate that overexpression of either KLF6 or NFκB-p65 enhances IFNγ–induced MCP1 expression compared to the pCI-neo-transfected control group (Fig. 6H). Importantly, cells stimulated with IFNγ following combined overexpression of KLF6 and NFκB-p65 robustly induced Mcp1 expression compared to any other group, suggesting that KLF6 co-operates with NFκB to enhance IFNγ-induced Mcp1 expression in RAW264.7 macrophages. Taken together, our results clearly indicate that KLF6 promotes IFNγ-induced NFκB transcriptional activity and co-operates with NFκB to regulate pro-inflammatory gene expression in myeloid cells.
Discussion
Our findings are the first to identify KLF6 as a significant transcriptional regulator in the context of intestinal inflammation, where it contributes to inflammatory macrophage activation and escape from inflammatory anergy. The key observations of this study are (1) KLF6 expression is elevated in diseased, colonic tissues and myeloid cells from IBD patients, and high KLF6 levels correlate with acutely involved areas of UC colon tissue; (2) experimental mouse models of IBD, including DSS-induced colitis and the TNFΔARE/+ model, reproduce the elevated KLF6 expression observed in IBD patients; (3) macrophage KLF6 promotes IFNγ-induced pro-inflammatory gene regulatory programs and diminishes IL-10-induced anti-inflammatory programs; (4) in vivo deletion of myeloid-specific KLF6 partially protects against DSS-induced colitis, suggesting that endogenous myeloid KLF6 contributes to macrophage pro-inflammatory functions and exacerbates intestinal inflammation; and (5) the mechanisms by which KLF6 regulates macrophage phenotype and function are two-pronged, acting to enhance NFκB-mediated gene expression as well as to inhibit STAT3-mediated expression of anti-inflammatory genes. Collectively, our findings indicate that KLF6 contributes to transcriptional re-programming of intestinal macrophages in response to cytokine signals in the inflamed IBD intestine.
Macrophages are well known to alter their activation states in response to changing environmental stimuli38. The spectrum of macrophage phenotypes and functions ranges from inflammation-anergic and related M2 macrophages, which promote tissue healing and repair processes, to pro-inflammatory M1 macrophages, which promote host defense by initiating immune responses against pathogens. Although M1 responses have evolved as a critical mechanism of host defense in response to pathogens, when left unchecked (as in the case of chronic inflammation, such as IBD), these responses can be extremely deleterious. The inflamed intestine is replete with pro-inflammatory cytokines, such as IFNγ, TNF, IL-1, and IL-6, which promote inflammation and tissue destruction, but also reinforce pro-inflammatory M1 macrophage activation. IBD patients produce high levels of pro-inflammatory cytokines and other soluble mediators, including nitric oxide (NO), a byproduct of L-arginine, ROS and NOS, cathepsins, and metalloproteinases12. Thus, once inflammation is established, environmental cues provide reinforcement for pathogenic, M1 activation.
Mounting evidence suggests that the local microenvironment within the inflamed, IBD intestines can result in a dynamic shift in macrophage polarization away from an inflammatory-anergic phenotype and towards an M1 phenotype, thus significantly contributing to the pathogenesis of IBD36. It is therefore critical to identify the molecular mechanisms underlying cytokine-induced M1 activation in the setting of chronic intestinal inflammation in order to design more effective and targeted therapeutic approaches. Our observations of increased KLF6 mRNA in colon tissue from UC and CD patients, as well as significantly elevated KLF6 mRNA in acutely inflamed biopsy tissue from UC patients (Fig. 1), suggests that locally high KLF6 levels correlate with worsened intestinal inflammation and tissue damage.
Given their role in directing lymphocyte phenotype and function, macrophages are at the epicenter of directing immune responses and are therefore an attractive target for therapeutic intervention. Pathogenic M1 polarization has been associated with numerous inflammatory conditions, including autoimmune diseases, atherosclerosis, and metabolic syndrome. Therefore, an enhanced understanding of the mechanisms by which macrophages are diverted away from the beneficial, protective M2 phenotypes will be relevant for many disease settings.
Transcriptional regulation is a central means of controlling macrophage activation. Several transcriptional regulators have been demonstrated to play a role in macrophage polarization, including NFκB, PPARs, and the STAT, AKT, and IRF families39. Previous studies have demonstrated that KLF2 and KLF4 have likewise been shown to regulate macrophage anti-inflammatory gene expression and function, although the role of these factors in macrophage-mediated intestinal inflammation remains unknown. Interestingly, our data shows that KLF6 functions in an opposing way to KLF2/4, by promoting pro-inflammatory M1 activation and diverting mucosal macrophages away from protective M2 activation. STAT1 and IRF5 also have demonstrated roles in promoting M1 phenotypes39, 40. Therefore, the potential crossregulation of STAT1, IRF5, and KLF6 represents an important area for future study. Our data show that IFNγ treatment of macrophages induces KLF6 expression (Fig. 6, S2), and IFNγ has been shown to signal in both STAT1-dependent and –independent manners41. Therefore, it is possible that STAT1 is upstream of KLF6, or that KLF6 is induced independently of STAT1.
Intriguingly, our studies show that regulation of macrophage activation by KLF6 occurs by two distinct pathways: cooperation with NFκB to induce pro-inflammatory gene expression, and inhibition of anti-inflammatory targets through STAT3 suppression (Fig. 6). Thus, KLF6 appears capable of modulating pro-and anti-inflammatory transcriptional regulatory molecules, leading to diverse outcomes for target gene expression. One mechanism underlying macrophage inflammatory anergy is blockade of pro-inflammatory NFκB signaling downstream of TLRs42, thus preventing NFκB translocation to the nucleus and subsequent transcription of pro-inflammatory genes. Given our observations of elevated KLF6 in the myeloid cells and intestinal tissue of IBD patients, we hypothesize that high levels of KLF6 may act to overcome blockade in NFκB-mediated signaling, thereby contributing to macrophage escape from inflammatory anergy.
Alterations in KLF expression and/or function have been associated with the pathogenesis of numerous human diseases, including bacterial infection/sepsis, cardiovascular disease, metabolic disorders and cancer23, 24. Interestingly, KLF6 has a known role as a tumor-suppressor and was shown to be inactivated in the majority of spontaneous and IBD-associated colorectal cancer (CRC) cases43. This finding further highlights the importance of each particular tissue microenvironment in determining the ultimate function of gene regulatory proteins in different contexts. Due to its robust expression in myeloid cells and its intersection with numerous other transcriptional regulators, KLF6 represents an attractive candidate for targeted therapeutic approaches for IBD and other conditions mediated by inflammatory macrophages.
The present study is first to identify KLF6 as a strong inducer of pro-inflammatory M1 activation in the context of chronic intestinal inflammation. Monocyte-derived macrophages from IBD patients express significantly elevated levels of pro-inflammatory cytokines and diminished anti-inflammatory molecules16, likely contributing to the inflammatory cytokine environment in the diseased tissue that reinforces M1 activation. Signaling downstream of KLF6 induces numerous pro-inflammatory gene targets, while simultaneously dampening the expression of anti-inflammatory targets; thereby, representing a central nexus point controlling many transcriptional targets. Thus, we propose that induction of KLF6, either in newly-recruited monocytes or in established tissue macrophages, participates in diverting macrophages away from protective, M2-like activation states and toward pathogenic, M1 activation. Given the central contribution of activated macrophages to chronic inflammation in IBD, this is likely an important mechanism leading to sustained tissue inflammation. Efforts at limiting the expression or functional activity of KLF6 represent promising areas for future development of biologics aimed at targeting innate immune mechanisms contributing to chronic intestinal inflammation.
Methods
Mice
Mac-KLF6-WT and Mac-KLF6-KO mice were generated by crossing Klf6 floxed mice with Lyz2cre mice expressing the lysozyme M promoter-driven Cre recombinase, as previously described28. Mice harboring two Klf6 floxed alleles and two Lyz2cre alleles were designated Mac-KLF6-KO, and Lyz2cre mice (harboring only two Cre alleles) were designated Mac-KLF6-WT. Lyz2cre and Klf6 genotyping have been previously described28. Mac-KLF6-KO mice display approximately 85–90% reduction in Klf6 expression in the myeloid compartment, as assessed by qPCR and Western blot28. TNFΔARE/+ mice were generated by breeding male mice heterozygous for the TNFΔARE mutant allele with TNF+/+ females, which results in 50% of offspring with posttranscriptional dysregulation of TNF and spontaneous intestinal inflammation. Wild-type TNF+/+ littermates were used as controls for these experiments. TNFΔARE/+ mice on the C57BL/6J background were generated as previously described 31,32 and propagated by breeding male mice heterozygous for the TNFΔARE mutant allele with TNF+/+ females, which results in 50% of offspring with posttranscriptional dysregulation of TNF and spontaneous intestinal inflammation. Wild-type TNF+/+ littermates were used as controls for these experiments. All mice were bred and maintained under special pathogen free (SPF) conditions, fed standard laboratory chow (Harlan Teklad, Indianapolis, IN), and kept on a 12h light/dark cycle. All procedures were approved by the IACUC at Case Western Reserve University and conformed to guidelines established by the American Association for Accreditation of Laboratory Animal Care.
Human tissue samples
Colonic biopsies were obtained from healthy adult volunteers or patients with diagnosed Crohn’s colitis or UC during endoscopy following informed consent. The control group consisted of individuals undergoing screening colonoscopy, who were included in the study only when endoscopy and pathology did not reveal any mucosal abnormalities. Endoscopies were performed at University Hospitals Case Medical Center, Cleveland, OH and at the Endoscopy Unit of the 1st Dept. of Internal Medicine, Propaedeutic, at Laikon Hospital, Athens, Greece. Specimens were maintained at 4°C for 1–2d in RNAlaterR Solution (AmbionR, Life Technologies, Austin, TX) until total RNA was extracted as described below. Peripheral blood mononuclear cells (PBMCs) were prepared from heparinized peripheral blood by density centrifugation, and cell viability was determined by trypan blue exclusion. Monocytes were isolated from PBMCs by magnetic bead separation as described below. Informed consent was obtained from all human subjects under protocols approved by the Institutional Review Board of University Hospitals Case Medical Center/Case Western Reserve University. This study was conducted in compliance with good clinical practice and according to the principles of the Declaration of Helsinki.
Confocal immunofluorescent microscopy
5µm tissue sections were cut from formalin-fixed, paraffin-embedded blocks. Sections were de-paraffinized and re-hydrated by sequential 4 min incubations in xylene and ethanol. Antigen retrieval was performed to unmask cross-linked epitopes by incubating sections for 20 min in boiling citrate buffer (Dako USA, Carpinteria, CA) and then cooling them to room temperature. Tissue was blocked with 10% chicken serum (Jackson ImmunoResearch, West Grove, PA) for 30 min, and then labeled with mouse anti-mouse CD68 (Novus Biologicals, Littleton, CO), rabbit anti-mouse KLF6 (Santa Cruz Biotechnology, Dallas, TX), or appropriate isotype controls for 1 hour at room temperature. Alexafluor 488- or 594-conjugated chicken anti-mouse or chicken anti-rabbit secondary antibodies (Life Technologies, Carlsbad, CA) were used to detect primary antibodies and Draq-5 (Axxora, San Diego, CA) was used as a nuclear marker. Images were collected using a 40× (NA 0.85) objective on an UltraVIEW Vox spinning disk confocal system (Perkin Elmer, Waltham, MA) attached to a Leica DMI 6000B inverted microscope.
DSS colitis model
8–10 week old male Mac-KLF6-WT and Mac-KLF6-KO mice were fed 3% DSS (TdB Consultancy AB, Uppsala, Sweden) for 5 days ad libitum in their drinking water. Mice had access to standard laboratory chow throughout the experiment and were assessed on days 0 (initiation of DSS), 1, 3, and 5 for body weight and intestinal bleeding (hemoccult assay) by an investigator blinded to mouse genotype. Following 5 days of DSS, mice were subjected to colonic endoscopy and euthanized for collection of colon tissues.
Histologic evaluation of colon inflammation
Full-length colons of experimental mice were removed, flushed of fecal contents, opened longitudinally, and placed in Bouin’s fixative for 24 hours. Tissues were embedded in paraffin, cut to 3 µm, and stained with H&E. Disease severity was evaluated by a trained pathologist (Wei Xin, M.D.) in a blinded fashion using an established histologic scoring system for colitis34. Images were obtained on an Axiophot microscope, captured on an Axiocam and assembled using Axiovision Release 4.5 (Carl Zeiss, Inc., Thornwood, NY).
Endoscopic evaluation of murine colitis
Experimental colitis following DSS treatment was assessed by a validated, reproducible endoscopic scoring system encompassing visual assessment of the extent and severity of colitis, as well as a numeric scoring system35. Briefly, following anesthetization with isoflurane, an Olympus URF-V flexible endoscope with attached camera (Olympus America, Center Valley, PA) was used to visualize transparency of the intestinal wall, mucosal bleeding, and focal lesions. Photos were acquired using narrow band imaging (NBI) to capture light of 390–445 nm and 530–550 nm, highlighting structures containing hemoglobin. Perianal inflammation (diarrhea, bloody discharge, rectal prolapse, granulation and fistulas) were observed prior to endoscopy procedure. A validated decimal-weighted scoring system35 was used to characterize endoscopic and perianal findings by a researcher blinded to mouse genotype. The system encompasses four parameters, each with a score of 0 to 3: perianal findings; wall transparency; intestinal bleeding; and focal lesions (edematous areas/ erosions/ ulcers).
Cell culture
RAW264.7 macrophages were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 10µg/ml streptomycin, and 2 mM glutamine in a humidified incubator (5% CO2 and 37°C). Mouse peritoneal macrophages were obtained by lavage 3 days post injection with 3% thioglycolate broth in 8–12 week old mice, as previously described26. Cells were cultured for short-term stimulation experiments in complete DMEM as described above. Mouse monocyte-derived macrophages were differentiated ex vivo by culturing CD14+ monocytes (isolated by negative selection) in DMEM supplemented with recombinant mouse M-CSF-1 for 7 days, as previously described28, and were then harvested and used for experiments. Human monocytes were isolated from unfractionated PBMCs by CD14 negative selection using magnetic beads (Miltenyi Biotec, San Diego, CA), and then cultured with recombinant human M-CSF (50ng/ml) ex vivo for 7 days to generate primary macrophages. All experiments involving human samples were approved by the Case Western Reserve University Institutional Review Board.
Transient transfection
Transfection of RAW264.7 cells was performed using Lipofectamine (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Efficiency of transfection of either pCl-neo (empty vector control) or pCl-neo-KLF6 (overexpressing KLF6) averaged 65–75% in all experiments. Transfected cells were stimulated in vitro with either IFNγ (100U/ml), IL-10 (10ng/ml), or PBS (control), then harvested and used for experiments.
Luciferase assays
RAW264.7 cells were transfected using the Lipofectamine® reagent (Life Technologies) according to the manufacturer’s instructions. Luciferase reporter plasmids driven by the NFκB concatemer (Promega) or STAT3 (Affymetrix, Inc) were transfected alone or together with plasmids encoding KLF6 or siKLF6. Cells were treated with IFNγ (100U/ml) or IL-10 (10ng/ml) for 16hrs, followed by measuring of Luciferase reporter activity. Data was normalized relative to the control (PBS-treated) group.
Chromatin immunoprecipitation
Chromatin immunoprecipitation experiments were performed using the EZ-Magna ChIP G kit (Millipore Corp., Billerica, MA) according to the manufacturer’s instructions. Briefly, peritoneal macrophages (PMs) from wild-type mice were isolated and stimulated ex vivo with IFNγ (100U/ml), IL-10 (10ng/ml), or PBS (control). Chromatin immunoprecipitations were performed using anti-KLF6 antibody (Santa Cruz Biotechnology, Inc) or IgG. Precipitated fragments were analyzed by real-time qPCR using primer sequences flanking the MCP1 promoter (−1363 to −1378) or SOCS3 promoter (−313 to −319).
Western blot
Western blots for KLF6 were performed using total cell lysates. Primary antibody against KLF6 (Santa Cruz Biotechnology, Inc.) was used at a 1:1000 dilution and polyclonal HRP-conjugated goat anti-rabbit IgG at 1:5000 dilution (Millipore) was used as a secondary antibody.
RNA extraction, Reverse-transcription, and Real-time quantitative PCR assay
Total RNA was isolated from tissue samples using Trizol (Life Technologies) or from macrophages using the High Pure RNA Isolation Kit (Roche Life Science, Indianapolis, IN). One microgram of total RNA was reverse-transcribed using M-MuLV reverse transcriptase (New England Biolabs, Ipswich, MA) in the presence of random hexamers and oligo(dT) primers. Real time quantitative PCR was performed using University SYBR Green PCR Master Mix on Applied Biosystems Step One Plus real-time PCR system using gene-specific primers.
Statistical analysis
Data are presented as mean ± SEM unless otherwise indicated. Two-tailed Student’s T-tests with Bonferroni’s corrections were performed to assess the significance of differences between groups. P values <0.05 were considered significant.
Supplementary Material
Acknowledgements
The authors thank Scott Howell, Minh Lam, Xiao-Ming Wang, and Becky Sebastian for their technical support and assistance.
This work was supported by grants from the NIH: HL097023, HL121193 and HL126626 (to GHM), DK056762 (to TTP), DK091222 and DK097948 (to TTP/FC), K01DK105138 (to WAG) and from the Crohn’s & Colitis Foundation of America: CDA329284 (to WAG) and RFA326877 (to CDS).
Abbreviations used in this paper
- AP-1
activator protein 1
- BCL
B cell lymphoma 2
- CD
Crohn’s disease
- DSS
dextran sulfate sodium
- IBD
Inflammatory bowel disease
- IFNγ
interferon gamma
- iNOS
inducible nitric oxide synthase
- IRF
interferon regulatory factor
- KLF
Kruppel-like Factor
- KO
knock-out
- MCP
monocyte chemotactic protein
- MCSF1
macrophage colony-stimulating factor 1
- MIP
macrophage inflammatory protein
- MLN
mesenteric lymph node
- MMP
matrix metalloproteinase
- NFκB
nuclear factor κB
- NOS
nitric oxide synthases
- PM
peritoneal macrophage
- PPAR
peroxisome proliferator-activated receptor
- PRR
pattern recognition receptor
- ROS
reactive oxygen species
- SBNO
strawberry notch homolog
- SOCS
suppressor of cytokine signaling
- STAT
signal transducer and activator of transcription
- TGF
transforming growth factor
- TLR
toll like receptor
- TNF
tumor necrosis factor
- Treg
T regulatory cell
- UC
ulcerative colitis
- WT
wild-type
Footnotes
Conflicts of interest: None
Author Contributions:
G.H.M., T.T.P. and W.A.G. conceived and designed the study.
G.H.M., W.A.G. S.O., D.D.,, G.D.K., L.D.M., and C.D.S. performed experiments.
G.H.M., T.T.P. F.C., W.A.G., S.O., D.D., and L.D.M. analyzed and interpreted the data.
G.B. and S.C. collected and provided human specimens.
W.A.G., G.H.M. and T.T.P. wrote and edited the manuscript that is approved by all authors.
References
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature reviews. Immunology. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee SH, Starkey PM, Gordon S. Quantitative analysis of total macrophage content in adult mouse tissues. Immunochemical studies with monoclonal antibody F4/80. The Journal of experimental medicine. 1985;161:475–489. doi: 10.1084/jem.161.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith PD, et al. Intestinal macrophages and response to microbial encroachment. Mucosal immunology. 2011;4:31–42. doi: 10.1038/mi.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qualls JE, Kaplan AM, van Rooijen N, Cohen DA. Suppression of experimental colitis by intestinal mononuclear phagocytes. Journal of leukocyte biology. 2006;80:802–815. doi: 10.1189/jlb.1205734. [DOI] [PubMed] [Google Scholar]
- 5.Hunter MM, et al. In vitro-derived alternatively activated macrophages reduce colonic inflammation in mice. Gastroenterology. 2010;138:1395–1405. doi: 10.1053/j.gastro.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 6.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Lin HH, et al. The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. The Journal of experimental medicine. 2005;201:1615–1625. doi: 10.1084/jem.20042307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nature immunology. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor symposia on quantitative biology. 1989;54 Pt 1:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Murray PJ, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smythies LE, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. The Journal of clinical investigation. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mowat AM, Bain CC. Mucosal macrophages in intestinal homeostasis and inflammation. Journal of innate immunity. 2011;3:550–564. doi: 10.1159/000329099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B, Alli R, Vogel P, Geiger TL. IL-10 modulates DSS-induced colitis through a macrophage-ROS-NO axis. Mucosal immunology. 2014;7:869–878. doi: 10.1038/mi.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeda K, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 15.Hontecillas R, et al. Immunoregulatory mechanisms of macrophage PPAR-gamma in mice with experimental inflammatory bowel disease. Mucosal immunology. 2011;4:304–313. doi: 10.1038/mi.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shouval DS, et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. 2014;40:706–719. doi: 10.1016/j.immuni.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dijkstra G, et al. Increased expression of inducible nitric oxide synthase in circulating monocytes from patients with active inflammatory bowel disease. Scandinavian journal of gastroenterology. 2002;37:546–554. doi: 10.1080/00365520252903099. [DOI] [PubMed] [Google Scholar]
- 18.Hausmann M, et al. Cathepsin D is up-regulated in inflammatory bowel disease macrophages. Clinical and experimental immunology. 2004;136:157–167. doi: 10.1111/j.1365-2249.2004.02420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinecker HC, et al. Enhanced secretion of tumour necrosis factor-alpha, IL-6, and IL-1 beta by isolated lamina propria mononuclear cells from patients with ulcerative colitis and Crohn's disease. Clinical and experimental immunology. 1993;94:174–181. doi: 10.1111/j.1365-2249.1993.tb05997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Targan SR, et al. A short-term study of chimeric monoclonal antibody cA2 to tumor necrosis factor alpha for Crohn's disease. Crohn's Disease cA2 Study Group. The New England journal of medicine. 1997;337:1029–1035. doi: 10.1056/NEJM199710093371502. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe N, et al. Elimination of local macrophages in intestine prevents chronic colitis in interleukin-10-deficient mice. Digestive diseases and sciences. 2003;48:408–414. doi: 10.1023/a:1021960401290. [DOI] [PubMed] [Google Scholar]
- 22.Scheinin T, Butler DM, Salway F, Scallon B, Feldmann M. Validation of the interleukin-10 knockout mouse model of colitis: antitumour necrosis factor-antibodies suppress the progression of colitis. Clinical and experimental immunology. 2003;133:38–43. doi: 10.1046/j.1365-2249.2003.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Z, Sun X, Icli B, Wara AK, Feinberg MW. Role of Kruppel-like factors in leukocyte development, function, and disease. Blood. 2010;116:4404–4414. doi: 10.1182/blood-2010-05-285353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiological reviews. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Kruppel-like transcription factors: a functional family. The international journal of biochemistry & cell biology. 2008;40:1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Mahabeleshwar GH, et al. The myeloid transcription factor KLF2 regulates the host response to polymicrobial infection and endotoxic shock. Immunity. 2011;34:715–728. doi: 10.1016/j.immuni.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma N, et al. Myeloid Kruppel-like factor 4 deficiency augments atherogenesis in ApoE−/− mice--brief report. Arteriosclerosis, thrombosis, and vascular biology. 2012;32:2836–2838. doi: 10.1161/ATVBAHA.112.300471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Date D, et al. Kruppel-like transcription factor 6 regulates inflammatory macrophage polarization. J Biol Chem. 2014;289:10318–10329. doi: 10.1074/jbc.M113.526749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palau N, et al. Genome-wide transcriptional analysis of T cell activation reveals differential gene expression associated with psoriasis. BMC genomics. 2013;14:825. doi: 10.1186/1471-2164-14-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bechmann LP, et al. Post-transcriptional activation of PPAR alpha by KLF6 in hepatic steatosis. Journal of hepatology. 2013;58:1000–1006. doi: 10.1016/j.jhep.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999 Mar;10(3):387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 32.Ho J, et al. A CD8+/CD103high T cell subset regulates TNF-mediated chronic murine ileitis. J Immunol. 2008;180:2573–2580. doi: 10.4049/jimmunol.180.4.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okayasu I, et al. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 34.Corridoni D, et al. Dysregulated NOD2 predisposes SAMP1/YitFc mice to chronic intestinal inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:16999–17004. doi: 10.1073/pnas.1311657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kodani T, et al. Flexible colonoscopy in mice to evaluate the severity of colitis and colorectal tumors using a validated endoscopic scoring system. Journal of visualized experiments : JoVE. 2013:e50843. doi: 10.3791/50843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bain CC, Mowat AM. Macrophages in intestinal homeostasis and inflammation. Immunological reviews. 2014;260:102–117. doi: 10.1111/imr.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Farrell AM, Liu Y, Moore KW, Mui AL. IL-10 inhibits macrophage activation and proliferation by distinct signaling mechanisms: evidence for Stat3-dependent and - independent pathways. The EMBO journal. 1998;17:1006–1018. doi: 10.1093/emboj/17.4.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills CD, Lenz LL, Ley K. Macrophages at the fork in the road to health or disease. Frontiers in immunology. 2015;6:59. doi: 10.3389/fimmu.2015.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nature reviews. Immunology. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 40.Ostuni R, Natoli G. Transcriptional control of macrophage diversity and specialization. European journal of immunology. 2011;41:2486–2490. doi: 10.1002/eji.201141706. [DOI] [PubMed] [Google Scholar]
- 41.Ramana CV, Gil MP, Schreiber RD, Stark GR. Stat1-dependent and - independent pathways in IFN-gamma-dependent signaling. Trends in immunology. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- 42.Smythies LE, et al. Inflammation anergy in human intestinal macrophages is due to Smad-induced IkappaBalpha expression and NF-kappaB inactivation. J Biol Chem. 2010;285:19593–19604. doi: 10.1074/jbc.M109.069955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reeves HL, et al. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology. 2004;126:1090–1103. doi: 10.1053/j.gastro.2004.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.