Abstract
Ticks play an important role in disease transmission as vectors for human and animal pathogens, including the Gram-negative pathogen Bartonella. Here, we evaluated the presence of Bartonella in ixodid ticks and domestic animals from Palestine. We tested 633 partly engorged ticks and 139 blood samples from domestic animals (dogs, sheep and camels) for Bartonella using ITS-PCR. Bartonella DNA was detected in 3.9% of the tested ticks. None of the ticks collected from sheep and goats were positive for Bartonella. Seventeen R. sanguineus ticks (17/391; 4.3%) collected from dogs were infected with B. rochalimae (n = 10), B. chomelii (n = 6), and B. koehlerae (n = 1). Four H. dromedarri ticks (4/63; 6.3%) obtained from camels were infected with B. bovis (n = 2) and B. rochalimae (n = 2). Among canine blood samples (n = 110), we found one asymptomatic female dog to be infected with B. rochalimae (0.9%). The detection of zoonotic Bartonella species in this study should raise awareness of these vector-borne diseases among physicians, veterinarians and public health workers and highlight the importance of surveillance and preventive measures in the region.
Keywords: bartonellosis, B. chomelii, B. rochalimae, B. bovis, Rhipicephalus, Hyalomma, Palestine
Introduction
Tick-borne diseases comprise a group of globally distributed and rapidly spreading illnesses caused by a range of pathogens. Molecular approaches make it possible to screen ticks for pathogens of veterinary and public health importance and perform detailed epidemiological studies (Sparagano et al., 1999). Bartonellosis is an infectious disease caused by bacteria from the genus Bartonella, which infect erythrocytes and endothelial cells in humans (Harms and Dehio, 2012). These pathogens are transmitted by biting arthropod vectors and infect a wide range of wild and domestic mammals, including rodents, cats, dogs, and cattle. In humans, Bartonella is responsible for emerging and reemerging diseases worldwide with presentations that range from subclinical or self-limiting infection to severe, life-threatening disease.
Different Bartonella species appear to be adapted to specific mammalian hosts (Breitschwerdt and Kordick, 2000; Vayssier-Taussat et al., 2010). For example, cats are the main reservoir for B. henselae, which causes cat-scratch disease. Cats are infected through the bites of cat fleas, while humans are directly infected through scratches or bites from an infected cat (Billeter et al., 2008).
By contrast, diverse species of Bartonella infect ruminants. B. bovis is the most commonly reported species in cattle, where it is associated with bovine endocarditis (Maillard et al., 2004; Erol et al., 2013). This species has been described in beef and dairy cattle worldwide, including North and South America, Italy, France, Israel, Taiwan and peninsular Malaysia (Martini et al., 2008; Cherry et al., 2009; Saisongkorh et al., 2009; Tsai et al., 2011; Bai et al., 2013; Rudoler et al., 2014; Kho et al., 2015). Another species, B. chomelii, has been isolated from cattle in France and Spain (Maillard et al., 2004; Antequera-Gomez et al., 2015). B. rochalimae infects humans, domestic animals and wild carnivores (Schaefer et al., 2012), so there has been much interest in its zoonotic potential (Eremeeva et al., 2007; Chomel et al., 2009).
Although transmission of Bartonella to humans typically occurs through traumatic contact with infected animals or by blood-sucking insect vectors such as fleas, lice and sand flies, this group of species has also been widely reported from ticks. The potential for involvement of ticks in transmission of Bartonella spp. has been suggested by several molecular and serological epidemiological studies (Billeter et al., 2008). Bartonella was first detected in questing ticks from the USA, including Ixodes pacificus, Dermacentor, and R. sanguineus (Chang et al., 2000). Additional surveys conducted in the Netherlands, France, Poland, and Austria have demonstrated the presence of Bartonella DNA in Ixodes ricinus ticks obtained from vegetation (Billeter et al., 2008). In France, the species B. henselae has been identified in Ixodes ricinus isolated from vegetation (Vayssier-Taussat et al., 2013), as well as from humans exposed to tick bites (Vayssier-Taussat et al., 2016).
Clinical studies have supported transmission of Bartonella by ticks to humans, as infections have occurred after tick bites without any known contact with other arthropods (Morozova et al., 2005; Billeter et al., 2008). Bartonella infection was reported in three patients with scalp eschar and neck lymphadenopathy following tick bites (Angelakis et al., 2010). A recent study by Vayssier-Taussat et al. (2016) identified potentially zoonotic Bartonella strains in symptomatic patients who reported tick bites. Natural co-infections with Bartonella species and other tick borne pathogens such as Babesia, Anaplasma, and Borrelia have also been demonstrated (Hofmeister et al., 1998; Angelakis et al., 2010). In particular, co-infections of Bartonella and Borrelia have been reported in humans from the USA and Europe (Billeter et al., 2008). These observations provide indirect evidence for tick-borne transmission, even in the absence of direct proof of tick vector competence for Bartonella (Cotte et al., 2008; Reis et al., 2011). Furthermore, experimental transmission studies using infected ticks and live susceptible animals support the role of ticks in the natural lifecycles of some Bartonella species (Cotte et al., 2008; Reis et al., 2011).
In Palestine, Bartonella DNA has been detected in 22% (64/289) of fleas collected from various animal hosts (dogs, cats and rodents) (Nasereddin et al., 2014). Several Bartonella species have been identified in Palestinian samples, including B. clarridgeiae, B. henselae, B. koehlerae, B. tribocorum, B. elizabethae, and B. rochalimae (Nasereddin et al., 2014).
In our previous studies, when we screened ticks from domestic animals, we identified several tick-borne pathogens, including Rickettsia from the spotted fever group, Babesia and Hepatozoon (Ereqat et al., 2016; Azmi et al., 2016). Given the potential role of ticks as a source of zoonotic Bartonella infection in humans (Vayssier-Taussat et al., 2016), we wished to determine whether ticks carry Bartonella in Palestine. More specifically, we set out to extend our previous surveys and to assess the presence of Bartonella in a set of previously studied ticks and in the blood samples collected from their animal hosts throughout Palestine.
Materials and Methods
Study Sites, Tick, and Animal Samples
A total of 633 hard ticks were collected during January to October 2014. The ticks were collected from dogs, sheep, goats, and camels in nine districts of Palestine (Hebron, Jenin, Jericho, Nablus, Qalqilia, Ramallah, Salfit, Tubas, and Tulkarem), located in three zones in the central, northern and southern regions of the country (Figure 1). Ticks were identified based on morphological characteristics (Feldman-Musham, 1954).
FIGURE 1.
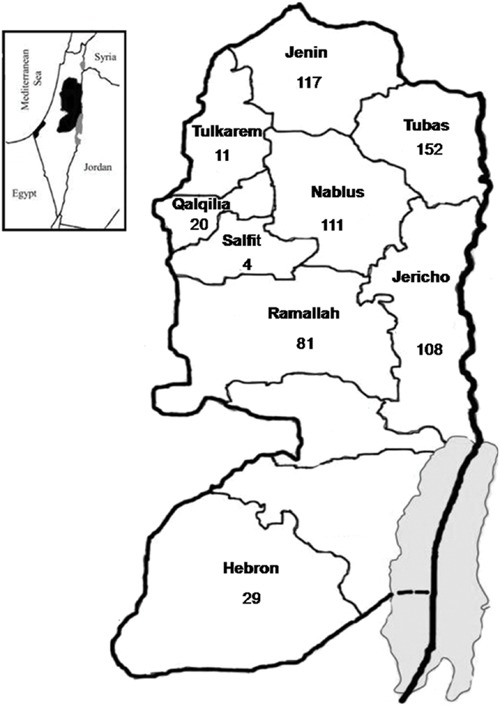
Distribution of Ixodid ticks collected from nine districts in Palestine from which Bartonella DNA was detected.
A total of 110 blood samples were collected from outdoor domestic dogs, with simultaneous tick collection from the locations mentioned above (17 from Hebron, 25 from Jenin, 18 from Jericho, three from Nablus, 21 from Ramallah, 13 from Salfit, and 13 from Tulkarem). All samples were collected in EDTA-anticoagulant tubes and stored at -20°C until further use. Study animals were selected irrespective of sex and age. In addition, 29 blood samples were obtained from camels (n = 19), sheep (n = 7), and goats (n = 3) from Jenin, Nablus, and Jericho, in January 2016. None of the animals showed clinical signs; all were apparently healthy at the time of sampling. The animal owners were verbally informed about the goals of this research and the sampling protocol. All owners gave their written informed consent to collect blood from their animals. The ethics committee at Al-Quds University approved the study.
DNA Extraction
DNA was extracted from each tick using a DNA extraction kit (QIAGEN GmbH, 40724 Hilden, Germany) following the manufacturer’s instructions. The eluted DNA (100 μl) was stored at -20°C until used as templates for PCR amplifications. DNA was extracted from whole blood (200 μl) following the QIAamp animal blood and Tissue Kit procedure (QIAGEN GmbH, Hilden, Germany), adjusted in 200 μl of Tris- EDTA (TE) buffer and stored at -20°C until further use.
Molecular Detection and Identification of Bartonella Species
For screening, conventional PCR was performed on all tick and blood samples (n = 772) targeting the Intergenic Transcribed Spacer (ITS) locus, using the following forward and reverse primers: (321s: 5′AGATGATGATCCCAAGCCTTCTGG and H493as: 5′-TGAACCTCCGACCTCACGCTTATC) as previously described (Maggi and Breitschwerdt, 2005; Gutierrez et al., 2014). PCR reactions were performed in 25-μl Syntezza PCR ready mix (Syntezza, Jerusalem), containing 1 μM of each set of primers and 5 μl of the extracted DNA. The thermal cycling procedure was as described previously (Norman et al., 1995; Renesto et al., 2001; Maggi and Breitschwerdt, 2005). Samples of PCR grade water were included as a negative (no-DNA) controls. To confirm amplicon identity, all PCR products from the positive samples underwent DNA sequencing; the nucleotide sequences were compared to those present in GenBank database using the Basic Alignment Search Tool (BLAST).1 Statistical analysis was done using the SPSS program v20.
Results
Bartonella DNA in Animal Blood
Among the canine blood samples (n = 110; 22 female: 88 males), one female dog sample was positive for Bartonella, yielding a PCR-amplified ITS sequence that showed 99% sequence identity to the reference sequence of B. rochalimae (FN645466.1). None of the blood samples from camels (n = 19), sheep (n = 7), and goats (n = 3) were positive for Bartonella.
Bartonella DNA in Ticks
A total of 633 hard ticks (292 female, 286 male, and 55 Nymph ticks) were obtained from 188 animals (137 dogs, 38 sheep, 10 camels, and three goats), residing in nine districts throughout Palestine. The geographic distribution of collected ticks was shown in Figure 1. All ticks representing three genera and seven species [Haemaphysalis parva (n = 43), Haemaphysalis adleri (n = 13), Rhipicephalus turanicus (n = 91), Rhipicephalus sanguineus (n = 391), Rhipicephalus bursa (n = 7), Rhipicephalus sp.(n = 20), Hyalomma dromedarii (n = 63), and Hyalomma impeltatum (n = 5)] were screened for Bartonella DNA.
Overall, 25 ticks (3.9%; 9 females, 11 males, and 5 nymphs) were positive for Bartonella DNA by ITS-PCR. Of these 21 ticks were collected from dogs and four from camels. None of the ticks from sheep or goats were positive for Bartonella (Table 1). Identification of Bartonella was successful for 21 positive samples (84%). The initial attempt to sequence four amplicons failed and no additional DNA samples were available. Seventeen R. sanguineus ticks were infected with B. rochalimae (n = 10), B. chomelii (n = 6), and B. koehlerae (n = 1). All these ticks were obtained from dogs.
Table 1.
The overall prevalence of Bartonella DNA in ticks species and their associated animal hosts.
| Ectoparasites | No. | pos. PCR | Infection rate | Animal host |
|---|---|---|---|---|
| Rhipicephalus turanicus | 91 | 0 | 0 | Dog, sheep |
| Rhipicephalus sanguineus | 391 | 20 | 5.1 | Dog, sheep, Goat, |
| Rhipicephalus bursa | 7 | 0 | 0 | Sheep |
| Rhipicephalus spp. | 20 | 1 | 5 | Dog |
| Haemaphysalis adleri | 13 | 0 | 0 | Dog, sheep |
| Haemaphysalis parva | 43 | 0 | 0 | Dog, sheep |
| Hyalomma dromedarii | 63 | 4 | 6.3 | Camel |
| Hyalomma impeltatum | 5 | 0 | 0 | Camel |
| Total ticks | 633 | 25 | 3.9 |
Four H. dromedarii ticks obtained from a single camel in Hebron and from two camels in Jericho were found to be infected with B. bovis (n = 2) and B. rochalimae (n = 2), respectively (Table 2). Bartonella species identification was based on the highest scoring BLAST hit on GenBank. The comparison of the PCR-amplified ITS sequences from the positive tick samples showed 97–99% sequence identity and 100% coverage when aligned against the reference sequences of B. bovis (KR733201.1), B. chomelii (KM215714.1), B. rochalimae (FN645466.1), and B. koehlerae (AF312490.1). Representative partial sequences of the 16S–23S ribosomal RNA intergenic spacer identified in the present study were deposited in GenBank under the following accession numbers: two B. rochalimae from a tick and its host (dog) (KX420619, KX420620), B. chomelii (KX420617), B. bovis (KX420618), and B. koehlerae (KX420616).
Table 2.
Molecular identification of Bartonella spp. collected from Ixodid ticks from different localities throughout Palestine.
| Bartonella spp. | Ectoparasites (n) | Site of collection |
|---|---|---|
| B. chomelii | Rhipicephalus sanguineus (6) | Hebron, Tubas |
| B. rochalimae | Rhipicephalus sanguineus (10), Hyalomma dromedarii (2) | Nablus, Jenin, Tubas, Ramallah, Jericho |
| B. bovis | Hyalomma dromedarii (2) | Hebron |
| B. koehlerae | Rhipicephalus sanguineus (1) | Jenin |
Discussion
Here, we report Bartonella DNA in ixodid ticks and blood samples from domestic animals from Palestine. The overall prevalence of Bartonella DNA in ticks (3.9%) was in agreement with previous screening undertaken worldwide including Czech Republic, United States, Italy and Thailand (Sanogo et al., 2003; Hercik et al., 2007; Billeter et al., 2008, 2012). Several Bartonella species have been identified in humans, animals, and their flea vectors in neighboring countries. However, Bartonella DNA was not detected in any of the Ixodid ticks examined in Israel and Egypt (Loftis et al., 2006; Harrus et al., 2011).
In our study, four Bartonella species were identified: B. rochalimae, B. chomelii, B. Bovis, and B. koehlerae. B. rochalimae was the predominant species among Bartonella-positive ticks (12 out of 25 samples; 48%). We examined seven hard tick species for Bartonella and found evidence of the pathogen in two of them: the brown dog tick, Rhipicephalus sanguineus and the camel tick, Hyalomma dromedarii. These findings represent the first detection of Bartonella in Ixodid ticks from the Middle East. Cases of human parasitism by the brown dog tick—the most widespread tick in the world—are well documented (Fernandez-Soto et al., 2006). Together, the exposure of animals to arthropod vectors and the proximity of infected animals to humans make some Bartonella species potential zoonotic agents.
People dealing with dogs (e.g., veterinarians, shepherds, dog owners and pet shop workers) appear to be at particular risk of exposure to R. sanguineus and its pathogens. We found Bartonella DNA in a single sample of dog blood, with high sequence identity to a reference sequence from B. rochalimae (99% DNA sequence identity; GenBank accession number FN645466.1). The dog that provided the sample was apparently healthy, showing no signs of bartonellosis. Detection of Bartonella in asymptomatic dogs has also been reported from Peru (Diniz et al., 2009). We identified B. rochalimae in two R. sanguineus ticks obtained from the same dog; both tick samples yielded sequences 100% identical to each other and to the sequence obtained from dog. Although the blood and tick samples were collected from dogs at the same time, Bartonella DNA was more prevalent in the dog ticks (5.4%) than in the dog blood (0.9%) suggesting that Bartonella be carried in partially engorged adult ticks. These findings provide highly suggestive molecular evidence that R. sanguineus ticks can act as vectors of animal-associated Bartonella infection in Palestine. Furthermore, although the role of dogs as source of human Bartonella infection remains unclear, we speculate that they may present a risk for zoonotic transmission similar to that seen with B. henselae in cat scratch disease (Chomel et al., 2006).
Recently, a study conducted in Israel confirmed the presence of a novel species of Bartonella in camelids, which has been named Bartonella dromedarii sp. nov (Rasis et al., 2014). In our study, we identified two species of Bartonella (B. rochalimae and B. bovis) in ticks obtained from camels, although we did not detect Bartonella DNA in blood samples from these camels. However, we cannot rule out bloodstream infections in the camels as the source of Bartonella in the ticks, as the blood samples were not taken at the same time as the ticks. Furthermore, even if camels are not reservoir hosts for these Bartonella species, they may have an important role as mechanical dispersers of infected ticks.
Our discovery of Bartonella in camel ticks from the species Hyalomma dromedarii is worrying because other species of hard ticks from the genus Hyalomma clearly bite humans (Psaroulaki et al., 2005; Bursali et al., 2011) and there are suggestions that H. dromedarii can do so too2. Other lines of evidence support the role of ticks in the natural cycles of some Bartonella species including those pathogenic for humans (Cotte et al., 2008; Reis et al., 2011; Liu and Bonnet, 2014). In particular, ticks can be infected in the larval or nymph stages by ingesting blood from an intermediate host carrying Bartonella. The pathogen can then survive in the midgut of ticks during molting and can be transmitted through feeding to an uninfected host.
In the present study, brown dog ticks were found to be infected with B. chomelii—a pathogen first isolated from French domestic cattle (Maillard et al., 2004) and found to be the most frequent Bartonella species infecting cattle grazing in Spain pastures (Antequera-Gomez et al., 2015). We also describe the first detection of B. koehlerae in a R. sanguineus tick obtained from dog. This species was first isolated from the blood of two pet cats in California (Droz et al., 1999). Since then, it has been detected in cats and their fleas in France, Israel and Palestine (Rolain et al., 2003; Gutierrez et al., 2013; Nasereddin et al., 2014), has been reported as causing endocarditis in humans and Boxer dogs in Israel (Avidor et al., 2004; Ohad et al., 2010). However, the presence of a microbial agent within a tick does not imply that the tick is a biological vector and might transmit it during the course of blood feeding (Telford and Wormser, 2010).
One of Bartonella -positive ticks we obtained from a dog harbored Rickettsia, a pathogen that is known to be tick-transmitted (Ereqat et al., 2016). This fits in with evidence from across the world that Bartonella is often found in ticks alongside well-known tick-transmitted organisms such as Anaplasma, Borrelia, and Rickettsia (Angelakis et al., 2010). Other studies supporting the hypothesis that Bartonella can be transmitted by ticks include a US report that dogs infected with Bartonella were also seropositive for Anaplasma phagocytophilum (MacDonald et al., 2004) and a human case study showing that patients infected with Borrelia burgdorferi after tick bites also carried Bartonella DNA in their blood (Podsiadly et al., 2003).
Conclusion
The detection of zoonotic Bartonella species in this study should increase the awareness of these vector-borne diseases among physicians, veterinarians and public health workers and highlight the importance of surveillance and preventive measures in the region. Additional epidemiologic surveys are required to enhance our understanding of the transmission dynamics of Bartonella in Palestine and in other parts of the Middle East.
Author Contributions
Conceived and designed the experiments: SE, AN, and ZA. Performed the experiments: SE, AA, and TZ. Analyzed the data: SE and AA-J. Wrote the first draft of the manuscript: SE. Directed, revised, and contributed to the writing of the manuscript: AN and V-TM. Final revision and approval of the manuscript to be published: SE, AN, V-TM, AA, AA-J, TZ, KA, and ZA.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by The Ministry of Foreign Affairs, The Hague, The Netherlands, through a grant to the Dutch (NVHU). The authors would like to thank the COST action TD1303 (EurNegVec) for their support. We thank Professor Mark Pallen for proof-reading and revising our manuscript.
Footnotes
References
- Angelakis E., Billeter S. A., Breitschwerdt E. B., Chomel B. B., Raoult D. (2010). Potential for tick-borne bartonelloses. Emerg. Infect. Dis 16 385–391. 10.3201/eid1603.081685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera-Gomez M. L., Lozano-Almendral L., Barandika J. F., Gonzalez-Martin-Nino R. M., Rodriguez-Moreno I., Garcia-Perez A. L., et al. (2015). Bartonella chomelii is the most frequent species infecting cattle grazing in communal mountain pastures in Spain. Appl. Environ. Microbiol. 81 623–629. 10.1128/AEM.03159-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor B., Graidy M., Efrat G., Leibowitz C., Shapira G., Schattner A., et al. (2004). Bartonella koehlerae, a new cat-associated agent of culture-negative human endocarditis. J. Clin. Microbiol. 42 3462–3468. 10.1128/JCM.42.8.3462-3468.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi K., Ereqat S., Nasereddin A., Al-Jawabreh A., Baneth G., Abdeen Z. (2016). Molecular detection of Theileria, Babesia, and Hepatozoon spp. in ixodid ticks from Palestine. Ticks Tick Borne Dis. 7 734–741. 10.1016/j.ttbdis.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Bai Y., Malania L., Alvarez Castillo D., Moran D., Boonmar S., Chanlun A., et al. (2013). Global distribution of Bartonella infections in domestic bovine and characterization of Bartonella bovis strains using multi-locus sequence typing. PLoS ONE 8:e80894 10.1371/journal.pone.0080894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter S. A., Miller M. K., Breitschwerdt E. B., Levy M. G. (2008). Detection of two Bartonella tamiae-like sequences in Amblyomma americanum (Acari: Ixodidae) using 16S-23S intergenic spacer region-specific primers. J. Med. Entomol. 45 176–179. 10.1603/0022-2585(2008)45[176:DOTBTS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Billeter S. A., Sangmaneedet S., Kosakewich R. C., Kosoy M. Y. (2012). Bartonella species in dogs and their ectoparasites from Khon Kaen Province, Thailand. Southeast Asian J. Trop. Med. Public Health 43 1186–1192. [PubMed] [Google Scholar]
- Breitschwerdt E. B., Kordick D. L. (2000). Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin. Microbiol. Rev. 13 428–438. 10.1128/CMR.13.3.428-438.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursali A., Tekin S., Keskin A., Ekici M., Dundar E. (2011). Species diversity of ixodid ticks feeding on humans in Amasya, Turkey: seasonal abundance and presence of Crimean-Congo hemorrhagic fever virus. J. Med. Entomol. 48 85–93. 10.1603/ME10034 [DOI] [PubMed] [Google Scholar]
- Chang C. C., Kasten R. W., Chomel B. B., Simpson D. C., Hew C. M., Kordick D. L., et al. (2000). Coyotes (Canis latrans) as the reservoir for a human pathogenic Bartonella sp.: molecular epidemiology of Bartonella vinsonii subsp. berkhoffii infection in coyotes from central coastal California. J. Clin. Microbiol. 38 4193–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry N. A., Maggi R. G., Cannedy A. L., Breitschwerdt E. B. (2009). Pcr detection of Bartonella bovis and Bartonella henselae in the blood of beef cattle. Vet. Microbiol. 135 308–312. 10.1016/j.vetmic.2008.09.063 [DOI] [PubMed] [Google Scholar]
- Chomel B. B., Boulouis H. J., Breitschwerdt E. B., Kasten R. W., Vayssier-Taussat M., Birtles R. J., et al. (2009). Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet. Res. 40:29 10.1051/vetres/2009011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomel B. B., Boulouis H. J., Maruyama S., Breitschwerdt E. B. (2006). Bartonella spp. in pets and effect on human health. Emerg. Infect. Dis. 12 389–394. 10.3201/eid1203.050931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotte V., Bonnet S., Le Rhun D., Le Naour E., Chauvin A., Boulouis H. J., et al. (2008). Transmission of Bartonella henselae by Ixodes ricinus. Emerg. Infect. Dis. 14 1074–1080. 10.3201/eid1407.071110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz P. P., Billeter S. A., Otranto D., De Caprariis D., Petanides T., Mylonakis M. E., et al. (2009). Molecular documentation of Bartonella infection in dogs in Greece and Italy. J. Clin. Microbiol. 47 1565–1567. 10.1128/JCM.00082-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droz S., Chi B., Horn E., Steigerwalt A. G., Whitney A. M., Brenner D. J. (1999). Bartonella koehlerae sp. nov., isolated from cats. J. Clin. Microbiol. 37 1117–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremeeva M. E., Gerns H. L., Lydy S. L., Goo J. S., Ryan E. T., Mathew S. S., et al. (2007). Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. New Engl. J. Med. 356 2381–2387. 10.1056/NEJMoa065987 [DOI] [PubMed] [Google Scholar]
- Ereqat S., Nasereddin A., Al-Jawabreh A., Azmi K., Harrus S., Mumcuoglu K., et al. (2016). Molecular Detection and Identification of Spotted Fever Group Rickettsiae in Ticks Collected from the West Bank, Palestinian Territories. PLoS Negl. Trop. Dis. 10: e0004348 10.1371/journal.pntd.0004348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erol E., Jackson C., Bai Y., Sells S., Locke S., Kosoy M. (2013). Bartonella bovis isolated from a cow with endocarditis. J. Vet. Diagn. Invest. 25 288–290. 10.1177/1040638713477408 [DOI] [PubMed] [Google Scholar]
- Feldman-Musham B. (1954). Revision of the genus Hyalomma. Bull. Res. Counc. Isr. 64 70–150. [Google Scholar]
- Fernandez-Soto P., Perez-Sanchez R., Alamo-Sanz R., Encinas-Grandes A. (2006). Spotted fever group rickettsiae in ticks feeding on humans in northwestern Spain: is Rickettsia conorii vanishing? Ann. N. Y. Acad. Sci. 1078 331–333. 10.1196/annals.1374.063 [DOI] [PubMed] [Google Scholar]
- Gutierrez R., Cohen L., Morick D., Mumcuoglu K. Y., Harrus S., Gottlieb Y. (2014). Identification of different Bartonella species in the cattle tail louse (Haematopinus quadripertusus) and in cattle blood. Appl. Environ. Microbiol. 80 5477–5483. 10.1128/AEM.01409-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R., Morick D., Gross I., Winkler R., Abdeen Z., Harrus S. (2013). Bartonellae in domestic and stray cats from Israel: comparison of bacterial cultures and high-resolution melt real-time Pcr as diagnostic methods. Vector Borne Zoonotic Dis. 13 857–864. 10.1089/vbz.2013.1308 [DOI] [PubMed] [Google Scholar]
- Harms A., Dehio C. (2012). Intruders below the radar: molecular pathogenesis of Bartonella spp. Clin. Microbiol. Rev. 25 42–78. 10.1128/CMR.05009-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrus S., Perlman-Avrahami A., Mumcuoglu K. Y., Morick D., Eyal O., Baneth G. (2011). Molecular detection of Ehrlichia canis, Anaplasma bovis, Anaplasma platys, Candidatus Midichloria mitochondrii and Babesia canis vogeli in ticks from Israel. Clin. Microbiol. Infect. 17 459–463. 10.1111/j.1469-0691.2010.03316.x [DOI] [PubMed] [Google Scholar]
- Hercik K., Hasova V., Janecek J., Branny P. (2007). Molecular evidence of Bartonella Dna in ixodid ticks in Czechia. Folia Microbiol. (Praha) 52 503–509. 10.1007/BF02932111 [DOI] [PubMed] [Google Scholar]
- Hofmeister E. K., Kolbert C. P., Abdulkarim A. S., Magera J. M., Hopkins M. K., Uhl J. R., et al. (1998). Cosegregation of a novel Bartonella species with Borrelia burgdorferi and Babesia microti in Peromyscus leucopus. J. Infect. Dis. 177 409–416. 10.1086/514201 [DOI] [PubMed] [Google Scholar]
- Kho K. L., Koh F. X., Jaafar T., Nizam Q. N., Tay S. T. (2015). Prevalence and molecular heterogeneity of Bartonella bovis in cattle and Haemaphysalis bispinosa ticks in Peninsular Malaysia. BMC Vet. Res. 11:153 10.1186/s12917-015-0470-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Y., Bonnet S. I. (2014). Hard tick factors implicated in pathogen transmission. PLoS Negl. Trop. Dis. 8:e2566 10.1371/journal.pntd.0002566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftis A. D., Reeves W. K., Szumlas D. E., Abbassy M. M., Helmy I. M., Moriarity J. R., et al. (2006). Rickettsial agents in Egyptian ticks collected from domestic animals. Exp. Appl. Acarol. 40 67–81. 10.1007/s10493-006-9025-2 [DOI] [PubMed] [Google Scholar]
- MacDonald K. A., Chomel B. B., Kittleson M. D., Kasten R. W., Thomas W. P., Pesavento P. (2004). A prospective study of canine infective endocarditis in northern California (1999-2001): emergence of Bartonella as a prevalent etiologic agent. J. Vet. Intern. Med. 18 56–64. [DOI] [PubMed] [Google Scholar]
- Maggi R. G., Breitschwerdt E. B. (2005). Potential limitations of the 16S-23S rrna intergenic region for molecular detection of Bartonella species. J. Clin. Microbiol. 43 1171–1176. 10.1128/JCM.43.3.1171-1176.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard R., Riegel P., Barrat F., Bouillin C., Thibault D., Gandoin C., et al. (2004). Bartonella chomelii sp. nov., isolated from French domestic cattle (Bos taurus). Int. J. Syst. Evol. Microbiol. 54 215–220. 10.1099/ijs.0.02770-0 [DOI] [PubMed] [Google Scholar]
- Martini M., Menandro M. L., Mondin A., Pasotto D., Mazzariol S., Lauzi S., et al. (2008). Detection of Bartonella bovis in a cattle herd in Italy. Vet. Rec. 162 58–59. 10.1136/vr.162.2.58 [DOI] [PubMed] [Google Scholar]
- Morozova O. V., Chernousova N., Morozov I. V. (2005). Detection of the Bartonella Dna by the method of nested Pcr in patients after tick bites in Novosibirsk region. Mol. Gen. Mikrobiol. Virusol. 4 14–17. [PubMed] [Google Scholar]
- Nasereddin A., Risheq A., Harrus S., Azmi K., Ereqat S., Baneth G., et al. (2014). Bartonella species in fleas from Palestinian territories: prevalence and genetic diversity. J. Vector Ecol. 39 261–270. 10.1111/jvec.12100 [DOI] [PubMed] [Google Scholar]
- Norman A. F., Regnery R., Jameson P., Greene C., Krause D. C. (1995). Differentiation of Bartonella-like isolates at the species level by Pcr-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 33 1797–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad D. G., Morick D., Avidor B., Harrus S. (2010). Molecular detection of Bartonella henselae and Bartonella koehlerae from aortic valves of Boxer dogs with infective endocarditis. Vet. Microbiol. 141 182–185. 10.1016/j.vetmic.2009.08.005 [DOI] [PubMed] [Google Scholar]
- Podsiadly E., Chmielewski T., Tylewska-Wierzbanowska S. (2003). Bartonella henselae and Borrelia burgdorferi infections of the central nervous system. Ann. N. Y. Acad. Sci. 990 404–406. 10.1111/j.1749-6632.2003.tb07400.x [DOI] [PubMed] [Google Scholar]
- Psaroulaki A., Germanakis A., Gikas A., Scoulica E., Tselentis Y. (2005). Simultaneous detection of “Rickettsia mongolotimonae” in a patient and in a tick in Greece. J. Clin. Microbiol. 43 3558–3559. 10.1128/JCM.43.7.3558-3559.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasis M., Rudoler N., Schwartz D., Giladi M. (2014). Bartonella dromedarii sp. nov. isolated from domesticated camels (Camelus dromedarius) in Israel. Vector Borne Zoonotic Dis. 14 775–782. 10.1089/vbz.2014.1663 [DOI] [PubMed] [Google Scholar]
- Reis C., Cote M., Le Rhun D., Lecuelle B., Levin M. L., Vayssier-Taussat M., et al. (2011). Vector competence of the tick Ixodes ricinus for transmission of Bartonella birtlesii. PLoS Negl. Trop. Dis. 5:e1186 10.1371/journal.pntd.0001186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renesto P., Gouvernet J., Drancourt M., Roux V., Raoult D. (2001). Use of rpoB gene analysis for detection and identification of Bartonella species. J. Clin. Microbiol. 39 430–437. 10.1128/JCM.39.2.430-437.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolain J. M., Franc M., Davoust B., Raoult D. (2003). Molecular detection of Bartonella quintana, B. koehlerae, B. henselae, B. clarridgeiae, Rickettsia felis, and Wolbachia pipientis in cat fleas, France. Emerg. Infect. Dis. 9 338–342. 10.3201/eid0903.020278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudoler N., Rasis M., Sharir B., Novikov A., Shapira G., Giladi M. (2014). First description of Bartonella bovis in cattle herds in Israel. Vet. Microbiol. 173 110–117. 10.1016/j.vetmic.2014.07.006 [DOI] [PubMed] [Google Scholar]
- Saisongkorh W., Barrassi L., Davoust B., De Broucker C. A., Raoult D., Rolain J. M. (2009). First isolation of Bartonella bovis from animals in French Guyana, South America. Clin. Microbiol. Infect. 15(Suppl. 2) 124–126. 10.1111/j.1469-0691.2008.02198.x [DOI] [PubMed] [Google Scholar]
- Sanogo Y. O., Zeaiter Z., Caruso G., Merola F., Shpynov S., Brouqui P., et al. (2003). Bartonella henselae in Ixodes ricinus ticks (Acari: Ixodida) removed from humans, Belluno province, Italy. Emerg. Infect. Dis. 9 329–332. 10.3201/eid0903.020133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer J. D., Moore G. M., Namekata M. S., Kasten R. W., Chomel B. B. (2012). Seroepidemiology of Bartonella infection in gray foxes from Texas. Vector Borne Zoonotic Dis. 12 428–430. 10.1089/vbz.2011.0805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparagano O. A., Allsopp M. T., Mank R. A., Rijpkema S. G., Figueroa J. V., Jongejan F. (1999). Molecular detection of pathogen Dna in ticks (Acari: Ixodidae): a review. Exp. Appl. Acarol. 23 929–960. 10.1023/A:1006313803979 [DOI] [PubMed] [Google Scholar]
- Telford S. R., III, Wormser G. P. (2010). Bartonella spp. transmission by ticks not established. Emerg. Infect. Dis. 16 379–384. 10.3201/eid1603.090443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Chomel B. B., Chang C. C., Kass P. H., Conrad P. A., Chuang S. T. (2011). Bartonella and Babesia infections in cattle and their ticks in Taiwan. Comp. Immunol. Microbiol. Infect. Dis. 34 179–187. 10.1016/j.cimid.2010.11.003 [DOI] [PubMed] [Google Scholar]
- Vayssier-Taussat M., Le Rhun D., Deng H. K., Biville F., Cescau S., Danchin A., et al. (2010). The Trw type Iv secretion system of Bartonella mediates host-specific adhesion to erythrocytes. PLoS Pathog. 6:e1000946 10.1371/journal.ppat.1000946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayssier-Taussat M., Moutailler S., Femenia F., Raymond P., Croce O., La Scola B., et al. (2016). Identification of novel zoonotic activity of Bartonella spp., France. Emerg. Infect. Dis. 22 457–462. 10.3201/eid2203.150269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayssier-Taussat M., Moutailler S., Michelet L., Devillers E., Bonnet S., Cheval J., et al. (2013). Next generation sequencing uncovers unexpected bacterial pathogens in ticks in western Europe. PLoS ONE 8:e81439 10.1371/journal.pone.0081439 [DOI] [PMC free article] [PubMed] [Google Scholar]


