Abstract
Trehalose and its metabolism have been demonstrated to play important roles in control of plant growth, development, and stress responses. However, direct genetic evidence supporting the functions of trehalose and its metabolism in defense response against pathogens is lacking. In the present study, genome-wide characterization of putative trehalose-related genes identified 11 SlTPSs for trehalose-6-phosphate synthase, 8 SlTPPs for trehalose-6-phosphate phosphatase and one SlTRE1 for trehalase in tomato genome. Nine SlTPSs, 4 SlTPPs, and SlTRE1 were selected for functional analyses to explore their involvement in tomato disease resistance. Some selected SlTPSs, SlTPPs, and SlTRE1 responded with distinct expression induction patterns to Botrytis cinerea and Pseudomonas syringae pv. tomato (Pst) DC3000 as well as to defense signaling hormones (e.g., salicylic acid, jasmonic acid, and a precursor of ethylene). Virus-induced gene silencing-mediated silencing of SlTPS3, SlTPS4, or SlTPS7 led to deregulation of ROS accumulation and attenuated the expression of defense-related genes upon pathogen infection and thus deteriorated the resistance against B. cinerea or Pst DC3000. By contrast, silencing of SlTPS5 or SlTPP2 led to an increased expression of the defense-related genes upon pathogen infection and conferred an increased resistance against Pst DC3000. Silencing of SlTPS3, SlTPS4, SlTPS5, SlTPS7, or SlTPP2 affected trehalose level in tomato plants with or without infection of B. cinerea or Pst DC3000. These results demonstrate that SlTPS3, SlTPS4, SlTPS5, SlTPS7, and SlTPP2 play roles in resistance against B. cinerea and Pst DC3000, implying the importance of trehalose and tis metabolism in regulation of defense response against pathogens in tomato.
Keywords: Trehalose, trehalose-6-phosphate synthase, trehalose-6-phosphate phosphatase (TPP), Botrytis cinerea, Pseudomonas syringae pv. tomato DC3000, disease resistance, defense response
Introduction
Trehalose (α-D-glucopyranosyl α-D-glucopyranoside) is a ubiquitously distributed non-reducing disaccharide (Elbein et al., 2003). The biosynthesis and degradation of trehalose in plants include three consecutive enzymatic steps. Firstly, trehalose-6-phosphate synthase (TPS) catalyzes the synthesis of trehalose-6-phosphate (T6P), which is subsequently dephosphorylated into trehalose by T-6-phosphate phosphatase (TPP). Furthermore, the synthesized trehalose can be hydrolyzed into two glucose monomers by the enzyme trehalase (TRE) (Schluepmann and Paulb, 2009). Biochemically, trehalose has been shown to be capable of stabilizing proteins and lipid membranes in cells and the trehalose metabolism is essentially required for some general metabolic pathways such as sugar status, carbon assimilation, biosynthesis, and degradation of starch in plants (Goddijn and van Dun, 1999; Paul et al., 2008; Lunn et al., 2014).
The TPSs and TPPs constitute two multi-gene families while the TRE is present as a single-copy gene in most of sequenced plant genomes (Lunn, 2007). For example, Arabidopsis contains 11 TPS genes (AtTPS1–AtTPS11) and 10 TPP genes (AtTPPA–AtTPPJ) (Leyman et al., 2001; Vandesteene et al., 2012) while rice has 11 TPS (OsTPS1–OsTPS11) and 11 TPP (OsTPP1–OsTPP11) (Ge et al., 2008; Zhang et al., 2011). Similar numbers of TPS and/or TPP genes were identified in wheat (Xie et al., 2015), maize (Henry et al., 2014; Zhou et al., 2014), poplar (Yang et al., 2012), and cotton (Mu et al., 2016). Plant TPSs can be divided into two groups with differences in structural features and biochemical activity. Group I TPSs contain both TPS and TPP domains and the Arabidopsis AtTPS1, AtTPS2, and AtTPS4 are active enzymes (Blazquez et al., 1998; Vandesteene et al., 2010; Delorge et al., 2015). Group II TPSs contain both TPS and TPP domains and most of them harbor conserved phosphatase domains (Vandesteene et al., 2010; Zhang et al., 2011). Whereas most of the Arabidopsis Class II TPSs are not active enzymes (Ramon et al., 2009), AtTPS6 and AtTPS11 were found to possess TPS or TPP activity (Chary et al., 2008; Singh et al., 2011). In addition, it was shown that the OsTPSs can form TPS complexes, which may potentially regulate T6P levels in plants (Zhang et al., 2011). By contrast, plant TPPs contain unique TPP domains with conserved phosphatase domains and all of them possess TPP activities (Shima et al., 2007).
Extensive genetic studies using loss-of-function and gain-of-function mutants have demonstrated that the trehalose metabolism plays critical roles in control of plant growth and development including embryo development, leaf morphology and senescence, and flowering (Satoh-Nagasawa et al., 2006; Gómez et al., 2010; Wingler et al., 2012; Nunes et al., 2013; Wahl et al., 2013) (for reviews, see Ramon and Rolland, 2007; Paul et al., 2008; Ponnu et al., 2011; Lunn et al., 2014; Tsai and Gazzarrini, 2014). Increasing evidence also supports that trehalose and its metabolism function in plant response to a number of unfavorable environmental conditions such as extreme temperatures, drought, salt and oxidative stresses (Iordachescu and Imai, 2008; Fernandez et al., 2012; Delorge et al., 2014; Lunn et al., 2014; Figueroa et al., 2016). For example, mutations in Arabidopsis AtTPS5 and AtTPPD impaired the tolerance to extreme temperatures and salt stress, respectively (Suzuki et al., 2008; Krasensky et al., 2014; Wang et al., 2016). By contrast, overexpression of AtTRE1 in Arabidopsis, OsTPS1 and OsTPP1 in rice, and heterologous TPS and TPP genes in transgenic plants confer improved abiotic stress tolerance (Garg et al., 2002; Jang et al., 2003; Pramanik and Imai, 2005; Karim et al., 2007; Miranda et al., 2007; Ge et al., 2008; Debast et al., 2011; Li et al., 2011; Van Houtte et al., 2013a). Thus, modulation of the endogenous trehalose metabolism is a promising strategy to improve stress tolerance in crop plants (Lunn et al., 2014).
There is also emerging evidence indicating that trehalose and its metabolism are involved in plant responses to biotic factors such as pathogenic microorganisms and herbivorous insects (Lunn et al., 2014). It was shown that exogenous trehalose acts as an elicitor of plant defense response (Bae et al., 2005) and can induce resistance in wheat plants against powdery mildew disease (Reignault et al., 2001; Renard-Merlier et al., 2007; Tayeh et al., 2014). Treatment with an inhibitor of trehalase, validamycin A, induced resistance to Fusarium wilt and late blight diseases, although exogenous trehalose did not confer resistance to powdery mildew disease (Ishikawa et al., 2005). Furthermore, expression of AtTPS11 and AtTRE in Arabidopsis plants was induced by infection with Tobacco mosaic virus (Golem and Culver, 2003) or Plasmodiophora brassica (Brodmann et al., 2002). Excess levels of trehalose accumulated in Arabidopsis roots after infection with a pathogenic nematode (Hofmann et al., 2010) or in citrus leaves infected with Xanthomonas citri subsp. citri (Piazza et al., 2015). Most recently, it was found that a Ralstonia solanacearum type III effector, ripTPS, is a functional TPS enzyme that elicits a hypersensitive response on tobacco (Poueymiro et al., 2014). However, genetic evidence originated from disease phenotype analysis of loss-of-function or gain-of-function mutants or transgenic lines is lacking to support the function of trehalose metabolism in pathogen resistance in plants. On the other hand, exogenous trehalose can also serve as a potential sign of dangers from infestation of herbivorous insects. For example, infestation of Arabidopsis and tomato plants by peach potato aphid led to accumulation of trehalose (Singh and Shah, 2012; Hodge et al., 2013) and mutation in Arabidopsis AtTPS11 impaired both the trehalose accumulation and resistance against aphids, suggesting that treahlose is an essential signal in the defense process (Singh et al., 2011).
The present study was aimed to explore the involvement of the trehalose metabolism in disease resistance against Botrytis cinerea, a necrotrophic fungal pathogen, and Pseudomonas syringae pv. tomato DC3000, a (hemi)biotrphic bacterial pathogen, in tomato. We identified 11 SlTPS, 8 SlTPP, and one SlTRE genes in tomato genome. Virus-induced gene silencing (VIGS)-based functional analyses revealed that VIGS-mediated silencing of SlTPS3, SlTPS4, or SlTPS7 deteriorated the resistance against B. cinerea and Pst DC3000, whereas silencing of SlTPS5 or SlTPP2 conferred an increased resistance against Pst DC3000. These findings demonstrate the importance of trehalose and its metabolic genes in regulation of defense response against pathogens in tomato.
Materials and Methods
Plant Growth and Treatments
Tomato (Solanum lycopersicum) cv. Suhong2003 was used for most of the experiments except that cultivar MicroTom was used in whole plant inoculation assays with B. cinerea. Growth of tomato plants and treatment with hormones were the same as previously described (Li et al., 2014b). Leaf samples were harvested at specific time points and stored at -80°C until use.
Characterization of SlTPS, SlTPP, and SlTRE1 Genes
Tomato genome database at the SOL Genomics Network1 (SGN) was searched using BlastP program with Arabidopsis AtTPSs, AtTPPs, and AtTRE1 as queries and the predicted nucleotide and amino acid sequences for SlTPSs, SlTPPs, and SlTRE1 were downloaded. Conserved TPS and TPP domains in the predicted SlTPS and SlTPP proteins were analyzed using the Conserved Domain Search program at NCBI website2 under default parameters and the Motif Scan program at MyHits website3 with the following parameters (hamap, pfam_fs, and pfam_Is). Putative ESTs or UniGenes and full-length cDNAs were searched against the tomato genome database and NCBI GenBank database, respectively, using predicted nucleotide sequences as queries. Phylogenetic trees for tomato, Arabidopsis and rice TPSs and TPPs were constructed using the neighbor-joining method of the MEGA6 program with the p-distance and complete deletion option parameters using a bootstrapping method with 1000 replicates.
VIGS Vector Construction and Agroinfiltration
Fragments of 300–400 bp, spanning partial 5′-UTR and coding sequences (Supplementary file 1), for selected SlTPSs, SlTPPs, and SlTRE1 were amplified by PCR with respective pairs of gene-specific primers (Supplementary Table S1). The amplified PCR products were digested with corresponding restriction enzymes (XbaI/XhoI or EcoRI/BamHI) and cloned into TRV2, yielding recombinant plasmids TRV-SlTPSs, TRV-SlTPPs, and TRV-SlTRE1. After confirmation by sequencing, the correct recombinant plasmids were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation and positive clones were selected by colony PCR. Cultivation of agrobacteria carrying different constructs of TRV-SlTPSs, TRV-SlTPPs and TRV-SlTRE1 and agroinfiltration for standard VIGS were carried out as described before (Li et al., 2014b). In all VIGS assays, a construct of TRV-PDS (Phytoene desaturase) was included as positive controls for silencing evaluation of the VIGS procedure (Liu et al., 2002).
Pathogen Inoculation and Disease Assays
Inoculation of tomato plants with B. cinerea was carried out using two different methods as described previously (Li et al., 2014b). Spore concentration in the inoculum was adjusted to 1 × 105 spores/mL. In detached leaf disease assays, leaves were collected from the second and third branches of 4-week-old plants and placed on wet cheesecloth in trays. After inoculation by dropping 5 μL of spore suspension on the surface of the detached leaves, the trays were covered with transparent plastic films to maintain high humidity. Lesion sizes were measured 4 days later. In whole plant disease assays, spore suspension was sprayed evenly on leaf surface of 4-week-old plants, which were then kept in high humidity in the growth room. Photos were taken at 4 days after inoculation. The inoculated leaves were harvested for gene expression and the determination of in planta fungal growth (Li et al., 2014b).
Plant inoculation with Pst DC3000 was carried out following previously described method (Li et al., 2014b). Briefly, plants were submerged into bacterial suspension (OD600 = 0.0002 in 10 mM MgCl2 with 0.04% Silwet l–77) and vacuum infiltrated under a -40 Kpa pressure for 1.5 min using a vacuum pump. The inoculated plants were kept in the growth room for growth with high humidity. Measurement of in planta bacterial growth was done as before (Li et al., 2014b).
RNA Extraction and qRT-PCR
Frozen leaf samples were homogenized in liquid nitrogen using a mortar and pestle. Total RNA was extracted using Trizol reagent (Invitrogen, Shanghai, China). First-strand cDNAs were synthesized using PrimeScript RT regent kit (TaKaRa, Dalian, China) and used for amplification of VIGS fragments and qRT-PCR analyses of gene expression. qRT-PCR was done with SYBR Premix Ex Taq (TaKaRa, Dalian, China) on a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA) and the conditions consisted of 40 cycles of denaturation at 95°C for 15 s, annealing at 55 or 60°C for 15 s and an extension at 72°C for 15 s. Dissociation curves were generated at the end of the PCR cycle to verify that a single product was amplified in the PCR reactions for each of the target genes using the software provided with the Bio-Rad System. Transcript levels of the target genes were normalized with the transcript level of a tomato Actin gene. Relative expression was calculated using 2-ΔΔCT method as described previously. Gene-specific primers used in qRT-PCR are listed in Supplementary Table S1.
Measurement of Trehalose Content
Measurement of trehalose content in tomato leaves was performed according to a previously described method (Jang et al., 2003; Ge et al., 2008). Briefly, leaf samples (2 g) were ground in liquid nitrogen and extracted in 20 ml boiling water for 10 min. The extract was centrifuged at 12,500g for 10 min and the supernatant was passed through a 0.45 μm filter. Trehalose content was determined by high-performance ion chromatography (DX500 HPIC system, Dionex 500, CA, USA). Commercial trehalose (Sigma, MO, USA) was used as a standard to calculate trehalose content in samples.
Detection of H2O2
Leaves collected at 0 and 24 h from B. cinerea-inoculated plants or at 0 and 48 h from Pst DC3000-inoculatd plants were used for detection of H2O2 accumulation by DAB staining as described before (Li et al., 2014b; Liu et al., 2014). Accumulation of H2O2 in stained leaves was visualized using a digital camera.
Experiment Design and Data Analysis
All experiments were independently repeated three times and three replicates were included in each of the independent experiments. At least 10 plants were used in each of independent experiments in whole plant inoculation assays with B. cinerea or with Pst DC3000 or leaves from 10 individual plants were collected for detached leaf inoculation assays with B. cinerea. Leaf samples were collected from three individual plants for analyses of H2O2 accumulation, trehalose content and gene expression. Data from three independent experiments were statistically analyzed according to the Student’s t-test and the probability of p < 0.05 was considered as significant difference.
Results
Characterization of SlTPS, SlTPP, and SlTRE1 Genes in Tomato
By Blastp searches against the tomato genome database using the characterized Arabidopsis AtTPSs, AtTPPs, and AtTRE1 as queries, we identified 11, 8, and 1 loci in tomato genome that were predicted to encode TPS, TPP, and TRE and designated as SlTPS1-11, SlTPP1-8, and SlTRE1, respectively (Table 1), based on their chromosomal locations.
Table 1.
Information on the SlTPS, SlTPP, and SlTRE genes and proteins.
| Family | Genes | Loci in SOL | Accessions in GenBank | ORF (bp) | Protein size and domains | UniGenes in SOL/cDNAs in GenBank | ||
|---|---|---|---|---|---|---|---|---|
| size (aa) | TPS | TPP | ||||||
| TPS | SlTPS1 | Solyc01g005210 | XP_004228746 | 2574 | 857 | Yes | Yes | SGN-U574042, SGN-U600459, SGN-U574043 |
| SlTPS2 | Solyc02g071590 | XP_010316884 | 2832 | 943 | Yes | Yes | – | |
| SlTPS3 | Solyc02g072150 | XP_004233035 | 2556 | 851 | Yes | Yes | SGN-U575044, SGN-U575051, SGN-U575049 | |
| SlTPS4 | Solyc04g025940 | XP_004237260 | 2574 | 857 | Yes | Yes | SGN-U576714, SGN-U567013 | |
| SlTPS5 | Solyc05g005750 | XP_004238680 | 2556 | 851 | Yes | Yes | SGN-U576715 | |
| SlTPS6 | Solyc07g006500 | XP_010323144 | 2631 | 876 | Yes | Yes | SGN-U576716, AB368491 | |
| SlTPS7 | Solyc07g055300 | XP_004243268 | 2577 | 858 | Yes | Yes | SGN-U585228, SGN-U599997 | |
| SlTPS8 | Solyc07g062140 | NP_001234879 | 2781 | 926 | Yes | Yes | SGN-U579539, SGN-U580026, EF151131 | |
| SlTPS9 | Solyc08g076650 | XP_004245918 | 2589 | 862 | Yes | Yes | SGN-U583981 | |
| SlTPS10 | Solyc10g007950 | XP_004248198 | 2574 | 857 | Yes | Yes | SGN-U584220, SGN-U600516 | |
| SlTPS11a | Solyc10g046770 | XM_010329326 | 735 | 244 | – | Yes | – | |
| TPP | SlTPP1 | Solyc03g007290 | XP_004234173 | 1011 | 336 | No | Yes | – |
| SlTPP2 | Solyc03g083960 | XP_010317997 | 1104 | 367 | No | Yes | SGN-U584704, AK319855, AK247068, AK322638 | |
| SlTPP3 | Solyc04g054930 | XP_004237406 | 1167 | 388 | No | Yes | SGN-U570949, AK320358 | |
| SlTPP4 | Solyc04g072920 | XP_004237894 | 1098 | 365 | No | Yes | SGN-U575865, AK321917 | |
| SlTPP5 | Solyc04g082550 | XP_004238632 | 882 | 293 | No | Yes | – | |
| SlTPP6 | Solyc05g051880 | XP_010321465 | 1047 | 348 | No | Yes | – | |
| SlTPP7 | Solyc06g060600 | XP_004242008 | 1020 | 339 | No | Yes | – | |
| SlTPP8 | Solyc08g079060 | XP_004245739 | 1161 | 386 | No | Yes | SGN-U584816, SGN-U584817, SGN-U568331 | |
| TRE | SlTRE1 | Solyc08g082860 | XP_004245478 | 1746 | 581 | – | – | SGN-U568010, AK320041 |
athe predicted ORFs seems incomplete for intact proteins.
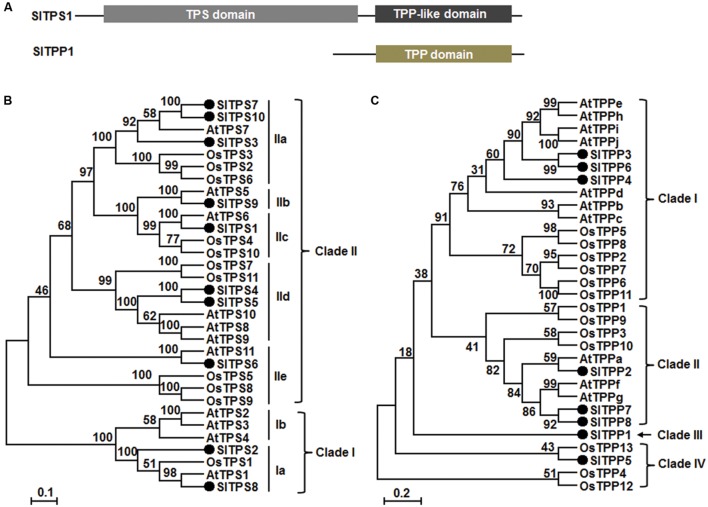
Among the 11 predicted SlTPSs, SlTPS1–SlTPS10 are complete TPSs containing both TPS and TPP-like domains (Figure 1A), but the predicted SlTPS11 is an incomplete TPS that only contains a partial TPP domain (Table 1). Nine of 11 SlTPS genes, accounting for 82% of the family, have available EST or full-length cDNAs (Table 1), indicating that these SlTPS genes are expressed normally in tomato plants. Phylogenetic tree analysis of the predicted protein sequences with Arabidopsis and rice TPSs indicated that the tomato SlTPSs can be classified into two main clades (Figure 1B). SlTPS2 and SlTPS8 belong to Clade I but both of them belong to Clade Ia, along with Arabidopsis AtTPS1 and rice OsTPS1 (Figure 1B). The remaining 8 SlTPSs, including SlTPS1, SlTPS3, SlTPS4, SlTPS5, SlTPS6, SlTPS7, SlTPS9, and SlTPS10, are members of Clade II (Figure 1B), which can be further classified into 5 subclades, Clade IIa-e (Yang et al., 2012; Henry et al., 2014).
FIGURE 1.
Structural features and phylogenetic tree of SlTPSs and SlTPPs with Arabidopsis and rice TPSs and TPPs. (A) Structures of SlTPS1 and SlTPP1. Conserved domains are indicated. (B,C) Phylogenetic tree of SlTPSs and SlTPPs. Phylogenetic trees were constructed by neighbor-joining method using MEGA program version 6.05. SlTPSs and SlTPPs in the trees are indicated by filled circles and different clades are labeled at right of the trees.
All of the 8 identified SlTPP proteins contain TPP domain but lack TPS domain (Table 1; Figure 1A). Four of these SlTPP genes including SlTPP2, SlTPP3, SlTPP4, and SlTPP8, accounting for 50% of the family, have available ESTs or full-length cDNAs (Table 1), indicating that these SlTPP genes are expressed in tomato plants. Phylogenetic tree analysis with Arabidopsis and rice TPPs revealed that SlTPPs can be classified into four clades (Figure 1C). Each of Clade I and Clade II harbors three SlTPPs (SlTPP3, SlTPP4, and SlTPP6 in Clade I and SlTPP2, SlTPP7 and SlTPP8 in Clade II) (Figure 1C). However, SlTPPs in Clade I and Clade II are closely clustered with Arabidopsis TPPs (Figure 1C). SlTPP5 was clustered with rice OsTPP13, forming Clade IV; however, SlTPP1 did not cluster with any of Arabidopsis and rice TPPs, becoming the only member in Clade III (Figure 1C). Together with the observations in Arabidopsis and maize (Vandesteene et al., 2012; Henry et al., 2014), the divergence of the SlTPPs proteins in the phylogenetic tree (Figure 1C) may imply that the SlTPP genes were evolved through duplication events after the monocot/dicot split.
Like that in Arabidopsis, rice and maize (Ge et al., 2008; Henry et al., 2014; Lunn et al., 2014), the tomato genome contains only one trehalase gene, SlTRE1 (Table 1). The SlTRE1 protein shows 53 and 57% of identity to Arabidopsis AtTRE1 and rice OsTRE1, respectively. One EST and one full-length cDNA that match to the predicted SlTRE1 sequence (Table 1) were identified in database, indicating SlTRE1 is also expressed normally in tomato plants.
Expression Patterns of Selected SlTPSs, SlTPPs, and SlTRE1 in Response to Pathogens and Defense Signaling Hormones
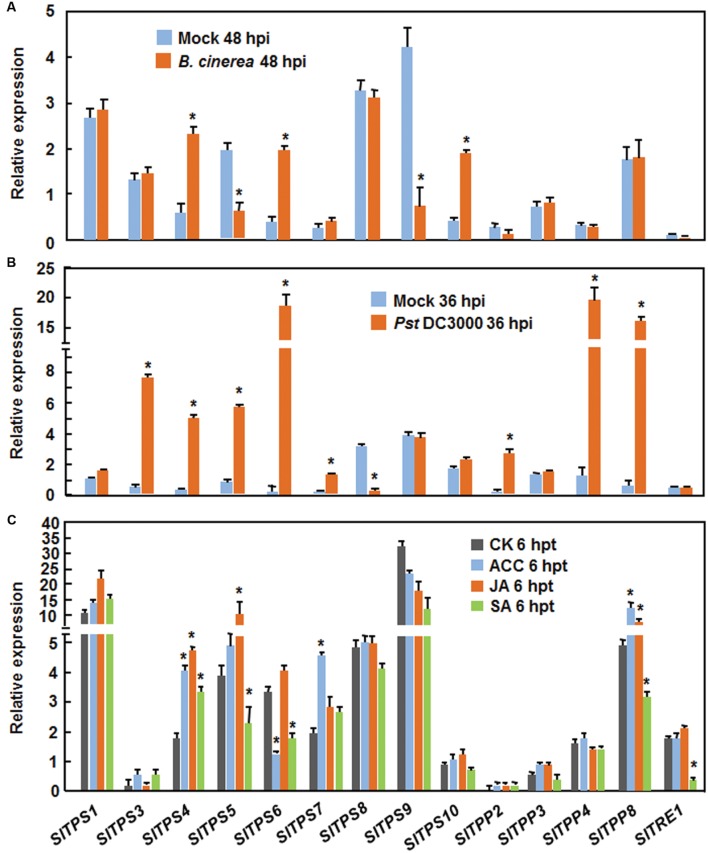
Nine SlTPSs (SlTPS1, SlTPS3, SlTPS4, SlTPS5, SlTPS6, SlTPS7, SlTPS8, SlTPS9, and SlTPS10), 4 SlTPPs (SlTPP2, SlTPP3, SlTPP4, and SlTPP8) and SlTRE1, which have EST or full-length cDNA supports (Table 1), were selected for further functional analysis. As a first step, we examined the expression of the selected SlTPS, SlTPS and SlTRE genes in tomato plants at 48 or 36 h after inoculation with B. cinerea or Pst DC3000, as the pathogens normally colonize and proliferate in the inoculated leaves at these time points (Li et al., 2014b, 2015; Zhang et al., 2014). At 48 h after inoculation with B. cinerea, the expression of SlTPS4, SlTPS6, and SlTPS10 was significantly upregulated, leading to 3.7∼6.3-fold increases, while the expression of SlTPS5 and SlTPS9 was markedly downregulated, resulting in 2.4- and 3.5-fold decrease, respectively, as compared with those in mock-inoculated plants (Figure 2A). Expression of other SlTPSs (SlTPS2, SlTPS3, SlTPS7, and SlTPS8), 4 SlTPPs and SlTRE1 was not affected by B. cinerea (Figure 2A). At 36 h after inoculation with Pst DC3000, the expression of SlTPS3, SlTPS4, SlTPS5, SlTPS6, SlTPS7, and SlTPP2, SlTPP4, and SlTPP8 was significantly upregulated, leading to 5.6∼91.1-fold increases, while the expression of SlTPS8 was markedly downregulated, resulting in 13.2-folds decrease, respectively, as compared with those in mock-inoculated plants (Figure 2B). Expression of SlTPS1, SlTPS9, SlTPS10, SlTPP3, and SlTRE1 was not affected by Pst DC3000 (Figure 2B). The responsiveness of these selected SlTPSs, SlTPPs, and SlTRE1 to defense signaling hormones such as salicylic acid (SA), methyl jasmonate (MeJA), and 1-amino cyclopropane-1-carboxylic acid (ACC, a precursor of ET) was also analyzed. As shown Figure 2C, the expression of SlTPS4, SlTPS5, SlTPS6, SlTPS7, SlTPP8, and SlTRE1 was affected by at least one of the defense signaling hormones at 6 h after treatment, while the expression of SlTPS1, SlTPS3, SlTPS8, SlTPS9, SlTPS10, SlTPP2, SlTPP3, and SlTPP4 was not affected by any of the defense signaling hormones. Among the genes whose expression was affected by defense signaling hormones, the expression of SlTPS4 was significantly upregulated by three defense signaling hormones (Figure 2C). In particular, SA suppressed the expression of SlTPS6 and SlTRE1 while JA induced the expression of SlTPS5 and SlTPP8 (Figure 2C). ACC induced the expression of SlTPS7 and SlTPP8 but suppressed the expression of SlTPS6 (Figure 2C). Taken together, these data indicate that some of the 14 selected SlTPS, SlTPP, and SlTRE1 genes responded with different expression patterns to infection of B. cinerea or Pst DC3000 and to at least one of the defense signaling hormones.
FIGURE 2.
Expression of selected SlPTSs, SlTPPs, and SlTRE1 in responses to infection with Botrytis cinerea or P. syringae pv. tomato DC3000 and to treatments with defense signaling hormones. (A) Expression of selected trehalose-related genes in response to B. cinerea. Four-week-old plants were inoculated by foliar spraying with spore suspension of B. cinerea or with same volume of buffer as a mock control and leaf samples were collected at 48 h after inoculation for analysis of gene expression. (B) Expression of selected trehalose-related genes in response to Pst DC3000. Four-week-old plants were inoculated by vacuum infiltration with suspension of Pst DC3000 or with 10 mM MgCl2 solution as a mock control and leaf samples were collected at 36 h after inoculation for analysis of gene expression. (C) Expression of selected trehalose-related genes in response to defense signaling hormones. Tomato plants were treated by foliar spraying of 100 μM SA, 100 μM MeJA, 100 μM ACC or similar volume of solution as a control and leaf samples were collected after 6 h for analysis of gene expression. Expression data were normalized with the value of a reference SlActin gene and relative expression was shown as folds of the SlActin expression level. Data presented are the means ± SD from three independent experiments and ∗ above the columns indicate significant differences at p < 0.05 level between the pathogen-inoculated or hormone-treated plants and the mock-inoculated/treated plants.
Silencing of 14 Selected SlTPSs, SlTPPs, and SlTRE1 Genes in Tomato
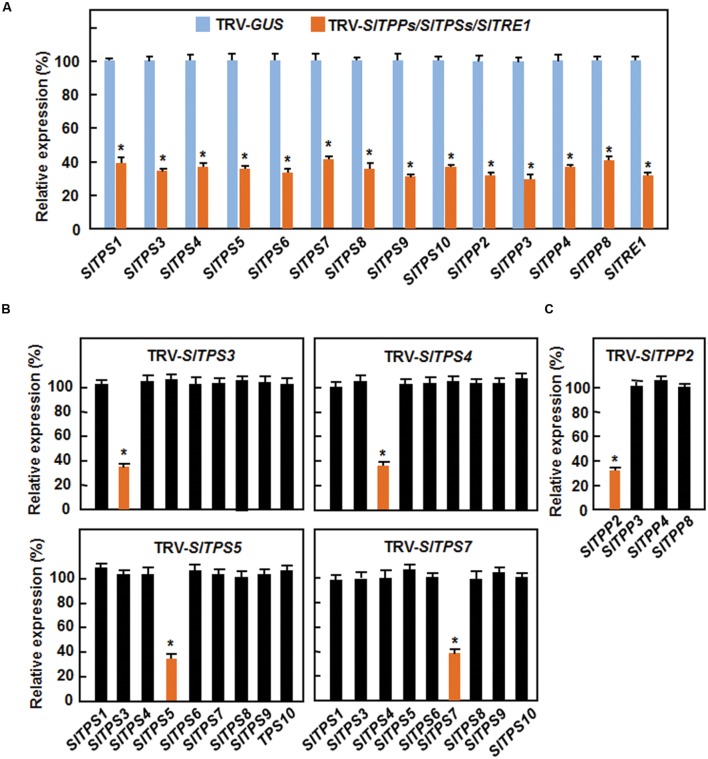
To explore the possible involvement of the trehalose-related genes in disease resistance, we manipulated the endogenous expression levels of each of the 14 selected SlTPS, SlTPP, and SlTRE1 genes by VIGS approach and examined their effects on disease resistance to B. cinerea or Pst DC3000. To do this, we first examined the silencing efficiency and specificity of the designed VIGS fragments for each of the selected SlTPS, SlTPP, and SlTRE1 genes. Standard VIGS protocol was applied to 2-week-old tomato plants (Liu et al., 2002; Li et al., 2014a,b) and the silencing efficiency was analyzed at 4 weeks after VIGS treatment. In our VIGS experiments, plants infiltrated with a TRV-PDS construct as positive controls started to display bleaching symptom on newly developed leaves at 10 days and >90% of the plants showed bleaching symptom at 4 weeks after VIGS infiltration. As shown in Figure 3A, the transcript levels for the target genes in corresponding TRV-SlTPSs-, TRV-SlTPPs-, or TRV-SlTRE1-infiltrated plants were 28–39% of those in TRV-GUS-infiltrated plants, indicating that the silencing efficiency for these trehalose-related genes was 61–72%. We also examined the silencing specificity of SlTPS3, SlTPS4, SlTPS5, SlTPS7, and SlTPP2, whose silencing led to altered resistance to B. cinerea or Pst DC3000 (see below), by comparing the transcript levels of the target gene and its relative family members in TRV-SlTPS3-, TRV-SlTPS4-, TRV-SlTPS5-, TRV-SlTPS7-, and TRV-SlTPP2-infiltrated plants. Compared with those in the TRV-GUS-infiltrated plants, the transcript levels for SlTPS3, SlTPS4, SlTPS5, SlTPS7, and SlTPP2 were significantly decreased in TRV-SlTPS3-, TRV-SlTPS4-, TRV-SlTPS5-, TRV-SlTPS7-, and TRV-SlTPP2-infiltrated plants, respectively, but the transcript levels of other family members were comparable (Figures 3B,C). These data demonstrate that silencing of SlTPS3, SlTPS4, SlTPS5, SlTPS7, or SlTPP2 only downregulated the expression of itself but did not affect the expression of other SlTPS or SlTPP genes in the same family.
FIGURE 3.
Silencing efficiency and specificity for selected SlTPS, SlTPP and SlTRE1 genes in VIGS-infiltrated plants. (A) Silencing efficiency for each of the selected trehalose-related genes in corresponding VIGS-infiltrated plants. (B,C) Silencing specificity for 4 SlTPS genes and for SlTPP2. Ten-day-old tomato plants were infiltrated with agrobacteria carrying TRV-SlTPSs/SlTPPs/SlTRE1 or TRV-GUS constructs and leaf samples were collected at 4 weeks after agroinfiltration. Transcript levels for the selected trehalose-related genes were analyzed by qRT-PCR using a tomato SlActin gene as an internal control. Expression levels of the selected trehalose-related genes in TRV-SlTPSs/SlTPPs/SlTRE1-infiltrated plants were shown as percentages of the levels in TRV-GUS-infiltrated plants. Data presented are the means ± SD from three independent experiments and ∗ above the columns indicate significant differences at p < 0.05 level between the TRV-SlTPSs/SlTPPs/SlTRE1-infiltrated and TRV-GUS-infiltrated plants.
During our studies, we noted that the SlTPS7- and SlTPS8-silenced plants displayed reduced plant heights, resulting in 25 and 33% of reduction at 4 weeks after VIGS infiltration, as compared with the TRV-GUS-infiltrated plants (Supplementary Figure S1). These results indicate that SlTPS7 and SlTPS8 may have functions in regulation of vegetative growth in tomato. However, silencing of each of other SlTPS (SlTPS1, SlTPS3, SlTPS4, SlTPS5, SlTPS6, SlTPS9, and SlTPS10), SlTPP (SlTPP2, SlTPP3, SlTPP4, and SlTPP8) and SlTRE1 genes did not affect vegetative growth of the silenced plants (data not shown).
Silencing of SlTPS3, SlTPS4, or SlTPS7 Led to Decreased Resistance to B. cinerea
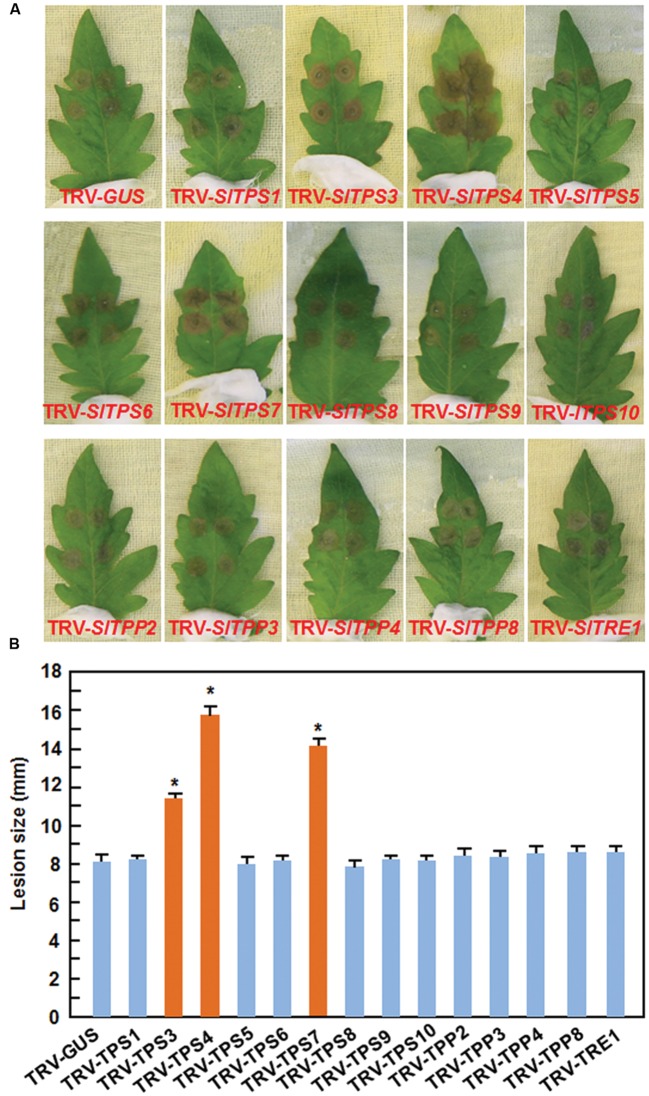
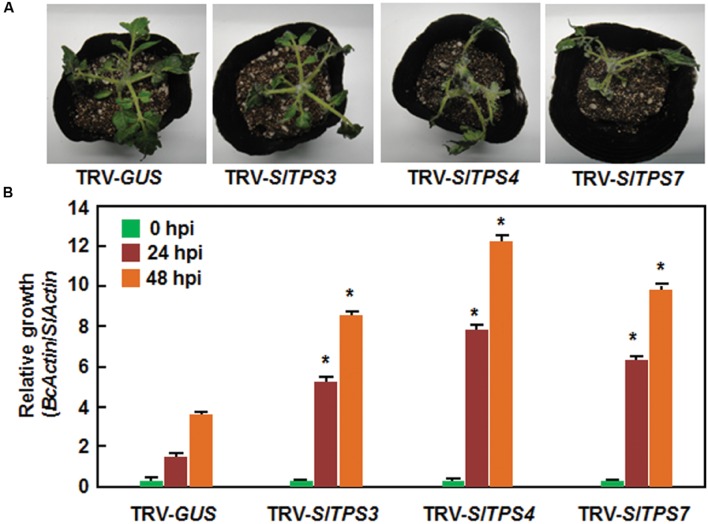
To examine the possible involvement of the selected SlTPS, SlTPP, and SlTRE1 genes in resistance to B. cinerea, a necrotrophic fungal pathogen, we used two different methods, detached leaf disease assays for fast evaluation and whole plant disease assays for confirmation, to compare the disease phenotype and in planta fungal growth in the TRV-SlTPS/SlTPP/SlTRE1-infiltrated plants with those in the TRV-GUS-infiltrated plants. In the detached leaf disease assays, typical small necrotic lesions were seen at 2 days post inoculation (dpi). At 3 dpi, sizes of the lesions on leaves from TRV-SlTPS1-, TRV- SlTPS5-, TRV-SlTPS6-, TRV-SlTPS8-, TRV-SlTPS9-, TRV-SlT PS10-, TRV-SlTPP2-, TRV-SlTPP3-, TRV-SlTPP4-, TRV-SlT PP8-, and TRV-SlTRE1-infiltrated plants were similar to that in the TRV-GUS-infiltrated plants (Figures 4A,B), indicating that SlTPS1, SlTPS5, SlTPS6, SlTPS8, SlTPS9, SlTPS10, SlTPP2, SlTPP3, SlTPP4, SlTPP8, and SlTRE1 may not be involved in resistance to B. cinerea. By contrast, sizes of the lesions on leaves from the TRV-SlTPS3-, TRV-SlTPS4-, and TRV-SlTPS7-infiltrated plants were significantly increased (Figure 4A), leading to 38, 97, and 75% of increases, respectively, than those in the TRV-GUS-infiltrated plants at 3 dpi (Figure 4B). To confirm this observation, we further evaluated the disease phenotype and measured in planta fungal growth of B. cinerea in the TRV-SlTPS3-, TRV-SlTPS4-, and TRV-SlTPS7-infiltrated plants using whole plant disease assays. As shown in Figure 5A, the TRV-SlTPS3-, TRV-SlTPS4-, and TRV-SlTPS7-infiltrated plants had larger necrotic areas and leaf maceration at 5 dpi, as compared with the TRV-GUS-infiltrated plants. Accordingly, in planta growth of B. cinerea, as represented by the transcript levels of the B. cinerea BcActinA gene, in leaf tissues of the TRV-SlTPS3-, TRV-SlTPS4-, and TRV-SlTPS7-infiltrated plants was significantly increased, showing three–four times higher than that in the TRV-GUS-infiltrated control plants at 24 and 48 hpi (Figure 5B). Taken together, these results demonstrate that silencing of SlTPS3, SlTPS4, or SlTPS7 deteriorated the resistance of tomato plants against B. cinerea and supported more growth of B. cinerea in the TRV-SlTPS3-, TRV-SlTPS4-, and TRV-SlTPS7-infiltrated plants.
FIGURE 4.
Silencing of SlTPS3, SlTPS4, and SlTPS7 led to decreased resistance against B. cinerea in detached leaf assays. Ten-day-old plants were infiltrated with agrobacteria carrying TRV-SlTPSs/SlTPPs/SlTRE1 or TRV-GUS constructs and leaves were collected at 4 weeks after agroinfiltration for disease assays with B. cinerea. (A) Disease symptom on representative leaves from the TRV-SlTPSs/SlTPPs/SlTRE1-infiltrated and TRV-GUS-infiltrated plants. (B) Size of lesions on leaves from the TRV-SlTPSs/SlTPPs/SlTRE1-infiltrated and TRV-GUS-infiltrated plants. Detached leaf disease assays were performed by dropping 5 μL of spore suspension onto the detached leaves and lesion sizes were measured 3 days after inoculation. At least 10 leaves from 10 individual TRV-SlTPSs/SlTPPs/SlTRE1- and TRV-GUS-infiltrated plants were used in each of three independent experiments. Similar results were obtained in independent experiments (A). Data presented in (B) are the means ± SD from three independent experiments and ∗ above the columns indicate significant differences at p < 0.05 level between the TRV-SlTPSs/SlTPSs/SlTRE1-infiltrated and TRV-GUS-infiltrated plants.
FIGURE 5.
Silencing of SlTPS3, SlTPS4, and SlTPS7 led to decreased resistance against B. cinerea in whole plant assays. (A) Disease phenotype of representative TRV-SlTPS3-, TRV-SlTPS4-, TRV-SlTPS7-, and TRV-GUS-infiltrated plants. Photos were taken at 4 days after inoculation. (B) In planta growth of B. cinerea in inoculated TRV-SlTPS3-, TRV-SlTPS4-, TRV-SlTPS7-, and TRV-GUS-infiltrated plants. Whole plant disease assays were done by foliar spraying with spore suspension at 4 weeks after VIGS infiltration. Transcript levels for B. cinerea BcActinA and tomato SlActin genes in B. cinerea-inoculated plants were analyzed using qRT-PCR and in planta relative growth of B. cinerea was shown as ratios of transcript levels of BcActinA/SlActin. Similar results were obtained in independent experiments (A) and data presented in (B) are the means ± SD from three independent experiments. ∗ above the columns indicate significant differences at p < 0.05 level between the TRV-SlTPS3/4/7-infiltrated and TRV-GUS-infiltrated plants.
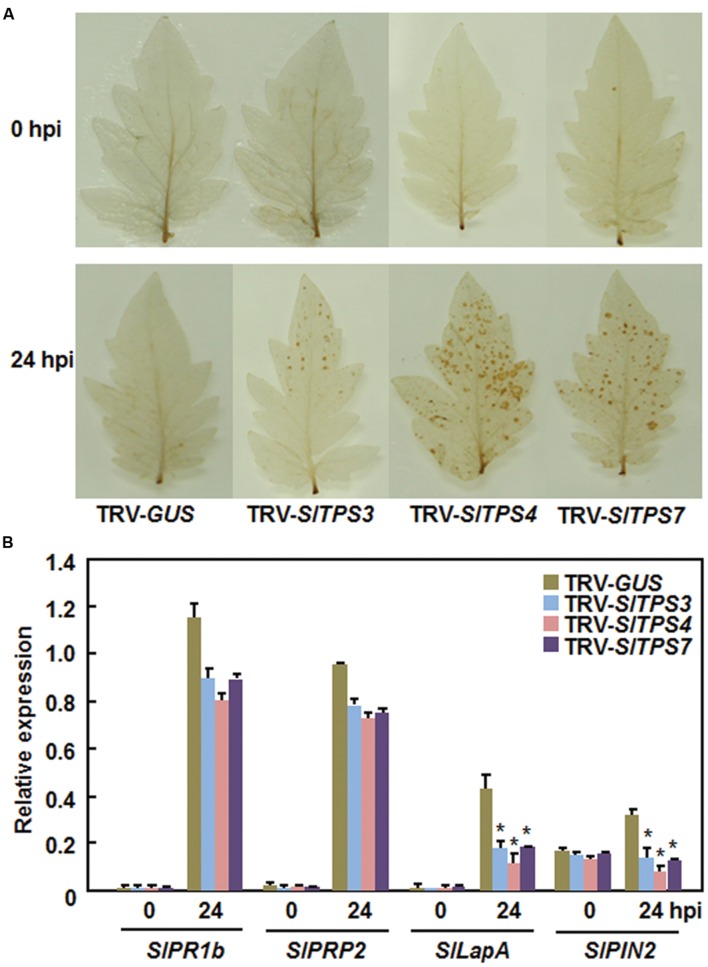
To explore the possible mechanism by which silencing of SlTPS3, SlTPS4, or SlTPS7 led to decreased resistance against B. cinerea, we analyzed and compared the defense responses including accumulation of reactive oxygen species (ROS) and expression of defense-related genes in the TRV-SlTPS3-, TRV-SlTPS4-, and TRV-SlTPS7-infiltrated plants before and after infection of B. cinerea. At 0 h, no accumulation of H2O2 was observed in the leaves from the TRV-SlTPS3-, TRV-SlTPS4-, or TRV-SlTPS7-infiltrated plants and the TRV-GUS-infiltrated plants (Figure 6A). However, significant accumulation of H2O2 was observed in leaves of the TRV-SlTPS3-, TRV-SlTPS4-, and TRV-SlTPS7-infiltrated plants, while only slight accumulation of H2O2 was detected in leaves of TRV-GUS-infiltrated plants, at 24 h after inoculation with B. cinerea (Figure 6A). Similarly, the expression of some selected SA signaling-responsive defense-related genes SlPR1b and SlPRP2 and JA/ET signaling-responsive defense-related genes SlLapA and SlPIN2 was comparable between the TRV-SlTPS3-, TRV-SlTPS4-, or TRV-SlTPS7-infiltrated plants and the TRV-GUS-infiltrated plants before infection of B. cinerea (Figure 6B). Although the expression of these SA signaling-responsive and JA/ET signaling-responsive defense-related genes was upregulated significantly by infection of B. cinerea; however, the expression levels of SlPR1b and SlPRP2 were slightly reduced while the expression levels of SlLapA and SlPIN2 were significantly decreased in TRV-SlTPS3-, TRV-SlTPS4-, and TRV-SlTPS7-infiltrated plants, as compared with those in the TRV-GUS-infiltrated plants, at 24 h (Figure 6B). Together, these data indicate that silencing of SlTPS3, SlTPS4, or SlTPS7 deregulated ROS accumulation and attenuated the expression of the JA/ET signaling-responsive defense-related genes upon infection of B. cinerea.
FIGURE 6.
Silencing of SlTPS3, SlTPS4, and SlTPS7 increased accumulation of H2O2 and decreased the expression levels of JA/ET signaling-responsive defense-related genes after infection with B. cinerea. Whole plant disease assays were done by foliar spraying with spore suspension at 4 weeks after VIGS infiltration and leaf samples were collected at 24 h after inoculation. (A) Accumulation of H2O2, as detected by DAB staining, in TRV- SlTPS3-, TRV-SlTPS4-, TRV-SlTPS7-, and TRV-GUS-infiltrated plants after infection of B. cinerea. (B) Expression patterns of selected defense-related genes in TRV-SlTPS3-, TRV-SlTPS4-, TRV-SlTPS7-, and TRV-GUS-infiltrated plants after infection of B. cinerea. Expression data for the selected defense-related genes in TRV-SlTPS3-, TRV-SlTPS4-, TRV-SlTPS7-, and TRV-GUS-infiltrated plants were normalized with the value of a reference SlActin gene and relative expression was shown as folds of the SlActin expression level. Similar results were obtained in independent experiments (A) and data presented in (B) are the means ± SD from three independent experiments. ∗ above the columns indicate significant differences at p < 0.05 level between the TRV-SlTPS3/4/7-infiltrated and TRV-GUS-infiltrated plants.
Silencing of SlTPS4 Decreased but Silencing of SlTPS5 or SlTPP2 Increased the Resistance against Pst DC3000
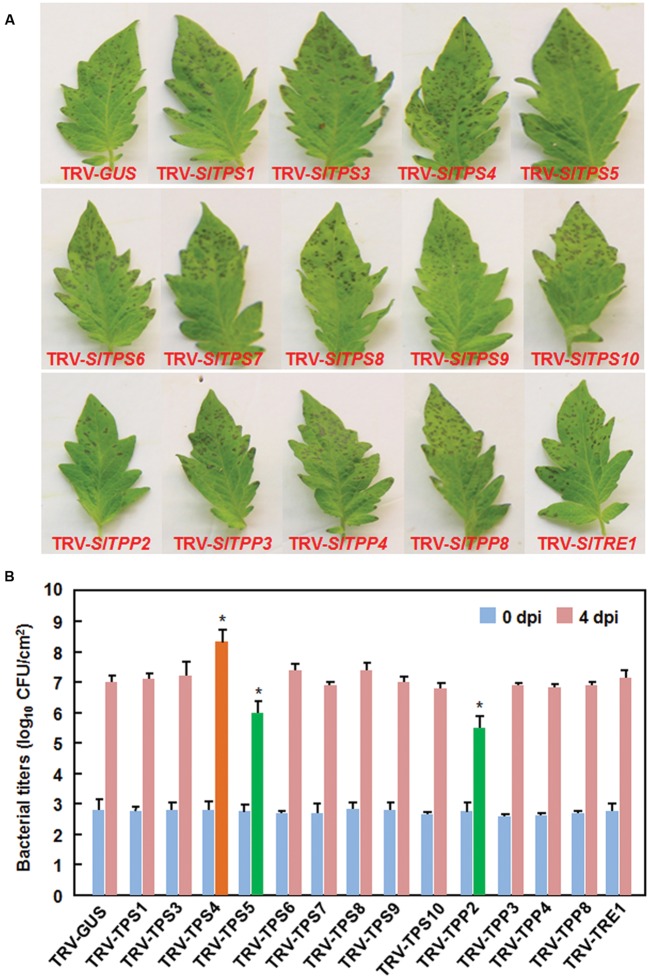
We next examined the possible involvement of the selected SlTPS, SlTPP, and SlTRE1 genes in resistance against Pst DC3000, a (hemi)biotrophic bacterial pathogen, by comparing the disease phenotype and in planta bacterial growth in the TRV-target SlTPS/SlTPP/SlTRE1-infiltrated plants with those in the TRV-GUS-infiltrated plants. At 3 dpi, the TRV-SlTPS4-infiltrated plants displayed more severe disease while the TRV-SlTPS5- and TRV-SlTPP2-infiltrated plants showed less severe disease, as compared with the TRV-GUS-infiltrated plants (Figure 7A). At 4 dpi, the bacterial population (2.24 × 108 colony-forming unit (cfu)/cm2 leaf tissues) in leaves of TRV-SlTPS4-infiltrated was 23.46 times higher than that in the TRV-GUS-infiltrated plants (9.55 × 106 cfu/cm2 leaf tissues). By contrast, the bacterial populations in leaves of the TRV-SlTPS5- and TRV-SlTPP2-infiltrated plants (1.07 × 106 cfu/cm2 leaf tissues and 3.63 × 105 cfu/cm2 leaf tissues, respectively) were 7.93 and 25.31 times less than that in the TRV-GUS-infiltrated plants, respectively, at 4 dpi (Figure 7B). Disease symptom on and bacterial growth in leaves from TRV-SlTPS1-, TRV-SlTPS3-, TRV-SlTPS6-, TRV-SlTPS7-, TRV-SlTPS8-, TRV-SlTPS9-, TRV-SlTPS10-, TRV-SlTPP3-, TRV-SlTPP4-, TRV-SlTPP8-, and TRV-SlTRE1-infiltrated plants were similar to those in the TRV-GUS-infiltrated plants (Figures 7A,B), indicating that SlTPS1, SlTPS3, SlTPS6, SlTPS7, SlTPS8, SlTPS9, SlTPS10, SlTPP3, SlTPP4, SlTPP8, and SlTRE1 may not be involved in resistance against Pst DC3000. These results indicate that silencing of SlTPS4 decreased the resistance while silencing of SlTPS5 or SlTPP2 increased the resistance against Pst DC3000 in tomato.
FIGURE 7.
Silencing of SlTPS4 decreased and silencing of SlTPS5 or SlTPP2 increased the resistance against P. syringae pv. tomato DC3000. Ten-day-old plants were infiltrated with agrobacteria carrying TRV-SlTPSs/SlTPPs/SlTRE1 or TRV-GUS constructs and disease assays were carried out at 4 weeks after agroinfiltration. The TRV-SlTPSs/SlTPPs/SlTRE1- and TRV-GUS-infiltrated plants were inoculated by vacuum infiltration with suspension of P. syringae pv. tomato DC3000. (A) Disease symptom on representative leaves from TRV-SlTPSs/SlTPPs/SlTRE1- and TRV-GUS-infiltrated plants at 4 days after inoculation with P. syringae pv. tomato DC3000. (B) Bacterial population in inoculated leaves of the TRV-SlTPSs/SlTPPs/SlTRE1- and TRV-GUS-infiltrated plants. Leaf samples were collected at 0 and 4 days after inoculation and bacterial population was measured. Similar results were obtained in independent experiments (A). Data presented in (B) are the means ± SD from three independent experiments and ∗ above the columns indicate significant differences at p < 0.05 level between the TRV-SlTPSs/SlTPSs/SlTRE1-infiltrated and TRV-GUS-infiltrated plants.
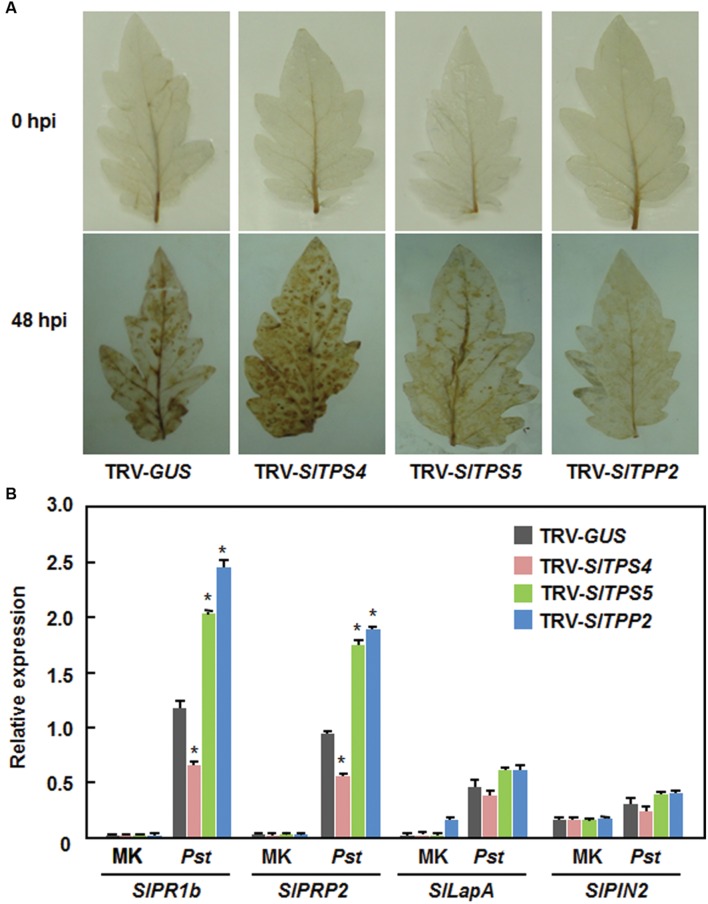
We also analyzed and compared the accumulation of ROS and expression of defense-related genes in the TRV-SlTPS4-, TRV-SlTPS5-, and TRV-SlTPP2-infiltrated plants before and after infection of Pst DC3000 to gain insights into the possible mechanism that silencing of SlTPS4, SlTPS5, or SlTPP2 affected the resistance against Pst DC3000. Before infection of Pst DC3000, no significant accumulation of H2O2 was seen in leaves of the TRV-SlTPS4-, TRV-SlTPS5-, TRV-SlTPP2-, and TRV-GUS-infiltrated plants (Figure 8A). However, at 3 dpi, significant accumulation of H2O2 was observed in leaves of the TRV-SlTPS4-infiltrated plants, while less accumulation of H2O2 in leaves of the TRV-SlTPS5- and TRV-TPP2-infiltrated plants was detected, as compared with that in leaves of the TRV-GUS-infiltrated plants (Figure 8A). Similarly, the expression of defense-related genes SlPR1b, SlPRP2, SlLapA, and SlPIN2 was comparable between the TRV-SlTPS4-, TRV-SlTPS5-, or TRV-SlTPP2-infiltrated plants and the TRV-GUS-infiltrated plants before infection of Pst DC3000 (Figure 8B). The expression levels of SlPR1b and SlPRP2 in the TRV-SlTPS4-infiltrated plants were decreased while the expression levels of these two defense-related genes in the TRV-SlTPS5- and TRV-SlTPP2-infiltrated plants were significantly increased, as compared with those in the TRV-GUS-infiltrated plants, at 2 dpi after inoculation with Pst DC3000 (Figure 8B). However, the expression levels of SlLapA and SlPIN2 in the TRV-SlTPS4-, TRV-SlTPS5-, or TRV-SlTPP2-infiltrated plants were not significantly affected, as compared with those in the TRV-GUS-infitlrated plants, at 2 dpi after inoculation with Pst DC3000 (Figure 8B). These data indicate that silencing of SlTPS4 attenuated while silencing of SlTPS5 or SlTPP2 strengthened the expression of the SA signaling-responsive defense-related genes upon infection of Pst DC3000.
FIGURE 8.
Silencing of SlTPS4, SlTPS5, and SlTPP2 affected H2O2 accumulation and expression of SA signaling-responsive defense-related genes after infection with P. syringae pv. tomato DC3000. Whole plant disease assays were done by vacuum infiltration with suspension of P. syringae pv. tomato DC3000 at 4 weeks after VIGS infiltration and leaf samples were collected at 24 h after inoculation. (A) Accumulation of H2O2, as detected by DAB staining, in TRV-SlTPS4-, TRV-SlTPS5-, TRV-SlTPP2-, and TRV-GUS-infiltrated plants after infection of Pst D3000. (B) Expression patterns of selected defense-related genes in TRV-SlTPS4-, TRV-SlTPS5-, TRV-SlTPP2-, and TRV-GUS-infiltrated plants after infection of Pst DC3000. Expression data for the selected defense-related genes in TRV-SlTPS4-, TRV-SlTPS5-, TRV-SlTPP2-, and TRV-GUS-infiltrated plants were normalized with the value of a reference SlActin gene and relative expression levels were shown as folds of the SlActin expression level. Similar results were obtained in independent experiments (A) and data presented in (B) are the means ± SD from three independent experiments. ∗ above the columns indicate significant differences at p < 0.05 level between the TRV-SlTPS4/SlTPS5/SlTPP2-infiltrated and TRV-GUS-infiltrated plants.
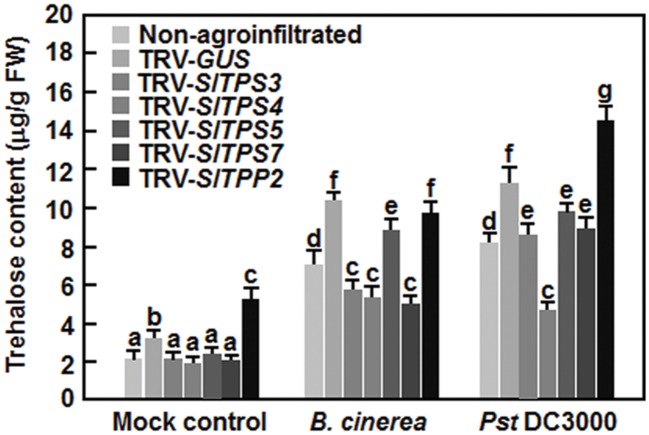
Silencing of SlTPS3, SlTPS4, SlTPS5, SlTPS7, or SlTPP2 Affected Trehalose Content in Tomato Plants with or without Pathogen Infection
To examine the possible involvement of trehalose in defense response, we analyzed the trehaolse contents in TRV-SlTPS3-, TRV-SlTPS4-, TRV-SlTPS5-, TRV-SlTPS7-, and TRV-SlTPP2-infiltrated plants with or without infection of B. cinerea or Pst DC3000. At 4 weeks after VIGS infiltration, trehalose content in TRV-GUS-infiltrated plants was 71% higher than that in non-agroinfiltrated plants (Figure 9), indicating that infiltrated agrobacteria and/or TRV affected trehalose content in tomato plants. Without pathogen infection, trehalose contents in TRV-SlTPS3-, TRV-SlTPS4-, TRV-SlTPS5-, and TRV-SlTPS7-silenced plants were decreased by 35∼45% while the trehalose content in TRV-SlTPP2-infiltrated plants was increased by 47%, as compared to that in TRV-GUS-silenced plants (Figure 9). At 48 h after inoculation, infection of B. cinerea or Pst DC3000 increased the trehalose contents in non-agroinfiltrated and TRV-GUS-infiltrated plants, leading to 1.9–2.5 and 2.2–3.1-folds of increases by B. cinerea and Pst DC3000, respectively (Figure 9). As compared with those in TRV-GUS-infiltrated plants, trehalose contents in TRV-SlTPS3-, TRV-SlTPS4-, and TRV-SlTPS7-infiltrated plants after infection of B. cinerea were decreased by 44–54% while trehalose content in TRV-SlTPS4-infiltrated plants after infection of Pst DC3000 was reduced by 58% (Figure 9). Trehalose content in TRV-SlTPP2-infiltrated plants was similar to that in TRV-GUS-infiltrated plants after infection of B. cinerea, whereas the content was increased by 27% after infection of Pst DC3000 (Figure 9). These data suggest that silencing of SlTPS3, SlTPS4, SlTPS5, SlTPS7, or SlTPP2 affected trehalose content in tomato plants with or without pathogen infection.
FIGURE 9.
Changes of trehalose contents in SlTPS3-, SlTPS4-, SlTPS5-, SlTPS7-, and SlTPP2-silenced plants with or without pathogen infection. Ten-day-old plants were infiltrated with agrobacteria carrying TRV-SlTPS3, TRV-SlTPS4, TRV-SlTPS5, TRV-SlTPS7, TRV-SlTPP2, or TRV-GUS construct and were inoculated with spore suspension of B. cinerea or bacterial suspension of Pst DC3000 at 4 weeks after VIGS infiltration. Leaf samples were collected for measurement of trehalose contents at 48 h after pathogen inoculation. Data presented are the means ± SD from three independent experiments and different letters above the columns indicate significant differences at p < 0.05 level.
Discussion
In the present study, we identified 11 SlTPS, 8 SlTPP, and 1 SlTRE1 genes in tomato (Table 1). The numbers of SlTPS, SlTPP, and SlTRE1 genes are similar to those observed in Arabidopsis (Leyman et al., 2001; Vandesteene et al., 2012), rice (Ge et al., 2008; Zhang et al., 2011), wheat (Xie et al., 2015), and maize (Henry et al., 2014; Zhou et al., 2014). This indicates that the SlTPS and SlTPP gene families in tomato are conserved with the TPS and TPP families in other plants, probably due to the evolution feature that at least the eudicot and many monocot TPP genes originate from whole-genome duplications (Vandesteene et al., 2012). Although the biological functions of TPSs, TPPs, and TRE1 in plant growth/development and abiotic stress response have been implicated, direct evidence supporting the roles of TPSs, TPPs, and TRE1 in plant disease resistance is lacking yet. Our VIGS-based functional analyses of 9 SlTPSs (82% of the family), 4 SlTPPs (50% of the family), and SlTRE1 revealed that silencing of SlTPS3, SlTPS4, SlTPS5, SlTPS7, or SlTPP2 affected the resistance against B. cinerea and Pst DC3000, two different pathogens with distinct infection styles. To our knowledge, these findings provide the first lines of evidence supporting the involvement of the trehalose-related genes in plant disease resistance.
In our VIGS assays, the silencing efficiency for individual target gene of the 14 selected SlTPSs, SlTPPs and SlTRE1 was estimated to be 61–72% (Figure 3A), which is similar to those observed in our previous studies (Li et al., 2014a,b; Liu et al., 2014; Zhang et al., 2014). Silencing specificity of SlTPS3, SlTPS4, SlTPS5, SlTPS7, or SlTPP2 (Figure 3B) demonstrates that the altered phenotypes in growth and disease resistance observed in the present study were the consequences of the silencing of specific individual SlTPS or SlTPP genes. Notably, we observed that silencing of either SlTPS7 or SlTPS8 led to inhibition of vegetative growth (Supplementary Figure S1), indicating that both SlTPS7 and SlTPS8 have functions in regulation of vegetative growth in tomato. SlTPS8 is phylogenetically closely related to Arabidopsis AtTPS1 (Figure 1B), which was shown to be essential for vegetative growth (van Dijken et al., 2004; Gómez et al., 2010). Therefore, it is likely that SlTPS8 and AtTPS1 have evolutionary conserved functions in vegetative growth of the Arabidopsis and tomato plants. In addition, AtTPS1 was also found to be essential for embryogenesis and flowering (Eastmond et al., 2002; van Dijken et al., 2004; Gómez et al., 2006, 2010; Wahl et al., 2013). The involvement of SlTPS8 in embryogenesis, flowering and other biological processes needs to be further investigated.
It was previously reported that expression of AtTPS11 was transiently induced by feeding of green peach aphids (Singh et al., 2011). However, some of the trehalose metabolic genes such as rice OsTPS1 responded with high level of expression by abiotic stress over a period of 3 days after treatment (Ge et al., 2008). Previous studies have shown that the expression of some of SlTPSs and SlTPPs can be induced by pathogen infection (Brodmann et al., 2002; Golem and Culver, 2003). Diverse spatiotemporal expression patterns were also observed for the 10 Arabidopsis AtTPP genes by analyzing promoter GUS/GFP lines (Vandesteene et al., 2012). The differential responsiveness of the selected SlTPS and SlTPP genes to infection of B. cinerea and Pst DC3000 and to defense signaling hormones indicates possible functional divergence among the SlTPSs and SlTPPs in disease resistance against B. cinerea and Pst DC3000. Moreover, we also noted that some of the SlTPS and SlTPP genes, which exhibited altered expression in pathogen-infected plants, did not affect the disease resistance to B. cinerea or Pst DC3000. This can be explained by a common phenomenon that induction of gene expression does not always correlate with an absolute requirement in defense response.
Previous studies have shown that pathogen-induced expression of trehalose-related genes can lead to trehalose accumulation (Brodmann et al., 2002; Hofmann et al., 2010; Piazza et al., 2015) and that transgenic expression of the trehalose metabolic genes can elevate the endogenous trehalose content (Jang et al., 2003; Ge et al., 2008; Singh et al., 2011; Wang et al., 2016). Similarly, we observed that infection of B. cinerea or Pst DC3000 as well as infiltration with agrobacteria harboring TRV construct induced the trehalose accumulation in tomato plants (Figure 9). Most of the Arabidopsis Class II TPSs are not active enzymes as revealed by yeast complementation assays (Ramon et al., 2009); however, overexpression of AtTPS11 and its cotton homologous gene GhTPS11 in transgenic Arabidopsis plants resulted in increased trehalose contents (Singh et al., 2011; Wang et al., 2016), implying that some of the Class II TPSs are active enzymes in planta that can catalyze trehalose metabolism. Silencing of SlTPS3, SlTPS4, SlTPS5, or SlTPS7, encoding for Class II TPSs, led to decreased trehalose content (Figure 9), indicating that SlTPS3, SlTPS4, SlTPS5, and SlTPS7 may be active trehalose metabolic enzymes in tomato. We noted that reduced pathogen-induced trehalose accumulation correlates with the decreased resistance in SlTPS3/4/7-silenced plants to B. cinerea and in SlTPS4-silenced plants to Pst DC3000 while increased pathogen-induced trehalose accumulation associates with enhanced resistance to Pst DC3000 in SlTPP2-silenced plants (Figures 4, 7, and 9). This is similar to the observation that Arabidopsis tps11 mutant plants displayed reduced resistance to aphids while the AtTPS11-overexpressing plants contain elevated trehalose content and exhibited increased resistance to aphids (Singh et al., 2011). Notably, silencing of SlTPS4 or SlTPS5 had opposite effect on resistance to Pst DC3000 (Figure 7). Possible explanations include that SlTPS4 and SlTPS5 have differential effects on the Pst DC3000-induced trehalose accumulation in SlTPS4/5-silenced plants (Figure 9), or other members of the Class II TPSs may complement the function of SlTPS5 in SlTPS5-silenced plants upon infection of Pst DC300 via a yet-unknown mechanism. It was reported that Arabidopsis AtTPPa and AtTPPg have redundant roles in leaf growth, root hair specification and energy-responses (Van Houtte et al., 2013a). Alternatively, it cannot be ruled out the possibility that altered T6P level due to the silencing of SlTPS5 in catalyzing the formation of T6P is responsible for resistance to Pst DC3000 in SlTPS5-silenced plants. Silencing of SlTPP2 led to increased trehalose content and enhanced resistance to Pst DC3000 (Figures 7 and 9). This is similar to the observation that mutations in some Arabidopsis TPP genes resulted in increased levels of T6P and trehalose (Vandesteene et al., 2012). In addition, the SlTPSs with functions in resistance contribute differentially to resistance against different pathogens. For example, SlTPS4 is required for resistance against both of B. cinerea and Pst DC3000 while SlTPS3 and SlTPS7 have functions in resistance against B. cinerea but not to Pst DC3000. Collectively, our data demonstrate an important role for trehalose and its metablic genes in resistance against different pathogens.
It was previously shown that trehalose is capable of protecting against damage from ROS such as hydroxyl radicals (Couee et al., 2006; Luo et al., 2008) and that overexpression of yeast TPS1 in tomato plants increased tolerance to oxidative stress (Cortina and Culianez-Macia, 2005). We observed that the SlTPS3/4/7-silenced plants accumulated excessive level of H2O2 after infection by B. cinerea or Pst DC3000 (Figures 6 and 8). ROS accumulated during the late stage may favor for the development of diseases caused by necrotrophic pathogens such as B. cinerea and (hemi)biotrophic pathogens like Pst DC3000 (Govrin and Levine, 2000; Govrin et al., 2006; Temme and Tudzynski, 2009; Ishiga et al., 2012; Mengiste, 2012). Thus, it is likely that deregulation of ROS accumulation caused by silencing of SlTPS3, SlTPS4, and SlTPS7 may be responsible for the decreased resistance against B. cinerea and Pst DC3000 in SlTPS3/4/7-silenced plants. On the other hand, the expression of SA signaling-responsive defense-related genes such as SlRP1b and SlRPP2 and JA/ET signaling-responsive defense-related genes SlLapA and SlPIN2 was attenuated in the SlTPS3-, SlTPS4-, and SlTPS7-silenced plants after infection of Pst DC3000 or B. cinerea, respectively (Figures 6 and 8). This may also be due to the reduced level of trehalose in the SlTPS3-, SlTPS4-, and SlTPS7-silenced plants (Figure 9), because exogenous trehalose was found to induce the expression of defense-related genes in wheat and citrus (Tayeh et al., 2014; Piazza et al., 2015). It is therefore likely that the reduced trehalose content may be responsible for deregulation of ROS accumulation and attenuated expression of defense-related genes in the SlTPS3-, SlTPS4-, and SlTPS7-silenced plants. However, this hypothesis cannot be used to explain the mechanism for the increased resistance against Pst DC3000 in the SlTPP2-silenced plants, which had elevated trehalose content but had decreased accumulation of H2O2 and upregulated expression of the defense-related genes after infection with Pst DC3000 (Figure 8). The facts that SA and JA affected the expression of SlTPS3, SlTPS4, SlTPS5, SlTPS7, and SlTPP2 (Figure 2C) and that silencing of these genes also affected the expression of pathogen-induced defense genes (Figures 6 and 8) may indicate that trehalose or its metabolism act downstream of the SA and JA. This can be verified further by testing whether SA or JA can rescue the reduced resistance phenotype in the SlTPS3/4/5/7- and SlTPP2-silenced plants.
It was previously reported that P. brassica-induced expression of AtTRE1 acts as a defense response to limit trehalose accumulation (Brodmann et al., 2002; Gravot et al., 2011) and overexpression of AtTRE1 improves drought stress tolerance in Arabidopsis (Van Houtte et al., 2013b). By contrast, the expression of SlTRE1 was not induced by both of B. cinerea and Pst DC3000 (Figure 2) and silencing of SlTRE1 did not affect the resistance against B. cinerea and Pst DC3000 (Figures 4, 5 and 7), indicating that SlTRE1 may not be involved in disease resistance against these two pathogens. Interestingly, B. cinerea Δtre1 mutant showed elevated trehalose content but showed similar pathogenicity to wild-type strain (Doehlemann et al., 2006). Thus, it is likely that TRE1 has limited function in tomato-B. cinerea interaction, although trehalose serves as a stress protectant and as a significant but not essential carbon source for conidial germination in B. cinerea (Doehlemann et al., 2006).
Author Contributions
Conceived and designed the experiments: FS and HZ. Performed the experiments: HZ, DL, LH, YH, SL, LT, YD, ZC, and LH. Analyzed the data: FS and HZ. Contributed to reagents/materials/analysis tools: HZ and DL. Wrote the paper: FS and HZ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National High-Tech R & D Program (No. 2012AA101505), the National Natural Science Foundation (No. 31272028), and the Research Fund for the Doctoral Program of Higher Education of China (20120101110070).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01176
References
- Bae H., Herman E., Bailey B., Bae H. J., Sicher R. (2005). Exogenous trehalose alters Arabidopsis transcripts involved in cell wall modification, abiotic stress, nitrogen metabolism, plant defense. Physiol. Plant. 125 114–126. 10.1111/j.1399-3054.2005.00537.x [DOI] [Google Scholar]
- Blazquez M. A., Santos E., Flores C. L., Martinez-Zapater J. M., Salinas J., Gancedo C. (1998). Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J. 13 685–689. 10.1046/j.1365-313X.1998.00063.x [DOI] [PubMed] [Google Scholar]
- Brodmann A., Schuller A., Ludwig-Muller J., Aeschbacher R. A., Wiemken A., Boller T., et al. (2002). Induction of trehalase in Arabidopsis plants infected with the trehalose-producing pathogen Plasmodiophora brassicae. Mol. Plant Microbe Interact. 15 693–700. 10.1094/MPMI.2002.15.7.693 [DOI] [PubMed] [Google Scholar]
- Chary S. N., Hicks G. R., Choi Y. G., Carter D., Raikhel N. V. (2008). Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol. 146 97–107. 10.1104/pp.107.107441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortina C., Culianez-Macia F. A. (2005). Tomato abiotic stress enhanced tolerance by trehalose biosynthesis. Plant Sci. 169 75–82. 10.1016/j.plantsci.2005.02.026 [DOI] [Google Scholar]
- Couee I., Sulmon C., Gouesbet G., El-Amrani A. (2006). Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J. Exp. Bot. 57 449–459. 10.1093/jxb/erj027 [DOI] [PubMed] [Google Scholar]
- Debast S., Nunes-Nesi A., Hajirezaei M. R., Hofmann J., Sonnewald U., Fernie A. R., et al. (2011). Altering trehalose-6-phosphate content in transgenic potato tubers affects tuber growth and alters responsiveness to hormones during sprouting. Plant Physiol. 156 1754–1771. 10.1104/pp.111.179903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorge I., Figueroa C. M., Feil R., Lunn J. E., Van Dijck P. (2015). Trehalose-6-phosphate synthase 1 is not the only active TPS in Arabidopsis thaliana. Biochem. J. 466 283–290. 10.1042/BJ20141322 [DOI] [PubMed] [Google Scholar]
- Delorge I., Janiak M., Carpentier S., Van Dijck P. (2014). Fine tuning of trehalose biosynthesis and hydrolysis as novel tools for the generation of abiotic stress tolerant plants. Front. Plant Sci. 5:147 10.3389/fpls.2014.00147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G., Berndt P., Hahn M. (2006). Trehalose metabolism is important for heat stress tolerance and spore germination of Botrytis cinerea. Microbiology 152 2625–2634. 10.1099/mic.0.29044-0 [DOI] [PubMed] [Google Scholar]
- Eastmond P. J., van Dijken A. J. H., Spielman M., Kerr A., Tissier A. F., Dickinson H. G., et al. (2002). Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J. 29 225–235. 10.1046/j.1365-313x.2002.01220.x [DOI] [PubMed] [Google Scholar]
- Elbein A. D., Pan Y. T., Pastuszak I., Carroll D. (2003). New insights on trehalose: a multifunctional molecule. Glycobiology 13 17R–27R. 10.1093/glycob/cwg047 [DOI] [PubMed] [Google Scholar]
- Fernandez O., Vandesteene L., Feil R., Baillieul F., Lunn J. E., Clement C. (2012). Trehalose metabolism is activated upon chilling in grapevine and might participate in Burkholderia phytofirmans induced chilling tolerance. Planta 236 355–369. 10.1007/s00425-012-1611-4 [DOI] [PubMed] [Google Scholar]
- Figueroa C. M., Feil R., Ishihara H., Watanabe M., Kölling K., Krause U., et al. (2016). Trehalose 6-phosphate coordinates organic and amino acid metabolism with carbon availability. Plant J. 85 410–423. 10.1111/tpj.13114 [DOI] [PubMed] [Google Scholar]
- Garg A. K., Kim J. K., Owens T. G., Ranwala A. P., Choi Y. D., Kochian L. V., et al. (2002). Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. U.S.A. 99 15898–15903. 10.1073/pnas.252637799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L. F., Chao D. Y., Shi M., Zhu M. Z., Gao J. P., Lin H. X. (2008). Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 228 191–201. 10.1007/s00425-008-0729-x [DOI] [PubMed] [Google Scholar]
- Goddijn O. J. M., van Dun K. (1999). Trehalose metabolism in plants. Trends Plant Sci. 4 315–319. 10.1016/S1360-1385(99)01446-6 [DOI] [PubMed] [Google Scholar]
- Golem S., Culver J. N. (2003). Tobacco mosaic virus induced alterations in the gene expression profile of Arabidopsis thaliana. Mol. Plant Microbe Interact. 16 681–688. 10.1094/MPMI.2003.16.8.681 [DOI] [PubMed] [Google Scholar]
- Gómez L. D., Baud S., Gilday A., Li Y., Graham I. A. (2006). Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J. 46 69–84. 10.1111/j.1365-313X.2006.02662.x [DOI] [PubMed] [Google Scholar]
- Gómez L. D., Gildaz A., Feil R., Lunn J. E., Graham I. A. (2010). AtTPS1 mediated trehalose-6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds stomatal guard cells. Plant J. 64 1–13. 10.1111/j.1365-313X.2010.04312.x [DOI] [PubMed] [Google Scholar]
- Govrin E. M., Levine A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10 751–757. 10.1016/S0960-9822(00)00560-1 [DOI] [PubMed] [Google Scholar]
- Govrin E. M., Rachmilevitch S., Tiwari B. S., Solomon M., Levine A. (2006). An elicitor from Botrytis cinerea induces the hypersensitive response in Arabidopsis thaliana and other plants and promotes the gray mold disease. Phytopathology 96 299–307. 10.1094/PHYTO-96-0299 [DOI] [PubMed] [Google Scholar]
- Gravot A., Grillet L., Wagner G., Jubault M., Lariagon C., Baron C., et al. (2011). Genetic and physiological analysis of the relationship between partial resistance to clubroot and tolerance to trehalose in Arabidopsis thaliana. New Phytol. 191 1083–1094. 10.1111/j.1469-8137.2011.03751.x [DOI] [PubMed] [Google Scholar]
- Henry C., Bledsoe S. W., Siekman A., Kollman A., Waters B. M., Feil R., et al. (2014). The trehalose pathway in maize: conservation and gene regulation in response to the diurnal cycle and extended darkness. J. Exp. Bot. 65 5959–5973. 10.1093/jxb/eru335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge S., Ward J. L., Beale M. H., Bennett M., Mansfield J. W., Powell G. (2013). Aphid-induced accumulation of trehalose in Arabidopsis thaliana is systemic and dependent upon aphid density. Planta 237 1057–1064. 10.1007/s00425-012-1826-4 [DOI] [PubMed] [Google Scholar]
- Hofmann J., El-Ashry A. N., Anwar S., Erban A., Kopka J., Grundler F. (2010). Metabolic profiling reveals local and systematic responses of host plants to nematode parasitism. Plant J. 62 1058–1071. 10.1111/j.1365-313X.2010.04217.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordachescu M., Imai R. (2008). Trehalose biosynthesis in response to abiotic stresses. J. Integr. Plant Biol. 50 1223–1229. 10.1111/j.1744-7909.2008.00736.x [DOI] [PubMed] [Google Scholar]
- Ishiga Y., Ishiga T., Wangdi T., Mysore K. S., Uppalapati S. R. (2012). NTRC and chloroplast-generated reactive oxygen species regulate Pseudomonas syringae pv. tomato disease development in tomato and Arabidopsis. Mol. Plant Microbe Interact. 25 294–306. 10.1094/MPMI-05-11-0130 [DOI] [PubMed] [Google Scholar]
- Ishikawa R., Shirouzu K., Nakashita H., Lee H. Y., Motoyama T., Yamaguchi I., et al. (2005). Foliar spray of validamycin A or validoxylamine a controls Tomato fusarium wilt. Phytopathology 95 1209–1216. 10.1094/PHYTO-95-1209 [DOI] [PubMed] [Google Scholar]
- Jang I. C., Oh S. J., Seo J. S., Choi W. B., Song S. I., Kin C. H., et al. (2003). Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth. Plant Physiol. 131 516–524. 10.1104/pp.007237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S., Aronsson H., Ericson H., Pirhonen M., Leyman B., Welin B., et al. (2007). Improved drought tolerance without undesired side effects in transgenic plants producing trehalose. Plant Mol. Biol. 64 371–386. 10.1007/s11103-007-9159-6 [DOI] [PubMed] [Google Scholar]
- Krasensky J., Broyart C., Rabanal F. A., Jonak C. (2014). The redox-sensitive chloroplast trehalose-6-phosphate phosphatase AtTPPD regulates salt stress tolerance. Antioxid. Redox Signal. 21 1289–1304. 10.1089/ars.2013.5693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman B., Van Dijck P., Thevelein J. M. (2001). An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana. Trends Plant Sci. 6 510–513. 10.1016/S1360-1385(01)02125-2 [DOI] [PubMed] [Google Scholar]
- Li H. W., Zang B. S., Deng X. W., Wang X. P. (2011). Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 234 1007–1018. 10.1007/s00425-011-1458-0 [DOI] [PubMed] [Google Scholar]
- Li X., Huang L., Hong Y., Zhang Y., Liu S., Li D., et al. (2015). Co-silencing of tomato S-adenosylhomocysteine hydrolase genes confers increased immunity against Pseudomonas syringae pv. tomato. Front. Plant Sci. 6:717 10.3389/fpls.2015.00717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Huang L., Zhang Y., Ouyang Z., Hong Y., Zhang H., et al. (2014a). Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. l 14:286 10.1186/s12870-014-0286-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang Y., Huang L., Ouyang Z., Hong Y., Zhang H., et al. (2014b). Tomato SlMKK2 and SlMKK4 contribute to disease resistance against Botrytis cinerea. BMC Plant Biol. 14:166 10.1186/1471-2229-14-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Ouyang Z., Zhang Y., Li X., Hong Y., Huang L., et al. (2014). Tomato NAC transcription factor SlSRN1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response. PLoS ONE 9:e102067 10.1371/journal.pone.0102067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31 777–786. 10.1046/j.1365-313X.2002.01394.x [DOI] [PubMed] [Google Scholar]
- Lunn J. E. (2007). Gene families and evolution of trehalose metabolism in plants. Funct. Plant Biol. 34 550–563. 10.1071/FP06315 [DOI] [PubMed] [Google Scholar]
- Lunn J. E., Delorge I., Figueroa C. M., Van Dijck P., Stitt M. (2014). Trehalose metabolism in plants. Plant J. 79 544–567. 10.1111/tpj.12509 [DOI] [PubMed] [Google Scholar]
- Luo Y., Li W. M., Wang W. (2008). Trehalose: protector of antioxidant enzymes or reactive oxygen species scavenger under heat stress? Environ. Exp. Bot. 63 378–384. 10.1016/j.envexpbot.2007.11.016 [DOI] [Google Scholar]
- Mengiste T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50 267–294. 10.1146/annurev-phyto-081211-172955 [DOI] [PubMed] [Google Scholar]
- Miranda J. A., Avonce N., Suárez R., Thevelein J. M., Van Dijck P., Iturriaga G. (2007). A bifunctional TPS-TPP enzyme from yeast confers tolerance to multiple and extreme abiotic-stress conditions in transgenic Arabidopsis. Planta 226 1411–1421. 10.1007/s00425-007-0579-y [DOI] [PubMed] [Google Scholar]
- Mu M., Lu X. K., Wang J. J., Wang D. L., Yin Z. J., Wang S., et al. (2016). Genome-wide Identification and analysis of the stress-resistance function of the TPS (Trehalose-6-Phosphate Synthase) gene family in cotton. BMC Genet. 17:54 10.1186/s12863-016-0360-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C., O’Hara L. E., Primavesi L. F., Delatte T. L., Schluepmann H., Somsen G. W., et al. (2013). The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation. Plant Physiol. 162 1720–1732. 10.1104/pp.113.220657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. J., Primavesi L. F., Jhurreea D., Zhang Y. (2008). Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 59 417–441. 10.1146/annurev.arplant.59.032607.092945 [DOI] [PubMed] [Google Scholar]
- Piazza A., Zimaro T., Garavaglia B. S., Ficarra F. A., Thomas L., Marondedze C., et al. (2015). The dual nature of trehalose in citrus canker disease: a virulence factor for Xanthomonas citri subsp. citri and a trigger for plant defence responses. J. Exp. Bot. 66 2795–2811. 10.1093/jxb/erv095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnu J., Wahl V., Schmid M. (2011). Trehalose-6-phosphate: connecting plant metabolism and development. Front. Plant Sci. 2:70 10.3389/fpls.2011.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro M., Cazalé A. C., François J. M., Parrou J. L., Peeters N., Genin S. (2014). A Ralstonia solanacearum type III effector directs the production of the plant signal metabolite trehalose-6-phosphate. MBio 5 e2065–14 10.1128/mBio.02065-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik M. H., Imai R. (2005). Functional identification of a trehalose 6-phosphate phosphatase gene that is involved in transient induction of trehalose biosynthesis during chilling stress in rice. Plant Mol. Biol. 58 751–762. 10.1007/s11103-005-7404-4 [DOI] [PubMed] [Google Scholar]
- Ramon M., De Smet I., Vandesteene L., Naudts M., Leyman B., Van Dijck P., et al. (2009). Extensive expression regulation and lack of heterologous enzymatic activity of the Class II trehalose metabolism proteins from Arabidopsis thaliana. Plant Cell Environ. 32 1015–1032. 10.1111/j.1365-3040.2009.01985.x [DOI] [PubMed] [Google Scholar]
- Ramon M., Rolland F. (2007). Plant development: introducing trehalose metabolism. Trends Plant Sci. 12 185–188. 10.1016/j.tplants.2007.03.007 [DOI] [PubMed] [Google Scholar]
- Reignault P., Cogan A., Muchembled J., Sahouri A. L. H., Durand R., Sancholle M. (2001). Trehalose induces resistance to powdery mildew in wheat. New Phytol. 149 519–529. 10.1094/PHYTO-07-13-0191-R [DOI] [PubMed] [Google Scholar]
- Renard-Merlier D., Randoux B., Nowak E., Farcy F., Durand R., Reiqnault P. (2007). Iodus 40, salicylic acid, heptanoyl salicylic acid and trehalose exhibit different efficacies and defence targets during a wheat/powdery mildew interaction. Phytochemistry 68 1156–1164. 10.1016/j.phytochem.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Satoh-Nagasawa N., Nagasawa N., Malcomber S., Sakai H., Jackson D. (2006). A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441 227–230. 10.1038/nature04725 [DOI] [PubMed] [Google Scholar]
- Schluepmann H., Paulb M. (2009). Trehalose metabolites in Arabidopsis-elusive, active and central. Arabidopsis Book 14:e0122 10.1199/tab.0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima S., Matsui H., Tahara S., Imai R. (2007). Biochemical characterization of rice trehalose-6-phosphate phosphatases supports distinctive functions of these plant enzymes. FEBS J. 274 1192–1201. 10.1111/j.1742-4658.2007.05658.x [DOI] [PubMed] [Google Scholar]
- Singh V., Louis J., Ayre B. G., Reese J. C., Pegadaraju V., Shah J. (2011). TREHALOSE PHOSPHATE SYNTHASE11-dependent trehalose metabolism promotes Arabidopsis thaliana defense against the phloem-feeding insect Myzus persicae. Plant J. 67 94–104. 10.1111/j.1365-313X.2011.04583.x [DOI] [PubMed] [Google Scholar]
- Singh V., Shah J. (2012). Tomato responds to green peach aphid infestation with the activation of trehalose metabolism and starch accumulation. Plant Signal. Behav. 7 605–607. 10.4161/psb.20066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Bajad S., Shuman J., Shulaev V., Mittle R. (2008). The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J. Biol. Chem. 283 9269–9275. 10.1074/jbc.M709187200 [DOI] [PubMed] [Google Scholar]
- Tayeh C., Randoux B., Vincent D., Bourdon N., Reignault P. (2014). Exogenous trehalose induces defenses in wheat before and during a biotic stress caused by powdery mildew. Phytopathology 104 293–305. 10.1094/PHYTO-07-13-0191-R [DOI] [PubMed] [Google Scholar]
- Temme N., Tudzynski P. (2009). Does Botrytis cinerea ignore H2O2-induced oxidative stress during infection? Characterization of Botrytis activator protein 1. Mol. Plant Microbe Interact. 22 987–998. 10.1094/MPMI-22-8-0987 [DOI] [PubMed] [Google Scholar]
- Tsai A. Y., Gazzarrini S. (2014). Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: the emerging picture. Front. Plant Sci. 5:119 10.3389/fpls.2014.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken A. J. H., Schluepmann H., Smeekens S. C. M. (2004). Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol. 135 969–977. 10.1104/pp.104.039743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houtte H., López-Galvis L., Vandesteene L., Beeckman T., Van Dijck P. (2013a). Redundant and non-redundant roles of the trehalose-6-phosphate phosphatases in leaf growth, root hair specification and energy-responses in Arabidopsis. Plant Signal. Behav. 8:e23209 10.4161/psb.23209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houtte H., Vandesteene L., López-Galvis L., Lemmens L., Kissel E., Carpentier S., et al. (2013b). Overexpression of the trehalase gene AtTRE1 leads to increased drought stress tolerance in Arabidopsis and is involved in abscisic acid-induced stomatal closure. Plant Physiol. 161 1158–1171. 10.1104/pp.112.211391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesteene L., López-Galvis L., Vanneste K., Feil R., Maere S., Lammens W., et al. (2012). Expansive evolution of the trehalose-6-phosphate phosphatase gene family in Arabidopsis. Plant Physiol. 160 884–896. 10.1104/pp.112.201400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesteene L., Ramon M., Le Roy K., Van Dijck P., Rolland F. (2010). A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in Arabidopsis. Mol. Plant 3 406–419. 10.1093/mp/ssp114 [DOI] [PubMed] [Google Scholar]
- Wahl V., Ponnu J., Schlereth A., Arrivault S., Langenecker T., Franke A., et al. (2013). Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339 704–707. 10.1126/science.1230406 [DOI] [PubMed] [Google Scholar]
- Wang C. L., Zhang S. C., Qi S. D., Zheng C. C., Wu C. A. (2016). Delayed germination of Arabidopsis seeds under chilling stress by overexpressing an abiotic stress inducible GhTPS11. Gene 575 206–212. 10.1016/j.gene.2015.08.056 [DOI] [PubMed] [Google Scholar]
- Wingler A., Delatte T. L., O’Hara L. E., Primavesi L. F., Jhurreea D., Paul M. J., et al. (2012). Trehalose 6-phosphate is required for the onset of leaf senescence associated with high carbon availability. Plant Physiol. 158 1241–1251. 10.1104/pp.111.191908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D. W., Wang X. N., Fu L. S., Sun J., Zheng W., Li Z. F. (2015). Identification of the trehalose-6-phosphate synthase gene family in winter wheat and expression analysis under conditions of freezing stress. J. Genet. 94 55–65. 10.1007/s12041-015-0495-z [DOI] [PubMed] [Google Scholar]
- Yang H. L., Liu Y. J., Wang C. L., Zeng Q. Y. (2012). Molecular evolution of trehalose-6-phosphate synthase (TPS) gene family in Populus, Arabidopsis and rice. PLoS ONE 7:e42438 10.1371/journal.pone.0042438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Li H., Li W., Deng X. W., Wang X. (2011). Analysis of trehalose-6-phosphate synthase (TPS) gene family suggests the formation of TPS complexes in rice. Plant Mol. Biol. 76 507–522. 10.1007/s11103-011-9781-1 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu B., Li X., Ouyang Z., Huang L., Hong Y., et al. (2014). The de novo biosynthesis of vitamin B6 is required for disease resistance against Botrytis cinerea in tomato. Mol. Plant Microbe Interact. 27 688–699. 10.1094/MPMI-01-14-0020-R [DOI] [PubMed] [Google Scholar]
- Zhou M. L., Zhang Q., Sun Z. M., Chen L. H., Liu B. X., Zhang K. X., et al. (2014). Trehalose metabolism-related genes in maize. J. Plant Growth Regul. 33 256–271. 10.1007/s00344-013-9368-y [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.