Abstract
Rabbits have been suggested as a zoonotic source of Hepatitis E virus. Phylogenetic analysis of HEV isolates from farmed, wild and pet rabbits in the Netherlands (23, 0, and 60 % respectively) showed them to be grouped amongst published rabbit HEV sequences and distinct from most human isolates. Dutch rabbits are unlikely to be a zoonotic source.
Electronic supplementary material
The online version of this article (doi:10.1007/s12560-016-9239-3) contains supplementary material, which is available to authorized users.
Keywords: Hepatitis E, HEV, Rabbits, Zoonosis
Introduction
Hepatitis E virus (HEV) incidence in the Netherlands seems to be on the rise (Koot et al. 2015). About two-thirds of the acute HEV cases in the Netherlands were suggested to be unrelated to travel to endemic countries (Herremans et al. 2007). Domestic pigs have been demonstrated to be a true reservoir (Bouwknegt et al. 2008) and seem to be a main source of infection (Van der Poel et al. 2001), but other species may play a role also (Meng 2010; Rutjes et al. 2014). Recently, rabbit HEV isolates were reported to be phylogenetically closely related to a particular human strain (Caruso et al. 2015; Izopet et al. 2012), suggesting rabbits as another potential source for zoonotic transmission. Since rabbits are widely kept as house pets and are also farmed for food and exist as an extensive wild population throughout the country, the consequences of possible zoonotic transmission of rabbit HEV may be far-reaching. We therefore investigated HEV in rabbits on petting farms, farmed rabbits and wild rabbits in the Netherlands to assess the likelihood of zoonotic transmission.
Materials and Methods
In 2013 faecal droppings were collected from 35 rabbits at 12 petting farms. Faecal samples and liver samples were collected from 10 farmed rabbits at slaughter. Faecal and liver samples (32 and 30, respectively) were collected from 32 wild rabbits shot by hunters at 11 locations throughout the Netherlands. Samples were stored at −20 °C until analysis. HEV RNA was isolated and Reverse Transcription-PCR for HEV was carried out to amplify a fragment of 70 base pairs using the method of Jothikumar et al. (2006) with adaptations. The RNA primers and probes used targeted the ORF 3 region (starting at bp 27). Primer and probe sequences were as follows: forward primer JVHEVF: 5′ GGTGGTTTCTGGGGTGAC 3′, reverse primer JVHEVR: 5′ AGGGGTTGGTTGGATGAA 3′ and probe JVHEVP: 5′ TGATTCTCAGCCCTTCGC 3′. The presence of a 70-bp-long fragment in the RT-PCR products was confirmed by gel electrophoresis (3 % agarose gel in Tris–borate buffer with EDTA, stained with SybrGold) (Online Resource 1).
RT-PCR amplification products of five samples showing the lowest Ct values (highest RNA concentrations) in the screening RT-PCR (see above) were amplified using a nested RT-PCR format targeting an ORF2 fragment of HEV using a method presented at a meeting of the Dutch Society for Clinical Virology, January 2012, Arnhem, The Netherlands (Online Resource 2). With this PCR, a product of 493 bp was obtained. The sequences of the rabbit isolates were subjected to phylogenetic analysis together with published sequences for human HEV from the Netherlands (healthy blood donors and liver patients) and for rabbit HEV and human HEV from other countries.
Results
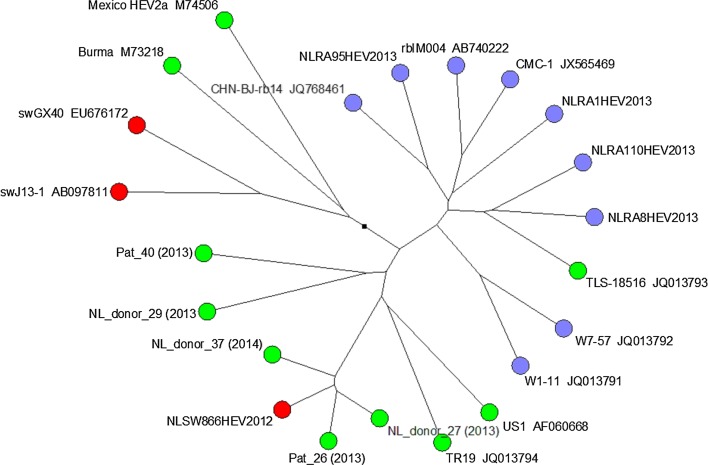
The prevalence of HEV was 8/35 (23 %) in petting farm rabbit droppings, 0/10 (0 %) in farmed rabbits, 5/32 (16 %) in wild rabbit faeces and 18/30 (60 %) in wild rabbit liver samples. Of the five wild rabbits with positive faecal samples, four of these also had positive liver samples. Phylogenetic analysis of PCR amplification products from two pets and two wild rabbits showed that they were located amongst sequences from rabbit isolates from other countries, as shown in Fig. 1. Sequences from Dutch blood donors and other human sequences from other regions do not cluster with the detected Dutch rabbit sequences except one. This concerns an earlier reported sequence of a human liver patient (TLS-18516 (Izopet et al. 2012)).
Fig. 1.
Phylogenetic tree of HEV ORF2 sequences detected in rabbits in the Netherlands (blue circles, labelled NLRA) related to HEV sequences of rabbits elsewhere (remaining blue circles), humans (green) and swine (red) reported in the literature
Discussion
To the authors’ knowledge, there is only one previous report of HEV in a pet rabbit. A house rabbit that had died suddenly was found to be HEV positive; the sequence similarity was determined to be 82.6–90.1 % with rabbit HEV and 71.4–89.5 % with human HEV (Caruso et al. 2015). HEV prevalence in farmed rabbits is reported to vary from 7 to 15 % as measured by HEV RNA in liver or faeces and 16–55 % as measured by the seroprevalence of HEV-specific antibodies (Cossaboom et al. 2011; Geng et al. 2011; Izopet et al. 2012). The small number of farmed rabbits sampled in the present study (n = 10) may have contributed to the lack of HEV-positive faecal samples. In a French study in which faeces from 20 groups of ten farmed rabbits were sampled, 13/20 of these groups were also negative for HEV RNA (Izopet et al. 2012). In the present study, we investigated viral RNA in faeces rather than seropositivity since the faecal–oral route is relevant to possible zoonotic infection. HEV prevalence presented here for wild rabbit livers (18/30 or 60 %) is higher than has been reported in France (47/205 or 23 %) (Izopet et al. 2012). This difference in prevalence may be due to differences in ecology, climate and geography of the two countries. However, differences in the numbers of samples or the methods and primers that were used may also contribute to this difference.
Phylogenetic analysis of PCR amplification products from pets and wild rabbits showed that they were located amongst sequences from rabbit isolates from other countries (Fig. 1). This location is distinct from published human HEV sequences including those from healthy blood donors and liver patients in the Netherlands but it is close to one isolate from a hospital patient in France (Izopet et al. 2012). These results are in agreement with papers that propose a classification of rabbit HEV as a distinct subtype of genotype 3, but this has not yet been adopted (Cossaboom et al. 2012; Smith et al. 2016; Vina-Rodriguez et al. 2015).
In conclusion, phylogenetic analysis of rabbit HEV from pets and wild rabbits showed them to be grouped amongst published rabbit HEV sequences and distinct from most human isolates. Rabbits in the Netherlands are therefore considered unlikely to be a source of zoonotic infection.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors are grateful to the petting zoos, hunters and rabbit abattoir for their cooperation in providing samples. The authors thank Gerdit Greve for technical assistance.
Financial Support Statement
This study was commissioned and supported by Utrecht University, the Dutch National Institute for Public Health and the Environment, and the Dutch Ministry of Agriculture and Foods.
Compliance with Ethical Standards
Conflict of interest
No conflicts of interest are declared.
Ethical Approval
All applicable international, national and institutional guidelines for the care and use of animals were followed.
References
- Bouwknegt M, Frankena K, Rutjes S, Wellenberg GJ, de Roda Husman AM, Van der Poel WHM, de Jong MCM. Basic reproduction ratio for hepatitis E virus transmission in swine by contact-exposure in one-to-one trials. Veterinary Research. 2008;39(5):40. doi: 10.1051/vetres:2008017. [DOI] [PubMed] [Google Scholar]
- Caruso C, Modesto P, Prato R, Scaglione FE, De Marco L, Bollo E, et al. Hepatitis E virus: First description in a pet house rabbit. A new transmission route for humans? Transboundary and Emerging Diseases. 2015;62:229–232. doi: 10.1111/tbed.12348. [DOI] [PubMed] [Google Scholar]
- Cossaboom CM, Cordoba L, Cao D, Ni YY, Meng XJ. Complete genome sequence of hepatitis E virus from rabbits in the United States. Journal of Virology. 2012;86(23):13124–13125. doi: 10.1128/JVI.02414-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossaboom CM, Córdoba L, Dryman B, Meng X-J. Hepatitis E virus in rabbits, Virginia, USA. Emerging Infectious Diseases. 2011;17:2047–2049. doi: 10.3201/eid1711.110428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Wang L, Wang X, Fu H, Bu Q, Zhu Y, Zhuang H. Study on prevalence and genotype of hepatitis E virus isolated from Rex Rabbits in Beijing, China. Journal of Viral Hepatitis. 2011;18:661–667. doi: 10.1111/j.1365-2893.2010.01341.x. [DOI] [PubMed] [Google Scholar]
- Herremans M, Vennema H, Bakker J, van der Veer B, Duizer E, Benne CA, Waar K, Hendrixks B, Schneeberger P, Blaauw G, Kooiman M, Koopmans MP. Swine-like hepatitis E viruses are a cause of unexplained hepatitis in the Netherlands. Journal of Viral Hepatitis. 2007;14:140–146. doi: 10.1111/j.1365-2893.2006.00786.x. [DOI] [PubMed] [Google Scholar]
- Izopet J, Dubois M, Bertagnoli S, Lhomme S, Marchandeau S, Boucher S, Kamar N, Abravanel F, Guérin JL. Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerging Infectious Diseases. 2012;18:1274–1281. doi: 10.3201/eid1808.120057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothikumar N, Cromeansa TL, Robertson BH, Meng XJ, Hill VR. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. Journal of Virological Methods. 2006;131:65–71. doi: 10.1016/j.jviromet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Koot H, Hogema BM, Koot M, Molier M, Zaaijer HL. Frequent hepatitis E in the Netherlands without traveling or immunosuppression. Journal of Clinical Virology. 2015;62:38–40. doi: 10.1016/j.jcv.2014.11.020. [DOI] [PubMed] [Google Scholar]
- Meng XJ. Hepatitis E virus: Animal reservoirs and zoonotic risk. Veterinary Microbiology. 2010;140:256–265. doi: 10.1016/j.vetmic.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjes SA, Bouwknegt M, van der Giessen JW, de Roda Husman AM, Reusken CBEM. Seroprevalence of hepatitis E virus in pigs from different farming systems in the Netherlands. Journal of Food Protection. 2014;77:640–642. doi: 10.4315/0362-028X.JFP-13-302. [DOI] [PubMed] [Google Scholar]
- Smith D, Simmonds P, Izopet J, Oliveira-Filho E, Ulrich R, Johne R, Koenig M, Jameel S, Harrison T, Meng X, Okamoto H, Van der Poel W, Purdy M. Proposed reference sequences for hepatitis E virus subtypes. Journal of General Virology. 2016;97(3):537–542. doi: 10.1099/jgv.0.000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Poel WHM, Verschoor F, van der Heide R, Herrera M-I, Vivo A, Kooreman M, de Roda Husman AM. Hepatitis E virus sequences in swine related to sequences in humans, the Netherlands. Emerging Infectious Diseases. 2001;7:970–976. doi: 10.3201/eid0706.010608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vina-Rodriguez A, Schlosser J, Becher D, Kaden V, Groschup MH, Eiden M. Hepatitis E virus genotype 3 diversity: A subtyping update and first detection of Genotype 3b in animals in Europe. Viruses. 2015;7:2704–2726. doi: 10.3390/v7052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.