Abstract
Background:
Previous studies have revealed that miR-26a-5p and miR-26b-5p act as tumour suppressors in various types of cancer tissues. Here, we aimed to investigate the functional roles of these miRNAs and to identify their regulatory targets in bladder cancer (BC).
Methods:
We performed functional assays in BC cells using transfection of mature microRNAs (miRNAs). In silico and luciferase reporter analyses were applied to identify target genes of these miRNAs. The overall survival (OS) of patients with BC was evaluated by the Kaplan–Meier method.
Results:
miR-26a-5p and miR-26b-5p were significantly downregulated in BC tissues. Restoration of these miRNAs inhibited cell migration and invasion in BC. The gene encoding procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (PLOD2), a collagen crosslinking enzyme, was directly regulated by miR-26a-5p and miR-26b-5p. Kaplan–Meier analysis revealed that patients with high PLOD2 expression had significantly shorter OS compared with those with low PLOD2 expression (P=0.0153).
Conclusions:
PLOD2, which is associated with the stiffness of the extracellular matrix, was directly regulated by miR-26a-5p and miR-26b-5p and may be a good prognostic marker in patients with BC.
Keywords: microRNA, miR-26a-5p, miR-26b-5p, PLOD2, bladder cancer, tumour suppressor
In developed countries, bladder cancer (BC) is the fifth most commonly diagnosed cancer (Siegel et al, 2012). Bladder cancer can be categorised as non-muscle-invasive BC (NMIBC) and muscle-invasive BC (MIBC); the 5-year survival rates for patients with MIBC and NMIBC are 60% and 90%, respectively (Zuiverloon et al, 2012). Nearly 80% of patients with distant metastases die in the first 5 years after diagnosis (Meeks et al, 2012). The molecular mechanisms of recurrence and metastasis in BC are not well understood. Previous studies have not yet identified effective chemotherapies for advanced BC (Bellmunt and Petrylak, 2012). Thus, although patients with advanced BC are generally treated with chemotherapy using gemcitabine and cisplatin, progression-free survival is short (De Santis et al, 2012). Most clinical trials evaluating chemotherapies or molecular-targeted therapeutics for advanced BC have shown limited benefits, and there are currently no effective second-line chemotherapies available (Ghosh et al, 2014). Therefore, novel prognostic markers and effective treatment strategies based on RNA network studies are needed to improve outcomes in patients with BC.
MicroRNAs (miRNAs) are small noncoding RNAs (19–22 bases in length) that regulate protein-coding genes by binding to the 3′-untranslated region (UTR) of the target mRNA, thereby inhibiting transcription (Bartel, 2004; Carthew and Sontheimer, 2009). Previous studies have shown that miRNAs are aberrantly expressed in various human cancers and have significant roles in human oncogenesis and metastasis (Di Leva and Croce, 2010). Therefore, detection of aberrantly expressed miRNAs is an important first step in the elucidation of miRNA-regulated oncogenic pathways.
Our laboratory has characterised the miRNA expression signatures of several human cancers and revealed that miR-26a-5p and miR-26b-5p are frequently downregulated in various types of cancer, suggesting that these miRNAs function as tumour suppressors by targeting multiple oncogenes (Fukumoto et al, 2015; Kato et al, 2015). However, the functional roles of these miRNAs in BC are unclear. The aim of the present study was to investigate the functional roles of miR-26a-5p and miR-26b-5p and to identify their molecular targets in BC cells. Our data demonstrated that miR-26a-5p and miR-26b-5p were significantly downregulated in clinical BC specimens and that transfection of BC cells with these miRNAs significantly inhibited cancer cell migration and invasion. In silico analyses suggested that the gene encoding procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (PLOD2), a collagen crosslinking enzyme, was a promising candidate target gene of these miRNAs. PLOD2 has been shown to be associated with extracellular matrix (ECM) stiffness and dysregulation of the ECM (Gilkes et al, 2014). The discovery of molecular targets regulated by tumour-suppressive miRNAs provides important insights into the potential mechanisms of BC oncogenesis and suggests novel therapeutic strategies for the treatment of BC.
Materials and methods
Clinical specimens and cell culture
The tissue specimens for quantitative real-time reverse transcription–polymerase chain reaction (qRT–PCR) were collected from patients with BC (n=69) who had received cystectomy (n=10) or transurethral resection of their bladder tumours (TURBT; n=59) at the Kagoshima University Hospital between 2003 and 2015. Normal bladder epithelia (n=23) were derived from patients with noncancerous disease. The specimens were staged according to the American Joint Committee on Cancer-Union Internationale Contre le Cancer tumour-node-metastasis (TNM) classification and histologically graded (Sobin and Compton, 2010). Our study was approved by the Bioethics Committee of the Kagoshima University; written prior informed consent and approval were obtained from all patients. Patient details and clinicopathological characteristics are summarised in Supplementary Table 1. The median follow-up of the patients was 45.8 months.
The human BC cell lines T24 and BOY were maintained in the recommended medium containing 10% foetal bovine serum, 50 μg ml−1 streptomycin, and 50 U ml−1 penicillin in a humidified atmosphere of 95% air/5% CO2 at 37 °C. Routine tests for mycoplasma infection were negative (Inoguchi et al, 2014; Itesako et al, 2014).
Tissue collection and RNA extraction
Tissues were immersed in RNAlater (Thermo Fisher Scientific, Waltham, MA, USA) and stored at −20 °C until RNA extraction. Total RNA, including miRNA, was extracted using a mirVana miRNA Isolation Kit (Thermo Fisher Scientific) following the manufacturer's protocol.
qRT–PCR
Stem-loop RT–PCR (TaqMan MicroRNA Assays; P/N: 000405 for miR-26a-5p, and P/N: 000407 for miR-26b-5p; Applied Biosystems, Foster City, CA, USA) was used to quantify miRNAs according to previously published conditions (Ichimi et al, 2009). TaqMan probes and primers for PLOD2 (P/N: Hs 01118190_m1; Applied Biosystems) were assay-on-demand gene expression products. We used human GUSB (P/N: Hs99999908_m1; Applied Biosystems) and RNU48 (P/N: 001006; Applied Biosystems) as internal controls, and the ΔCt method was used to calculate the fold changes relative to the expression levels of internal controls.
Transfection with mature miRNA and siRNA
As described elsewhere (Ichimi et al, 2009; Yoshino et al, 2011), T24 and BOY cells were transfected with Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) and Opti-MEM (Thermo Fisher Scientific) with 10 nM mature miRNA. Mature miRNA and negative-control miRNA (Applied Biosystems) were used in gain-of-function experiments, whereas PLOD2 small interfering RNA (siRNA) (cat. nos HSS108124 and HSS108125; Thermo Fisher Scientific) and negative-control siRNA (D-001810-10; Thermo Fisher Scientific) were used in loss-of-function experiments.
Cell proliferation, migration, and invasion assays
T24 and BOY cells were transfected with 10 nM miRNA or siRNA by reverse transfection. Cells were seeded in 96-well plates at 3 × 103 cells per well for XTT assays. After 72 h, cell proliferation was determined using a Cell Proliferation Kit II (Roche Diagnostics GmbH, Mannheim, Germany) as described previously (Tatarano et al, 2011; Kojima et al, 2012; Nohata et al, 2013). Cell migration activity was evaluated with wound healing assays. Cells were plated in 6-well plates at 2 × 105 cells per well, and after 48 h of transfection, the cell monolayer was scraped using a P-20 micropipette tip. The initial gap length (0 h) and the residual gap length 24 h after wounding were calculated from photomicrographs as described (Tatarano et al, 2011; Kojima et al, 2012; Nohata et al, 2013). Cell invasion assays were performed using modified Boyden chambers consisting of Transwell precoated Matrigel membrane filter inserts with 8-mm pores in 24-well tissue culture plates (BD Biosciences, Bedford, MA, USA). At 72 h after transfection, cells were plated in 24-well plates at 1 × 105 cells per well. Minimum essential medium containing 10% foetal bovine serum in the lower chamber served as the chemoattractant, as described previously (Tatarano et al, 2011; Kojima et al, 2012; Nohata et al, 2013). All experiments were performed in triplicate.
Western blot analysis
Cells were harvested 72 h after transfection, and lysates were prepared. Fifty micrograms of protein was separated by NuPAGE on 4–12% bis-tris gels (Invitrogen, Carlsbad, CA, USA) and transferred to PVDF membranes. Immunoblotting was carried out with diluted anti-PLOD2 antibodies (1 : 200; 21214-1-AP; Proteintech, Chicago, IL, USA) and anti-GAPDH antibodies (MAB374; Chemicon, Temecula, CA, USA). Specific complexes were visualised with an Echo-chemiluminescence Detection System (GE Healthcare, Little Chalfont, UK) as described previously (Yoshino et al, 2011; Kinoshita et al, 2013).
Genome-wide gene expression analysis and in silico analysis for the identification of genes regulated by miR-26a-5p and miR-26b-5p
In silico analysis was used to identify target genes of miR-26a-5p and miR-26b-5p. To obtain candidate target genes regulated by miR-26a-5p and miR-26b-5p, we used the TargetScan database Release 6.2 (http://www.targetscan.org). Additionally, the Gene Expression Omnibus database (accession numbers: GSE11783 and GSE31684) was used to identify upregulated genes in BC specimens.
Plasmid construction and dual-luciferase reporter assays
Partial wild-type sequences of the 3′-UTR of PLOD2 or sequences with deletion of the miR-26a-5p and miR-26b-5p target sites (positions 905–912 and 1188–1194 of the PLOD2 3′-UTR) were inserted between the XhoI and PmeI restriction sites in the 3′-UTR of the PLOD2 gene in the psiCHECK-2 vector (C8021; Promega, Madison, WI, USA). The protocol for vector construction was described previously (Chiyomaru et al, 2010; Kinoshita et al, 2013). The procedure for dual-luciferase reporter assay was described previously (Yoshino et al, 2013a).
Statistical analysis
The relationships between two groups were analysed using Mann–Whitney U-tests. The relationships between three variables and numerical values were analysed using Bonferroni-adjusted Mann–Whitney U-tests. Overall survival (OS) in patients with BC was evaluated by the Kaplan–Meier method. Patients were divided into two groups according to the median value of PLOD2 expression, and the differences between the two groups were evaluated by log-rank tests. Multivariable analysis was evaluated by the Cox proportional hazards model. All analyses were carried out using Expert StatView software, version 5.0 (SAS Institute, Cary, NC, USA).
Results
Expression levels of miR-26a-5p and miR-26b-5p in BC
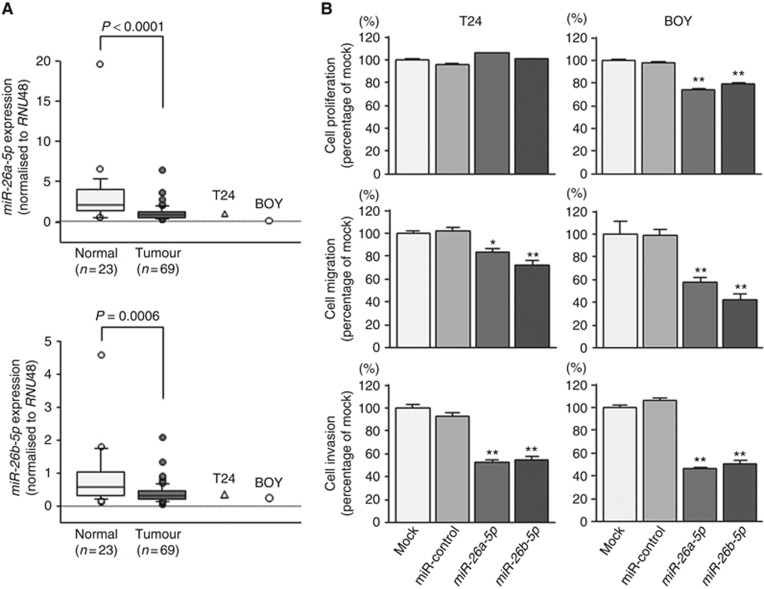
First, we evaluated the expression levels of miR-26a-5p and miR-26b-5p in BC tissues (n=69), and normal bladder epithelia (n=23). The expression levels of miR-26a-5p and miR-26b-5p were significantly reduced in tumour tissues compared with those in normal bladder epithelia (P<0.0001 and P=0.0006, respectively; Figure 1A). There were no significant relationships between any of the clinicopathological parameters (i.e., tumour stage, grade, and survival rate) and the expression levels of miR-26a-5p and miR-26b-5p (data not shown).
Figure 1.
(A) Expression levels of miR-26a-5p and miR-26b-5p. Quantitative real-time reverse transcription–polymerase chain reaction showed that the expression levels of miR-26a-5p and miR-26b-5p were significantly lower in BC tissues and BC cell lines compared with that in non-BC tissues (P<0.0001 and P=0.0006, respectively). (B) Effects of miR-26a-5p and miR-26b-5p transfection on the functionality of BC cell lines. The XTT assay showed that cell proliferation was inhibited in miR-26a-5p/26b-5p-transfected BOY cells compared with that in mock or miR-control transfectants. However, cancer cell proliferation was not exhibited in miR-26a-5p/26b-5p-transfected T24 cells compared with that in mock or miR-control transfectants. Migration and invasion assays demonstrated that cell migration and invasion were significantly inhibited in miR-26a-5p- and miR-26b-5p-transfected cells in comparison with those in mock- or miR-control-transfected cells. *P=0.002; **P<0.0001.
Effects of miR-26a-5p and miR-26b-5p restoration on cell proliferation, migration, and invasion activities in BC cell lines
We performed gain-of-function studies using miR-26a-5p or miR-26b-5p transfected T24 and BOY cells to investigate the functional roles of these miRNAs. XTT assays showed that miR-26a-5p and miR-26b-5p transfection inhibited cancer cell proliferation in BOY cells compared with that in mock or miR-control transfectants (Figure 1B). Moreover, migration assays demonstrated that cell migration activity was significantly inhibited in miR-26a-5p and miR-26b-5p transfectants in comparison with that in mock or miR-control transfectants (Figure 1B). Finally, Matrigel invasion assays demonstrated that cell invasion activity was significantly inhibited in miR-26a-5p and miR-26b-5p transfectants in comparison with that in mock or miR-control transfectants (Figure 1B). These data suggested that miR-26a-5p and miR-26b-5p functioned as tumour suppressors via inhibition of cell migration and invasion in BC.
Identification of molecular pathways modulated by miR-26a/b and putative target genes in BC cells
Next, in silico analysis was used to gain additional insights into the molecular mechanisms and pathways regulated by tumour-suppressive miR-26a-5p and miR-26b-5p in BC cells. Candidate miR-26-regulated genes were identified using TargetScan database Release 6.2 (http://www.targetscan.org). Among these candidate genes, we selected 112 genes that had two or more conserved sites (Supplementary Table 2). We applied the Gene Expression Omnibus database (accession numbers: GSE11783 and GSE31684) to identify upregulated genes in BC specimens and subsequently selected the 28 genes listed in Table 1. For knockdown studies, it is necessary to select highly expressed genes. Evaluation of the expression levels of the top 6 genes in Table 1 by qRT–PCR showed that LOXL2 and PLOD2 were adequately expressed in the examined cell lines (Supplementary Figure 1). In this report, we have focused on PLOD2; studies of LOXL2 are ongoing in our laboratory.
Table 1. Putative candidate of target genes.
|
TargetScan |
Gene expression omnibus (GSE11783+GSE31684) |
||||||
|---|---|---|---|---|---|---|---|
| Entrez gene ID | Gene symbol | Description | Genomic location | Number of conserved sites | Number of poorly conserved sites | Fold change | P-value |
| 647309 | GEMC1 | Geminin coiled-coil domain-containing protein 1 | 3q28 | 2 | 0 | 10.139 | 9.420E−05 |
| 3759 | KCNJ2 | Potassium inwardly rectifying channel, subfamily J, member 2 | 17q24.3 | 2 | 0 | 6.435 | 5.670E−05 |
| 4017 | LOXL2 | Lysyl oxidase-like 2 | 8p21.3 | 2 | 0 | 4.706 | 5.316E−05 |
| 5352 | PLOD2 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | 3q24 | 2 | 0 | 4.616 | 1.135E−04 |
| 170692 | ADAMTS18 | ADAM metallopeptidase with thrombospondin type 1 motif, 18 | 16q23 | 2 | 0 | 4.186 | 4.376E−05 |
| 55117 | SLC6A15 | Solute carrier family 6 (neutral amino-acid transporter), member 15 | 12q21.3 | 2 | 0 | 4.119 | 1.640E−04 |
| 144455 | E2F7 | E2F transcription factor 7 | 12q21.2 | 2 | 0 | 3.726 | 5.316E−05 |
| 23657 | SLC7A11 | Solute carrier family 7 (anionic amino-acid transporter light chain, xc system), member 11 | 4q28.3 | 2 | 2 | 3.381 | 1.003E−04 |
| 84206 | MEX3B | Mex-3 homologue B (Caenorhabditis elegans) | 15q25.2 | 2 | 0 | 3.293 | 9.231E−03 |
| 1993 | ELAVL2 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) | 9p21 | 3 | 0 | 3.237 | 7.790E−04 |
| 10847 | SRCAP | Snf2-related CREBBP activator protein | 16p11.2 | 2 | 0 | 2.818 | 8.308E−05 |
| 7468 | WHSC1 | Wolf–Hirschhorn syndrome candidate 1 | 4p16.3 | 2 | 0 | 2.590 | 4.670E−05 |
| 9120 | SLC16A6 | Solute carrier family 16, member 6 (monocarboxylic acid transporter 7) | 17q24.2 | 2 | 0 | 2.431 | 1.285E−04 |
| 54620 | FBXL19 | F-box and leucine-rich repeat protein 19 | 16p11.2 | 2 | 0 | 2.429 | 1.405E−03 |
| 10622 | POLR3G | Polymerase (RNA) III (DNA directed) polypeptide G (32 kDa) | 5q14.3 | 2 | 2 | 2.281 | 1.642E−03 |
| 64768 | IPPK | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase | 9q22.31 | 2 | 0 | 2.223 | 1.067E−04 |
| 23276 | KLHL18 | Kelch-like 18 (Drosophila) | 3p21.31 | 2 | 1 | 1.854 | 4.670E−05 |
| 124801 | LSM12 | LSM12 homologue (Saccharomyces cerevisiae) | 17q21.31 | 2 | 0 | 1.795 | 1.003E−04 |
| 54726 | OTUD4 | OTU domain containing 4 | 4q31.21 | 2 | 1 | 1.589 | 2.975E−04 |
| 2182 | ACSL4 | Acyl-CoA synthetase long-chain family member 4 | Xq22.3-q23 | 2 | 0 | 1.589 | 8.447E−03 |
| 51701 | NLK | Nemo-like kinase | 17q11.2 | 2 | 1 | 1.548 | 1.405E−03 |
| 200424 | TET3 | Tet oncogene family member 3 | 2p13.1 | 3 | 1 | 1.456 | 1.421E−02 |
| 26973 | CHORDC1 | Cysteine and histidine-rich domain (CHORD) containing 1 | 11q14.3 | 2 | 1 | 1.435 | 7.383E−03 |
| 23633 | KPNA6 | Karyopherin α6 (importin α7) | 1p35.1 | 2 | 1 | 1.413 | 7.373E−04 |
| 85403 | EAF1 | ELL-associated factor 1 | 3p25.1 | 2 | 1 | 1.372 | 1.148Ev02 |
| 27 | ABL2 | v-abl Abelson murine leukaemia viral oncogene homologue 2 | 1q25.2 | 3 | 2 | 1.371 | 3.487E−03 |
| 23367 | LARP1 | La ribonucleoprotein domain family, member 1 | 5q33.2 | 2 | 1 | 1.299 | 2.057E−02 |
| 114885 | OSBPL11 | Oxysterol binding protein-like 11 | 3q21 | 2 | 0 | 1.289 | 1.749E−02 |
PLOD2 was directly regulated by miR-26a-5p and miR-26b-5p in BC cells
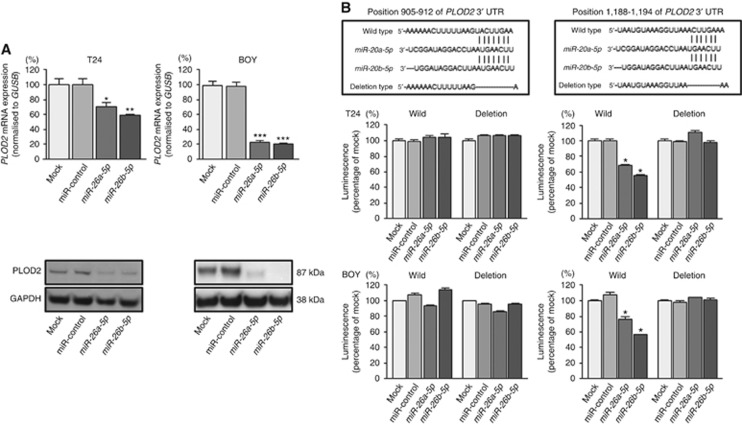
We performed qRT–PCR analysis and western blot analyses to confirm that restoration of miR-26a-5p and miR-26b-5p resulted in downregulation of PLOD2 in T24 and BOY cells. PLOD2 mRNA and protein levels were significantly reduced in miR-26a-5p and miR-26b-5p transfectants in comparison with those in mock or miR-control transfectants (Figure 2A).
Figure 2.
Direct regulation of PLOD2 by miR-26a-5p and miR-26b-5p.(A) The expression of PLOD2 was significantly repressed in miR-26a-5p/26b-5p transfectants in comparison with that in mock or miR-control transfectants. GUSB was used as an internal control. *P=0.0109; **P=0.0016; ***P<0.0001. The expression of PLOD2 protein was significantly repressed in miR-26a-5p/26b-5p transfectants in comparison with that in mock or miR-control transfectants. GAPDH was used for a loading control. (B) miR-26a-5p/26b-5p binding sites at positions 1188–1194 in the 3′-UTR of PLOD2 mRNA. Dual-luciferase reporter assays using vectors encoding putative miR-26a-5p/26b-5p target sites for wild-type or deleted regions. Normalised data were calculated as ratios of Renilla/firefly luciferase activities. The luminescence intensity was significantly reduced by co-transfection with miR-26a-5p/26b-5p and the vector carrying the wild-type sequence for positions 1188–1194 in the 3′-UTR of PLOD2 mRNA, whereas transfection with the deletion vector and the wild-type sequence for positions 905–912 in the 3′-UTR of PLOD2 mRNA blocked the decrease in luminescence. *P<0.0001.
We then performed dual-luciferase reporter assays in T24 and BOY cells to determine whether PLOD2 was directly regulated by these miRNAs. The TargetScan database predicted that there were two binding sites for miR-26a-5p and miR-26b-5p at positions 905–912 and 1188–1194 in the PLOD2 3′-UTR. We used vectors encoding the partial wild-type sequence of the 3′-UTR of PLOD2, including the predicted miR-26a-5p/miR-26b-5p target sites. We found that the luminescence intensity was significantly reduced by co-transfection with miR-26a-5p or miR-26b-5p and the vector carrying the wild-type sequences at positions 1188–1194 of the PLOD2 3′-UTR, whereas transfection with the deletion vector (in which the binding site had been removed) and the wild-type sequence at positions 905–912 of the PLOD2 3′-UTR blocked the decrease in luminescence (Figure 2B). These data suggested that miR-26a-5p/26b-5p bound directly to specific sites at positions 1188–1194 of the PLOD2 3′-UTR.
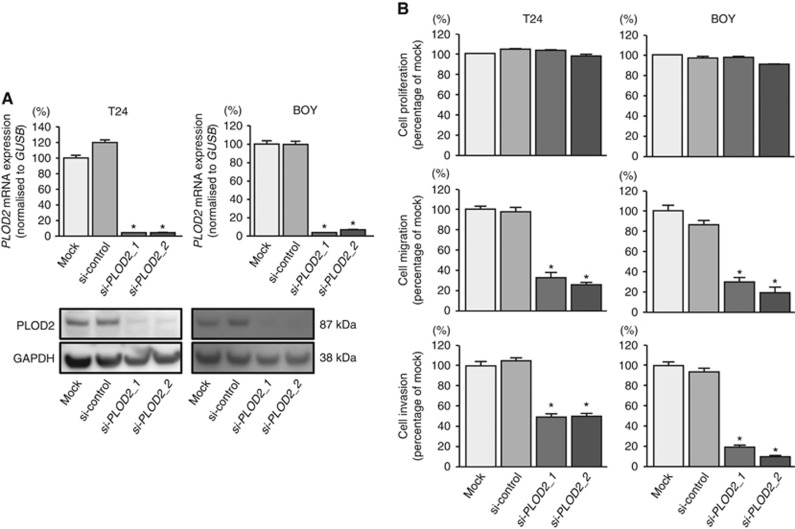
Effects of PLOD2 knockdown on cell proliferation, migration, and invasion in BC cell lines
To investigate the functional role of PLOD2 in BC cells, we performed loss-of-function studies using cells transfected with two si-PLOD2 constructs (si-PLOD2-1 and si-PLOD2-2). We evaluated the knockdown efficiency of si-PLOD2 transfection in T24 and BOY cells. Quantitative real-time reverse transcription–polymerase chain reaction analysis and western blot analysis indicated that these siRNAs effectively downregulated PLOD2 mRNA and protein expression in both cell lines (Figure 3A). XTT assays demonstrated that cell proliferation was not inhibited in si-PLOD2 transfectants in comparison with that in mock or miR-control transfectants (Figure 3B). In contrast, migration assays demonstrated that cell migration activity was significantly inhibited in si-PLOD2 transfectants in comparison with that in mock or miR-control transfectants (Figure 3B). Matrigel invasion assays demonstrated that cell invasion activity was significantly inhibited in si-PLOD2 transfectants in comparison with that in mock or miR-control transfectants (Figure 3B).
Figure 3.
Effects of si-PLOD2 transfection on BC cell lines. We evaluated the knockdown efficiency of si-PLOD2-1 and si-PLOD2-2 transfection in T24 and BOY cells. (A) Quantitative real-time reverse transcription–polymerase chain reaction analysis indicated that these siRNAs effectively downregulated PLOD2 expression in both cell lines. *P<0.0001. Western blot analyses indicated that these siRNAs effectively downregulated PLOD2 protein expression in both cell lines. (B) XTT assays demonstrated that cell proliferation was not inhibited in si-PLOD2-transfected cells in comparison with that in mock- or miR-control-transfected cells. Migration and Matrigel invasion assays demonstrated that cell migration and invasion were significantly inhibited in si-PLOD2-transfected cells in comparison with that in mock- or miR-control-transfected cells. *P<0.0001.
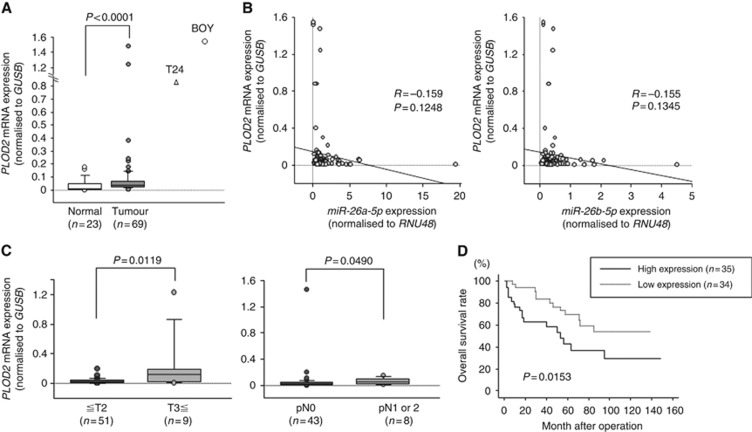
Expression levels of PLOD2 in BC specimens
Quantitative real-time reverse transcription–polymerase chain reaction analysis showed that PLOD2 expression was significantly upregulated in BC specimens (n=69) compared with that in normal specimens (n=23; P<0.0001; Figure 4A). Spearman's rank test showed the trend towards significance about inverse correlations between the expression levels of miR-26a-5p/miR-26b-5p and PLOD2 (P=0.1248, R=−0.159 and P=0.1345, R=−0.155, respectively; Figure 4B). The expression level of PLOD2 was significantly higher in pT3 and pT4 specimens (P=0.0119) and in BCs with positive lymph node invasion (P=0.0490) compared with that in their counterparts (Figure 4C). However, we could not find the positive correlations in terms of other clinicopathological parameters (Supplementary Figure 2). Moreover, Kaplan–Meier analysis showed that patients in the high PLOD2 expression group exhibited significantly shorter OS than patients in the low PLOD2 expression group (P=0.0153; Figure 4D). When we looked at the muscle-invasive cancer group, there was a trend towards significance between PLOD2 and survival (P=0.0571). Our cohort might be too small to find a positive correlations in the muscle-invasive cancer groups. There was also a trend towards significance of PLOD2 expression with OS after controlling for pathologic stage, age, and gender in a multivariable analysis (P=0.0545) (Supplementary Table 4).
Figure 4.
Expression level of PLOD2 in BC clinical specimens. (A) The expression level of PLOD2 was significantly upregulated in BC tissues in comparison with that in normal bladder tissues. GUSB was used for normalisation (P=0.0001). (B) Spearman's rank test showed the trend towards significance about inverse correlations between the expression levels of miR-26a-5p/miR-26b-5p and PLOD2 (P=0.1248, R=−0.159 and P=0.1345, R=−0.155, respectively). (C) To determine whether the levels of PLOD2 mRNA in tumour tissues were correlated with clinicopathological parameters, we analysed the expression level of the PLOD2 gene in human tumour samples. PLOD2 expression was significantly increased in samples from patients with T3 stage disease as compared with that in samples from patients with T2 stage disease or lower (P=0.0119). Additionally, PLOD2 expression was significantly increased in pN1 or pN2 samples compared with that in pN0 samples (P=0.0490). (D) Kaplan–Meier survival plots for high and low expression groups for PLOD2, as determined for 69 patients. Overall survival was significantly prolonged in patients with low PLOD2 expression compared with that in patients with high expression (P=0.0153). The median follow-up of the patients was 45.8 months.
Discussion
Although many studies have shown that miRNA regulatory mechanisms can be disrupted by the aberrant expression of tumour-suppressive or oncogenic miRNAs in cancer cells, each miRNA targets different genes and cancer pathways. Therefore, identification of tumour-suppressive miRNAs and the molecular pathways mediated by these miRNAs is important to improve our understanding of cancer mechanisms. We have previously reported that tumour-suppressive miRNAs mediate novel molecular targets and pathways in some types of cancers (Yoshino et al, 2013b; Matsushita et al, 2015). Among them, we found that some miRNAs contribute to BC development, progression, and metastasis (Yoshino et al, 2013c).
In this study, we focused on miR-26a/b because these miRNAs have been shown to have strong anticancer effects in prostate cancer, head and neck carcinoma, and papillary thyroid carcinoma (Fukumoto et al, 2015; Kato et al, 2015; You et al, 2015) through regulation of genes associated with the ECM and cell cycle regulation. Other researchers have also reported that miR-26a delivery prevents the progression of hepatocellular carcinoma, highlighting its potential therapeutic applications (Kota et al, 2009). In addition, c-MYC has been reported to suppress miR-26a and miR-26b (Chang et al, 2008). Because miR-26a-5p and miR-26b-5p may regulate different molecular targets in several cancers, elucidation of the molecular mechanisms of aberrant miR-26a-5p and miR-26b-5p expression in each type of cancer is important. In our study, these miRNAs were significantly downregulated in clinical BC specimens compared with those in normal bladder tissue, suggesting that these miRNAs may function as tumour suppressors in BC. Moreover, cell function assays revealed that cell migration and invasion were inhibited in BC cells transfected with these miRNAs.
During our miR-26a/b target analyses, we used in silico analysis with the TargetScan database to obtain candidate miR-26-regulated genes. Among these candidate genes, we identified 112 genes having two or more conserved sites. In addition, we applied the Gene Expression Omnibus database to identify upregulated genes in BC clinical specimens. From this analysis, we selected 28 genes for further analysis. Molecular target searches suggested that PLOD2, a collagen crosslinking enzyme, was a promising candidate target regulated by these miRNAs in BC cells.
PLOD2 is a collagen-modifying enzyme, similar to prolyl 4-hydroxylase α-subunit (P4HA) isoform 1, P4HA2, and LOX. Procollagens are post-translationally modified within the cisternae of the endoplasmic reticulum by P4HA1–3 and PLOD1-3 lysyl hydroxylase enzymes. Collagen, similar to elastics, fibronectins, laminins, and proteoglycans, is an ECM protein that regulates tissue homeostasis, organ development, inflammation, and diseases such as cancer. Therefore, aberrant expression of PLOD2 results in ECM disruption, promoting cell migration and invasion. A recent study showed that high expression of PLOD2 mRNA in breast cancer was associated with poorer disease-free survival of 159 patients (Gilkes et al, 2013). Another report in breast cancer showed that the expression levels of miR-26a and miR-26b are decreased by oestrogen stimulation and that forced expression of miR-26a or miR-26b negatively regulates oestrogen-stimulated breast cancer cell growth both in vitro and in vivo. Additionally, oestrogen-dependent c-MYC expression suppresses miR-26a and miR-26b expression. Interestingly, screening of oestrogen-responsive genes predicted to be targeted by miR-26 led to identification of PLOD2 and other targets (Tan et al, 2014). In our study, PLOD2 knockdown BC cells inhibited cell migration and invasion. Importantly, high expression of PLOD2 was associated with poorer OS. Therefore, we speculated that miR-26a/b downregulation triggered upregulation of PLOD2, contributing to the poor outcomes observed in patients with BC. However, despite the observed correlations between PLOD2 and survival, our data did not show any correlations between miR-26 and survival. In our speculation, this observation may be explained by the fact that multiple microRNAs would target PLOD2. Therefore, it is no wonder that no correlations were found between individual miRNAs and the survival in BC patients. We also could not find any correlations between PLOD2 mRNA expression and the recurrence status in the BC patients (Supplementary Figure 2). Our cohort included patients who had undergone different treatment modalities such as radical cystectomy or TURBT with or without adjuvant bacillus calmette guerin instillation into bladder. Thus, these complicated background may affect the results.
Furthermore, we investigated the molecular mechanisms of PLOD2 downstream signalling using si-PLOD2 transfectants, as shown in Table 2. Our results showed that hexokinase 2 (HK2) was a potential target of PLOD2 signalling in BC cells (Supplementary Table 3). In rapidly growing tumour tissue, hypoxia-inducible factor (HIF)-1 helps hypoxic tumour cells to shift toward the glycolytic pathway from the more efficient oxidative phosphorylation pathway to maintain energy production (the Warburg effect). Therefore, hypoxic cells tend to consume more glucose to meet their energy needs. Hypoxia-inducible factor 1 mediates this metabolic conversion through the induction of enzymes involved in the glycolysis pathway, such as HK2 and glucose transporters (Masoud and Li, 2015). Among these, our recent study showed that the tumour-suppressive miR-143/145 cluster directly regulates HK2 in renal cell carcinoma cells (Yoshino et al, 2013a). Although we did not demonstrate a correlation between hypoxia and miR-26a/b or PLOD2 in this study, it is plausible that hypoxia might inhibit miR-26a/b, leading to increased expression of PLOD2 and HK2. Therefore, further studies are necessary.
Table 2. Significantly enriched pathways (downstream genes of PLOD2).
| KEGG pathway entry number | Annotation | Number of genes | P-value | Genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4010 | MAPK signalling pathway | 16 | 8.14E−04 | IL1B | GADD45A | FGF2 | DDIT3 | FLNA | IL1A | DUSP6 | DUSP5 | MECOM |
| CASP3 | KRAS | RPS6KA3 | ARRB1 | GADD45B | PAK2 | DUSP22 | ||||||
| 5200 | Pathways in cancer | 14 | 1.56E−02 | CYCS | FGF2 | ITGA6 | ITGA3 | TPM3 | TCEB1 | LAMC1 | MECOM | CASP3 |
| WNT5B | KRAS | RALA | TRAF1 | CBLB | ||||||||
| 4141 | Protein processing in endoplasmic reticulum | 14 | 8.33E−05 | UBE2J1 | HYOU1 | HSPA5 | DNAJA2 | DNAJB11 | DDIT3 | UBE2E1 | DNAJC1 | SEC24A |
| DERL2 | MAN1A2 | CAPN2 | HERPUD1 | PDIA4 | ||||||||
| 4144 | Endocytosis | 9 | 3.67E−02 | ITCH | CAV1 | PIP5KL1 | RAB31 | SMAP2 | ARRB1 | EPS15 | ERBB3 | CBLB |
| 5152 | Tuberculosis | 9 | 2.53E−02 | IL1B | CYCS | RIPK2 | CREB1 | IL1A | CASP3 | CEBPG | CORO1A | HLA-DMB |
| 4120 | Ubiquitin-mediated proteolysis | 9 | 7.81E−03 | CDC26 | UBE2J1 | PRPF19 | CDC20 | ITCH | UBE2E1 | TCEB1 | ERCC8 | CBLB |
| 4910 | Insulin signalling pathway | 9 | 7.78E−03 | EXOC7 | HK2 | RPS6KB1 | RHEB | HKDC1 | KRAS | PPP1R3C | PYGL | CBLB |
| 5010 | Alzheimer's disease | 8 | 4.15E−02 | IL1B | CYCS | APH1A | CAPN2 | NDUFB3 | PLCB3 | CASP3 | COX7A2L | |
| 5323 | Rheumatoid arthritis | 8 | 1.75E−03 | IL1B | ATP6V1C1 | ATP6V1A | CCL20 | IL1A | CSF1 | CCL5 | HLA-DMB | |
| 3008 | Ribosome biogenesis in eukaryotes | 8 | 8.44E−04 | UTP18 | NOL6 | NOP56 | NXT1 | DKC1 | WDR43 | UTP14A | RPP40 | |
| 4115 | p53 signalling pathway | 8 | 6.70E−04 | CYCS | GADD45A | SESN2 | CASP3 | STEAP3 | CCNG1 | GADD45B | PMAIP1 | |
| 4110 | Cell cycle | 7 | 3.68E−02 | CDC26 | GADD45A | CDC20 | CDC25C | MAD2L1 | ORC1 | GADD45B | ||
| 4012 | ErbB signalling pathway | 7 | 9.20E−03 | NRG1 | RPS6KB1 | NRG2 | KRAS | PAK2 | ERBB3 | CBLB | ||
| 4640 | Haematopoietic cell lineage | 7 | 7.55E−03 | IL1B | ITGA6 | ITGA3 | IL1A | CSF1 | CD9 | IL7 | ||
| 520 | Amino sugar and nucleotide sugar metabolism | 7 | 8.07E−04 | HK2 | UAP1 | UGDH | GNPNAT1 | GNPDA1 | HKDC1 | GFPT1 | ||
| 970 | Aminoacyl-tRNA biosynthesis | 6 | 1.23E−03 | IARS | FBXO17 | GARS | MTFMT | YARS | SARS | |||
| 5020 | Prion diseases | 6 | 6.56E−04 | IL1B | HSPA5 | IL1A | LAMC1 | PRNP | CCL5 | |||
| 5120 | Epithelial cell signaling in Helicobacter pylori infection | 5 | 3.82E−02 | JAM2 | ATP6V1C1 | ATP6V1A | CASP3 | CCL5 | ||||
| 3018 | RNA degradation | 5 | 3.74E−02 | DCPS | PAN3 | ENO3 | CNOT6 | LSM5 | ||||
| 5416 | Viral myocarditis | 5 | 3.35E−02 | CYCS | CAV1 | CASP3 | EIF4G2 | HLA-DMB | ||||
| 10 | Glycolysis/gluconeogenesis | 5 | 3.29E−02 | HK2 | ENO3 | HKDC1 | PGAM1 | PGAM4 | ||||
| 4150 | mTOR signalling pathway | 5 | 2.01E−02 | RPS6KB1 | RHEB | EIF4B | CAB39L | RPS6KA3 | ||||
| 500 | Starch and sucrose metabolism | 5 | 1.98E−02 | HK2 | ENPP1 | UGDH | HKDC1 | PYGL | ||||
| 4130 | SNARE interactions in vesicular transport | 4 | 2.60E−02 | VAMP1 | VAMP4 | STX3 | BET1 | |||||
| 260 | Glycine, serine and threonine metabolism | 4 | 2.09E−02 | CTH | PHGDH | PSAT1 | PSPH | |||||
| 910 | Nitrogen metabolism | 3 | 4.18E−02 | ASNS | CTH | CA13 | ||||||
| 524 | Butirosin and neomycin biosynthesis | 2 | 2.52E−02 | HK2 | HKDC1 | |||||||
Abbreviations: KEGG=Kyoto Encyclopedia of Genes and Genomes; MAPK=mitogen-activated protein kinase; mTOR=mammalian target of rapamycin; PLOD2=procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2; SNARE=soluble N-ethylmaleimide-sensitive factor attached protein receptor.
Investigation of the molecular mechanisms downstream of PLOD2 signalling also identified integrin A3 (ITGA3). Integrins comprise a large family of cell receptors for ECM proteins and ligands, such as fibronectin and collagen on other cells. Therefore, aberrant expression of extracellular proteins triggers intracellular signalling events, which may lead to cancer metastasis and invasion (Givant-Horwitz et al, 2005). Based on our study, PLOD2-induced ECM fibrosis may contribute to aberrant intracellular signalling. Our recent study showed that the tumour-suppressive miR-223 directly regulates ITGA3/ITGB1 in prostate cancer (Kurozumi et al, 2016), suggesting that miR-26a/b may also regulate ITGA3 through PLOD2 regulation in BC. Because there is no evidence of direct connections among PLOD2, HK2, and integrin, further studies are necessary.
Conclusions
Downregulation of miR-26a-5p and miR-26b-5p was frequently observed in BC cells, and both of these miRNAs significantly inhibited cancer cell migration and invasion. PLOD2 was directly regulated by the tumour suppressors miR-26a-5p and miR-26b-5p and may be a good prognostic marker for survival in patients with BC. Recent studies have shown that aberrant expression of ECM components contributes to cancer cell invasion and metastasis. PLOD2 functions as a collagen crosslinking enzyme is associated with ECM stiffness. This is the first report demonstrating the positive correlation between PLOD2 and survival in clinical BC specimens. The discovery of molecular targets mediated by tumour-suppressive miRNAs provides important insights into the potential mechanisms of BC metastasis.
Acknowledgments
This study was supported by the KAKENHI (C), 25462490.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bellmunt J, Petrylak DP (2012) New therapeutic challenges in advanced bladder cancer. Semin Oncol 39: 598–607. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT (2008) Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N, Nakagawa M (2010) miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer 102: 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santis M, Bellmunt J, Mead G, Kerst JM, Leahy M, Maroto P, Gil T, Marreaud S, Daugaard G, Skoneczna I, Collette S, Lorent J, de Wit R, Sylvester R (2012) Randomized phase II/III trial assessing gemcitabine/carboplatin and methotrexate/carboplatin/vinblastine in patients with advanced urothelial cancer who are unfit for cisplatin-based chemotherapy: EORTC study 30986. J Clin Oncol 30: 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva G, Croce CM (2010) Roles of small RNAs in tumor formation. Trends Mol Med 16: 257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto I, Hanazawa T, Kinoshita T, Kikkawa N, Koshizuka K, Goto Y, Nishikawa R, Chiyomaru T, Enokida H, Nakagawa M, Okamoto Y, Seki N (2015) MicroRNA expression signature of oral squamous cell carcinoma: functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br J Cancer 112: 891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Brancato SJ, Agarwal PK, Apolo AB (2014) Targeted therapies in urothelial carcinoma. Curr Opin Oncol 26: 305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Bajpai S, Wong CC, Chaturvedi P, Hubbi ME, Wirtz D, Semenza GL (2013) Procollagen lysyl hydroxylase 2 is essential for hypoxia-induced breast cancer metastasis. Mol Cancer Res 11: 456–466. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gilkes DM, Semenza GL, Wirtz D (2014) Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer 14: 430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givant-Horwitz V, Davidson B, Reich R (2005) Laminin-induced signaling in tumor cells. Cancer Lett 223: 1–10. [DOI] [PubMed] [Google Scholar]
- Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, Tsujimoto G, Nakagawa M, Seki N (2009) Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer 125: 345–352. [DOI] [PubMed] [Google Scholar]
- Inoguchi S, Seki N, Chiyomaru T, Ishihara T, Matsushita R, Mataki H, Itesako T, Tatarano S, Yoshino H, Goto Y, Nishikawa R, Nakagawa M, Enokida H (2014) Tumour-suppressive microRNA-24-1 inhibits cancer cell proliferation through targeting FOXM1 in bladder cancer. FEBS Lett 588: 3170–3179. [DOI] [PubMed] [Google Scholar]
- Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, Yonezawa T, Nohata N, Kinoshita T, Nakagawa M, Enokida H (2014) The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS One 9: e84311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Goto Y, Matsushita R, Kurozumi A, Fukumoto I, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa T, Seki N (2015) MicroRNA-26a/b directly regulate La-related protein 1 and inhibit cancer cell invasion in prostate cancer. Int J Oncol 47: 710–718. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Nohata N, Hanazawa T, Kikkawa N, Yamamoto N, Yoshino H, Itesako T, Enokida H, Nakagawa M, Okamoto Y, Seki N (2013) Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. Br J Cancer 109: 2636–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M, Seki N (2012) Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer 106: 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT (2009) Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurozumi A, Goto Y, Matsushita R, Fukumoto I, Kato M, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa T, Seki N (2016) Tumor-suppressive microRNA-223 inhibits cancer cell migration and invasion by targeting ITGA3/ITGB1 signaling in prostate cancer. Cancer Sci 107: 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoud GN, Li W (2015) HIF-1alpha pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B 5: 378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita R, Seki N, Chiyomaru T, Inoguchi S, Ishihara T, Goto Y, Nishikawa R, Mataki H, Tatarano S, Itesako T, Nakagawa M, Enokida H (2015) Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br J Cancer 113: 282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks JJ, Bellmunt J, Bochner BH, Clarke NW, Daneshmand S, Galsky MD, Hahn NM, Lerner SP, Mason M, Powles T, Sternberg CN, Sonpavde G (2012) A systematic review of neoadjuvant and adjuvant chemotherapy for muscle-invasive bladder cancer. Eur Urol 62: 523–533. [DOI] [PubMed] [Google Scholar]
- Nohata N, Hanazawa T, Kinoshita T, Inamine A, Kikkawa N, Itesako T, Yoshino H, Enokida H, Nakagawa M, Okamoto Y, Seki N (2013) Tumour-suppressive microRNA-874 contributes to cell proliferation through targeting of histone deacetylase 1 in head and neck squamous cell carcinoma. Br J Cancer 108: 1648–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62: 10–29. [DOI] [PubMed] [Google Scholar]
- Sobin LH, Compton CC (2010) TNM seventh edition: what's new, what's changed: communication from the International Union Against Cancer and the American Joint Committee on Cancer. Cancer 116: 5336–5339. [DOI] [PubMed] [Google Scholar]
- Tan S, Ding K, Li R, Zhang W, Li G, Kong X, Qian P, Lobie PE, Zhu T (2014) Identification of miR-26 as a key mediator of estrogen stimulated cell proliferation by targeting CHD1, GREB1 and KPNA2. Breast Cancer Res 16: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarano S, Chiyomaru T, Kawakami K, Enokida H, Yoshino H, Hidaka H, Yamasaki T, Kawahara K, Nishiyama K, Seki N, Nakagawa M (2011) miR-218 on the genomic loss region of chromosome 4p15.31 functions as a tumor suppressor in bladder cancer. Int J Oncol 39: 13–21. [DOI] [PubMed] [Google Scholar]
- Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N, Nakagawa M (2011) The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer 104: 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino H, Enokida H, Itesako T, Kojima S, Kinoshita T, Tatarano S, Chiyomaru T, Nakagawa M, Seki N (2013. a) Tumor-suppressive microRNA-143/145 cluster targets hexokinase-2 in renal cell carcinoma. Cancer Sci 104: 1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino H, Enokida H, Itesako T, Tatarano S, Kinoshita T, Fuse M, Kojima S, Nakagawa M, Seki N (2013. b) Epithelial–mesenchymal transition-related microRNA-200s regulate molecular targets and pathways in renal cell carcinoma. J Hum Genet 58: 508–516. [DOI] [PubMed] [Google Scholar]
- Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H (2013. c) Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol 10: 396–404. [DOI] [PubMed] [Google Scholar]
- You H, Lin H, Zhang Z (2015) CKS2 in human cancers: clinical roles and current perspectives (Review). Mol Clin Oncol 3: 459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuiverloon TC, Nieuweboer AJ, Vekony H, Kirkels WJ, Bangma CH, Zwarthoff EC (2012) Markers predicting response to bacillus Calmette-Guerin immunotherapy in high-risk bladder cancer patients: a systematic review. Eur Urol 61: 128–145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.