Abstract
Background:
The management of locally advanced pancreatic cancer (LAPC) patients remains controversial. Better discrimination for overall survival (OS) at diagnosis is needed. We address this issue by developing and validating a prognostic nomogram and a score for OS in LAPC (PROLAP).
Methods:
Analyses were derived from 442 LAPC patients enrolled in the LAP07 trial. The prognostic ability of 30 baseline parameters was evaluated using univariate and multivariate Cox regression analyses. Performance assessment and internal validation of the final model were done with Harrell's C-index, calibration plot and bootstrap sample procedures. On the basis of the final model, a prognostic nomogram and a score were developed, and externally validated in 106 consecutive LAPC patients treated in Besançon Hospital, France.
Results:
Age, pain, tumour size, albumin and CA 19-9 were independent prognostic factors for OS. The final model had good calibration, acceptable discrimination (C-index=0.60) and robust internal validity. The PROLAP score has the potential to delineate three different prognosis groups with median OS of 15.4, 11.7 and 8.5 months (log-rank P<0.0001). The score ability to discriminate OS was externally confirmed in 63 (59%) patients with complete clinical data derived from a data set of 106 consecutive LAPC patients; median OS of 18.3, 14.1 and 7.6 months for the three groups (log-rank P<0.0001).
Conclusions:
The PROLAP nomogram and score can accurately predict OS before initiation of induction chemotherapy in LAPC-untreated patients. They may help to optimise clinical trials design and might offer the opportunity to define risk-adapted strategies for LAPC management in the future.
Keywords: prognostic score, nomogram, overall survival, locally advanced pancreatic cancer, chemotherapy
Pancreatic cancer (PC) is one of the leading causes of cancer-related mortality worldwide, with 5-year relative survival of only 8%. The predicted death rates have progressively increased over the past years (Siegel et al, 2016). This unfavourable trend emphasises the importance of giving priority to research in PC prevention and treatment. Number of deaths by PC in 2014 in the European Union (EU) has been estimated to 82 300 cases equally distributed in men and women. By analogy, a similar prediction has been reported for women with breast cancer (N=89 300) (Malvezzi et al, 2014).
Most PCs are unresectable due to the presence of either distant metastases or locoregional metastases, including vascular invasion (locally advanced PC: LAPC). At the time of diagnosis, up to 35% of patients with PC present with locally advanced disease (Philip, 2011). The management of those patients is still controversial, specifically the role of radiotherapy remains uncertain (Huguet et al, 2007; Krishnan et al, 2007; Chauffert et al, 2008; Loehrer et al, 2011; Philip, 2011; Hajj and Goodman, 2015).
Currently, there is a lack of staging system and consensus regarding specific risk profile for overall survival (OS) in LAPC, which can lead to confusion in research strategies development and potentially inappropriate management of patients. Given this aspect and the current interest in precision medicine, there is a need for prognostic tools to optimise patients' selection for appropriate treatment therapy in order to achieve more personalised management. Specifically, more accurate prediction of OS at diagnosis may help to ensure well-balanced arms in clinical trials and to a better management patient care in the future. Ultimately, such a tool may lead to improve patient's health-related quality of life (HRQoL), chemotherapy (CT) cost-effectiveness ratio, and design of future clinical trials.
Nomograms have been increasingly used for survival prediction in different cancer settings as an alternative to traditional TNM staging system or a new standard (Balachandran et al, 2015). Unfortunately, no single study evaluating a nomogram for survival prediction in LAPC patients is available.
We aimed to develop and validate a new prognosis model, nomogram and score for OS using a broad spectrum of parameters currently available at baseline. This model was developed based on a large cohort of LAPC patients enrolled in the phase III LAP07 trial and validated in patients encountered in daily clinical practice.
Patients and Methods
Patients
The development cohort was obtained from an international, multicentre, randomised phase III LAP07 trial (ClinicalTrials.gov NCT00634725). The complete design and trial procedures of the LAP07 study was previously described (Hammel et al, 2016). Briefly, the LAP07 study assessed whether chemoradiotherapy (CRT) administered to LAPC patients whose tumour has been controlled after 4 months of gemcitabine alone or plus erlotinib induction CT increases OS compared with continuation of the same CT. In this trial, CRT did not increase OS compared with the continuation of CT alone. Demographics, cancer history, clinicopathological, biological and radiological parameters at baseline as well as treatment outcomes were collected.
The external validation cohort included 106 consecutive LAPC patients treated at the University Hospital of Besançon, France between January 2003 and December 2013.
Main eligibility and exclusion criteria along with treatment and procedures for the development and validation cohorts are specified in Supplementary Appendix A1.
Follow-up
In the development cohort, patients were observed at 2, 4, 7, 9 and 11 months and every 2 months thereafter. At each follow-up visit, a detailed clinical history and a complete physical examination were carried out. Database was locked on 23 February 2014.
In the validation cohort, patients were treated using standard clinical protocols. A radiological evaluation was performed every 3 months. Database was locked on 13 November 2015.
Statistical analysis
The primary analysis was performed on the LAP07 intent to treat population. Mean (s.d.) values and frequency (percentage) were provided for the description of continuous and categorical variables, respectively. Means and proportions were compared using Student's t-test and χ2-test (or Fisher's exact test, if appropriate), respectively.
In the development cohort, OS was calculated from the date of study randomisation to the date of death from any cause. Patients last known to be alive were censored at the time of their last follow-up assessment (Bonnetain et al, 2014). In the external validation cohort, the starting point for OS was the date of diagnosis of LAPC. OS was estimated using the Kaplan–Meier method and described using median or rate at specific time points with 95% confidence intervals (95% CI). Follow-up was calculated using a reverse Kaplan–Meier estimation when feasible (Schemper and Smith, 1996).
Cox-proportional-hazard models were performed to estimate the hazard ratio (HR) and 95% CI for factors associated with OS. The association of the baseline parameters with OS was first assessed using univariate Cox analyses, and then those with P<0.10 were entered into a final multivariate Cox regression model. Significantly skewed continuous variables, such as neutrophil count, total bilirubin, alkaline phosphatase, aspartate transaminase, alanine aminotransaminase, lactate dehydrogenase, albumin and cancer antigen 19.9 (CA 19-9), were normalised by logarithmic transformation. Hazard proportionality was checked by plotting log-minus-log survival curves and by the cumulative martingale process plots. Log-linearity assumption for continuous variables was also checked graphically by making a categorical variable. For continuous variables involved in the final multivariate model, a fractional polynomial method was applied to validate the transformation used and to assess the relationship between predictors and survival. In the absence of clinical hypothesis and/or documentation in the literature, no interaction term was considered in the model construction.
Accuracy of the final model was verified regarding two parameters: discrimination and calibration. The predictive value and the discrimination ability (that is, the ability to separate patients with different prognosis) of the final model were evaluated with the Harrell's concordance index (C-index) (Harrell et al, 1996). One thousand random samples of the population were used to derive 95% CI for the C-statistic. Calibration and goodness of fit refers to the ability to provide unbiased survival predictions in groups of similar patients. These were assessed by visual examination of calibration plot and tested with an extension of the Hosmer–Lemeshow test for survival data. Internal validation of the final multivariate model was performed with a bootstrap sample procedure.
The final model was used to derive a nomogram allowing the estimation of median and individual post-diagnosis OS probabilities at 6, 12, 24 and 48 months. For estimation at population level, a prognostic score was constructed and weighted with β-coefficients estimations in the final model, and normalised between 0 and 5. To give a reasonable spread of risk, we chose to work on two and three prognostic risk groups. In such way, different cut points for the risk score from the development set were explored. These were based on the optimal cut point (Hothorn and Lausen, 2003) and Cox's (Cox, 1957) methods for the two- and three-risk groups approach, respectively. Median and tertiles were also considered. The discrimination abilities of the different prognostic scores produced were assessed with the Harrell's concordance index (C-index) by considering risk groups classification.
Patients' baseline key characteristics were compared between prognostic risk groups using Fisher-exact test and the Kruskal–Walis test for categorical and quantitative parameters, respectively.
To explore the performance of the final model, sensitivity analyses were performed by forcing the treatment assigned at first randomisation (gemcitabine vs gemcitabine plus erlotinib) to enter into the model and by including baseline histological grade and systolic blood pressure; two parameters that were not initially selected in the multivariate analysis process due to their high rate of missing data. To assess potential bias arising from missing data for parameters involved in the multivariate final model on their significance (P-value) and estimates (β and its standard error), a multiple imputation procedure with a Markov chain Monte Carlo method was performed using SAS MI and MIANALYZE procedure.
The final multivariate and the prognostic score discrimination abilities were confirmed in an external validation cohort and evaluated with the Harrell's concordance index (C-index) with survival predictions from the model and from risk groups classification, respectively. To identify risk groups and to determine their survival profile, the same development cohort-derived risk predictive algorithm was applied.
All analyses were performed using SAS version 9.3 (SAS Institute) and R software version 2.15.2 (R Development Core Team; http://www.r-project.org). Values of P<0.05 were considered statistically significant and all tests were two-sided. Details on the interpretation of important statistical concepts are given in Supplementary Appendix A2.
Results
Patient characteristics
A total of 442 LAPC patients treated in the LAP07 trial between February 2008 and December 2011 formed the development cohort. The median follow-up time was 34.3 months (95% CI, 27.6–43.7). Overall, 106 LAPC patients treated in the University Hospital of Besançon between January 2003 and December 2013 were included in the external validation cohort. Patient baseline characteristics according to the cohort set are summarised in Table 1. As expected, given its daily clinical nature the validation cohort differed from the LAP07 population with respect to, age, histological grade, WHO-PS, tumour size and treatment regimens.
Table 1. Baseline characteristics according to the cohort set.
| Development set cohort–LAP07 trial (N=442) | External validation set cohort–University Hospital of Besançon cohort (N=106) | P | |
|---|---|---|---|
| Age, yearsa | 63.5±9.6 | 67.1±10.2 | 0.0007 |
| Gender, N (%) | |||
| Male | 228 (52%) | 57 (54%) | |
| Female | 214 (48%) | 49 (46%) | 0.7456 |
| Localisation, N (%) | |||
| Head/head and body | 302 (69%) | 71 (70%) | |
| Other (body and/or tail) | 138 (31%) | 31 (30%) | 0.9059 |
| Unknown | 2 | 4 | |
| Grading, N (%) | |||
| Well differentiated | 107 (48%) | 9 (26%) | |
| Moderately differentiated | 75 (34%) | 17 (50%) | |
| Poorly differentiated | 41 (18%) | 8 (24%) | 0.05556 |
| Missing | 219 | 72 | |
| Lymph nodes, N (%) | |||
| Yes | 179 (41%) | — | |
| No | 258 (59%) | — | |
| Missing | 5 | — | |
| WHO-PS, N (%) | |||
| 0 | 197 (46%) | 31 (30%) | |
| 1 | 203 (47%) | 59 (56%) | |
| 2 | 31 (7%) | 14 (13%) | |
| 3 | 0 (0%) | 1(1%) | 0.004892 |
| Missing | 11 | 1 | |
| RECIST tumour size (mm)a | 44.0±18.0 | 38.3±14.7 | 0.00385 |
| Missing | 20 | 9 | |
| RECIST tumour size (mm), N (%) | |||
| <30 | 66 (16%) | 22 (23%) | |
| 30–50 | 226 (54%) | 56 (58%) | |
| ⩾50 | 130 (31%) | 19 (20%) | 0.04676 |
| Missing | 20 | 9 | |
| Vascular invasion, N (%) | |||
| Superior mesenteric artery | 144 (33%) | — | |
| Coeliac trunk | 188 (43%) | — | |
| Hepatic artery | 25 (6%) | — | |
| No | 79 (18%) | — | |
| Missing | 6 | — | |
| Median follow-up time, months (95% CI) | 34.3 (27.6–43.7) | All patients were followed until death (maximum time observed=76.6) except five patients alive with a median follow-up equal to 10 |
Abbreviations: CI=confidence interval; NA=not available; PS=performance status; WHO=World Health Organization. Bold values refer to significant P-values reflecting statistical significant difference for the parameter considered between the 2 cohorts.
Plus–minus values are means±s.d.
Prognostic factors for OS
Only 8 out of 30 evaluated baseline parameters were associated with OS in the univariate Cox analyses with a P<0.1. These were age, histological grade, performance status (the World Health Organization scale), systolic blood pressure, pain, albumin level, tumour size and CA 19-9 (Table 2). The multivariate Cox analysis (N=358) showed five independent risk factors for OS: age; pain; albumin; tumour size; and CA 19-9 (Table 3).
Table 2. Univariate analysis of factors associated with overall survival.
| Number of patients | Number of events | HR | 95% CI | P | |
|---|---|---|---|---|---|
|
Demographic parameters | |||||
| Age at inclusion, years | 442 | 379 | 1.013 | 1.003–1.024 | 0.0148 |
| Sex | |||||
| Male | 228 | 198 | 1 | — | — |
| Female | 214 | 181 | 0.915 | 0.748–1.120 | 0.3897 |
|
Pathological parameters | |||||
| Primary tumour site | |||||
| Head–head and body | 302 | 260 | 1 | ||
| Body and/or tail | 138 | 117 | 0.951 | 0.764–1.183 | 0.6519 |
| Unknown | 2 | ||||
| Histological grade | |||||
| Well differentiated | 107 | 89 | 1 | — | — |
| Moderately differentiated | 75 | 62 | 1.106 | 0.799–1.530 | — |
| Poorly differentiated | 41 | 40 | 1.939 | 1.331–2.825 | 0.0021 |
| Missing | 219 | ||||
| Regional lymph node invasion | |||||
| No | 258 | 222 | 1 | — | — |
| Yes | 179 | 152 | 0.954 | 0.776–1.174 | 0.6584 |
| Missing | 5 | — | — | — | — |
| Arterial involvement | |||||
| No | 10 | 7 | 1 | — | — |
| Yes | 430 | 370 | 0.635 | 0.299–1.345 | 0.2352 |
| Missing | 2 | — | — | — | — |
|
Clinical parameters | |||||
| PS (WHO) | |||||
| 0 | 197 | 162 | 1 | — | — |
| 1 | 203 | 179 | 1.252 | 1.012–1.550 | — |
| 2 | 31 | 30 | 1.731 | 1.171–2.558 | 0.0092 |
| Missing | 11 | — | — | — | — |
| Systolic blood pressure, mmHg | 252 | 222 | 1.011 | 1.003 to 1.019 | 0.0048 |
| Diastolic blood pressure, mmHg | 260 | 222 | 1.006 | 0.997–1.015 | 0.2176 |
| Diarrhoea | |||||
| No | 408 | 351 | 1 | — | — |
| Yes | 31 | 27 | 0.763 | 0.516–1.129 | 0.1763 |
| Missing | 3 | — | — | — | — |
| Pain | |||||
| No | 186 | 155 | 1 | — | — |
| Yes | 253 | 223 | 1.199 | 0.976–1.472 | 0.0834 |
| Missing | 3 | — | — | — | — |
| Jaundice | |||||
| No | 387 | 334 | 1 | — | — |
| Yes | 51 | 44 | 0.971 | 0.709–1.331 | 0.8550 |
| Missing | 4 | — | — | — | — |
| Body Mass Index, kg m−2 | 434 | 372 | 0.993 | 0.968–1.019 | 0.5831 |
| Body mass index, kg m−2 | |||||
| Normal weight (18.5–25) | 249 | 216 | 1 | ||
| Underweight <18.5 | 37 | 32 | 1.041 | 0.718–1.510 | |
| Overweight (25–30) | 114 | 97 | 0.979 | 0.770–1.244 | |
| Obese⩾30 | 34 | 27 | 0.944 | 0.632–1.409 | 0.9822 |
| Missing | 8 | ||||
| Percentage of weight loss | 385 | 329 | 1.009 | 0.993–1.025 | 0.2653 |
|
Biological parameters | |||||
| Haematological function | |||||
| Neutrophils, mm3 (log-value) | 437 | 374 | 1.473 | 0.818–2.654 | 0.1973 |
| Haemoglobin, g dl−1 | 442 | 379 | 0.954 | 0.887–1.025 | 0.1976 |
| Platelets, mm3 | 438 | 376 | 1.000 | 1.000–1.000 | 0.1442 |
| Biochemical | |||||
| Creatinine, μmol l−1 | 440 | 377 | 0.996 | 0.990–1.002 | 0.2153 |
| Creatinine clearance rate, ml min−1 | 313 | 263 | 1.000 | 0.996–1.005 | 0.9169 |
| Potassium, mmol l−1 | 422 | 363 | 0.985 | 0.784–1.237 | 0.8950 |
| Total bilirubin, μmol l−1 (log-value) | 440 | 377 | 1.028 | 0.769–1.375 | 0.8517 |
| Alkaline phosphatase, UI l−1 (log-value) | 432 | 371 | 1.147 | 0.789–1.667 | 0.4727 |
| Aspartate transaminase, UI l−1 (log-value) | 438 | 375 | 1.297 | 0.832–2.021 | 0.2510 |
| Alanine aminotransferase, UI l−1 (log-value) | 436 | 373 | 0.866 | 0.623–1.205 | 0.3930 |
| Lactate dehydrogenase, UI l−1 (log-value) | 313 | 268 | 1.089 | 0.599–1.983 | 0.7792 |
| Albumin, g l−1 | 393 | 336 | 0.963 | 0.944–0.982 | 0.0002 |
| Proteinuria, g l−1 | 313 | 272 | 0.988 | 0.968–1.007 | 0.2152 |
| Hypercalcemia, mmol l−1 | 347 | 301 | 1.885 | 0.755–4.707 | 0.1748 |
| CA 19-9, UI (log-value) | 411 | 359 | 1.171 | 1.057–1.296 | 0.0025 |
|
Radiological parameters | |||||
| Tumour size, mm | 422 | 362 | 1.008 | 1.002–1.015 | 0.0081 |
Abbreviations: CI=confidence interval; HR=hazard ratio; PS=performance status; WHO=World Health Organization. Bold values refer to P-values <0.1 highlighting parameters involved in the final multivariate construction as described in the statistical section.
Table 3. Multivariate analysis of factors associated with overall survival (N=358).
| Number of patients | Number of death | HR | 95% CI | P | Internal validation BCA HR 95% | Max scores in nomogram | |
|---|---|---|---|---|---|---|---|
| Age at diagnosis, years | 358 | 307 | 1.013 | 1.000–1.025 | 0.0478 | 1.000–1.025 | 43 |
| Pain | |||||||
| No | 155 | 130 | 1 | – | – | ||
| Yes | 203 | 177 | 1.292 | 1.023–1.632 | 0.0317 | 1.046–1.657 | 16 |
| Albumin, g l−1 | 358 | 307 | 0.963 | 0.942–0.984 | 0.0006 | 0.942–0.988 | 96 |
| Tumour size, mm | 358 | 307 | 1.008 | 1.001–1.015 | 0.0214 | 1.000–1.016 | 100 |
| CA 19-9, UI (log-value) | 358 | 307 | 1.172 | 1.047–1.311 | 0.0056 | 1.039–1.306 | 80 |
Abbreviations: BCA=accelerated bootstrap confidence interval; CI=confidence interval; HR=hazard ratio. Bold values reflect that P-values are all significant <0.05.
Performance assessment and internal validation of the final model
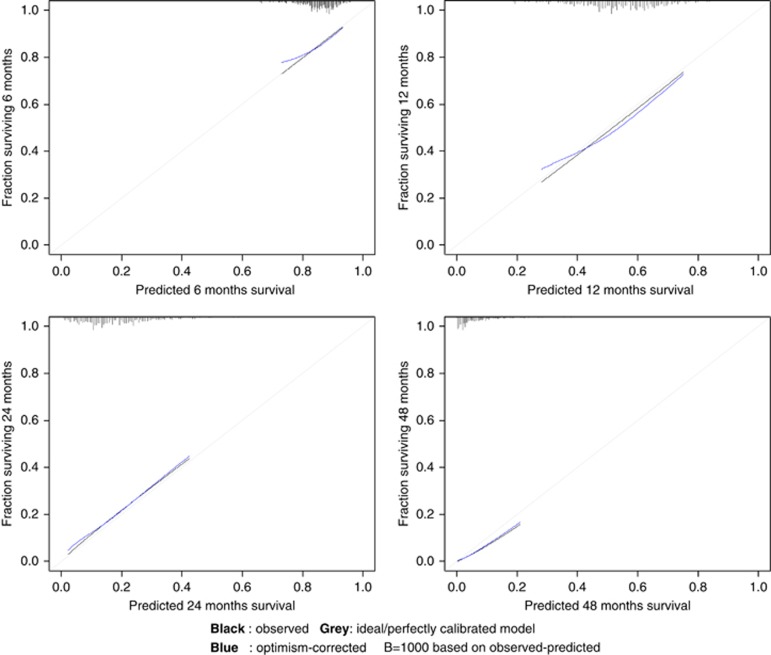
The multivariate model exhibited acceptable discrimination ability (C-index 0.60; 95% bootstrap percentile CI, 0.57–0.64) and a good calibration at 6, 12, 24 and 48 months (P=0.99, P=0.55, P=0.30, and P=0.08, respectively). The calibration plots showed an optimal agreement between model prediction and actual observation for predicting OS probability at 6, 12, 24, and 48 months (Figure 1).
Figure 1.
Calibration plots at 6, 12, 24 and 48 months for the final multivariate model. Vertical axis is the observed proportion of patients surviving at time of interest.
In the internal validation, uncertainties around HR measured with the bootstrapping procedure (Table 3) reflected the robustness of the final model.
Sensitivity analyses
In the sensitivity analysis, the performance of the final model was investigated by forcing the allocated treatment (gemcitabine vs gemcitabine plus erlotinib). Associations remained unchanged (Supplementary Appendix Table A1A).
After adjustment of the final model for histological grading and systolic blood pressure, main associations of the final multivariate model remained unchanged except for CA 19-9 (Supplementary Appendix Table A1B).
The final multivariate model was based on 358 (81.0%) patients (complete-subject analysis) for whom the five parameters were available. A multiple imputation analysis based on 1000 imputed data sets for all the variables provided similar results to the complete-subject analysis in term of significance for the association (P-value) and for coefficient estimations (β and SE) (Supplementary Appendix Table A1C).
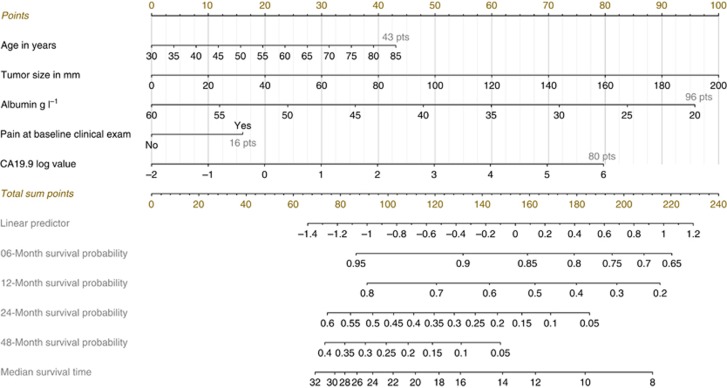
Prognostic nomogram for OS
Nomogram integrating all significant independent factors for OS was built (Figure 2). An illustration and details on the nomogram development are provided in Supplementary Appendix A3.
Figure 2.
Prognostic nomogram to predict individual overall survival probability in patients with locally advanced pancreatic cancer. Points are assigned for each risk factor by drawing a line upward from the corresponding values to the ‘point' line. The total sum of points for four risk factors is plotted on the ‘total points' line. A line is drawn down to read the corresponding predictions of 6-, 12-, 24- and 48-month-survival probability and median survival time. For example, a patient with the following characteristics–63 years old (26 points), tumour measurement equal to 105 mm (53 points), albumin at 40.9 g l−1 (46 points), CA 19-9 at 89 (log-value=1.95; 39 points) and the presence of pain at baseline clinical exam (16 point) will have a total points of 180, which corresponds to 6-, 12-, 24- and 48-month-survival probabilities of 80% (95% CI 71–86), 40% (95% CI 25–55), 7% (95% CI 2–17) and 1% (95% CI 0–4) and a predicted median survival time of 10.5 months (95% CI 8.75–13.5). An online web-based application smartphone compatible was developed that provides individualised survival estimates from the nomogram: http://www.umqvc.org/en/tool/prolap.html.
Prognostic score for death-risk stratification
A PROLAP (PRognostic score for Overall survival in LAPC) score ranging from 0 to 5 based on the final model was calculated for each patient. All factors included in the score were weighted according to the β-parameter estimation issued from the final Cox model. The contribution of each individual factor was summed to generate a raw score that was finally applied to normalisation between 0 and 5. This score construction and equation are provided as Supplementary Appendix A3.
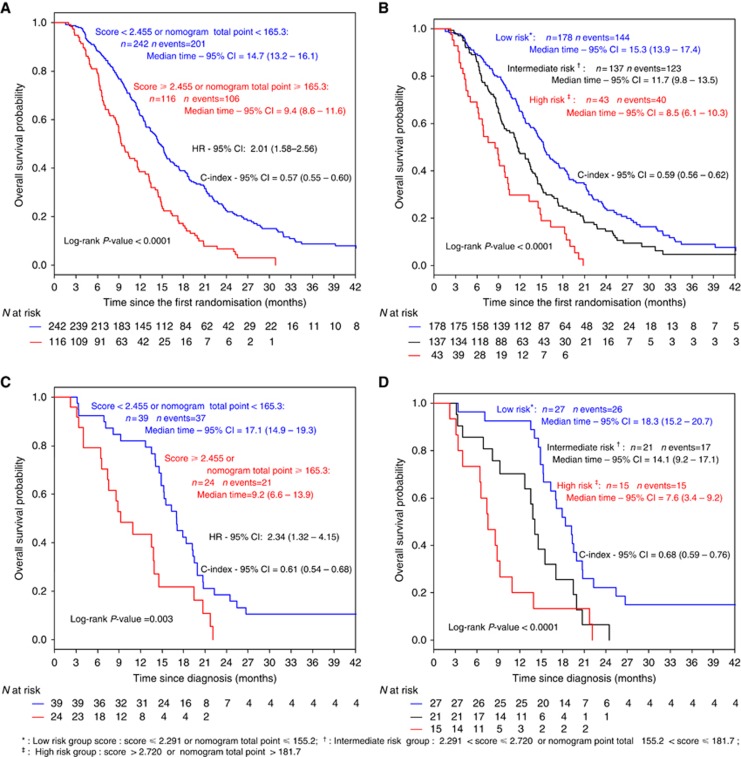
On the basis of an optimal cut point determined by Hothorn and Lausen method (Supplementary Appendix Figure A1) patients were categorised into two-risk groups with significantly different prognostic profiles (Figure 3A): a low-risk group (N=242, score<2.455) and a high-risk group (N=116, score ⩾2.455, HR=2.01 95% CI, 1.58–2.56; P<0.0001). The median OS was 14.7 and 9.4 months for the low-risk group and the high-risk group, respectively (P<0.0001).
Figure 3.
Kaplan–Meier curves of overall survival for (A, B) the development set cohort and (C, D) the external validation set cohort according to prognostic score group for the Hothorn & Lausen optimal cut point (A, C) and Cox's methods (B, D) for the two- and three-risk groups approach, respectively. An online web-based smartphone compatible application was developed that provides risk-group classification for OS from the PROLAP score: http://www.umqvc.org/en/tool/prolap.html.
Considering that the prognostic score assumes a normal distribution (mean=2.27; s.d.=0.39) for the three-risk groups approach, Cox demonstrated that the loss of information from grouping would be minimised by selecting cut points equal to 2.291 and 2.720. Such grouping achieved a clear separation of the Kaplan–Meier curves (Figure 3B). The median OS was 15.3, 11.7 and 8.5 months for the low, intermediate and the high-risk groups, respectively (P<0.0001).
Results for classification of patient's into two- and three-risk groups related to the median and tertiles approach are provided in Supplementary Appendix Figure A2. These, however, were not selected as the main result given the low efficiency of this approach.
Baseline characteristics of patients according to the risk groups
For the two-risk group approach, in the high-risk group, 75% of patients presented with pain, were older, with moderate albumin level and had tumour of large size. In contrast, in the low-risk group, 48% of patients reported pain, were younger, had normal albumin levels and had tumour of small size (Supplementary Appendix Table A1A). A similar trend across groups was observed in the three-risk groups approach (Supplementary Appendix Table A1B).
Agreement between nomogram prediction and the PROLAP score
Given that the prognostic nomogram and score were built from the same multivariate final model parameters, they can be connected. Characterisation of risk groups with the total number of points derived from the nomogram is provided in the Kaplan–Meir curves (Figure 3A and B).
Predictive value of treatments on OS according to the risk groups
In the two- and three-risk group approaches, there were no differential effects on OS of the gemcitabine-erlotinib combination across groups. In addition, no significant OS difference was observed in favour of CRT over CT across groups.
Interestingly, a total of 121 (68%) low-risk, 85 (62%) intermediate-risk and 19 (44%) high-risk patients reached the second randomisation (P=0.0027). A similar significant trend was also observed in the two-risk group analysis. Detailed results are provided in Supplementary Appendix A4.
Validation of the discriminatory ability of the PROLAP score
A total of 63 (59%) of patients from the external validation cohort had all the information required to be eligible for the score calculation using five baseline parameters (pain, CA 19-9, albumin, tumour size and age). Similar characteristics at diagnosis and outcomes were observed between eligible and non-eligible patients (Supplementary Appendix Table A2).
Overall, the final multivariate model identified in the development set exhibited a better discrimination ability in the validation cohort (C-index 0.68; 95% bootstrap percentile CI, 0.59–0.77).
In the two-risk group approach, the discrimination ability of the risk score algorithm developed in the main analysis was externally confirmed, with median OS estimated to 17.1 months for the low-risk group and 9.2 months for the high-risk group (HR=2.3; 95% CI, 1.3 to 4.2; log-rank P=0.003; Figure 3C) and with a better C-index estimation (0.61; 95% CI 0.54 to 0.68) than in the development cohort.
Similarly in the three-risk group approach, the discrimination ability of the risk score was externally confirmed, with median OS estimated to 18.3 months for the low-risk group, 14.1 months for the intermediate-risk group and 7.6 months for the high-risk group (log-rank P<0.0001; Figure 3D) and with a better C-index estimation (0.68; 95% CI 0.59–0.76) than in the development cohort.
Discussion
Currently, there is no efficient prognostic staging system that could help to estimate OS at diagnosis in LAPC patients. On the basis of the largest phase III clinical trial of LAPC, we established a novel easy-to-use survival prediction model built on five key parameters (age, albumin, tumour size, CA 19-9 and pain) and provided a prognostic nomogram and score.
Nomograms are an important component of modern medical decision-making (Balachandran et al, 2015). The main goal of such staging systems is to provide the most accurate predictions for specified end points. In this way, parameters to consider should be easily available and measurable. The proposed nomogram fulfils these requirements and produces individual survival estimations in LAPC patients. The baseline prognostic score can be considered as a complementary tool for clinical practice by producing a classification of the patients into well-identified death-risk populations. We have developed an open-access web site smartphone compatible to automatically compute the prognostic nomogram and score estimations. (http://www.umqvc.org/en/tool/prolap.html).
The present work revealed a considerable heterogeneity among LAPC patients regarding their OS-risk profiles. The recognition of risk groups with different prognostic outcomes may offer the opportunity to define risk-adapted strategies for LAPC management in the future. Indeed, while our analysis was based on OS, but not on progression-free survival, we accurately identified patients more likely not to be eligible for the second randomisation. Overall, 32% of the low-risk, 38% of the intermediate-risk and 56% of the high-risk patients failed to reach the second randomisation due to progression.
Our study supports the role of the nomogram and/or the PROLAP score as tools to be used both for patient selection and/or for patients' stratification in future clinical trials. These may help to reduce heterogeneity among treatment arms in terms of risk profile and thus better estimate the treatment effect. Indeed, if patients from the risk groups identified are distributed unequally among arms this difference can lead to a distortion in the outcome analysis. It seems therefore reasonable to recommend pre-planned subgroup analyses according to the level of such a score in future trials.
It is worth noting that two of the five factors included in our model, pain and albumin are objective surrogates for measurement of HRQoL (Burris et al, 1997). The importance of these parameters in the model and the recognition of HRQoL as an additional prognostic value for OS in metastatic disease (Gourgou-Bourgade et al, 2013) and in other tumour types (Diouf et al, 2014; Fiteni et al, 2016; Paquette et al, 2016) suggest that HRQoL needs to be evaluated in the future prognostic models for LAPC.
Our study has several strengths. The score was constructed in the framework of a large international multicentre trial with a broad spectrum of parameters available at diagnosis. Moreover, discrimination, calibration and internal validation underlined satisfactory performance and validity of the model. We built our model in a rigorous methodological framework respecting a recent recommended checklist for nomograms (Balachandran et al, 2015) and providing a transparent reporting of the multivariate model as suggested in the TRIPOD statement (Collins et al, 2015). The internal validation ensured a reliable performance for patients similar to those of the development cohort. Our prognostic score was fully externally replicated in the ‘real-world' patients encountered in daily clinical practice: different centres; clinicians; treatments; and periods. This external validation population is quite different to the development set and underlines the robustness of our results in the clinical setting. A validation in a larger cohort of patients will be performed with the International Pancreas Database Program ARCAD metabase, which is currently under development. Finally, the five parameters identified in our study reflect the general condition of the patient and disease activity and therefore provide clinically relevant information in LAPC. These factors are admitted prognostic factors in LAPC patients by clinicians and accordingly enhance the relevance of the tools developed.
The present study has also several limitations. The role of pain status could be further refined by its assessment at diagnosis before administration of analgesic or by documenting whether the patient was receiving opioids or not at the time of assessment. In case of baseline cholestasis, the CA 19-9 level should be better estimated after biliary drainage whenever possible in order to minimise overestimation. Despite the broad spectrum of parameters analysed and the acceptable performance of the score, we recognise that additional potentially relevant variables such as a neutrophil to lymphocyte ratio could not be assessed and merit future consideration (Proctor et al, 2012; Stotz et al, 2013; Goldstein et al, 2015) given that novel biologic insights have recently been validated through the pancreatic genome project (Bailey et al, 2016).
In conclusion, we present here a novel OS prediction model based on five key independent prognostic factors at the time of diagnosis. This allowed us to develop and externally validate the first prognostic staging system for OS in LAPC and then to provide functional tools at individual (nomogram) and population (score) levels: http://www.umqvc.org/en/tool/prolap.html. In this study, we revealed considerable heterogeneity among LAPC patients regarding their OS-risk profiles with the recognition of clearly different risk groups. The prognostic nomogram and the score proposed may help to optimise clinical trials design and might offer the opportunity to define risk-adapted strategies for LAPC management in the future.
Acknowledgments
We thank Florian Limousin (graphic designer and independent web developer in Pearlweb society) for the web development application support. The LAP07 trial was supported in part by a grant from Roche. Data from the INTERNATIONAL INTERGROUP STUDY LAP07 (France: GERCOR, FFCD & FNCLCC (PRODIGE)-SFRO; Belgium: BGDO; Australia: AGITG; Sweden: Nordic Group).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
PA and TA have received honoraria and consulting fee from Roche. PH has received consulting fee from Roche. FB has received honoraria and research funding from Roche. The remaining authors declare no conflict of interest.
Supplementary Material
References
- Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, Miller DK, Christ AN, Bruxner TJC, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grützmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM Australian Pancreatic Cancer Genome Initiative Munzy D, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey U-MH, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM (2016) Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531: 47–52. [DOI] [PubMed] [Google Scholar]
- Balachandran VP, Gonen M, Smith JJ, DeMatteo RP (2015) Nomograms in oncology: more than meets the eye. Lancet Oncol 16: e173–e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnetain F, Bonsing B, Conroy T, Dousseau A, Glimelius B, Haustermans K, Lacaine F, Van Laethem JL, Aparicio T, Aust D, Bassi C, Berger V, Chamorey E, Chibaudel B, Dahan L, De Gramont A, Delpero JR, Dervenis C, Ducreux M, Gal J, Gerber E, Ghaneh P, Hammel P, Hendlisz A, Jooste V, Labianca R, Latouche A, Lutz M, Macarulla T, Malka D, Mauer M, Mitry E, Neoptolemos J, Pessaux P, Sauvanet A, Tabernero J, Taieb J, van Tienhoven G, Gourgou-Bourgade S, Bellera C, Mathoulin-Pélissier S, Collette L (2014) Guidelines for time-to-event end-point definitions in trials for pancreatic cancer. Results of the DATECAN initiative (Definition for the Assessment of Time-to-event End-points in CANcer trials). Eur J Cancer 50: 2983–2993. [DOI] [PubMed] [Google Scholar]
- Burris HA, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, Cripps MC, Portenoy RK, Storniolo AM, Tarassoff P, Nelson R, Dorr FA, Stephens CD, Von Hoff DD (1997) Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol 15: 2403–2413. [DOI] [PubMed] [Google Scholar]
- Chauffert B, Mornex F, Bonnetain F, Rougier P, Mariette C, Bouché O, Bosset JF, Aparicio T, Mineur L, Azzedine A, Hammel P, Butel J, Stremsdoerfer N, Maingon P, Bedenne L (2008) Phase III trial comparing intensive induction chemoradiotherapy (60Gy, infusional 5-FU and intermittent cisplatin) followed by maintenance gemcitabine with gemcitabine alone for locally advanced unresectable pancreatic cancer. Definitive results of the 2000-01 FFCD/SFRO study. Ann Oncol 19: 1592–1599. [DOI] [PubMed] [Google Scholar]
- Collins GS, Reitsma JB, Altman DG, Moons KGM (2015) Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Br J Cancer 112: 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR (1957) Note on Grouping. J Am Stat Assoc 52: 543–547. [Google Scholar]
- Diouf M, Chibaudel B, Filleron T, Tournigand C, Hug de Larauze M, Garcia-Larnicol M-L, Dumont S, Louvet C, Perez-Staub N, Hadengue A, de Gramont A, Bonnetain F (2014) Could baseline health-related quality of life (QoL) predict overall survival in metastatic colorectal cancer? The results of the GERCOR OPTIMOX 1 study. Health Qual Life Outcomes 12: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiteni F, Vernerey D, Bonnetain F, Vaylet F, Sennélart H, Trédaniel J, Moro-Sibilot D, Herman D, Laizé H, Masson P, Derollez M, Clément-Duchêne C, Milleron B, Morin F, Zalcman G, Quoix E, Westeel V (2016) Prognostic value of health-related quality of life for overall survival in elderly non-small-cell lung cancer patients. Eur J Cancer Oxf Engl 1990 52: 120–128. [DOI] [PubMed] [Google Scholar]
- Goldstein D, El-Maraghi RH, Hammel P, Heinemann V, Kunzmann V, Sastre J, Scheithauer W, Siena S, Tabernero J, Teixeira L, Tortora G, Van Laethem J-L, Young R, Penenberg DN, Lu B, Romano A, Von Hoff DD (2015) nab-Paclitaxel plus gemcitabine for metastatic pancreatic cancer: long-term survival from a phase III trial. J Natl Cancer Inst e-pub ahead of print 31 Jan 2015 doi:10.1093/jnci/dju413. [DOI] [PubMed]
- Gourgou-Bourgade S, Bascoul-Mollevi C, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, Adenis A, Raoul J-L, Boige V, Bérille J, Conroy T (2013) Impact of FOLFIRINOX compared with gemcitabine on quality of life in patients with metastatic pancreatic cancer: results from the PRODIGE 4/ACCORD 11 randomized trial. J Clin Oncol 31: 23–29. [DOI] [PubMed] [Google Scholar]
- Hajj C, Goodman KA (2015) Role of radiotherapy and newer techniques in the treatment of GI cancers. J Clin Oncol 33: 1737–1744. [DOI] [PubMed] [Google Scholar]
- Hammel P, Huguet F, van Laethem J-L, Goldstein D, Glimelius B, Artru P, Borbath I, Bouché O, Shannon J, André T, Mineur L, Chibaudel B, Bonnetain F, Louvet C (2016) Effect of chemoradiotherapy vs chemotherapy on survival in patients with locally advanced pancreatic cancer controlled after 4 months of gemcitabine with or without erlotinib: the LAP07 Randomized Clinical Trial. JAMA 315: 1844–1853. [DOI] [PubMed] [Google Scholar]
- Harrell FE Jr, Lee KL, Mark DB (1996) Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 15: 361–387. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Lausen B (2003) On the exact distribution of maximally selected rank statistics. Comput Stat Data Anal 43: 121–137. [Google Scholar]
- Huguet F, André T, Hammel P, Artru P, Balosso J, Selle F, Deniaud-Alexandre E, Ruszniewski P, Touboul E, Labianca R, de Gramont A, Louvet C (2007) Impact of chemoradiotherapy after disease control with chemotherapy in locally advanced pancreatic adenocarcinoma in GERCOR phase II and III studies. J Clin Oncol 25: 326–331. [DOI] [PubMed] [Google Scholar]
- Krishnan S, Rana V, Janjan NA, Varadhachary GR, Abbruzzese JL, Das P, Delclos ME, Gould MS, Evans DB, Wolff RA, Crane CH (2007) Induction chemotherapy selects patients with locally advanced, unresectable pancreatic cancer for optimal benefit from consolidative chemoradiation therapy. Cancer 110: 47–55. [DOI] [PubMed] [Google Scholar]
- Loehrer PJ, Feng Y, Cardenes H, Wagner L, Brell JM, Cella D, Flynn P, Ramanathan RK, Crane CH, Alberts SR, Benson AB (2011) Gemcitabine alone versus gemcitabine plus radiotherapy in patients with locally advanced pancreatic cancer: an Eastern Cooperative Oncology Group trial. J Clin Oncol 29: 4105–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E (2014) European cancer mortality predictions for the year 2014. Ann Oncol 25(8): 1650–1656. [DOI] [PubMed] [Google Scholar]
- Paquette B, Vernerey D, Chauffert B, Dabakuyo S, Feuvret L, Taillandier L, Frappaz D, Taillia H, Schott R, Ducray F, Fabbro M, Tennevet I, Ghiringhelli F, Guillamo J-S, Durando X, Castera D, Frenay M, Campello C, Dalban C, Skrzypski J, Chinot O, Anota A, Bonnetain F (2016) Prognostic value of health-related quality of life for death risk stratification in patients with unresectable glioblastoma. Cancer Med e-pub ahead of print 1 June 2016 doi:10.1002/cam4.734. [DOI] [PMC free article] [PubMed]
- Philip PA (2011) Locally advanced pancreatic cancer: where should we go from here? J Clin Oncol 29: 4066–4068. [DOI] [PubMed] [Google Scholar]
- Proctor MJ, McMillan DC, Morrison DS, Fletcher CD, Horgan PG, Clarke SJ (2012) A derived neutrophil to lymphocyte ratio predicts survival in patients with cancer. Br J Cancer 107: 695–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schemper M, Smith TL (1996) A note on quantifying follow-up in studies of failure time. Control Clin Trials 17: 343–346. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66: 7–30. [DOI] [PubMed] [Google Scholar]
- Stotz M, Gerger A, Eisner F, Szkandera J, Loibner H, Ress AL, Kornprat P, AlZoughbi W, Zoughbi WA, Seggewies FS, Lackner C, Stojakovic T, Samonigg H, Hoefler G, Pichler M (2013) Increased neutrophil-lymphocyte ratio is a poor prognostic factor in patients with primary operable and inoperable pancreatic cancer. Br J Cancer 109: 416–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.