Abstract
Different viruses have been identified as etiologic agents of respiratory tract infections, including severe cases. Among these, human rhinoviruses (HRVs) and human enteroviruses (HEVs) are recognized as leading causes. The present study describes the molecular epidemiology of HRVs and HEVs in Senegal over a 3-year surveillance period. From January 2012 to December 2014, nasopharyngeal and oropharyngeal swabs specimen were collected from patients with influenza-like illness (ILI). A real-time reverse transcription polymerase chain reaction was performed for HRV and HEV detection using the RV16 kit. Two regions were targeted for the molecular characterization of RVs: 5′ untranslated region (5′UTR) and viral protein 4/viral protein 2 (VP4/VP2) transition region. For enteroviruses (EVs) phylogeny, VP1 gene was targeted. A total of 4,194 samples were collected. Children up to 5 years accounted for 52.9%. Among them, 1,415 (33.7%) were positive for HRV, 857 (20.4%) for HEV, and 437 cases of dual infections HRV/HEV. HRVs and HEVs were identified significantly in children aged 5 years or less. Only cough and vomiting signs were observed with significant association with viral infection. Both viruses co-circulated all year long with a marked increase of activity during rainy and cold period. All HRV types circulate in Senegal. HRV-A and C groups were the most common. HEV serotyping identified coxsackie B viruses (CBV) only. VP1 region revealed different CBV (CBV1, CBV2, CBV3, CBV4, and CBV5), echoviruses, coxsackieviruses A4–like strains and a poliovirus 2. The results suggest strong year-round respiratory picornavirus activity in children up to 5 years of age. Molecular studies identified a wide variety of RVs along with diverse EVs in samples from patients with ILI.
Introduction
Acute respiratory tract infections (RTIs) are a leading cause of morbidity and mortality worldwide, particularly in children and the elderly.1–5 About 1.9 million child deaths occur each year with 70% of these in Africa and southeast Asia.2 Many etiological studies show that viruses are the main cause of RTIs.6 Indeed, a wide variety of viruses have been identified as the etiologic agents of RTIs including particularly severe cases.
Among these, human rhinoviruses (HRVs) and human enteroviruses (HEVs) are recognized as leading causes of RTIs7–10 and are responsible for 60.5% of microbiologically unexplained acute RTI diagnosed in hospitalized pediatric cases.10
HRVs and HEVs belong to the Picornaviridae family and exhibit high genomic similarity.11 However, HEVs and HRVs differ in terms of their preferred growth temperature, sensitivity to pH, and cell tropism.
Senegal has had a National Influenza Center since 1974; this latter has been part of the World Health Organization (WHO) Global Influenza Surveillance Network since 1996.12 The purpose of this influenza surveillance has traditionally been the early detection of influenza epidemics in the community, the identification of predominant circulating strains of influenza virus, and the issuing of public health recommendations. A previous study showed that only 13.4% of specimens from patients with influenza-like illness (ILI) collected between 1996 and 2009 were confirmed as influenza positive, suggesting the existence of other etiologies in most cases.13 Thus, surveillance was extended to cover other respiratory viruses in 2012.14
The aim of the present study was to describe the epidemiology and molecular characteristics of respiratory picornaviruses (EVs and RVs) in Senegal over a 4-year surveillance period.
Materials and Methods
Samples and data collection.
From January 2012 to December 2014, in collaboration with the epidemiology unit of the institute Pasteur de Dakar, nasopharyngeal and oropharyngeal swabs specimen were collected from consenting ILI patients. Once collected, the swabs were placed in 2-mL cryovials containing viral transport medium (Universal Transport Medium; COPAN Diagnostics Inc., Murrieta, CA) and transported under a controlled temperature (4°C) to the laboratory. The ILI was defined according to the Centers for Disease Control (CDC) case definition: sudden onset of fever (≥ 38°C) with a cough or sore throat lasting less than 3 days. Each sample was accompanied by a case report form containing basic epidemiologic information and a description of the symptoms before presentation. On arrival at the laboratory, the specimens were processed immediately for virus isolation/detection, identification, and characterization. Aliquots of each sample were also stored at −80°C for additional analysis.
Case reports were entered into an Epi Info database (CDC, Atlanta, GA) and merged with laboratory data.
RNA extraction and detection of respiratory viruses.
Nucleic acid extraction and a two-step real-time reverse transcription polymerase chain reaction was performed as previously described using the CFX96TM Real-time PCR system (Bio-Rad, Singapore).14 The RV16 Kit (Seegene, Seoul, Korea) used allows differentiation of RVs from other EVs.
Enterovirus isolation and serotyping.
Human epidermoid carcinoma (Hep2c) and human rhabdomyosarcoma (RD) cell lines were used for enterovirus isolation. All cell lines were donated by the CDC for the polioviruses surveillance program. Nasopharyngeal samples from patient positive for enterovirus were inoculated onto both cell lines. Temporal repartition (weeks and years) of samples collection criterion have also been taken into account in the sample selection.
Each sample (200 μL) was treated with antibiotics (penicillin/streptomycin, 0.5%), filtered through a 0.22-μm filter, and added to a cell culture tube previously prepared with a suspension of 105 cells in a final volume of 1 mL medium. Tubes were incubated at 37°C in an atmosphere containing 5% CO2 conditions. The inoculated cells were examined daily for any cytopathic effect (CPE). Samples showing CPEs were stored at −20°C pending serotyping. Tubes containing cells showing no CPE after 7 days were frozen, thawed, re-passaged, and examined for a further 7 days.
Supernatants from all CPEs-positive tubes either on RD or Hep2c tubes were harvested for the viral serotyping using the antiserum pool containing horse typing antisera against HEV serotypes. Serotyping was performed according to the WHO Polio Laboratory Manual protocol.15 In brief, after a titration step, equal amount at the half maximal inhibitory concentration dilution of the isolate was mixed with different combinations of the antisera pools and incubated for 3 hours at 37°C before the addition of Hep2c cells. Cultures were observed daily for 5 days, and characteristics CPE were noted. Antisera combinations that inhibited CPEs facilitated virus identification.
Molecular studies.
For sequencing, we basically took into account three criteria to select samples: a positive result in real-time PCR, distribution of positive samples per year and week, and the cycle threshold values detection. Indeed, only samples positive at the first catcher melting temperature analysis point level (which means an important viral load) were targeted. Unfortunately, many samples showed no amplification or poor-quality sequences. Indeed, only 46% samples were successfully amplified and sequenced for RV and for 66% EVs. The low sensitivity of conventional PCR compared with real-time PCR or nonspecific amplifications could be the cause of these failures.
For RVs two regions were targeted to enable the molecular characterization: the 5′ untranslated region (5′UTR) region and the viral protein 4/viral protein 2 (VP4/VP2) transition region were used to identify infecting HRV serotypes.16
To amplify the 5′UTR region, a first round PCR targeting a 913-nucleotide fragment was performed using reverse primer SRHI1 (GCATCIGGYARYTTCCACCACCANCC) and forward primer DK001 (CAAGCACTTCTGTTTCCC).17,18 In brief, 10.7 μL H2O ribonuclease (RNase) free, 6 μL of reaction buffer 5×, 1.2 μL of deoxyribonucleotide triphosphates (dNTPs) mix (10 mM each), 1.8 μL of each primer (diluted at 10 μM), 3 μL of Q solution, five units of taq DNA polymerase (Qiagen, Hilden, Germany), and 5 μL of cDNA were mixed in a PCR tube. After denaturation for 15 minutes at 95°C, the PCR was performed as follows: 40 cycles at 95°C for 30 seconds, 55°C for 60 seconds, and 72°C for 60 seconds, followed by an extension step at 72°C for 10 minutes and a 4°C hold.
A semi-nested PCR targeting a 550-nucleotide fragment was then performed using a new inner sense primer (SRHI2: 5′-GGGACCAACTACTTTGGGTGTCCGTGT-3′) and 1 μL of the negative PCR products as templates.
PCR products were run on a 1% agarose gel along with appropriated molecular weight markers (100 bp ladder; New England Biolabs, Ipswich, MA). The gels were then stained with ethidium bromide (0.5 μg/mL) before visualization under ultraviolet light. Positive samples were sequenced by Beckman Coulter Services (Hope End Takeley, United Kingdom) using the Sanger method. Data in fast adaptive shrinkage thresholding algorithm format were then sent for analysis.
The VP4/VP2 region was amplified using the couple of primers VP4/VP2 IS (5′-ACCRACTACTTTGGGTGTCCGTG-3′) and VP4/VP2 IAS (5′ TCWGGHARYTTCCAMCACCANCC-3′). The reaction mixture contained 12.3 μL H2O RNase free, 2 μL of reaction buffer 10×, 1.6 μL of dNTPs mixed (2.5 mM each), 1 μL of 10 μM of each primer, 0.5 units of taq DNA polymerase, and 10 μL of cDNA. The thermal cycling conditions were as follows: 5 minutes at 95°C for initial denaturation, 40 cycles of 15 seconds at 95°C for denaturation, 30 seconds at 55°C for annealing, 1 minute at 70°C for extension, and 15 minutes at 72°C for a final extension step. The targeted amplicon (541 bp) was visualized as described above, and positive samples were sent for sequencing.
AMTH and GDCL primers, described elsewhere,19 were used for EVs serotypes characterization. The latter allows the amplification of a 1,491-bp fragment that encompasses the whole VP1 gene. The reaction mixture (50 μL of volume) contained 5 μL of cDNAs, 34.3 μL H2O RNase free, 5 μL of reaction buffer 10×, 1.2 μL of dNTPs mix (10 mM), 2 μL of each primer (10 μM), and 2.5 units of taq DNA polymerase. The thermocycler profile was as follows: 15 minutes at 95°C, followed by 40 cycles of 95°C for 30 seconds, 55°C for 30 seconds, 60°C for 2 minutes, and 15 minutes at 60°C for final extension. Positive samples (1,500-bp amplicon) were bidirectionally sequenced as described for RVs.
Sequence analysis and multiple sequence comparison.
Sequences were aligned against related sequences retrieved from GenBank, and searches for sequence similarities was carried out using the Basic Local Alignment Search Tool (Blastn) from National Center for Biotechnology Information BLAST web portal. Multiple alignments were edited using the BioEdit Sequence Alignment Editor.20 Phylogenetic trees were generated using the MEGA version 5 for constructing maximum likelihood tree using the Tamura–Nei evolutionary model with neighbor-joining using 100 bootstrap replicates,21 with bootstrap values ≥ 70.
Statistical analysis.
We compared the distribution of RV and EV infection cases in the different age groups to verify whether the associated rates were statistically supported. The Fisher's exact test was used, and a P value < 0.05 was considered statistically significant. We used the 0–5 years age group as the reference group. The R.3.0.1 tool was used to perform the analyses.
Ethical considerations.
This study is a component of the 4S network syndromic surveillance.14 Principles of the 4S network were approved by the Ministry of Health in its guidelines for influenza surveillance policy, finalized with the support of Pasteur Institute in Dakar and the Strengthening Influenza Sentinel Surveillance in Africa project funded by the WHO. The protocol and oral consent were determined as routine surveillance activity by the Senegalese National Ethics committee and the steering committee for 4S network, an entity representing Ministry of Health, Institut Pasteur Dakar, WHO, and clinicians in compliance with all applicable national regulations governing the protection of human subjects. Data were collected anonymously in an objective of surveillance and applicable to molecular epidemiology studies on the detected pathogen. The information provided to participants was an informal description of the study. Respiratory specimens were collected only after informed consent was granted, verbally, to local health-care workers by the patients or parents in the case of minors. Oral consent was documented in the patient form with two questions about received information and about oral consent. If patients refuse to participate, no specimen was taken. For the surveillance activities, written consent is judged not necessary by the Senegalese National Ethics Committee, which has also previously approved the work of the National Influenza Center. Collections of nonsensitive data or an observation from normal care in which participants remain anonymous do not require ethics committee review. The patients included in this study were of all ages and consulted the sentinel sites due to influenza-like symptoms; the patients, or parents in the case of minors, accepted the tests for respiratory viruses largely because these tests were free and safe.
Results
Demographic and clinical characteristics of the enrolled patients.
From January 2012 to December 2014, a total of 4,194 samples were collected from patients presenting with ILI at the different sentinel sites in Senegal: 753 (17.9%) in 2012, 1,519 (36.2%) in 2013, and 1,922 (45.8%) in 2014 (Table 1). Ages ranged from 1 month to 95 years. The mean age was 10 years 5 months and the median age was 4 years. The male-to-female ratio was about 0.97 (2,054 [49%] males and 2,121 [50.6%] females). Children up to 5 years of age accounted for 52.9% (2,218/4,194) of ILI cases, followed by 5–10 years age group with 10.7% (447/4,194). Patients over 50 years of age represented only 2.7% (113/4,194) of enrolled patients. Age was not documented for 8.5% (356/4,194) of patients.
Table 1.
Demographical characteristics and symptoms
| Characteristics | 2012 | 2013 | 2014 | Total |
|---|---|---|---|---|
| (N = 753) | (N = 1,519) | (N = 1,922) | (N = 4,194) | |
| Gender, n (%) | ||||
| Male | 378 (50.2) | 744 (49.0) | 932 (48.5) | 2,054 (49.0) |
| Female | 371 (49.3) | 767 (50.5) | 983 (51.1) | 2,121 (50.6) |
| Missing | 4 (0.5) | 8 (0.5) | 7 (0.4) | 19 (0.4) |
| Age group (years), n (%) | ||||
| 0–5 | 520 (69.1) | 763 (50.2) | 935 (48.6) | 2,218 (52.9) |
| 5–10 | 78 (10.4) | 162 (10.7) | 207 (10.8) | 447 (10.7) |
| 10–15 | 44 (5.8) | 85 (5.6) | 122 (6.3) | 251 (6.0) |
| 15–25 | 40 (5.3) | 122 (8.0) | 233 (12.1) | 395 (9.4) |
| 25–50 | 30 (4.0) | 120 (7.9) | 264 (13.7) | 414 (9.9) |
| ≥ 50 | 10 (1.3) | 18 (1.2) | 85 (4.4) | 113 (2.7) |
| Missing | 31 (4.1) | 249 (16.4) | 76 (3.9) | 356 (8.5) |
| Clinical signs, n (%) | ||||
| Myalgia | 85 (11.3) | 342 (22.5) | 307 (16.0) | 734 (17.5) |
| Fever | 709 (94.2) | 1,362 (89.7) | 1,856 (96.6) | 3,927 (93.6) |
| Cough | 561 (74.5) | 1,174 (77.3) | 1,663 (86.5) | 3,398 (81.0) |
| Vomiting | 96 (12.7) | 35 (2.3) | 87 (4.5) | 218 (5.2) |
| Diarrhea | 70 (9.3) | 32 (2.1) | 44 (2.3) | 146 (3.5) |
| headache | 55 (7.3) | 183 (12.0) | 264 (13.7) | 502 (12.0) |
| Dyspnea | 15 (2.0) | 24 (1.6) | 85 (4.4) | 124 (3.0) |
| Rhinitis | 542 (72.0) | 1,048 (69.0) | 1,622 (84.4) | 3,212 (76.6) |
| Pharyngitis | 670 (89.0) | 812 (53.5) | 1,619 (84.2) | 3,101 (73.9) |
Regarding clinical symptoms, fever was the primary inclusion criterion although it was not always reported on the data form (93.6%; 3,927/4,194). Cough (81%; 3,398/4,194), rhinitis (76.6%; 3,212/4,194), and pharyngitis (73.9%; 3,101/4,194) were also common symptoms. Myalgia (17.5%; 734/4,194), headache (12%; 502/4,194), and dyspnea (3%; 124/4,194) were also reported. Enteric symptoms as vomiting (5.2%; 218/4,194), diarrhea (3.5%; 146/4,194), and abdominal pains have been noted in some patients.
Patients and viral infections.
Of the 4,194 ILI patients specimens collected and tested using real-time PCR, 1,415 (33.7%) were positive for HRV and 857 (20.4%) for HEV. The detection rate of both HRV and HEV over the study period was highest in 2013 (42.8% for HRV and 33.4% for HEV). A total of 437 cases (10.4%; 437/4,194) of dual infections HRV/HEV were also observed.
HRVs and HEVs were identified significantly more often in children of 5 years or less of age, 52.9% and 52.1% of viruses detected, respectively, a statistically significant finding (P < 0.05 for both) (Table 2). However, the detection rates in the other groups were also high (e.g., 31.9% of HRV detection for > 50 years old). The gender distribution of the HRV- and HEV-positive patients was not significantly different from the negative patients. Taking into account the clinical symptoms and viral infections, only cough (P < 0.005 for both viruses) and vomiting (P = 0.012 for HRV and 0.0004 for HEV) signs were observed with a statistical significant association with viral infection. We also noted that among the 108 cases of diarrhea reported, 48 (44.4%) were infected by HRV.
Table 2.
Detection rates of human rhinovirus and human enterovirus infection in patients with influenza-like illness per year from 2012 to 2014 in Senegal and comparison of the distribution in different age groups
| Years | 2012 | 2013 | 2014 | Total | P values | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | 753 | 1,519 | 1,922 | 4,194 | HRV | HEV | ||||
| Pathogen | Rhinovirus | Enterovirus | Rhinovirus | Enterovirus | Rhinovirus | Enterovirus | Rhinovirus | Enterovirus | ||
| Positivity (per year) | 31.5 | 15.9 | 42.8 | 33.4 | 27.4 | 12.0 | 33.7 | 20.4 | ||
| Positive | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | ||
| Sex male | 128 (54) | 62 (51.7) | 342 (52.5) | 257 (50.7) | 258 (48.9) | 118 (51.3) | 728 (51.4) | 437 (51) | 0.16 | 0.49 |
| Age group (years) | ||||||||||
| 0–5 | 176 (74.3) | 82 (68.3) | 310 (47.6) | 235 (46.3) | 262 (49.7) | 130 (56.5) | 748 (52.9) | 447 (52.1) | – | – |
| 5–10 | 22 (9.3) | 14 (11.7) | 66 (10.1) | 49 (9.7) | 43 (8.2) | 17 (7.4) | 131 (9.2) | 80 (9.3) | 0.02 | 0.03 |
| 10–15 | 14 (5.9) | 8 (6.7) | 29 (4.4) | 35 (6.9) | 22 (4.2) | 6 (2.6) | 65 (4.6) | 49 (5.7) | ||
| 15–25 | 14 (5.9) | 13 (10.8) | 58 (8.9) | 39 (7.7) | 49 (9.3) | 16 (6.9) | 121 (8.5) | 68 (7.9) | ||
| 25–50 | 8 (3.4) | 2 (1.7) | 52 (8) | 47 (9.2) | 85 (16.1) | 23 (10) | 145 (10.2) | 72 (8.4) | ||
| ≥ 50 | 3 (1.2) | 1 (0.8) | 11 (1.7) | 5 (1) | 22 (4.2) | 8 (3.5) | 36 (2.5) | 14 (1.6) | ||
| Missing | 0 (0) | 0 (0) | 125 (19.2) | 97 (19.1) | 44 (8.3) | 30 (13) | 169 (11.9) | 127 (14.8) | ||
| Total | 237 (100) | 120 (100) | 651 (100) | 507 (100) | 527 (100) | 230 (100) | 1,415 (100) | 857 (100) | ||
P values in bold are considered statistically significant.
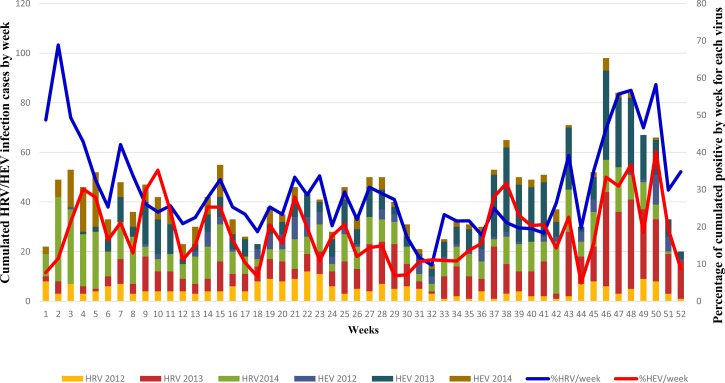
To evaluate the temporal distribution pattern of HRV and HEV species throughout the year during the study period (2012–2014), Figure 1 presents cumulative viral detection data on a weekly basis for each virus. We noted that both viruses co-circulated all year long with detection peaks of different amplitudes. However, for both viruses, there was a marked increase in the activity during last weeks (around weeks 38 and 39 for HEV and week 42 for HRV with major peaks occurring at around weeks 46–49). This increase in activity is also extended to the first weeks of the following year. It is important to note that July to October corresponds to the rainy season and the end and beginning of years are the cold periods in Senegal.
Figure 1.
Cumulated number of cases of human rhinovirus (HRV) and human enterovirus (HEV) per week over the 2012–2014 period in Senegal.
Phylogenetic analyses of HRV and HEV.
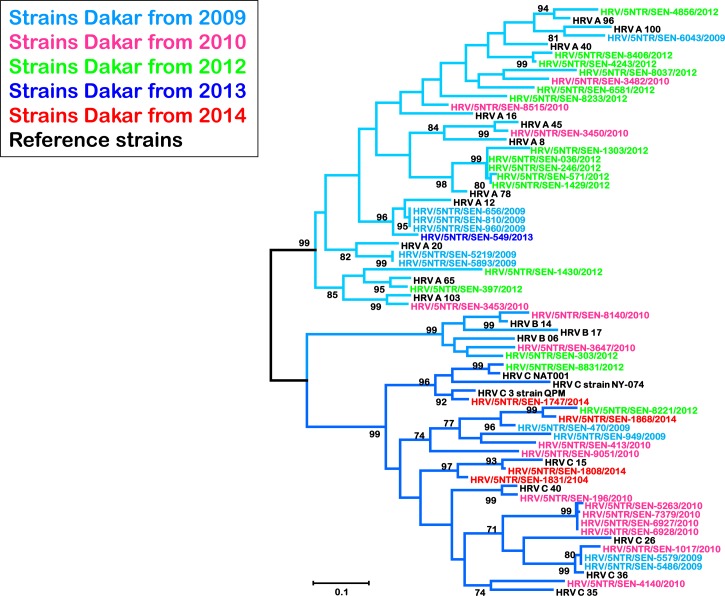
When examining the 5′UTR region, we included sequences that were previously obtained with RV strains from 2009 to 2010. Then a total of 46 sequences from 2009 to 2014 were obtained (Figure 2 ): 10 from 2009, 15 from 2010, 16 from 2012, 1 from 2013, and 4 from 2014. Maximum likelihood phylogenetic comparison of clinical specimens with international reference sequences identified all known HRV groups in Senegal. Of these clinical specimens, randomly selected HRV-A group was the most common (52.2%; 24/46), followed by HRV-C (41.3%; 19/46). Only three strains (6.5%; 3/46) grouped with B species.
Figure 2.
Phylogenetic analysis of human rhinovirus (HRV) isolates from Senegal between 2009 and 2014 based on the 5′ untranslated region noncoding region using the neighbor-joining method with 1,000 bootstrap replicates with MEGA 5 version. Senegal isolates are highlighted in different colors for each year and reference strains from GenBank are in black. Only bootstrap values over 70 are shown. The branches under the same subtype (HRV A, B, or C) are labeled with different blue color nuances.
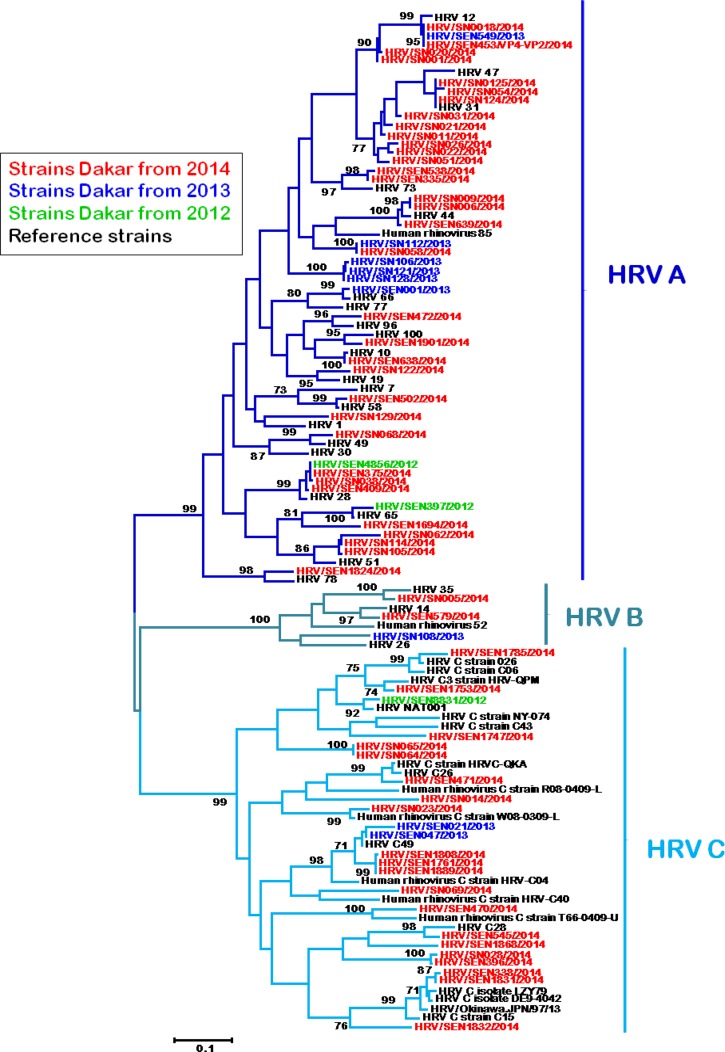
The VP4/VP2 region, which allowed more accurate species identification, was successfully amplified and sequenced from 68 clinical samples. Phylogenetic analysis confirmed that all three HRV subtypes were circulating within the study population, with a pattern (Figure 3 ) similar to the one obtained from the 5′UTR region. Again, type HRV-A (61.8%; 42/68) followed by HRV-C type (33.8%; 23/68) were largely predominant among the randomly selected HRV-positive clinical samples, while only three cases (4.4%) of HRV-B infection was detected. This dominance was even more evident if we considered sequences from 2014 only (in red), year in which we performed more exhaustive sequencing. Regarding the antigenic diversity, the HRV strains from Senegal were closely related to 43 reference serotypes corresponding to 21 HRV-A, 4 HRV-B, and 19 HRV-C strains. Most of Senegalese HRV-C strains were related closely to previously identified QPM, QCE, NAT001, NY-074, C 26, LZY79 HRV-C reference strains, which were reported as Ca subspecies with a HRV-A-like 5′UTR; the remainder assigned as HRV-Cc.22 HRV-B strains from Senegal seem genetically closer to HRV-B14, B26, and B35 reference strains.
Figure 3.
Phylogenetic analysis of human rhinovirus (HRV) isolates from Senegal between 2012 and 2014 based on the viral protein 4/viral protein 2 (VP4/VP2) transition coding region using the neighbor-joining method with 1,000 bootstrap replicates with MEGA 5 version. Senegal isolates are highlighted in different colors for each year and reference strains from GenBank are in black. Only bootstrap values over 70 are shown. The branches under the same subtype (HRV A, B, and C) are with different blue color nuances.
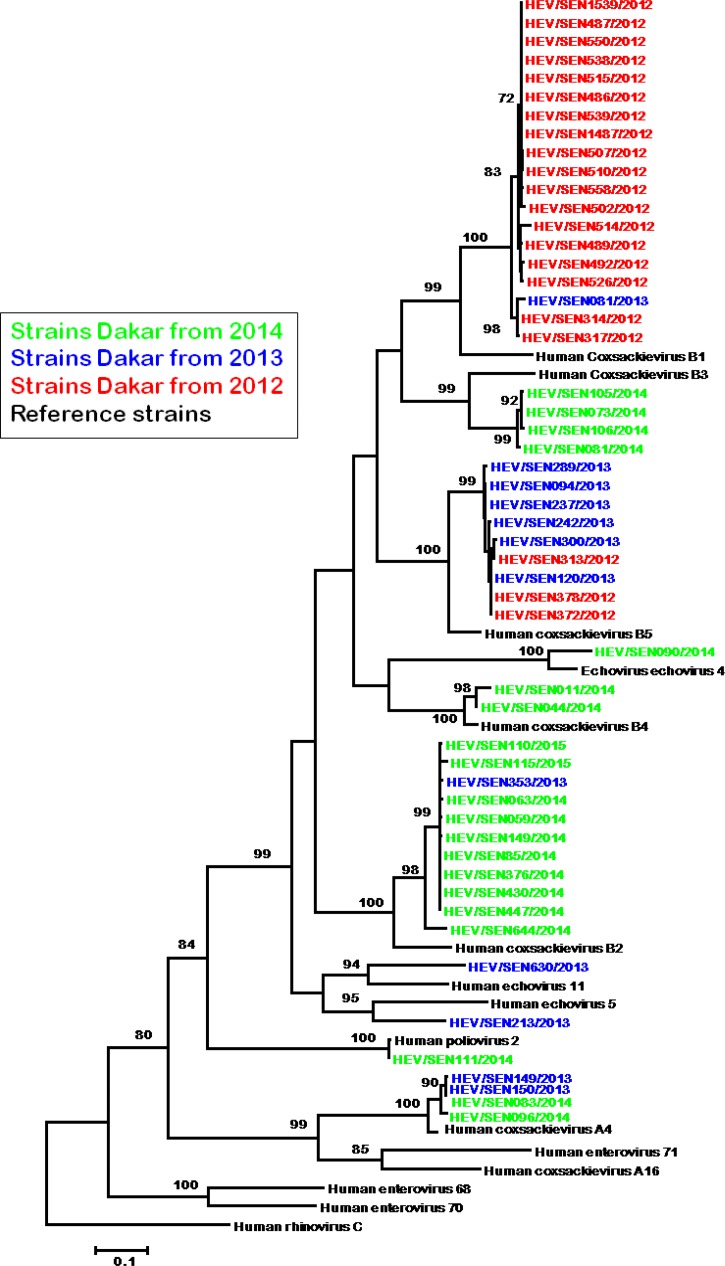
For HEV, a total of 53 strains have been isolated from samples and sequenced successfully. Isolates were all obtained after inoculation on Hep2c cell line, no isolation was achieved with RD cell line. Of these 53 strains, 21 were from 2012, 12 from 2013, and 20 from 2014. Serotyping identified 45 coxsackie B viruses (CBV) and eight isolates that were untypeable using available antisera pools.
Phylogenetic analysis based on the VP1 gene (Figure 4 ) revealed that 19 isolates from Senegal clustered with CBV1, 11 clustered with CBV2, four with CBV3, two with CBV4, and nine with CBV5. CVB1 serotypes were most often isolated from specimens collected in 2012 (18 of 19 isolates), CVB5 from specimens collected in 2013 (six of nine isolates), and CVB2 mostly from specimens collected in 2014 (10 of 11 isolates). Analysis of the VP1 sequence revealed that among the eight serologically untypeable isolates, three were echoviruses genetically closer to the reference serotypes echovirus 4, 5, and 11; four were strongly grouped with the coxsackieviruses A4. The remaining strain clustered with the poliovirus type 2.
Figure 4.
Phylogenetic analysis of human enterovirus (HEV) isolates from Senegal between 2009 and 2014 based on the P1 coding region using the neighbor-joining method with 1,000 bootstrap replicates with MEGA 5 version. Senegal isolates are highlighted in different colors for each year and reference strains from GenBank are in black. Only bootstrap values over 70 are shown.
Discussion
This study reports the first extensive laboratory surveillance of respiratory picornaviruses (HRV and HEV) in ILI patients in Senegal. The study was based on a 3 years' period of surveillance of outpatients. The findings showed that HRVs/HEVs are the most common pathogens present in the nasopharynx of patients with ILI in Senegal across all seasons. Indeed 51% of patients presented with HRV/HEV infection.
The detection rates of HRV and HEV were 34.2% and 20.8%, respectively. RV was the most common respiratory pathogen present in all age groups.23 In a previous study in elderly, Nicholson and others (1997) showed that RVs were responsible for a greater disease burden (activities restriction, duration of illness) than influenza viruses; HRVs were detected in 52% of cases.24 Greenberg also found that RV was the most prevalent pathogen (121 isolates; 53%) detected in 231 cases of upper respiratory infection.25 In a large study in outpatients with ILI from the United States, HRVs/HEVs were the most commonly detected viruses (after influenza), with an overall detection rate of 21%.26 The findings of these different studies are in broad agreement with those presented herein.
However, a recent large study in children and young adults from Latin America revealed a much lower detection rate than the present study.9 Indeed, HRV and HEV were identified in 16% (548/3,375) and 3% (84/3,375) of ILI cases, respectively. A study from Gabon, reported that 12.3% (128/1,041) of ILI cases were infected with HEV and 7.8% (81/1,041) with HRV.27 In two other studies conducted on ILI patients, in South Korea and Philippines, Noh and others and Otomaru and others detected 4.3% (86/1,983) and 7.5% (153/2,031) of RV infection, respectively.28,29 No HEV infection was reported in the study from Korea. These discrepancies in the rate of detection, beyond the technical approaches, may be due to geographical differences in virus burden, differences in subject recruitment strategies (outpatients or hospitalized patients), or to the periods during which samples were collected.
Considering the two regions (5′UTR and VP4/VP2), HRV-A and HRV-C species accounted for the majority of the HRV subtyped (57.9% and 36.8%, respectively), with a variety of serotypes (24 for HRV-A and 17 for HRV-C). Only four HRV-B serotypes were found. Despite the low number of HRV strains studied, these results are in line with those published in studies from other areas (Europe, North America, and east Asia), and all show similar HRV distributions.30–35 Overall, different genotypes of RV appear to circulate simultaneously within a given period and geographic area with predominance of types A and C.22,36,37
Among HEVs characterized herein, CBV (B1, B2, B3, B4, and B5) were predominant (45/53). The temporal distribution of these strains suggests a predominant circulating strain of CBV that can change from one year to another. Results also show the presence of echovirus 4, 5, and 11 serotypes. Four strains were apparently coxsackieviruses A4–like strains. One strain shows proximity with the poliovirus 2 serotype. The child of 16 months age had probably been immunized (oral polio vaccine) just before the nasopharyngeal swab. This diversity of respiratory HEV in ILI patients can be improved with a more exhaustive number of characterized strains. Indeed, recent studies show that EVs associated with respiratory infections are highly diverse.9,38,39
HRVs and HEVs were detected more frequently in children aged < 5 year (35.4% and 21.2%, respectively). These results are in concordance with those of other studies, which concluded that children of 5 years of age or younger had a higher risk of infection by HRV and HEV,9,40 although the risk is slighter lower for HEV. Feikin and others (2013) studied Kenyan children under 5 years of age, who had severe acute respiratory infection (SARI).41 They found that HRV/HEV was the primary etiological agent, with a prevalence of 50% among patients. In China, Zhang and others (2013) reported 54% of child SARI cases harbored HRV/HEV.42 However, particularly for RV infections, it is reported that the infection rate is highest during the early years and decreases with age, probably due to the immunity induced by cumulative exposure to different serotypes.43
However, the presence of a virus in the nasopharynx of a child does not necessarily mean that it is the etiological agent of the ILI; indeed, it may only represent a coincidental upper airway infection, an asymptomatic carrier state, or prolonged shedding of a pathogen that caused a previous infection. This may be particularly important in the case of RVs because a number of epidemiological studies have shown that they are present in the respiratory secretions of 12–22% of asymptomatic subjects.32,44,45
HRV and HEV showed a similar profile with respect to seasonality. Indeed both viruses were endemic throughout the 3-year period, with epidemic peaks showing greater amplitudes during rainy and cold periods in Senegal. These observations agree with those reported in studies performed elsewhere in the world.46,47 A more recent study from Latin America shows that HRV and HEV circulate regularly all year long, but show greater activity during the rainy season.9 Findings from temperate regions show an increase in HRV/HEV activity during the summer,48 peaking from July until the beginning of the fall, followed by a slow decline. However, monitoring must continue for several years if we are to provide a more detailed analysis of the seasonality of these viruses in a sub-Saharan context.
During data analysis, we observed some limitations in our study. First, only a small number of strains were subtyped for both viruses (HRV and HEV). Indeed, the sequencing results do not reflect the full spectrum of HRV/HEV strains that may be circulating in ILI patients in Senegal, and may be biased toward samples with a high viral load; and especially for HRV, systematic type identification of positive samples would allow to pick up seasonal differences among the individual HRV species in Senegal, as reported in studies conducted in other geographical areas.49–51
In conclusion, the results of the present study suggest strong year-round respiratory picornavirus activity in children up to 5 years of age in Senegal. Molecular studies identified a wide variety of RVs along with CBV in samples from patients with ILI. The next step will be to measure the burden of these viruses among children in Senegal; such studies should include asymptomatic controls SARI cases, and provide information on disease outcome, atypical clinical signs, duration of symptoms, and treatment. Data regarding viral load, shedding, and other possible etiologies (e.g., bacterial and other viruses) would also enable a more thorough assessment of the viral effective disease (or symptom) causality.
Acknowledgments
This study would not have been possible without the excellent support from all the health-care workers of the 4S network who contributed, every day, to the surveillance network. We convey our special thanks to Kathleen Victoir from the International Network of Pasteur Institutes for her unwavering support to the 4S network. We acknowledge the Senegalese Ministry of Health for their help in implementing the 4S network. We also thank Cheikh Loucoubar (G4 BBS, IPD) for his great help for statistical analyses, and Kader Ndiaye, Aichatou Diouf Fall, Pape Amadou Vieux Mbathio Diop, and Ameth Fall for their helpful advices in enteroviruses isolation and serotyping. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
Footnotes
Financial support: This work was supported by the Department of Health and Human Service (DHSS) via the International Network of Pasteur Institutes and Pasteur Institute of Dakar.
Authors' addresses: Amary Fall, Ndongo Dia, Ousmane Kébé, Davy E. Kiori, El Hadj Abdoul Khadir Cissé, Sara Sy, Deborah Goudiaby, Ousmane Madiagne Diop, and Mbayame Ndiaye Niang, Medical Virology Unit, Institut Pasteur de Dakar, Senegal, E-mails: amary022@hotmail.com, ndia@pasteur.sn, bigouze87@live.fr, dekiori@pasteur.sn, akacisse@pasteur.sn, sarasy16@outlook.fr, dgoudiaby@pasteur.sn, diopo@who.int, and niang@pasteur.sn. Fatoumata Diene Sarr and Vincent Richard, Epidemiology Unit, Institut Pasteur de Dakar, Senegal, E-mails: fdsarr@yahoo.fr and virc@hotmail.fr.
Reprint requests: Mbayame N. Niang, Institut Pasteur de Dakar, 36 Avenue Pasteur, BP 220, Dakar, Senegal, E-mail: niang@pasteur.sn, Tel: 00 221 839 92 22.
References
- 1.Sloots TP, Whiley DM, Lambert SB, Nissen MD. Emerging respiratory agents: new viruses for old diseases? J Clin Virol. 2008;42:233–243. doi: 10.1016/j.jcv.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams BG, Gouws E, Boschi-Pinto C, Bryce J, Dye C. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis. 2002;2:25–32. doi: 10.1016/s1473-3099(01)00170-0. [DOI] [PubMed] [Google Scholar]
- 3.Louie JK, Yagi S, Nelson FA, Kiang D, Glaser CA, Rosenberg J, Cahill CK, Schnurr DP. Rhinovirus outbreak in a long term care facility for elderly persons associated with unusually high mortality. Clin Infect Dis. 2005;41:262–265. doi: 10.1086/430915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longtin J, Winter AL, Heng D, Marchand-Austin A, Eshaghi A, Patel S, Jamieson F, Weir E, Low DE, Gubbay JB. Severe human rhinovirus outbreak associated with fatalities in a long-term care facility in Ontario, Canada. J Am Geriatr Soc. 2010;58:2036–2038. doi: 10.1111/j.1532-5415.2010.03091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pierangeli A, Scagnolari C, Selvaggi C, Verzaro S, Spina MT, Bresciani E, Antonelli G, Bertazzoni G. Rhinovirus frequently detected in elderly adults attending an emergency department. J Med Virol. 2011;83:2043–2047. doi: 10.1002/jmv.22205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev. 2010;23:74–98. doi: 10.1128/CMR.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jartti T, Lehtinen P, Vuorinen T, Osterback R, van den Hoogen B, Osterhaus AD, Ruuskanen O. Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg Infect Dis. 2004;10:1095–1101. doi: 10.3201/eid1006.030629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacques J, Bouscambert-Duchamp M, Moret H, Carquin J, Brodard V, Lina B, Motte J, Andréoletti L. Association of respiratory picornaviruses with acute bronchiolitis in French infants. J Clin Virol. 2006;35:463–466. doi: 10.1016/j.jcv.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Garcia J, Espejo V, Nelson M, Sovero M, Villaran MV, Gomez J, Barrantes M, Sanchez F, Comach G, Arango AE, Aguayo N, de Rivera IL, Chicaiza W, Jimenez M, Aleman W, Rodriguez F, Gonzales MS, Kochel TJ, Halsey ES. Human rhinoviruses and enteroviruses in influenza-like illness in Latin America. Virol J. 2013;10:305. doi: 10.1186/1743-422X-10-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renois F, Lévêque N, Deliège PG, Fichel C, Bouin A, Abely M, N'guyen Y, Andréoletti L. Enteroviruses as major cause of microbiologically unexplained acute respiratory tract infections in hospitalized pediatric patients. J Infect. 2013;66:494–502. doi: 10.1016/j.jinf.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden FG. Rhinovirus and the lower respiratory tract. Rev Med Virol. 2004;14:17–31. doi: 10.1002/rmv.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization National Influenza Centres. 2010. http://www.who.int/csr/disease/influenza/centres/en/index.html Available at. Accessed March 12, 2010.
- 13.Niang MN, Dosseh A, Ndiaye K, Sagna M, Gregory V, Goudiaby D, Hay A, Diop OM. Sentinel surveillance for influenza in Senegal, 1996–2009. J Infect Dis. 2012;206((Suppl 1)):S129–S135. doi: 10.1093/infdis/jis576. [DOI] [PubMed] [Google Scholar]
- 14.Dia N, Diene Sarr F, Thiam D, Faye Sarr T, Espié E, OmarBa I, Coly M, Niang M, Richard V, 4S Network Group Influenza-like illnesses in Senegal: not only focus on influenza viruses. PLoS One. 2014;9:e93227. doi: 10.1371/journal.pone.0093227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO . Polio Laboratory Manual. Geneva, Switzerland: Department of Immunization, Vaccines and Biologicals, World Health Organization; 2004. [Google Scholar]
- 16.McIntyre CL, Knowles NJ, Simmonds P. Proposals for the classification of human rhinovirus species A, B and C into genotypically assigned types. J Gen Virol. 2013;94:1791–1806. doi: 10.1099/vir.0.053686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savolainen C, Mulders MN, Hovi T. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res. 2002;85:41–46. doi: 10.1016/s0168-1702(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 18.Kiang D, Yagi S, Kantardjieff KA, Kim EJ, Louie JK, Schnurr DP. Molecular characterization of a variant rhinovirus from an outbreak associated with uncommonly high mortality. J Clin Virol. 2007;38:227–237. doi: 10.1016/j.jcv.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 19.Bessaud M, Jegouic S, Joffret ML, Barge C, Balanant J, Gouandjika-Vasilache I, Delpeyroux F. Characterization of the genome of human enteroviruses: design of generic primers for amplification and sequencing of different regions of the viral genome. J Virol Methods. 2008;149:277–284. doi: 10.1016/j.jviromet.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 20.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 21.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang T, Wang W, Bessaud M, Ren P, Sheng J, Yan H, Zhang J, Lin X, Wang Y, Delpeyroux F, Deubel V. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One. 2009;4:e6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knipe DM, Howley PM. Fields Virology. 5th edition. Philadelphia, PA: Lippincott Williams and Wilkins; 2007. [Google Scholar]
- 24.Nicholson KG, Kent J, Hammersley V, Cancio E. Acute viral infections of upper respiratory tract in elderly people living in the community: comparative, prospective, population based study of disease burden. BMJ. 1997;315:1060–1064. doi: 10.1136/bmj.315.7115.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenberg SB. Viral respiratory infections in elderly patients and patients with chronic obstructive pulmonary disease. Am J Med. 2002;112((Suppl 6A)):28S–32S. doi: 10.1016/S0002-9343(01)01061-0. [Review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fowlkes A, Giorgi A, Erdman D, Temte J, Goodin K, Di Lonardo S, Sun Y, Martin K, Feist M, Linz R, Boulton R, Bancroft E, McHugh L, Lojo J, Filbert K, Finelli L, IISP Working Group Viruses associated with acute respiratory infections and influenza-like illness among outpatients from the Influenza Incidence Surveillance Project, 2010–2011. J Infect Dis. 2014;209:1715–1725. doi: 10.1093/infdis/jit806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lekana-Douki SE, Nkoghe D, Drosten C, Ngoungou EB, Drexler JF, Leroy EM. Viral etiology and seasonality of influenza-like illness in Gabon, March 2010 to June 2011. BMC Infect Dis. 2014;14:373. doi: 10.1186/1471-2334-14-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noh JY, Song JY, Cheong HJ, Choi WS, Lee J, Lee JS, Wie SH, Jeong HW, Kim YK, Choi SH, Han SB, So BH, Kim H, Kim WJ. Laboratory surveillance of influenza-like illness in seven teaching hospitals, South Korea: 2011–2012 season. PLoS One. 2013;8:e64295. doi: 10.1371/journal.pone.0064295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otomaru H, Kamigaki T, Tamaki R, Opinion J, Santo A, Daya E, Okamoto M, Saito M, Tallo V, Lupisan S, Suzuki A, Oshitani H. Influenza and other respiratory viruses detected by influenza-like illness surveillance in Leyte Island, the Philippines, 2010–2013. PLoS One. 2015;10:e0123755. doi: 10.1371/journal.pone.0123755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang Z, Gonzalez R, Wang Z, Xiao Y, Chen L, Li T, Vernet G, Paranhos-Baccalà G, Jin Q, Wang J. Human rhinoviruses in Chinese adults with acute respiratory tract infection. J Infect. 2010;61:289–298. doi: 10.1016/j.jinf.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwane MK, Prill MM, Lu X, Miller EK, Edwards KM, Hall CB, Griffin MR, Staat MA, Anderson LJ, Williams JV, Weinberg GA, Ali A, Szilagyi PG, Zhu Y, Erdman DD. Human rhinovirus species associated with hospitalizations for acute respiratory illness in young US children. J Infect Dis. 2011;204:1702–1710. doi: 10.1093/infdis/jir634. [DOI] [PubMed] [Google Scholar]
- 32.Lee WM, Lemanske RF, Jr, Evans MD, Vang F, Pappas T, Gangnon R, Jackson DJ, Gern JE. Human rhinovirus species and season of infection determine illness severity. Am J Respir Crit Care Med. 2012;186:886–891. doi: 10.1164/rccm.201202-0330OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daleno C, Piralla A, Scala A, Senatore L, Principi N, Esposito S. Phylogenetic analysis of human rhinovirus isolates collected from otherwise healthy children with community-acquired pneumonia during five successive years. PLoS One. 2013;8:e80614. doi: 10.1371/journal.pone.0080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piralla A, Lilleri D, Sarasini A, Marchi A, Zecca M, Stronati M, Baldanti F, Gerna G. Human rhinovirus and human respiratory enterovirus (EV68 and EV104) infections in hospitalized patients in Italy, 2008–2009. Diagn Microbiol Infect Dis. 2012;73:162–167. doi: 10.1016/j.diagmicrobio.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 35.Calvo C, Casas I, García-García ML, Pozo F, Reyes N, Cruz N, García-Cuenllas L, Pérez-Breña P. Role of rhinovirus C respiratory infections in sick and healthy children in Spain. Pediatr Infect Dis J. 2010;29:717–720. doi: 10.1097/INF.0b013e3181d7a708. [DOI] [PubMed] [Google Scholar]
- 36.Esposito S, Daleno C, Tagliabue C, Scala A, Tenconi R, Borzani I, Fossali E, Pelucchi C, Piralla A, Principi N. Impact of rhinoviruses on pediatric community-acquired pneumonia. Eur J Clin Microbiol Infect Dis. 2012;31:1637–1645. doi: 10.1007/s10096-011-1487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mermond S, Zurawski V, D'Ortenzio E, Driscoll AJ, DeLuca AN, Deloria-Knoll M, Moïsi JC, Murdoch DR, Missotte I, Besson-Leaud L, Chevalier C, Debarnot V, Feray F, Noireterre S, Duparc B, Fresnais F, O'Connor O, Dupont-Rouzeyrol M, Levine OS. Lower respiratory infections among hospitalized children in New Caledonia: a pilot study for the Pneumonia Etiology Research for Child Health project. Clin Infect Dis. 2012;54((Suppl 2)):S180–S189. doi: 10.1093/cid/cir1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puenpa J, Mauleekoonphairoj J, Linsuwanon P, Suwannakarn K, Chieochansin T, Korkong S, Theamboonlers A, Poovorawan Y. Prevalence and characterization of enterovirus infections among pediatric patients with hand foot mouth disease, herpangina and influenza like illness in Thailand, 2012. PLoS One. 2014;9:e98888. doi: 10.1371/journal.pone.0098888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tapparel C, Siegrist F, Petty TJ, Kaiser L. Picornavirus and enterovirus diversity with associated human diseases. Infect Genet Evol. 2013;14:282–293. doi: 10.1016/j.meegid.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 40.Wisdom A, Leitch EC, Gaunt E, Harvala H, Simmonds P. Screening respiratory samples for detection of human rhinoviruses (HRVs) and enteroviruses: comprehensive VP4-VP2 typing reveals high incidence and genetic diversity of HRV species C. J Clin Microbiol. 2009;47:3958–3967. doi: 10.1128/JCM.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feikin DR, Njenga MK, Bigogo G, Aura B, Aol G, Audi A, Jagero G, Muluare PO, Gikunju S, Nderitu L, Winchell JM, Schneider E, Erdman DD, Oberste MS, Katz MA, Breiman RF. Viral and bacterial causes of severe acute respiratory illness among children aged less than 5 years in a high malaria prevalence area of western Kenya, 2007–2010. Pediatr Infect Dis J. 2013;32:e14–e19. doi: 10.1097/INF.0b013e31826fd39b. [DOI] [PubMed] [Google Scholar]
- 42.Zhang C, Zhu N, Xie Z, Lu R, He B, Liu C, Ma X, Tan W. Viral etiology and clinical profiles of children with severe acute respiratory infections in China. PLoS One. 2013;8:e72606. doi: 10.1371/journal.pone.0072606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wald TG, Shult P, Krause P, Miller BA, Drinka P, Gravenstein SA. A rhinovirus outbreak among residents of a long-term care facility. Ann Intern Med. 1995;123:588–593. doi: 10.7326/0003-4819-123-8-199510150-00004. [DOI] [PubMed] [Google Scholar]
- 44.van Benten I, Koopman L, Niesters B, Hop W, van Middelkoop B, de Waal L, van Drunen K, Osterhaus A, Neijens H, Fokkens W. Predominance of rhinovirus in the nose of symptomatic and asymptomatic infants. Pediatr Allergy Immunol. 2003;14:363–370. doi: 10.1034/j.1399-3038.2003.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright PF, Deatly AM, Karron RA, Belshe RB, Shi JR, Gruber WC, Zhu Y, Randolph VB. Comparison of results of detection of rhinovirus by PCR and viral culture in human nasal wash specimens from subjects with and without clinical symptoms of respiratory illness. J Clin Microbiol. 2007;45:2126–2129. doi: 10.1128/JCM.02553-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monto AS. Epidemiology of viral respiratory infections. Am J Med. 2002;112((Suppl 6A)):4S–12S. doi: 10.1016/s0002-9343(01)01058-0. [DOI] [PubMed] [Google Scholar]
- 47.Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pallansch MA, Roos R. Enteroviruses: polioviruses, coxsackiviruses, echoviruses, and newer enterovirus. In: Knipe DM, Howley PM, editors. Fields Virology. 5th edition. vol. 1. Philadelphia, PA: Lippincott Williams and Wilkins; 2007. pp. 839–893. [Google Scholar]
- 49.Kennett ML, Birch CJ, Lewis FA, Yung AP, Locarnini SA, Gust ID. Enterovirus type 71 infection in Melbourne. Bull World Health Organ. 1974;51:609–615. [PMC free article] [PubMed] [Google Scholar]
- 50.Miller EK, Edwards KM, Weinberg GA, Iwane MK, Griffin MR, Hall CB, Zhu Y, Szilagyi PG, Morin LL, Heil LH, Lu X, Williams JV. New Vaccine Surveillance Network A novel group of rhinoviruses is associated with asthma hospitalizations. J Allergy Clin Immunol. 2009;123:98–104.e1. doi: 10.1016/j.jaci.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin Y, Yuan XH, Xie ZP, Gao HC, Song JR, Zhang RF, Xu ZQ, Zheng LS, Hou YD, Duan ZJ. Prevalence and clinical characterization of a newly identified human rhinovirus C species in children with acute respiratory tract infections. J Clin Microbiol. 2009;47:2895–2900. doi: 10.1128/JCM.00745-09. [DOI] [PMC free article] [PubMed] [Google Scholar]