Abstract
Because O blood group has been associated with more severe cholera infections, it has been hypothesized that cholera toxin (CT) may bind non-O blood group antigens of the intestinal mucosae, thereby preventing efficient interaction with target GM1 gangliosides required for uptake of the toxin and activation of cyclic adenosine monophosphate (cAMP) signaling in target epithelia. Herein, we show that after exposure to CT, human enteroids expressing O blood group exhibited marked increase in cAMP relative to cells derived from blood group A individuals. Likewise, using CRISPR/Cas9 engineering, a functional group O line (HT-29-A−/−) was generated from a parent group A HT-29 line. CT stimulated robust cAMP responses in HT-29-A−/− cells relative to HT-29 cells. These findings provide a direct molecular link between blood group O expression and differential cellular responses to CT, recapitulating clinical and epidemiologic observations.
Cholera is a leading cause of epidemic diarrhea worldwide with estimates of 3–5 million cases occurring annually.1 The voluminous watery diarrhea characteristic of cholera is caused by cholera toxin (CT), the principal virulence determinant of Vibrio cholerae. CT is an AB5 toxin consisting of an active A subunit and a pentameric B subunit (CT-B), responsible for binding to its cognate receptor, GM1 ganglioside, on the surface of enterocytes. Endocytosis and eventual release of the A subunit leads to adenosine diphosphate–ribosylation of the intracellular guanine nucleotide protein, Gsα. Inhibition of GTPase activity of Gsα promotes activation of adenylate cyclase resulting in increase in intracellular cyclic adenosine monophosphate (cAMP) levels. This increase in cAMP activates protein kinase A–mediated phosphorylation of the cystic fibrosis transmembrane receptor, enhancing chloride secretion into the intestinal lumen while reducing sodium uptake by inhibition of sodium/hydrogen ion exchange via NHE3. Combined, the net electrolyte loss leads to profound secretory diarrhea characteristic of cholera.2,3
Similar to other important enteric pathogens,4,5 blood group antigens appear to be an important determinant of the outcome of infections by V. cholerae. Blood group O individuals tend to develop more severe disease after infection with either classical or El Tor strains.6–8 Strengthening this observation, group O individuals are underrepresented in the Ganges delta where cholera infections are highly endemic.8
However, the reasons for the association between blood group O and severe illness have not been thoroughly elucidated. Hypothetically, differential binding of CT to non-O blood group gastrointestinal glycans may impair efficient interaction of CT with cognate GM1 receptors.9,10 Indeed, CT has been shown to bind blood group antigens.9,11,12 Although the affinity of these interactions relative to CT binding to GM1 is relatively weak,13,14 it has been suggested that more rapid dissociation of CT from O-antigen glycans9 could permit accelerated binding to cognate GM1 gangliosides receptors in blood group O individuals.
Human blood groups are determined by allelic variations in a glycosyltransferase that in blood group B adds terminal galactose residues to the core H (blood group O) antigen, whereas the blood group A enzyme adds N-acetylgalactosamine (GalNac). Blood group O is determined by a premature stop codon within the glycosyltransferase gene yielding a truncated allele, resulting in complete loss of enzymatic activity and undecorated core H antigens.
To investigate the direct impact of blood group expression on CT activation of target epithelia, we chose two novel enteric model systems. First, we examined the ability of CT to stimulate cAMP responses in enteroids derived from gastrointestinal stem cells obtained from individuals with either blood group O or A. In addition, we compared the effect of CT on transformed HT-29 gastrointestinal epithelial cells derived from an individual with blood group A to CRISPR/Cas9 engineered cells in which the glycosyltransferase was disrupted yielding isogenic HT-29-A−/− cells expressing the core H (blood group O) antigen.
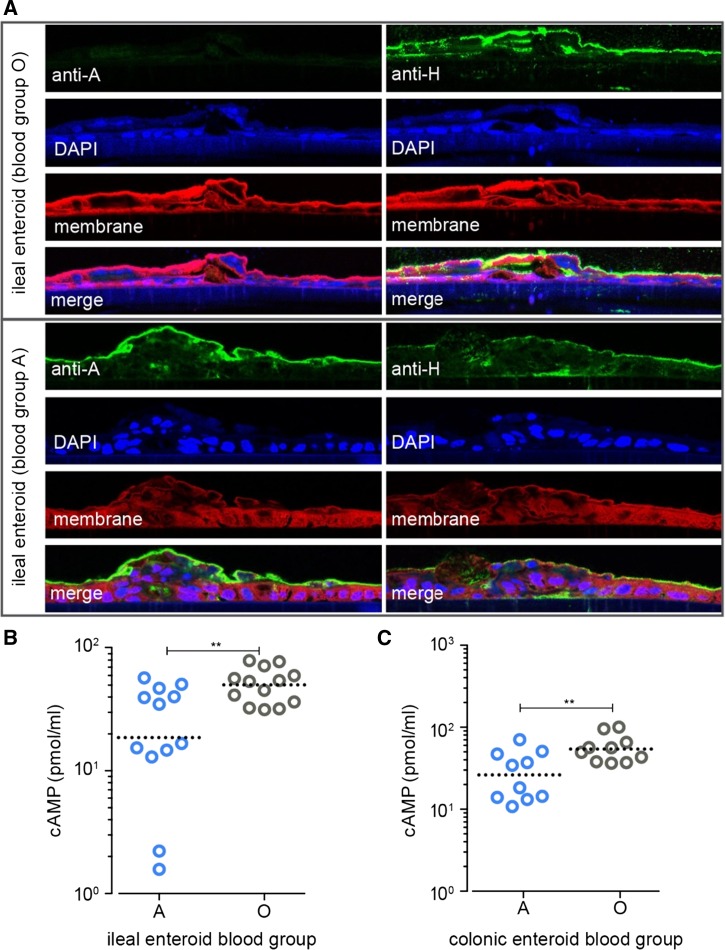
Enteroids can recapitulate many features of human intestinal epithelium including generation of enterochromaffin cells, goblet cells, and enterocytes.15 In addition, we found that enteroids appropriately express blood group antigens on the surface of enterocytes (Figure 1A ). We therefore used enteroids derived from ileal or colonic biopsies from individuals belonging to different blood groups to examine CT-mediated activation of cAMP. Interestingly, intestinal cells derived from blood group O individuals exhibited consistently higher levels of cAMP (P value = 0.0096) relative to cells expressing the blood group A (Figure 1B). Although cholera is thought to be an ileal infection, we also found that cells derived from colonic biopsies from blood group O individuals exhibited more robust cAMP response on exposure to CT (P value = 0.016, Figure 1C). These data provide additional in vitro evidence to support an association between O-blood group expression and differential responses to CT.
Figure 1.
(A) Confocal immunofluorescence images of enteroids grown on transwells demonstrating the presence of H antigen (group O) in ileal enteroids derived from a blood group O individual (top panel), or blood group A subject (bottom panel). (B) Cyclic adenosine monophosphate (cAMP) production after overnight stimulation of ileal enteroids with cholera toxin (CT, 0.1 μg/mL). Data for both blood groups represent composite data from two different subjects (**P value = 0.0096) (C) cAMP production in colonic enteroids from two blood group A and two blood group O subjects after CT stimulation as described in B (**P value = 0.016). Statistical calculations in B, C performed using two-tailed, Mann–Whitney U comparisons.
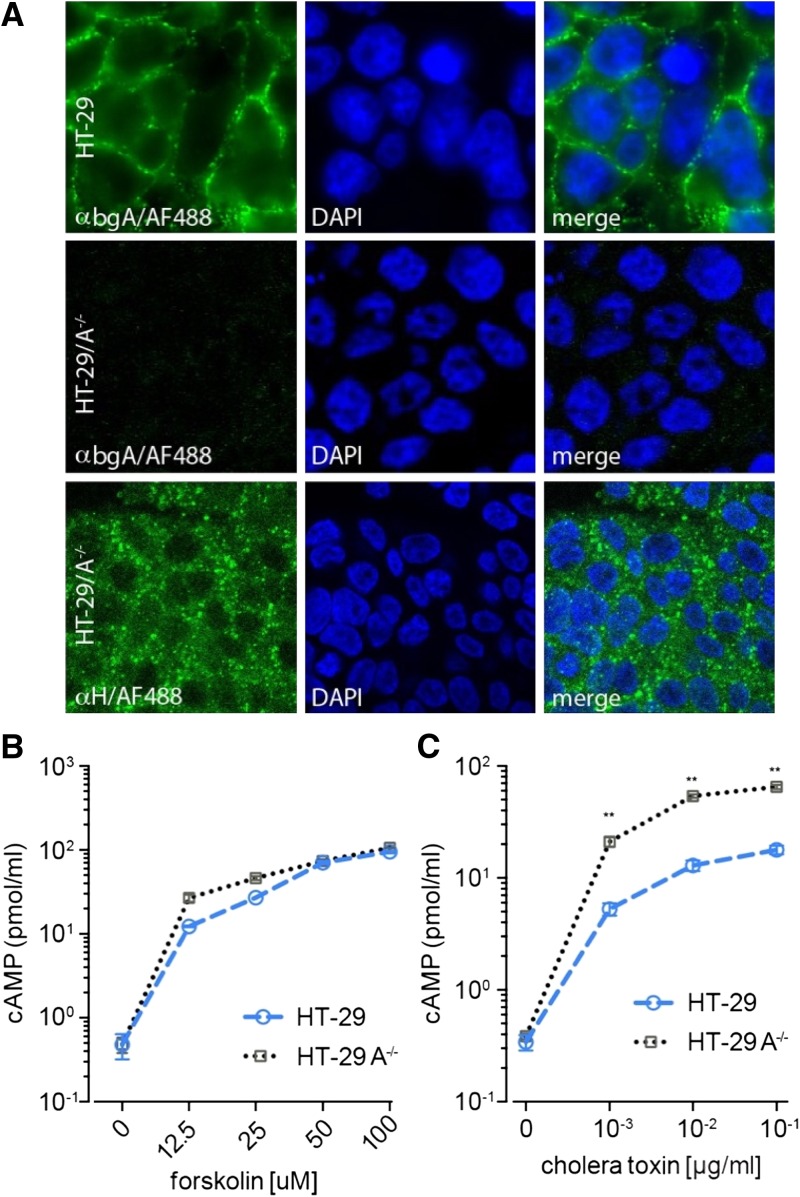
Furthermore, to examine more directly the specific impact of blood group expression on CT activation of target epithelia, we compared the responses of parental HT-29 cells (blood group A) to isogenic H-antigen expressing CRISPR/Cas9 engineered HT-29-A−/− cells (effectively, blood group O; Figure 2A ). Although both the parental and mutant cell lines responded to forskolin, a diterpene activator of adenylate cyclase (Figure 2B), CT stimulation of cAMP production in HT-29-A−/− cells consistently exceeded that observed in the parental A blood group–expressing cells (range: 3.7- to 4.2-fold increase; Figure 2C).
Figure 2.
(A) Confocal immunofluorescence images demonstrating the presence of group A antigen expressed on the surface of parental HT-29 cells (first row), but not the CRISPR engineered HT-29-A−/− cells which lack the α1-3-N-acetylgalactosaminyltransferase required to form the A blood group (second row), but which retain expression of the core H (blood group O) antigen (third row). (B) Parental HT-29 (blue symbols), and the engineered HT-29-A−/− cells (grey) exhibit similar response to the adenylate cyclase agonist forskolin. (C) HT-29 A/A− cells exhibit enhanced cyclic adenosine monophosphate (cAMP) responses to cholera toxin (CT) compared with parent HT-29 cells. (**P = 0.002, two-tailed Mann–Whitney U comparisons).
Collectively, these in vitro data obtained using two different enteric systems strongly suggest that blood group O expression by gastrointestinal epithelia permits an accelerated response to CT, the principal effector molecule of V. cholerae responsible for cholera diarrheal illness. Interestingly, the data obtained using the enteroids derived from multiple individuals expressing different blood groups offer striking parallels to clinical and epidemiologic associations between O-blood group and the risk for severe cholera. Our findings suggest that O-blood group enterocytes respond vigorously to CT stimulation, potentially providing a direct cellular link to disease severity. Moreover, the enhanced responses to CT observed in cells engineered to express only the core-H (blood group O) antigen suggest that this phenomenon is not simply a genetic association between O blood group factors that segregate by blood type, but a direct effect of expression of the unmodified core-H glycan.
These findings could not be explained by differences in direct binding of CT to the respective epithelial cells as we did not observe a strict association between overall CT binding to epithelial cells and blood group. We were unable to detect significant differences in either the pattern (Supplemental Figure 1A) or the amount of CT bound to the surface of HT-29 and HT-29-A−/− cells (Supplemental Figure 1B). Likewise, in both A and O blood group–derived enteroids, we observed similar colocalization of labeled CT-B subunit and the major secreted mucin, MUC2 (Supplemental Figure 1C). The studies reported here were limited by the lack of available cells from blood group B individuals for comparison. Although we were unable to discern a clear difference in binding of CT to cells expressing the different blood group antigens, our experiments were not designed to detect a more rapid dissociation of CT from blood group O glycans, which could accelerate binding to its cognate GM1 ganglioside receptor.9 Nevertheless, they highlight the advantages of these newly available tools in the dissection of clinically important pathogen–host interactions.
In summary, we have established the utility of two model enteric systems, to study the effects of a major virulence effector, CT. In particular, we are able to show that cellular responses to CT are enhanced in cells that express the O blood group, offering interesting parallels to the earlier epidemiologic and clinical observations that have associated blood group O with severe cholera.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kelly Monroe, Naomi Sonnek, and Emily Vivio for their assistance in acquiring the enteroid cell lines used in this study.
Disclaimer: The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID, NIDDK, NIH, or the VA.
Footnotes
Financial support: This work was supported by funding from the Department of Veterans Affairs(grant 5I01BX001469); grant R01AI89894 from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID); CTSA grant UL1 TR000448 from the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH), and the Digestive Diseases Research Core Center at Washington University School of Medicine; grant P30 DK52574 from the National Institute of Diabetes and Digestive and Kidney Diseases. Matthew A. Ciorba was supported by grants DK100737, DK089016, and DK109384 from the NIDDK, AI095776 from the NIAID, and a Crohn's and Colitis Foundation Senior Research Award.
Authors' addresses: F. Matthew Kuhlmann, Pardeep Kumar, and Qingwei Luo, Division of Infectious Diseases, Department of Internal Medicine, Washington University School of Medicine, Saint Louis, MO, E-mails: fkuhlman@dom.wustl.edu, pradeepkumar.19@gmail.com, and qluo@dom.wustl.edu. Srikanth Santhanam, Division of Gastroenterology, Department of Internal Medicine, Washington University School of Medicine, Saint Louis, MO, E-mail: ssanthan@dom.wustl.edu. Matthew A. Ciorba, Division of Gastroenterology, Department of Internal Medicine, Washington University School of Medicine, Saint Louis, MO, and Molecular Microbiology and Microbial Pathogenesis Program, Division of Biology and Biomedical Sciences, Washington University School of Medicine, Saint Louis, MO, E-mail: mciorba@dom.wustl.edu. James M. Fleckenstein, Division of Infectious Diseases, Department of Internal Medicine, Washington University School of Medicine, Saint Louis, MO, John Cochran Division, Veterans Affairs Medical Center, Saint Louis, MO, and Molecular Microbiology and Microbial Pathogenesis Program, Division of Biology and Biomedical Sciences, Washington University School of Medicine, Saint Louis, MO, E-mail: jflecken@dom.wustl.edu.
References
- 1.Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB. Cholera. Lancet. 2012;379:2466–2476. doi: 10.1016/S0140-6736(12)60436-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fleckenstein JM, Hardwidge PR, Munson GP, Rasko DA, Sommerfelt H, Steinsland H. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect. 2010;12:89–98. doi: 10.1016/j.micinf.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopic S, Geibel JP. Toxin mediated diarrhea in the 21 century: the pathophysiology of intestinal ion transport in the course of ETEC, V. cholerae and rotavirus infection. Toxins (Basel) 2010;2:2132–2157. doi: 10.3390/toxins2082132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anstee DJ. The relationship between blood groups and disease. Blood. 2010;115:4635–4643. doi: 10.1182/blood-2010-01-261859. [DOI] [PubMed] [Google Scholar]
- 5.Boren T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892–1895. doi: 10.1126/science.8018146. [DOI] [PubMed] [Google Scholar]
- 6.Glass RI, Holmgren J, Haley CE, Khan MR, Svennerholm AM, Stoll BJ, Belayet Hossain KM, Black RE, Yunus M, Barua D. Predisposition for cholera of individuals with O blood group. Possible evolutionary significance. Am J Epidemiol. 1985;121:791–796. doi: 10.1093/oxfordjournals.aje.a114050. [DOI] [PubMed] [Google Scholar]
- 7.Harris JB, Khan AI, LaRocque RC, Dorer DJ, Chowdhury F, Faruque AS, Sack DA, Ryan ET, Qadri F, Calderwood SB. Blood group, immunity, and risk of infection with Vibrio cholerae in an area of endemicity. Infect Immun. 2005;73:7422–7427. doi: 10.1128/IAI.73.11.7422-7427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlsson EK, Harris JB, Tabrizi S, Rahman A, Shlyakhter I, Patterson N, O'Dushlaine C, Schaffner SF, Gupta S, Chowdhury F, Sheikh A, Shin OS, Ellis C, Becker CE, Stuart LM, Calderwood SB, Ryan ET, Qadri F, Sabeti PC, Larocque RC. Natural selection in a Bangladeshi population from the cholera-endemic Ganges River Delta. Sci Transl Med. 2013;5:192ra86. doi: 10.1126/scitranslmed.3006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heggelund JE, Haugen E, Lygren B, Mackenzie A, Holmner A, Vasile F, Reina JJ, Bernardi A, Krengel U. Both El Tor and classical cholera toxin bind blood group determinants. Biochem Biophys Res Commun. 2012;418:731–735. doi: 10.1016/j.bbrc.2012.01.089. [DOI] [PubMed] [Google Scholar]
- 10.Holmner A, Mackenzie A, Krengel U. Molecular basis of cholera blood-group dependence and implications for a world characterized by climate change. FEBS Lett. 2010;584:2548–2555. doi: 10.1016/j.febslet.2010.03.050. [DOI] [PubMed] [Google Scholar]
- 11.Holmner A, Lebens M, Teneberg S, Angstrom J, Okvist M, Krengel U. Novel binding site identified in a hybrid between cholera toxin and heat-labile enterotoxin: 1.9 A crystal structure reveals the details. Structure. 2004;12:1655–1667. doi: 10.1016/j.str.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 12.Vasile F, Reina JJ, Potenza D, Heggelund JE, Mackenzie A, Krengel U, Bernardi A. Comprehensive analysis of blood group antigen binding to classical and El Tor cholera toxin B-pentamers by NMR. Glycobiology. 2014;24:766–778. doi: 10.1093/glycob/cwu040. [DOI] [PubMed] [Google Scholar]
- 13.Cuatrecasas P. Gangliosides and membrane receptors for cholera toxin. Biochemistry. 1973;12:3558–3566. doi: 10.1021/bi00742a032. [DOI] [PubMed] [Google Scholar]
- 14.Hansson HA, Holmgren J, Svennerholm L. Ultrastructural localization of cell membrane GM1 ganglioside by cholera toxin. Proc Natl Acad Sci USA. 1977;74:3782–3786. doi: 10.1073/pnas.74.9.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VanDussen KL, Marinshaw JM, Shaikh N, Miyoshi H, Moon C, Tarr PI, Ciorba MA, Stappenbeck TS. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2015;64:911–920. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taki T, Kibayashi K. A simple ABO genotyping by PCR using sequence-specific primers with mismatched nucleotides. Leg Med (Tokyo) 2014;16:168–172. doi: 10.1016/j.legalmed.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.