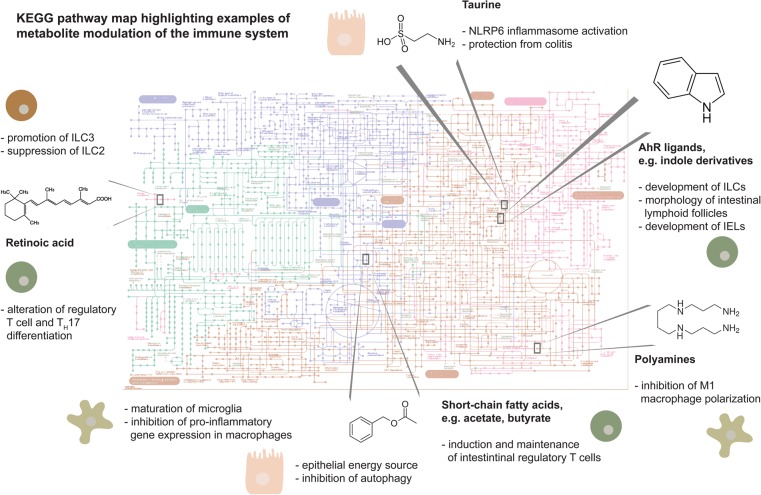
In this review, Levy et al. highlight examples of how microbiota-modulated metabolites control the development, differentiation, and activity of the immune system and classify them into functional categories that illustrate the spectrum of ways by which microbial metabolites influence host physiology.
Keywords: immune, metabolites, microbiome
Abstract
The mammalian intestine harbors one of the largest microbial densities on Earth, necessitating the implementation of control mechanisms by which the host evaluates the state of microbial colonization and reacts to deviations from homeostasis. While microbial recognition by the innate immune system has been firmly established as an efficient means by which the host evaluates microbial presence, recent work has uncovered a central role for bacterial metabolites in the orchestration of the host immune response. In this review, we highlight examples of how microbiota-modulated metabolites control the development, differentiation, and activity of the immune system and classify them into functional categories that illustrate the spectrum of ways by which microbial metabolites influence host physiology. A comprehensive understanding of how microbiota-derived metabolites shape the human immune system is critical for the rational design of therapies for microbiota-driven diseases.
The mammalian intestine harbors a dense and complex microbial community, termed the microbiota. The commensal microbiome and, in particular, the dense and diverse microbial community inhabiting the gastrointestinal tract play a pivotal role in the maintenance of organismal homeostasis and stable physiology. Research over the last decade has highlighted the concept that the eukaryotic host and its microbiota, rather than existing in isolation, compose a complex meta-organism termed the “holobiont,” jointly regulating multiple aspects of mammalian physiology, including immune system development, metabolism, and nervous system function (Sommer and Backhed 2013). The interaction between the microbiota and the host is most prominent at mucosal surfaces, which provide an interface between the mostly microbe-free host, the microbiota, and the environment. At the same time, the presence of the microbial community within the eukaryotic host necessitates the development of a sophisticated immune system that controls and maintains a beneficial symbiosis of the holobiont through the continuous communication between the microbiome and the host.
The recognition of microbial patterns by the innate immune system initiates a signaling cascade downstream from pattern recognition receptors, which triggers an anti-microbial immune response. At the same time, signals downstream from microbial pattern recognition can also result in the induction of mechanisms involved in the maintenance of tolerance.
Deficiency of innate immune pathways leads to alterations of the microbial community, termed dysbiosis, which in some cases result in disease pathogenesis (Rakoff-Nahoum et al. 2004; Petnicki-Ocwieja et al. 2009; Elinav et al. 2011; Thaiss et al. 2014). Different microbiome configurations produce, modulate, and degrade a large array of small molecules (referred to here as “metabolites”), thereby providing a functional complementation to the metabolic capacities of the host; e.g., complex proteins and carbohydrates that cannot be degraded by the host can be metabolized by the microbial community (Fischbach and Sonnenburg 2011; Nicholson et al. 2012). One additional important and recently recognized function of this complex network of microbially produced, modulated, and degraded metabolites relates to the intimate communication of the microbiota with its eukaryotic host, mediated by metabolite signaling through a series of innate immune receptors. In addition, metabolites can be a form of bacterial signaling such as quorum sensing and can drive changes in the composition and function of the microbiota (Bassler and Losick 2006).
It has thus become increasingly apparent that the microbiota and its metabolites are important orchestrators of host physiology and pathophysiology through the control of a large range of metabolic, inflammatory, and even behavioral processes (Hsiao et al. 2013; Tang et al. 2013). Here, we provide an overview of the molecular relationship between microbiota, metabolites, and the host immune system and highlight prominent examples of how metabolites contribute to health and disease of the holobiont (Fig. 1). Rather than categorizing metabolites by chemical groups, we focus on commonalities with respect to their functional impact on the immune system with the goal of illustrating common themes unifying the currently known microbiota–metabolite–immune cell interactions.
Figure 1.
Chart of metabolic pathways with highlighted examples of microbiome-derived or -modulated pathways involved in modulation of the immune response. (ILCs) Innate lymphoid cells; (IELs) intraepithelial lymphocytes. The pathway map was obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/pathway/map01100) (Kanehisa and Goto 2000; Kanehisa et al. 2016).
Metabolites as drivers of immune system development and differentiation
One of the most striking examples of the importance of the microbiota for the immune system is the observation that germ-free mice, which are raised in the absence of any microbial colonization, feature a profoundly underdeveloped immune system (Smith et al. 2007), which can be largely reversed by adult conventionalization with a species-specific microbiota (Chung et al. 2012). While the molecular mechanisms by which the microbiome controls immune system ontogeny are still largely unknown, several examples have been found that highlight the involvement of specific bacterial-derived metabolites in the regulation of immune cell development and differentiation (Fig. 1).
Short chain fatty acids (SCFAs)
Among the most abundant molecules produced by gut bacteria are SCFAs, which have been found recently to control multiple aspects of human immunity and metabolism (Morrison and Preston 2016). Soluble dietary fibers and nondigestible carbohydrates such as cellulose are an integral component of the human diet. While humans lack the enzymes to degrade such polysaccharides, anaerobic commensal bacteria in the cecum and large intestine can ferment these fibers. Dietary fiber fermentation results in the generation of SCFAs—namely, acetate, propionate, and butyrate, which can be detected by the host through the intracellular receptor PPARγ; the surface proteins GPR41 and GPR43; and the butyrate receptor GPR109a (Brown et al. 2003; Alex et al. 2013). Acetate and propionate are produced mainly by Bacteroidetes, while butyrate is primarily produced by the Firmicutes phylum (Hoverstad and Midtvedt 1986; Macfarlane and Macfarlane 2003).
Administration of SCFAs leads to alterations in hematopoiesis, resulting in an enhanced myeloid output due to elevated numbers of myeloid precursors (Balmer et al. 2014; Khosravi et al. 2014). This myeloid skew promotes clearance of systemic infection (Balmer et al. 2014; Khosravi et al. 2014) and ameliorates allergic reactions (Trompette et al. 2014). SFCAs also influence myeloid cells post-hematopoiesis, as has been described for microglia in the brain. The cells feature abnormalities in morphology and function in the absence of the microbiota, which can be partially restored by SCFA supplementation (Erny et al. 2015). These examples illustrate that part of the impact that the microbiota exerts on host immune system development is mediated through microbiota-modulated metabolites.
Apart from these examples of the systemic effects of microbiota-derived metabolites that influence molecular and cellular processes far from the intestinal tract, the microbiota also modulates tissue-level immune system maturation through its profound impact on local metabolite levels.
Tryptophan metabolites
One prominent example of how the microbiota impacts tissue-level immune maturation is the microbial metabolism of tryptophan. It was shown that commensal Lactobacilli use tryptophan as an energy source to produce ligands of the aryl hydrocarbon receptor (AhR), such as the metabolite indole-3-aldehyde (Zelante et al. 2013). AhR is a ligand-activated transcription factor critically important for the organogenesis of intestinal lymphoid follicles (ILFs). Immune cells expressing AhR include RORγt+ group 3 innate lymphoid cells (ILC3s) that are involved in ILF genesis, and AhR expression on ILC3s is functionally required for their expansion (Kiss et al. 2011). In addition, AhR-induced IL-22 production by ILCs drives the secretion of the anti-microbial peptides lipocalin-2, S100A8, and S100A9, which protect from pathogenic infection by Candida albicans (Zelante et al. 2013). Similarly, mice lacking AhR are more susceptible to infection with Citrobacter rodentium and Listeria monocytogenes (Shi et al. 2007; Qiu et al. 2012).
In addition to its role in the function of ILCs, AhR was also found to be necessary for the maintenance of the epithelial barrier and the homeostasis of intraepithelial lymphocytes (IELs) (Li et al. 2011; Lee et al. 2012). When AhR-deficient mice were subjected to DSS-induced colitis, they featured enhanced disease susceptibility compared with wild-type mice, which was not apparent when mice were fed a synthetic diet depleted of dietary AhR ligands. The transfer of functional IELs to AhR-deficient mice resulted in ameliorated disease and enhanced recovery (Li et al. 2011; Lee et al. 2012).
Retinoic acid (RA)
In addition to the orchestration of immune system maturation, metabolites fine-tune immune responses at the level of post-developmental differentiation. The vitamin A lipid metabolite RA was shown to regulate the balance between proinflammatory and anti-inflammatory immune responses. RA deficiency affects both the composition of the microbiota and immune system function. Mice deficient in RA harbor reduced numbers of segmented filamentous bacteria (SFB), which might contribute to the decreased number of T helper 17 (TH17) cells present in vitamin A-deficient mice (Gaboriau-Routhiau et al. 2009; Cha et al. 2010).
In the steady state, RA has an important role in the maintenance of immune homeostasis in the intestine, as it can promote both regulatory T (Treg) cell development through TGF-β and the production of IgA by B cells (Mora et al. 2006; Coombes et al. 2007; Mucida et al. 2007; Sun et al. 2007). RA mediates the expansion of Treg cells and the suppression of TH17 lineage (Bergstrom et al. 2010; Hall et al. 2011b) through induction of histone acetylation at the FoxP3 promoter, the master regulator of Treg cell identity (Kang et al. 2007). DCs from the lamina propria of the small intestine can promote CD4+ T cell conversion to Tregs in a manner dependent on TGF-β and RA (Sun et al. 2007). This capability of DCs is influenced by the microbiota, as feeding of mice with Bifidobacterium infantis results in elevated number of DCs capable of synthesizing RA as well as FoxP3+ cells in the lamina propria (Konieczna et al. 2013).
Paradoxically, during inflammation, RA is also involved in eliciting proinflammatory CD4+ T-cell responses to infection (Hall et al. 2011a). Accordingly, mice fed a diet deficient in vitamin A were reported to feature inhibited TH17 cell differentiation in the lamina propria of the small intestine (Cha et al. 2010). RA was also shown to be important for the expression of gut-homing molecules on immune cells. In the absence of TLR signaling in MyD88-deficient mice, intestinal DCs express low levels of retinal dehydrogenases, a critical enzyme for RA biosynthesis, and are impaired in their ability to induce gut-homing lymphocytes (Iwata et al. 2004; Mora et al. 2006; Wang et al. 2011; Bakdash et al. 2015).
The impact of vitamin A even reaches beyond T-cell immunity. In the absence of vitamin A, the numbers of ILC3s are strongly diminished, while ILC2 cells and their immune program became more dominant (Spencer et al. 2014). A similar phenomenon can be observed in adaptive lymphocytes, where TH2 cells expand under vitamin A-deficient conditions on the expense of TH1 and TH17 immunity (Pino-Lagos et al. 2011). Together, these studies highlight the power of a diet-derived and microbiota-modulated metabolite in directing a specific type of immune response.
Other vitamins can likewise participate in the post-developmental fate decisions of leukocytes. For instance, vitamin D profoundly affects T-cell activation (von Essen et al. 2010). Several studies have linked vitamin D deficiency to inflammatory bowel disease susceptibility (Sun 2010), and vitamin D receptor expression is significantly lower in IBD- and colitis-associated colon cancer patients (Abreu et al. 2004; Wada et al. 2009).
The link between the gut microbiota and vitamins is particularly apparent in the case of vitamins belonging to B and K groups, as, in their case, the host is unable to perform the biosynthetic reactions and depends on members of the commensal microbiota (Roth et al. 1996; Hill 1997; Martens et al. 2002). For instance, the gut commensals Bifidobacterium and Lactobacillus can synthesize the B9 vitamin folate (Noda et al. 1994; Pompei et al. 2007; Strozzi and Mogna 2008; Kleerebezem and Vaughan 2009), which was shown to have a critical role in the maintenance of Treg cells in the small intestine (Kunisawa et al. 2012). Vitamin B12 deficiency results in a decreased number of lymphocytes and suppressed NK cell activity (Tamura et al. 1999). The recent discovery of vitamin metabolite presentation to mucosa-associated invariant T (MAIT) cells by the MHC class I-like molecule MR1 (Kjer-Nielsen et al. 2012; Le Bourhis et al. 2013) suggested a direct link between microbial vitamin metabolism and immune cell priming.
Metabolites as regulators of symbiosis
The quintessential task of mutualistic host–microbiota interactions at mucosal surfaces is the preservation of tissue homeostasis. Over the last decade, several microbial molecules have been identified that aid in the orchestration of the tightrope walk between immunity and tolerance at the mucosal–bacterial interface (Fig. 1). A common strategy by several such molecules is the promotion of an anti-inflammatory response. The commensal Bacteroides fragilis produces surface polysaccharide A (PSA) that suppresses the production of the proinflammatory IL-17 and promotes expression of IL-10 by CD4+ T cells through binding to TLR2 (Mazmanian et al. 2008; Round and Mazmanian 2010). It was first demonstrated that monocolonization of germ-free mice with B. fragilis modulates CD4+ T-cell homeostasis and cytokine production in a PSA-dependent manner. PSA presented by DCs activates cognate CD4+ T cells (Mazmanian et al. 2005). Later, it was shown that PSA of B. fragilis dampens Helicobacter hepaticus-driven intestinal inflammation and likewise prevents colitis induced by the chemical compound TNBS and that a mutant of B. fragilis lacking PSA could not prevent inflammation (Mazmanian et al. 2008). The recognition of PSA by TLR2 specifically expressed on Treg cells induces their activation, which in turn leads to suppression of the inflammatory response. This circuit is required for the colonization of B. fragilis (Round et al. 2011), potentially providing an example for active niche construction by a commensal bacterium through modulating the host immune response.
Such metabolite-induced feedback loops might emerge as a common theme in the regulation of a stable microbial colonization by the immune system. This is best illustrated by the epithelial innate immune sensor NLRP6, a member of the NOD-like receptor family, which is involved in viral recognition as well as inflammasome formation (Levy et al. 2015a; Wang et al. 2015a). The inflammasome pathway is influenced by the microbiota-modulated metabolites taurine, histamine, and spermine, thereby regulating the level of epithelial IL-18 production, anti-microbial peptide secretion, and intestinal community composition (Levy et al. 2015b). Thus, the metabolic activity of the microbiome is sensed by the immune system, and this sensing is translated into an anti-microbial response aimed at maintaining a stable colonization. If this pathway is disturbed in genetically modified mice, dysbiosis arises, leading to the manifestation of intestinal autoinflammation (Elinav et al. 2011), enhanced susceptibility to enteric infection (Wlodarska et al. 2014), intestinal tumorigenesis (Hu et al. 2013), and liver inflammation (Henao-Mejia et al. 2012).
In addition to the amino acids that modulate NLRP6 signaling, other amino acids are involved in the maintenance of intestinal homeostasis. It was shown that protein malnutrition (and specifically tryptophan depletion) alters the severity of intestinal inflammation (Hashimoto et al. 2012). Angiotensin-converting enzyme 2 (ACE2) controls the expression of the neutral amino acid transporter in the intestine. Ace2−/− mice feature altered gut microbial composition, and these mice develop severe colitis after induction of epithelial damage by DSS. Transplantation of the gut microbiota to germ-free mice transfers the inflammatory phenotype and susceptibility to colitis, while a tryptophan-rich diet reverses microbial composition in this model (Hashimoto et al. 2012).
Similar to Ace2−/− mice, mice lacking indoleamine 2,3-dioxygenase (IDO1) display altered microbial composition as well as aberrations in metabolic pathways of both the microbiota and the host (Puccetti and Grohmann 2007; Zelante et al. 2013). IDO1 activity was shown to inhibit IL-10, which, in a model of atherosclerosis, leads to ameliorated disease (Metghalchi et al. 2015). Ido1−/− mice showed blooming of Lactobacilli similar to mice on a tryptophan-rich diet, which resulted in an accumulation of the AhR ligand indole-3-aledhyde. Moreover, tryptophan catabolism through IDO1 affects the differentiation of CD4+ T cells that secrete IL-17, which is modulated by the microbiota (Huengsberg et al. 1998; Romani et al. 2008; Favre et al. 2010).
In addition to tryptophan, the microbiota is pivotal for the regulation of intestinal levels of the amino acid arginine, which in turn exerts a modulatory effect on the immune system. Germ-free mice feature elevated levels of arginine, indicating that commensal bacteria are involved in the metabolism of arginine to downstream derivatives, including polyamines (Matsumoto et al. 2012; Mardinoglu et al. 2015). Correspondingly, polyamine levels are strongly diminished in the absence of the microbiome (Matsumoto et al. 2012). Polyamines in turn exert their immunomodulatory effect on various cell types, including macrophages and epithelial cells, where they contribute to suppressed inflammation (Kibe et al. 2014; Levy et al. 2015b).
Although arginine is primarily metabolized in the liver, immune cells can also serve as an extrahepatic source for arginase-1 (Arg1) activity during infection and inflammation (Raber et al. 2012; Wynn et al. 2013; Thomas and Mattila 2014; Murray 2015). Myeloid cell Arg1 has immunosuppressive capacities (Rodriguez et al. 2004; Munder et al. 2006), and, recently, Arg1 was shown to perform a cell-intrinsic role in the regulation of ILC2 metabolism and type 2 inflammation in the lung (Monticelli et al. 2016).
A further example of an anti-inflammatory molecule produced by the microbiota is provided by SCFAs, which have been briefly discussed above in the context of their impact on hematopoiesis, indicating that certain metabolites may perform a multitude of functions at several layers of immune regulation. The anti-inflammatory effect of SCFAs has been well characterized in both immune cells and epithelial cells. The intestinal microbiota, through the production of SCFAs, can suppress inflammation through several distinct mechanisms. Germ-free mice as well as colonized mice treated with acetate show ameliorated severity of DSS-induced colitis, and this beneficial effect is dependent on the receptor GPR43. Consequently, Gpr43−/− mice feature elevated severity of inflammation compared with wild-type mice (Maslowski et al. 2009). As in the case of immune system development, SFCAs exert their symbiosis-promoting effects both locally at the tissue level and systemically. Circulating SCFAs can suppress the inflammatory response in the lungs in a GPR41-dependent manner. A low-fiber diet increased the severity of allergic airway inflammation, while propionate administration protected from airway inflammation with reduced amounts of IL-4, IL-5, IL-13, and IL-17 in the lung (Trompette et al. 2014).
Another major function of SCFAs in dampening the inflammatory response is the regulation of colonic Treg homeostasis (Atarashi et al. 2011, 2013; Smith et al. 2013). SCFAs were shown to selectively expand Tregs in the large intestine. Feeding of germ-free mice with SCFAs increased the numbers of FoxP3+ Treg cells to a level similar to that observed in conventionalized mice. Propionate administration increased the expression of FoxP3 and IL-10 in colonic Treg cells (Smith et al. 2013). It was further shown that intestinal Tregs express GPR43 and that, in Gpr43−/− mice, the expansion of Tregs by SCFAs was abrogated (Smith et al. 2013). Interestingly, a mixture of Clostridia species, which are a prominent source of SCFA production, induced the development of Tregs and the production of the anti-inflammatory cytokine IL-10 (Atarashi et al. 2011, 2013). Mechanistically, Clostridia species cooperatively produce SCFAs that elicit a TGF-β response by epithelial cells. SCFAs also facilitate peripheral extrathymic generation of Tregs through a different mechanism (Furusawa et al. 2013; Smith et al. 2013). Butyrate epigenetically regulates gene expression through inhibition of histone deacetylases (HDACs), resulting in enhanced histone acetylation in the noncoding sequences of the FoxP3 locus (Arpaia et al. 2013; Smith et al. 2013). Consequently, butyrate-treated CD4+ T cells depicted increased histone H3 acetylation of the FoxP3 promoter region and at the intragenic enhancer element conserved noncoding DNA sequence 1 (CNS1) (Arpaia et al. 2013; Furusawa et al. 2013; Smith et al. 2013).
The role of butyrate as a HDAC inhibitor was also shown to contribute to the suppression of inflammatory response by intestinal macrophages (Chang et al. 2014). Treatment of bone marrow-derived macrophages with butyrate induced hyporesponsiveness to LPS stimulation and suppressed the proinflammatory cytokines IL-6 and IL-12p40 due to the HDAC inhibitory activity of butyrate and H3K9ac deposition on the Il6 and Il12b loci (Chang et al. 2014). Taken together, these results highlight the central importance of SCFAs in the communication between the microbiome and the host immune system.
Conclusions
The study of metabolites as messengers between the microbiota and the immune system has initiated a paradigm change in our understanding of host–microbial interactions. The increasing mechanistic knowledge about how microbiota metabolism shapes the physiology of its host impacts at least three areas of fundamental importance to both the conceptualization of host–microbiota interactions and its translation to clinical applications.
First, the discoveries made in the field of microbiome research over the last decade have challenged our model of how the immune system recognizes and eliminates micro-organisms upon contact with the host. The notion that microbial recognition by pattern recognition receptors of the innate immune system inevitably leads to the initiation of an immune and inflammatory response that aims at achieving eradication of the microbial trigger cannot possibly apply to the situation of colonization by commensal microbes. As such, the mechanisms of an immune response to pathogenic infection, studied in detail for almost a century, might represent the exception rather than the rule with regard to microbial handling by the immune system. The conceptual difficulty in the distinction between commensal colonization and pathogenic infection by the immune system lies in the fact that the ligands of all known pattern recognition receptors are conserved molecular structures that are essential for microbial existence and therefore shared across the spectrum of pathogenic and nonpathogenic micro-organisms. To reconcile these apparently opposing concepts, microbial viability, localization, and virulence factor expression have been suggested as hallmarks of pathogen-induced immune responses (Sander et al. 2011; Kamada et al. 2013). The discovery of bacterial metabolites as modulators of immune responses adds an important facet to our understanding of the distinction between mutualism and pathogenicity at the host–microbial interface. By sensing microbial metabolites, the immune system may evaluate microbial activity rather than the mere presence or absence of micro-organisms at a given location. The metabolite profile characteristic of a pathogenic invasion likely differs from the one characteristic of homeostasis, and thus this change in metabolite composition and concentrations may be sensed by the immune system as a deviation from homeostatic set points to initiate an appropriate inflammatory response.
Second, despite the rapid progress over the last decade in the identification of bacterial drivers of human diseases, the selective eradication of disease-associated micro-organisms from the intestinal microbial community remains a major challenge. Currently, antibiotics and fecal microbiota transplantation are the only clinically approved approaches to targeting a disease-causing microbiota. The use of antibiotics in the treatment of aberrant host–microbial interactions resembles the use of chemotherapy in cancer treatment in that it removes the causative agent of a disease at the cost of major disruptions in the endogenous cellular organization. Similarly, the effectiveness of fecal microbiota transplantation is based on the replacement of the entire intestinal community—a radical intervention resulting in an often unpredictable outcome. The identification of specific microbiota-derived metabolites and their effect on the host immune system has provided mechanistic insights that are much more tangible than the association of disease phenotypes with microbial taxa in large data set collections. On the one hand, metabolites are ideally suited as biomarkers for disease development, as their detection is usually affordable and scalable and may precede the onset of clinically manifest disease symptoms. On the other hand, the causal involvement of microbiota-derived metabolites in the molecular etiology of human disease presents direct opportunities for the rational development of new patient-tailored therapies. Beneficial metabolites can be administered with typically easy-to-control pharmacokinetics and without the danger of inadvertent immune responses to the drug. In addition, disease-associated metabolites can be targeted by pharmacological or nutritional intervention, potentially without strong interference from the homeostatic metabolism. The first proof of principal studies have included microbial targeting of the TMAO metabolism in the treatment of atherosclerosis (Wang et al. 2015b), and future research is warranted to further exploit such metabolite-based therapeutic options.
Third, the microbiome harbors a sheer endless reservoir of metabolic capabilities, which has led to the suggestion that, while the cataloging of microbial taxa that colonize mammalian mucosal surfaces may reach saturation within the next decade of microbiome research, the assessment of the entirety of the chemical complexity encoded by the microbiome is still in its infancy. This is interesting with regard to the pharmacologic opportunities mentioned above but may also help to better define a fundamental aspect of host–microbiota interactions; namely, the evolutionary teleology of microbial niche construction through modulation of the host immune system. The initial goal at early stages of the Human Microbiome Project had been to define a core microbiome shared by all individuals, motivated by the assumption that coevolution between the microbiota and its host has yielded an indispensable canon of bacteria characteristic of the intestinal community in the human gut (Turnbaugh et al. 2007). At later states, however, this notion has been replaced by the realization that the immense interindividual variability in the composition of the microbiota does not allow for the definition of a shared “core” on the taxonomic level (The Human Microbiome Project Consortium 2012). With the study of metabolites as common outputs of bacterial metabolic function, the notion of a functional core can be revisited. It is conceivable that different taxonomic groups of bacteria have evolved to perform similar metabolic functions and are thus classifiable by the type of metabolites that they produce and, consequently, the type of physiologic response that they elicit in the host. One may speculate that, despite the vast taxonomic diversity and variability that has been documented in cataloging studies of various human microbiomes to date, the members of the commensal microbiota can be classified into a limited variety of functional subsets with respect to their metabolite profile (Thaiss and Elinav 2014). Therefore, distinct groups of bacteria with niche-specific immunomodulatory activity might have evolved, employing similar metabolic pathways and metabolite signatures for the maintenance of homeostatic colonization. Such a level of understanding regarding the molecular details of colonization mechanisms and immune cell–microbiota cross-talk would open up a new area in this young field of study, resulting in numerous possibilities for the therapeutic exploitation of basic discoveries.
Acknowledgments
We thank the members of the Elinav laboratory for fruitful discussions. We apologize to investigators whose relevant work was not included in this review owing to space constraints. C.A.T. received a Boehringer Ingelheim Fonds PhD Fellowship. E.E. is supported by Yael and Rami Ungar, Israel; the Gurwin Family Fund for Scientific Research; the Leona M. and Harry B. Helmsley Charitable Trust; Crown Endowment Fund for Immunological Research; the estate of Jack Gitlitz; the estate of Lydia Hershkovich; the Benoziyo Endowment Fund for the Advancement of Science; the Adelis Foundation; John L. and Vera Schwartz; Pacific Palisades; Alan Markovitz, Canada; Cynthia Adelson, Canada; Centre National de la Recherche Scientifique (CNRS); the estate of Samuel and Alwyn J. Weber; Mr. and Mrs. Donald L. Schwarz, Sherman Oaks; grants funded by the European Research Council; the Kenneth Rainin Foundation; the German-Israel Binational Foundation; the Israel Science Foundation; the Minerva Foundation; the Rising Tide foundation; and the Alon Foundation scholar award. E.E. is the incumbent of the Rina Gudinski Career Development Chair and a senior fellow of the Canadian Institute For Advanced Research (CIFAR).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.284091.116.
References
- Abreu MT, Kantorovich V, Vasiliauskas EA, Gruntmanis U, Matuk R, Daigle K, Chen S, Zehnder D, Lin YC, Yang H, et al. 2004. Measurement of vitamin D levels in inflammatory bowel disease patients reveals a subset of Crohn's disease patients with elevated 1,25-dihydroxyvitamin D and low bone mineral density. Gut 53: 1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alex S, Lange K, Amolo T, Grinstead JS, Haakonsson AK, Szalowska E, Koppen A, Mudde K, Haenen D, Al-Lahham S, et al. 2013. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor γ. Mol Cell Biol 33: 1303–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504: 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. 2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500: 232–236. [DOI] [PubMed] [Google Scholar]
- Bakdash G, Vogelpoel LT, van Capel TM, Kapsenberg ML, de Jong EC. 2015. c. Mucosal Immunol 8: 265–278. [DOI] [PubMed] [Google Scholar]
- Balmer ML, Schurch CM, Saito Y, Geuking MB, Li H, Cuenca M, Kovtonyuk LV, McCoy KD, Hapfelmeier S, Ochsenbein AF, et al. 2014. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J Immunol 193: 5273–5283. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Losick R. 2006. Bacterially speaking. Cell 125: 237–246. [DOI] [PubMed] [Google Scholar]
- Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, et al. 2010. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog 6: e1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, et al. 2003. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem 278: 11312–11319. [DOI] [PubMed] [Google Scholar]
- Cha HR, Chang SY, Chang JH, Kim JO, Yang JY, Kim CH, Kweon MN. 2010. Downregulation of Th17 cells in the small intestine by disruption of gut flora in the absence of retinoic acid. J Immunol 184: 6799–6806. [DOI] [PubMed] [Google Scholar]
- Chang PV, Hao L, Offermanns S, Medzhitov R. 2014. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci 111: 2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, et al. 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149: 1578–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. 2007. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J Exp Med 204: 1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145: 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, et al. 2015. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 18: 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre D, Mold J, Hunt PW, Kanwar B, Loke P, Seu L, Barbour JD, Lowe MM, Jayawardene A, Aweeka F, et al. 2010. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med 2: 32ra36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL. 2011. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10: 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504: 446–450. [DOI] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31: 677–689. [DOI] [PubMed] [Google Scholar]
- Hall JA, Cannons JL, Grainger JR, Dos Santos LM, Hand TW, Naik S, Wohlfert EA, Chou DB, Oldenhove G, Robinson M, et al. 2011a. Essential role for retinoic acid in the promotion of CD4+ T cell effector responses via retinoic acid receptor α. Immunity 34: 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Grainger JR, Spencer SP, Belkaid Y. 2011b. The role of retinoic acid in tolerance and immunity. Immunity 35: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, et al. 2012. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 487: 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, et al. 2012. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482: 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MJ. 1997. Intestinal flora and endogenous vitamin synthesis. Eur J Cancer Prev 6: S43–S45. [DOI] [PubMed] [Google Scholar]
- Hoverstad T, Midtvedt T. 1986. Short-chain fatty acids in germfree mice and rats. J Nutr 116: 1772–1776. [DOI] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. 2013. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155: 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Elinav E, Huber S, Strowig T, Hao L, Hafemann A, Jin C, Wunderlich C, Wunderlich T, Eisenbarth SC, et al. 2013. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci 110: 9862–9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huengsberg M, Winer JB, Gompels M, Round R, Ross J, Shahmanesh M. 1998. Serum kynurenine-to-tryptophan ratio increases with progressive disease in HIV-infected patients. Clin Chem 44: 858–862. [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. 2004. Retinoic acid imprints gut-homing specificity on T cells. Immunity 21: 527–538. [DOI] [PubMed] [Google Scholar]
- Kamada N, Chen GY, Inohara N, Nunez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14: 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44: D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Lim HW, Andrisani OM, Broxmeyer HE, Kim CH. 2007. Vitamin A metabolites induce gut-homing FoxP3+ regulatory T cells. J Immunol 179: 3724–3733. [DOI] [PubMed] [Google Scholar]
- Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, Mazmanian SK. 2014. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 15: 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibe R, Kurihara S, Sakai Y, Suzuki H, Ooga T, Sawaki E, Muramatsu K, Nakamura A, Yamashita A, Kitada Y, et al. 2014. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci Rep 4: 4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. 2011. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334: 1561–1565. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, et al. 2012. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491: 717–723. [DOI] [PubMed] [Google Scholar]
- Kleerebezem M, Vaughan EE. 2009. Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Annu Rev Microbiol 63: 269–290. [DOI] [PubMed] [Google Scholar]
- Konieczna P, Ferstl R, Ziegler M, Frei R, Nehrbass D, Lauener RP, Akdis CA, O'Mahony L. 2013. Immunomodulation by Bifidobacterium infantis 35624 in the murine lamina propria requires retinoic acid-dependent and independent mechanisms. PLoS One 8: e62617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisawa J, Hashimoto E, Ishikawa I, Kiyono H. 2012. A pivotal role of vitamin B9 in the maintenance of regulatory T cells in vitro and in vivo. PLoS One 7: e32094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourhis L, Mburu YK, Lantz O. 2013. MAIT cells, surveyors of a new class of antigen: development and functions. Curr Opin Immunol 25: 174–180. [DOI] [PubMed] [Google Scholar]
- Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, et al. 2012. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol 13: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Katz MN, Suez J, Elinav E. 2015a. Inflammasomes and the microbiota—partners in the preservation of mucosal homeostasis. Semin Immunopathol 37: 39–46. [DOI] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, et al. 2015b. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163: 1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, Wilhelm C, Veldhoen M. 2011. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147: 629–640. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. 2003. Regulation of short-chain fatty acid production. Proc Nutr Soc 62: 67–72. [DOI] [PubMed] [Google Scholar]
- Mardinoglu A, Shoaie S, Bergentall M, Ghaffari P, Zhang C, Larsson E, Backhed F, Nielsen J. 2015. The gut microbiota modulates host amino acid and glutathione metabolism in mice. Mol Syst Biol 11: 834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, Barg H, Warren MJ, Jahn D. 2002. Microbial production of vitamin B12. Appl Microbiol Biotechnol 58: 275–285. [DOI] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461: 1282–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E, Koga Y, Benno Y. 2012. Impact of intestinal microbiota on intestinal luminal metabolome. Sci Rep 2: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122: 107–118. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453: 620–625. [DOI] [PubMed] [Google Scholar]
- Metghalchi S, Ponnuswamy P, Simon T, Haddad Y, Laurans L, Clement M, Dalloz M, Romain M, Esposito B, Koropoulis V, et al. 2015. Indoleamine 2,3-dioxygenase fine-tunes immune homeostasis in atherosclerosis and colitis through repression of interleukin-10 production. Cell Metab 22: 460–471. [DOI] [PubMed] [Google Scholar]
- Monticelli LA, Buck MD, Flamar AL, Saenz SA, Tait Wojno ED, Yudanin NA, Osborne LC, Hepworth MR, Tran SV, Rodewald HR, et al. 2016. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat Immunol 17: 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora JR, Iwata M, Eksteen B, Song SY, Junt T, Senman B, Otipoby KL, Yokota A, Takeuchi H, Ricciardi-Castagnoli P, et al. 2006. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science 314: 1157–1160. [DOI] [PubMed] [Google Scholar]
- Morrison DJ, Preston T. 2016. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. 2007. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 317: 256–260. [DOI] [PubMed] [Google Scholar]
- Munder M, Schneider H, Luckner C, Giese T, Langhans CD, Fuentes JM, Kropf P, Mueller I, Kolb A, Modolell M, et al. 2006. Suppression of T-cell functions by human granulocyte arginase. Blood 108: 1627–1634. [DOI] [PubMed] [Google Scholar]
- Murray PJ. 2015. Amino acid auxotrophy as a system of immunological control nodes. Nat Immunol 17: 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science 336: 1262–1267. [DOI] [PubMed] [Google Scholar]
- Noda H, Akasaka N, Ohsugi M. 1994. Biotin production by bifidobacteria. J Nutr Sci Vitaminol 40: 181–188. [DOI] [PubMed] [Google Scholar]
- Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, Kobayashi KS. 2009. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci 106: 15813–15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino-Lagos K, Guo Y, Brown C, Alexander MP, Elgueta R, Bennett KA, De Vries V, Nowak E, Blomhoff R, Sockanathan S, et al. 2011. A retinoic acid-dependent checkpoint in the development of CD4+ T cell-mediated immunity. J Exp Med 208: 1767–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompei A, Cordisco L, Amaretti A, Zanoni S, Matteuzzi D, Rossi M. 2007. Folate production by bifidobacteria as a potential probiotic property. Appl Environ Microbiol 73: 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccetti P, Grohmann U. 2007. IDO and regulatory T cells: a role for reverse signalling and non-canonical NF-κB activation. Nat Rev Immunol 7: 817–823. [DOI] [PubMed] [Google Scholar]
- Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, Zhou L. 2012. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raber P, Ochoa AC, Rodriguez PC. 2012. Metabolism of L-arginine by myeloid-derived suppressor cells in cancer: mechanisms of T cell suppression and therapeutic perspectives. Immunol Invest 41: 614–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118: 229–241. [DOI] [PubMed] [Google Scholar]
- Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, Delgado A, Correa P, Brayer J, Sotomayor EM, et al. 2004. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res 64: 5839–5849. [DOI] [PubMed] [Google Scholar]
- Romani L, Fallarino F, De Luca A, Montagnoli C, D'Angelo C, Zelante T, Vacca C, Bistoni F, Fioretti MC, Grohmann U, et al. 2008. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature 451: 211–215. [DOI] [PubMed] [Google Scholar]
- Roth JR, Lawrence JG, Bobik TA. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol 50: 137–181. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. 2010. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci 107: 12204–12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332: 974–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Muller M, Blander JM. 2011. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature 474: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LZ, Faith NG, Nakayama Y, Suresh M, Steinberg H, Czuprynski CJ. 2007. The aryl hydrocarbon receptor is required for optimal resistance to Listeria monocytogenes infection in mice. J Immunol 179: 6952–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, McCoy KD, Macpherson AJ. 2007. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol 19: 59–69. [DOI] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Backhed F. 2013. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 11: 227–238. [DOI] [PubMed] [Google Scholar]
- Spencer SP, Wilhelm C, Yang Q, Hall JA, Bouladoux N, Boyd A, Nutman TB, Urban JF Jr, Wang J, Ramalingam TR, et al. 2014. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science 343: 432–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strozzi GP, Mogna L. 2008. Quantification of folic acid in human feces after administration of Bifidobacterium probiotic strains. Journal of clinical gastroenterology 42: S179–S184. [DOI] [PubMed] [Google Scholar]
- Sun J. 2010. Vitamin D and mucosal immune function. Curr Opin Gastroenterol 26: 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. 2007. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med 204: 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura J, Kubota K, Murakami H, Sawamura M, Matsushima T, Tamura T, Saitoh T, Kurabayshi H, Naruse T. 1999. Immunomodulation by vitamin B12: augmentation of CD8+ T lymphocytes and natural killer (NK) cell activity in vitamin B12-deficient patients by methyl-B12 treatment. Clin Exp Immunol 116: 28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. 2013. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 368: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss CA, Elinav E. 2014. Exploring new horizons in microbiome research. Cell Host Microbe 15: 662–667. [DOI] [PubMed] [Google Scholar]
- Thaiss CA, Levy M, Suez J, Elinav E. 2014. The interplay between the innate immune system and the microbiota. Curr Opin Immunol 26: 41–48. [DOI] [PubMed] [Google Scholar]
- Thomas AC, Mattila JT. 2014. ‘Of mice and men’: arginine metabolism in macrophages. Front Immunol 5: 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, et al. 2014. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 20: 159–166. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007. The human microbiome project. Nature 449: 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Essen MR, Kongsbak M, Schjerling P, Olgaard K, Odum N, Geisler C. 2010. Vitamin D controls T cell antigen receptor signaling and activation of human T cells. Nat Immunol 11: 344–349. [DOI] [PubMed] [Google Scholar]
- Wada K, Tanaka H, Maeda K, Inoue T, Noda E, Amano R, Kubo N, Muguruma K, Yamada N, Yashiro M, et al. 2009. Vitamin D receptor expression is associated with colon cancer in ulcerative colitis. Oncol Rep 22: 1021–1025. [DOI] [PubMed] [Google Scholar]
- Wang S, Villablanca EJ, De Calisto J, Gomes DC, Nguyen DD, Mizoguchi E, Kagan JC, Reinecker HC, Hacohen N, Nagler C, et al. 2011. MyD88-dependent TLR1/2 signals educate dendritic cells with gut-specific imprinting properties. J Immunol 187: 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhu S, Yang L, Cui S, Pan W, Jackson R, Zheng Y, Rongvaux A, Sun Q, Yang G, et al. 2015a. Nlrp6 regulates intestinal antiviral innate immunity. Science 350: 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, Gu X, Huang Y, Zamanian-Daryoush M, Culley MK, et al. 2015b. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163: 1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, et al. 2014. NLRP6 inflammasome orchestrates the colonic host–microbial interface by regulating goblet cell mucus secretion. Cell 156: 1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, Pollard JW. 2013. Macrophage biology in development, homeostasis and disease. Nature 496: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi-Benedetti C, Fallarino F, et al. 2013. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39: 372–385. [DOI] [PubMed] [Google Scholar]