Yang et al. report the crystal structures of the SETD2 catalytic domain bound to H3K36M or H3K36I peptides with SAH. In the complex structure, the catalytic SET domain adopts an open conformation, with the K36M/I peptide snuggly positioned in a newly formed substrate channel.
Keywords: oncohistone; SETD2 methyltransferase; crystal structure, epigenetic regulation
Abstract
High-frequency point mutations of genes encoding histones have been identified recently as novel drivers in a number of tumors. Specifically, the H3K36M/I mutations were shown to be oncogenic in chondroblastomas and undifferentiated sarcomas by inhibiting H3K36 methyltransferases, including SETD2. Here we report the crystal structures of the SETD2 catalytic domain bound to H3K36M or H3K36I peptides with SAH (S-adenosylhomocysteine). In the complex structure, the catalytic domain adopts an open conformation, with the K36M/I peptide snuggly positioned in a newly formed substrate channel. Our structural and biochemical data reveal the molecular basis underying oncohistone recognition by and inhibition of SETD2.
Histone post-translational modifications (PTMs) are linked to tumorigenesis, mostly via dysfunction of their regulators (e.g., readers, writers, and erasers) that are frequently mutated in tumors (Dawson and Kouzarides 2012). Recently, high-frequency mutations in genes encoding histones themselves, rather than the histone regulators, were identified in a number of cancer types. Exome sequencing studies have identified recurrent hot spot missense mutations in histone H3. Notably, these mutations are located at or adjacent to H3 lysine residues that undergo acetylation and/or methylation. For example, the H3K27M mutation was identified in the majority of pediatric diffuse intrinsic pontine gliomas (Schwartzentruber et al. 2012; Wu et al. 2012), and the H3K36M mutation was found to occur predominantly in chondroblastomas (Behjati et al. 2013) and rarely in other cancer types such as head and neck squamous cell carcinoma and colorectal cancer (Shah et al. 2014). In addition, the H3K36-neighboring G34 mutations, such as G34R/V and G34W/L, have been detected in pediatric non-brain stem gliomas (Schwartzentruber et al. 2012; Wu et al. 2012) and giant cell tumors of the bone (Behjati et al. 2013), respectively.
Biochemical and cellular studies showed that H3K27M reduced global H3K27 methylation in vitro and in vivo by inhibiting the methyltransferase activity of polycomb-repressive complex 2 (PRC2) (Chan et al. 2013; Lewis et al. 2013; Justin et al. 2016). Recently, we and others identified a similar “poisoning” mechanism underlying H3K36M-driven tumorigenesis involving the inactivation of H3K36 methyltransferases (Fang et al. 2016; Lu et al. 2016). H3K36 can be methylated by enzymes such as NSD1/2/3, ASH1L, and SETD2 (Wagner and Carpenter 2012). Among them, SETD2 serves as the major H3K36 methyltransferase that is able to generate the trimethylated H3K36 from the unmethylated, monomethylated, or dimethylated states in vitro and in cells (Edmunds et al. 2008; Hu et al. 2010). SETD2 plays important roles in cellular processes such as transcription elongation (Yoh et al. 2008), RNA splicing (de Almeida et al. 2011; Kim et al. 2011), and DNA damage repair (Li et al. 2013; Pai et al. 2014). However, due to the lack of peptide-bound structures, the molecular basis underlying oncogenic histone H3 recognition by SETD2 remains unknown.
Here, we report the crystal structure of SETD2 catalytic domain bound to the H3K36M/I peptide and SAH (S-adenosylhomocysteine). In the complex structure, the catalytic SET domain undergoes dramatic conformational change to accommodate the histone H3 peptide. The molecular insights gained from this work shed light on mechanisms of action of other H3K36 methyltransferase family members.
Results and Discussion
Overall structure of the SETD2–H3K36M–SAH complex
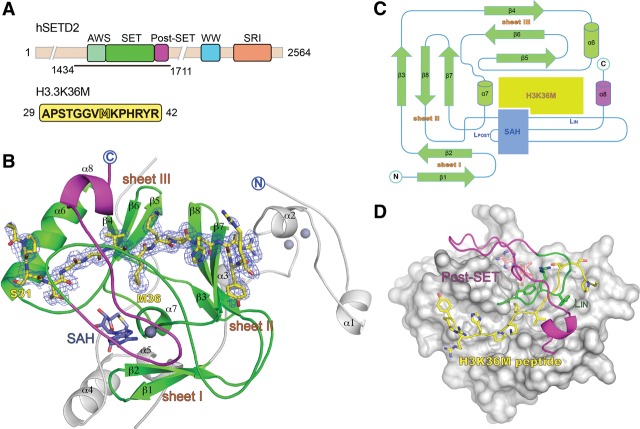
To decipher the molecular basis for H3K36M recognition by SETD2, we solved the ternary crystal structure of the human SETD2 catalytic domain (SETD2CD, amino acids 1434–1711) (Fig. 1A) bound to SAH and mutant histone H3.329–42K36M at 2.05 Å as well as a SAH-bound binary structure at 2.4 Å (Supplemental Table S1). The SETD2CD is composed of an N-terminal AWS zinc finger motif, a center SET domain, and a C-terminal Post–SET motif (Fig. 1A). The SET domain is characteristic of three discrete β sheets (I, β1–β2; II, β3–β8–β7; III, β4–β6–β5) that are arranged in a triangular shape (Fig. 1B,C). The SAH is located at the pocket formed by loops Lβ1β2, Lα7β7, and LPOST. Based on the electron densities, we could clearly trace histone H3.3 A29–R42 at the peptide-binding channel of the SET domain with the M36 long side chain pointing to SAH at the active center (Fig. 1B). Upon complex formation, the C-terminal fragments of SETD2CD (loop LIN and Post–SET) form a “knot”-like structure to cover the N-terminal part of the H3 peptide and stabilize its recognition (Fig. 1D). We calculated a buried solvent-accessible surface (SAS) area of 1270 Å2, which accounts for up to 60% of the SAS area of the H3 peptide.
Figure 1.
Overall structure of the SETD2CD–H3K36M–SAH ternary complex. (A) The domain architecture of human SETD2 (hSETD2) and the sequence of histone H3K36M peptide used for crystallization. (B) Overall structure of SETD2CD bound to the H3K36M peptide and SAH. SETD2CD is colored in light gray for the AWS motif, green for the SET domain, and magenta for the Post–SET motif. The H3K36M peptide is shown as yellow sticks covered by the simulated annealing Fo–Fc omit map countered at the 2.5 σ level. (C) The topology of SETD2CD SET–Post–SET motifs in the open state (the AWS motif was omitted for clarity). The newly formed C-terminal helix α8 is colored magenta. The loops LPOST and LIN mentioned in the text are labeled. The H3K36M peptide and SAH-binding sites are shaded in lime green and slate blue, respectively. (D) Covering of the H3K36M peptide by the C-terminal segments of SETD2CD. The Post–SET motif (magenta) and loop LIN (green) are represented by a cartoon. The H3K36M peptide is shown as yellow sticks. The rest of SETD2CD is represented as a gray surface.
Conformational change trigged by histone peptide binding
The structural alignment of the ternary complex with the SAH-bound binary structures reported previously (Zheng et al. 2012) as well as the one solved in this study revealed dramatic structural changes of the LIN–Post–SET fragment upon H3 binding (Supplemental Movie S1). First, the LIN loop connecting the SET domain and the Post–SET motif undergoes stepwise conformational changes following H3 peptide binding (Fig. 2A in stereo view, closed in salmon, half-open in green, and open in magenta). In the closed state (Protein Data Bank [PDB] code: 4H12), the LIN loop takes on a crouched conformation and inserts its residue, R1670, into the active center. Particularly, R1670 overlaps with H3K36M and occupies the K36 access pocket, therefore inhibiting the enzymatic activity of SETD2. Our binary structure captured a half-open state of SETD2CD in which R1670 flips outwards from the active center by ∼8.3 Å, thus priming the pocket for peptide entrance. In the open state, loop LIN adopts an extended conformation with R1670 further shifted ∼4.5 Å away, which allows proper docking of the H3 peptide in the substrate channel. Interestingly, the autoinhibitory LIN loop in the closed state turned out to facilitate H3 peptide binding in the open state (Fig. 1D; detailed later), suggesting dualistic function of the LIN loop at different stages of the enzymatic cycle. A second conformational change trigged by H3 peptide binding is the ordering of an invisible C-terminal loop-α8 fragment (1692–1703) of Post–SET that stretches over the substrate channel (Fig. 2A). Notably, a short α helix (α8) is induced at the C terminus to interact with α6 of the SET domain through hydrophobic contacts, further fastening H3 binding (Fig. 2B). In the presence of Pr-SNF (N-propyl sinefungin), a synthetic SAM (S-adenosyl methionine) analog, an open-state SETD2CD (PDB code: 4FMU) has been captured in the absence of bound peptide (Zheng et al. 2012). Structural superimposition revealed a number of conformation differences between the Pr-SNF-bound and peptide-bound “open” states of SETD2CD, which involves structural adjustments of the LIN loop and the ordering of the Post–SET C-terminal fragment in the latter case as an adaptation to peptide binding (Supplemental Fig. S1). In addition, structural and sequence alignment of SETD2CD with other H3K36 methyltransferases (An et al. 2011; Qiao et al. 2011) revealed that C211 of NSD1 (PDB code:3OOI) and S2259 of ASH1L (PDB code: 3OPE) are well superimposed with R1670 of SETD2 in its closed form (Fig. 2C,D), which suggests similar conformational change-driven regulatory mechanisms shared among H3K36 methyltransferases.
Figure 2.

Conformational change of SETD2CD and its comparison with other H3K36 methyltransferases. (A) Structure alignment of SETD2CD in closed (salmon), half-open (green), and open (magenta) states in stereo view. R1670 residues in three states are highlighted as sticks. Distances between the Cα of R1670 residues are labeled. Note the newly formed helix α8 that stacks against α6 in the open state. (B) Hydrophobic cluster formation involving helices α8 (magenta) and α6 (gray) around histone H3 V35 (yellow). I1614 is from β6, and Y1671 is from loop LIN. (C) Superposition of the SET domains among SETD2 (closed [blue] and open [magenta]), ASH1L (cyan), and NSD1 (orange). The conserved autoinhibitory residues are shown as sticks. The H3K36M peptide is colored yellow. (D) Sequence alignment of the LIN –Post–SET segments among SETD2 paralogs. Conserved residues are shaded violet. The flexible region that becomes visible in the peptide-bound complex is underlined.
Details of H3 peptide recognition by the SETD2 catalytic domain
Interaction analysis by LigPlot (Laskowski and Swindells 2011) revealed 14 direct and eight water-mediated hydrogen-bonding pairs as well as 15 pairs of hydrophobic contacts between SETD2CD and the H3K36M peptide (Supplemental Fig. S2). Notably, site-specific recognition of histone H3 by SETD2CD is determined by extensive interactions involving residues A29–P30–S31–T32–G33–G34–V35 and K37–P38–H39–R40–Y41–R42 flanking position 36. The small, noncharged A29–V35 fragment is located at a positive channel and partly buried; meanwhile, the K37–R42 fragment is rich in basic and bulky residues and docked into an exposed acidic groove (Fig. 3A). Specifically, the main chain of H3 S31–K37 is sandwiched by loops Lα6β5 and LIN of SETD2CD, and stabilized by a “pseudo-β-sheet” formation (Fig. 3B, panel i); furthermore, the H3 H39–Y41 main chain forms two hydrogen-bonding pairs (H39H3–T1637SETD2 and Y41H3–E1636SETD2) with E1636–T1637 side chains, thus completing the H3 main chain-mediated hydrogen-bonding network (Fig. 3B, panel ii). In addition, E1636 contributes to H3R40 recognition by forming charge-stabilized hydrogen-bonding pairs mediated by their side chains (Fig. 3B, panel ii). Moreover, G33–M36 is embraced by hydrophobic clusters formed by residues F1589, Y1604, F1606, F1668, and Y1671 around H3 G33–V35 (Fig. 3B, panel iii) as well as Y1579, M1607, F1664, and Y1666 around H3K36M (Fig. 3B, panel iv).
Figure 3.

Details for H3K36M peptide recognition by SETD2CD. (A) Electrostatic surface view of the SETD2CD ternary complex. Electrostatic potential is expressed as a spectrum ranging from −6 kT/e (red) to +6 kT/e (blue). The H3K36M peptide is shown as yellow sticks. (B, panel i) Sandwiching the H3 peptide by “pseudo-β-sheet” formation. The loops Lα6β5 and LIN of SETD2CD are colored blue, and the H3K36M peptide is colored yellow. Hydrogen bonds are depicted as magenta dashes. Arrows denote peptide direction from the N termini to the C termini. (Panel ii) Hydrogen bonding between H39–Y41 of the H3 peptide and residues E1636 and T1637 of SETD2. (Panels iii,iv) Hydrophobic contacts around H3 G33–V35 (panel iii) and H3 M36 (panel iv). (C) In vitro methyltransferase assays of wild-type and mutant SETD2CD with HeLa mononucleosome as substrate. Enzymatic activity was quantified by scintillation, and error bars represent standard deviation of three repeats.
In order to validate the functional importance of the above-mentioned residues, we next generated corresponding alanine mutations of SETD2CD and conducted in vitro methyltransferase assays. As summarized in Figure 3C, mutation of hydrophobic residues around H3K36M at the active center disrupted the activity almost completely, suggesting their essential role in H3K36 substrate recognition and/or catalysis. F1668A and Y1671A in loop LIN and F1589A in α6 displayed 30%–60% residual activities, underscoring their roles in H3 G33–V35 recognition. Interestingly, Y1604A, E1636A, and T1637A mutants showed elevated enzymatic activities compared with wild-type protein. Y1604 is part of the Lα6β5 loop and stacks against H3G33 from one side, whereas E1636 and T1637 participate in H3 H39–Y41 hydrogen bonding. As these mutations are located away from the active center, it is conceivable that the resultant interaction loss may be beneficial for more efficient enzymatic activities, possibly by promoting product release.
Structural basis for cis inhibition of SETD2 activity by H3G34 mutants
Histone H3G34 mutations (G34R/V/W/L) have been detected in brain (Schwartzentruber et al. 2012; Wu et al. 2012) and bone (Behjati et al. 2013) tumors. In the complex structure, the G33–G34 step is fully buried and threads through a narrow tunnel of the SETD2CD substrate channel (Fig. 4A), which is reminiscent of H3 G33–G34 recognition by the H3K36 demethylase KDM2A (Cheng et al. 2014). The inner wall of the H3 G33–G34 tunnel is formed by aromatic rings of Y1604, F1668, and Y1671 as well as main chains of Q1669–G1672 within loop LIN (Fig. 4B). The dimension of the tunnel is highly restrictive, being perfect for accommodating side chain-free glycine residues. Conceivably, mutation of H3G34 into any other bulkier residues will severely block histone H3 binding (Supplemental Fig. S3A–C) and thereafter abolish subsequent H3K36 methylation by SETD2 as shown previously (Lewis et al. 2013). This may serve as one fundamental molecular mechanism underlying the oncogenic activity of histone H3G34R/V/W/L mutants.
Figure 4.
The H3 G33–G34-accomodating tunnel of SETD2. (A) Burial of the G33–G34 fragment of H3 peptide (yellow) in the SETD2CD substrate channel. SETD2CD is shown as half-transparent surface (gray). (B) Close-up view of H3 G33–G34 (yellow) surrounded by an aromatic residue cluster of SETD2CD (blue). Sequence (C) and structural (D) alignments of G33–G34 tunnel residues among SETD2, NSD1/2/3, and ASH1L. The structural superimposition is based on the “closed state” structures of SETD2 (PDB code: 4H12) (blue), NSD1 (PDB code: 3OOI) (cyan), and ASH1L (PDB code: 3OPE) (salmon). (E) Encapsulation of H3 G33–G34 (yellow) by a modeled NSD1/2/3 tunnel. Modeled tunnel residues are shown as cyan sticks.
Sequence alignment showed that the tunnel-forming residues of SETD2 are essentially conserved in other H3K36 methyltransferases, including NSD1/2/3 and ASH1L (Fig. 4C). In principle, an aromatic and planar feature is conserved at position Y1604 (Y, F, and H), which stacks against the T32–G33 amide plane, and a hydrophobic feature is conserved at positions F1668 (F, L, and F) and Y1671 (Y, L, and F), which encapsulate the side chain-free H3 G33–G34. These tunnel residues are well aligned in the peptide-free state (Fig. 4D), and structural modeling suggested that mutating Y1604/F1668/Y1671 to F/L/L of NSD1/2/3 can well restrict H3 G33–G34 in a narrow tunnel (Fig. 4E; Supplemental Fig. S3D,E). Collectively, these results indicate that other H3K36 methyltransferase family members may be similarly inhibited by H3G34 oncogenic mutations.
Structural basis for trans inhibition of SETD2 activity by H3K36M/I mutants
Histone H3K36M is a high-frequency oncogenic mutation identified in chondroblastoma (Behjati et al. 2013). Our ternary structure in this study revealed registration and snug insertion of the K36M side chain into the active center of SETD2CD, where the S-methyl thioether group of K36M points to SAH—the product form of SAM after methyl transfer (Fig. 5A, panel i). The K36M side chain is confined in a hydrophobic K36 access pocket formed by residues Y1579, M1607, F1664, and Y1666 with sound shape complementarity (Supplemental Fig. S4A, in stereo view). These residues are well conserved in NSD1/2/3 and ASH1L (Supplemental Fig. S5), suggesting similar K36M preference by the lysine access pocket. Besides hydrophobic contacts, the K36M side chain stacks against the aromatic ring of Y1666 and is further stabilized by sulfur–aromatic (Valley et al. 2012) as well as CH–π interactions (Fig. 5B, panel i; Brandl et al. 2001). These features will conceivably promote SETD2CD-H3K36M association and thus sequester and inhibit SETD2 activity in trans to block global H3K36 methylation.
Figure 5.

The effect of the H3K36M/I mutation on SETD2 methyltransferase activity. (A) Cutaway view of the SETD2CD active pocket with bound K36M (panel i) and K36I (panel ii) of the ternary complexes. SETD2CD is shown as a gray surface. K36M/I and SAH are depicted as yellow and green sticks, respectively, and are covered by the simulated annealing Fo–Fc omit map countered at the 3.0 σ level. (B) Stacking interactions of SETD2 Y1666 with H3K36M (panel i) and K36I (panel iii). (Panel ii) Superposition of M36 (yellow) and I36 (blue) of H3. (Panel iv) The modeled K36L mutant introduced steric clash between K36L and Y1666, as denoted by red plates (arrow). (C) Peptide inhibition methyltransferase assays of SETD2CD, with mononucleosome as the substrate in the presence of 50 μM denoted H3 peptides. Enzymatic activity was quantified by scintillation, and error bars represent standard deviation of three repeats.
Systematic amino acid substitution studies at H3K27 showed that only the K-to-I mutation displayed an inhibitory effect on PRC2 similar to that of K-to-M (Lewis et al. 2013), and we recently observed a similar oncogenic effect of H3K36I in cell culture models as well as the identification of the K36I mutation in pediatric undifferentiated sarcoma tumors (Lu et al. 2016). Interestingly, in an effort to obtain additional complex structures using wild-type, K36 methylated (me1/2/3), or other K36 mutated (K36I, K36R, K36L, and K36Q) H3.329–42 peptides, we were able to get only H3.329–42K36I-bound SETD2CD crystal under essentially the same crystallization condition as that of H3.329–42K36M, indicating that K36M and K36I are better ligands for complex formation. In the new ternary structure determined at 1.5 Å (Supplemental Table S1), K36I is positioned in the same K36M access hydrophobic pocket (Fig. 5A, panel ii). Strikingly, K36I overlapped perfectly with K36M in positions Cα, Cβ, Cγ1 (Cγ in M36), and Cδ (Sδ in M36) (Fig. 5B, panel ii). The Cγ2 atom of I36 is branched out to occupy a spare space near the entrance of the hydrophobic pocket (Supplemental Fig. S4B,C). Although lacking the sulfur–aromatic interaction by nature, K36I recognition is notably contributed by hydrophobic encapsulation of its side chain as well as CH–π interactions with Y1666 (Fig. 5B, panel iii). Despite a high similarity to isoleucine, leucine substitution of H3K36 in our modeling study revealed a direct steric clash of K36L Cδ2-methyl with Y1666 due to deeper burial of the branched Cδ groups (Fig. 5B, panel iv), which explains the incapability of K36L to be an oncogenic mutation. Conceivably, other H3K36 mutants may introduce a similar steric clash or lack sufficient interactions within the K36-binding pocket and thus be less competent to inhibit the H3K36 methylation by SETD2.
We next performed methyltransferase inhibition assays with wild-type mononucleosome as a substrate in the presence of different H3K36 peptides. As summarized in Figure 5C, compared with the peptide-free control, the enzymatic activity of SETD2 is severely reduced in the presence of histone H326–46 K36M and K36I peptides; in contrast, addition of H3K36L, H3K36Q, H3K36R, or H3K36me3 peptide has a minimal effect on SETD2 activity. Taken together, our biochemical and structural studies suggest that H3K36M and H3K36I are preferred nonreactive ligands for SETD2 and likely its structurally conserved paralogs (Fig. 2C) to inhibit their enzymatic activities, therefore leading to global reduction of H3K36 methylation in cells in the process of tumorigenesis.
In summary, we report here the first peptide-bound structures of SETD2CD in the open state, thus shedding new light on H3K36 methyltransferases with regard to substrate recognition and enzymatic regulation. Our current work along with two recent structural studies on H3K27M recognition by the PRC2 complex (Justin et al. 2016) and H3K9M recognition by G9a (Jayaram et al. 2016) collectively emphasize the role of K to M among other oncohistone mutations in promoting loss of specific lysine methylation through recognition by and inhibition of SET domain methyltransferases. Given the importance of histone methylation in health and disease, the molecular mechanisms uncovered here not only help to elucidate the etiology associated with aberrant histone modification but also pave the way for new initiatives in the treatment of diseases linked to oncohistone mutation.
Materials and methods
Protein production and crystallographic studies
Wild-type and mutant human SETD2CD (residues 1434–1711) were recombinantly produced in Escherichia coli and purified as His-SUMO-tagged proteins. Crystallization was performed via vapor diffusion method. Diffraction data were collected at Shanghai Synchrotron Radiation Facility beamline BL17U under cryo conditions and processed with the HKL2000 software packages. The structures were solved by molecular replacement using the MolRep program (Vagin and Teplyakov 2010), with the free SETD2 SET domain structure (PDB code: 4H12) as the search model. All structures were refined using PHENIX (Adams et al. 2010) with iterative manual model building with COOT (Emsley and Cowtan 2004). Detailed structural refinement statistics are in Supplemental Table S1.
In vitro methyltransferase assay
Radiometric filter assay was used to measure the enzymatic activity of SETD2CD and its mutants. The mononucleosome samples were prepared from HeLa cells.
Detailed descriptions about the Materials and Methods are in the Supplemental Material.
Accession codes
The coordinates and structure factors for the SETD2–H3K36M–SAH, SETD2–H3K36I–SAH, and SETD2–SAH structures have been deposited under accession codes 5JJY, 5JLB, and 5JLE, respectively.
Supplementary Material
Acknowledgments
We thank the staff at beamline BL17U of the Shanghai Synchrotron Radiation Facility and Dr. S. Fan at Tsinghua Center for Structural Biology for assistance in data collection. We thank the Center of Biomedical Analysis in Tsinghua University for providing isotope facilities. This work was supported by grants from the Ministry of Science and Technology of China (2016YFA0500700 and 2015CB910503) and the Tsinghua University Initiative Scientific Research Program to H.L., and funds from Rockefeller University and National Institutes of Health grant (P01CA196539) to C.D.A. C.L. is the Kandarian Family Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2195-14). H.L. conceived the study. S.Y. designed and performed the experiments with help from X.Z. and C.L. under the guidance of H.L. G.-M.L. and C.D.A. provided critical comments, and H.L. and S.Y. wrote the manuscript with input from the other authors. All authors reviewed the results and approved the final version of the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.284323.116.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Yeo KJ, Jeon YH, Song JJ. 2011. Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. J Biol Chem 286: 8369–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H, et al. 2013. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 45: 1479–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl M, Weiss MS, Jabs A, Suhnel J, Hilgenfeld R. 2001. C-H…π-interactions in proteins. J Mol Biol 307: 357–377. [DOI] [PubMed] [Google Scholar]
- Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, et al. 2013. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Cheung P, Kuo AJ, Yukl ET, Wilmot CM, Gozani O, Patel DJ. 2014. A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36. Genes Dev 28: 1758–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. 2012. Cancer epigenetics: from mechanism to therapy. Cell 150: 12–27. [DOI] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. 2011. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol 18: 977–983. [DOI] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. 2008. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 27: 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fang D, Gan H, Lee JH, Han J, Wang Z, Riester SM, Jin L, Chen J, Zhou H, Wang J, et al. 2016. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352: 1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Sun XJ, Zhang YL, Kuang Y, Hu CQ, Wu WL, Shen SH, Du TT, Li H, He F, et al. 2010. Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proc Natl Acad Sci 107: 2956–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram H, Hoelper D, Jain SU, Cantone N, Lundgren SM, Poy F, Allis CD, Cummings R, Bellon S, Lewis PW. 2016. S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3. Proc Natl Acad Sci 113: 6182–6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, De Marco V, Haire LF, Walker PA, Reinberg D, et al. 2016. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun 7: 11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Fong N, Erickson B, Bentley DL. 2011. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci 108: 13564–13569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Swindells MB. 2011. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model 51: 2778–2786. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM. 2013. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell 153: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Jain SU, Hoelper D, Bechet D, Molden RC, Ran L, Murphy D, Venneti S, Hameed M, Pawel BR, et al. 2016. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352: 844–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CC, Deegan RS, Subramanian L, Gal C, Sarkar S, Blaikley EJ, Walker C, Hulme L, Bernhard E, Codlin S, et al. 2014. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat Commun 5: 4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Li Y, Chen Z, Wang M, Reinberg D, Xu RM. 2011. The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J Biol Chem 286: 8361–8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231. [DOI] [PubMed] [Google Scholar]
- Shah MA, Denton EL, Arrowsmith CH, Lupien M, Schapira M. 2014. A global assessment of cancer genomic alterations in epigenetic mechanisms. Epigenetics Chromatin 7: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 66: 22–25. [DOI] [PubMed] [Google Scholar]
- Valley CC, Cembran A, Perlmutter JD, Lewis AK, Labello NP, Gao J, Sachs JN. 2012. The methionine-aromatic motif plays a unique role in stabilizing protein structure. J Biol Chem 287: 34979–34991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. 2012. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. 2012. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44: 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Lucas JS, Jones KA. 2008. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev 22: 3422–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Ibanez G, Wu H, Blum G, Zeng H, Dong A, Li F, Hajian T, Allali-Hassani A, Amaya MF, et al. 2012. Sinefungin derivatives as inhibitors and structure probes of protein lysine methyltransferase SETD2. J Am Chem Soc 134: 18004–18014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.