Here, Lim et al. report a new version of TAIL-seq (mRNA TAIL-seq [mTAIL-seq]) with enhanced sequencing depth for mRNAs (by ∼1000-fold compared with the previous version). Using their new methodology, the authors investigated mRNA tailing in Drosophila oocytes and embryos and demonstrated a relationship between poly(A) tail length and translational efficiency during egg activation.
Keywords: poly(A) tail, TAIL-seq, cytoplasmic polyadenylation, oogenesis, egg activation, Drosophila
Abstract
Eukaryotic mRNAs are subject to multiple types of tailing that critically influence mRNA stability and translatability. To investigate RNA tails at the genomic scale, we previously developed TAIL-seq, but its low sensitivity precluded its application to biological materials of minute quantity. In this study, we report a new version of TAIL-seq (mRNA TAIL-seq [mTAIL-seq]) with enhanced sequencing depth for mRNAs (by ∼1000-fold compared with the previous version). The improved method allows us to investigate the regulation of poly(A) tails in Drosophila oocytes and embryos. We found that maternal mRNAs are polyadenylated mainly during late oogenesis, prior to fertilization, and that further modulation occurs upon egg activation. Wispy, a noncanonical poly(A) polymerase, adenylates the vast majority of maternal mRNAs, with a few intriguing exceptions such as ribosomal protein transcripts. By comparing mTAIL-seq data with ribosome profiling data, we found a strong coupling between poly(A) tail length and translational efficiency during egg activation. Our data suggest that regulation of poly(A) tails in oocytes shapes the translatomic landscape of embryos, thereby directing the onset of animal development. By virtue of the high sensitivity, low cost, technical robustness, and broad accessibility, mTAIL-seq will be a potent tool to improve our understanding of mRNA tailing in diverse biological systems.
Eukaryotic mRNA experiences multiple types of modifications at its 3′ end throughout its life cycle. Newly synthesized mRNA is known to acquire a long poly(A) tail (up to ∼250 nucleotides [nt]) through canonical polyadenylation coupled to transcription, which facilitates mRNA export from the nucleus (Wahle and Keller 1996). In the cytoplasm, poly(A) tails are associated with cytoplasmic poly(A)-binding proteins (PABPCs) that stabilize mRNA by acting as a safeguard against multiple decay machineries and promote protein synthesis (Eliseeva et al. 2013; Norbury 2013). Once mRNA is deadenylated to a certain threshold (∼25 nt), PABPC is released, and mRNA becomes translationally inactive and prone to degradation. Poly(A) tails can be elongated post-transcriptionally by noncanonical poly(A) polymerases, which can play a particularly important role in cells with limited transcriptional activity (Weill et al. 2012; D'Ambrogio et al. 2013). For instance, in neuronal synapses and oocytes, deadenylated mRNAs are not degraded but are stored in a dormant state until protein production is required (Weill et al. 2012; D'Ambrogio et al. 2013). In response to cellular signals during meiotic maturation or synaptic stimulation, some deadenylated mRNAs are known to undergo cytoplasmic polyadenylation, which enhances translation, promoting rapid accumulation of protein independently of transcription. In such conditions, poly(A) tail length serves as a determining factor in translational reactivation.
Cytoplasmic polyadenylation has been mainly studied with maternal mRNAs stored in oocytes and early embryos of Drosophila, Xenopus, and mice (Salles et al. 1994; Mendez and Richter 2001; Richter 2007). Previous analyses showed that cell cycle genes and embryo patterning factors are regulated via polyadenylation, which is critical for normal development. In vertebrates, the cytoplasmic polyadenylation element (CPE) is enriched in the 3′ untranslated regions (UTRs) of target mRNAs (Pique et al. 2008). GLD-2 (also known as TUTase2 or PAPD4 in mammals) is a noncanonical poly(A) polymerase and acts on mRNAs containing CPEs through the interaction with CPE-binding protein (CPEB) (Kwak et al. 2004; Norbury 2013). In Drosophila, a GLD-2 homolog, Wispy, is known to be required for poly(A) tail elongation (Benoit et al. 2008; Cui et al. 2008, 2013).
Our current knowledge of cytoplasmic polyadenylation is mainly from analyses of individual genes using Northern blotting-based or RT–PCR-based methods (Salles et al. 1999). Previous attempts at global analysis had to rely on microarrays because current sequencing platforms cannot sequence long homopolymers (Whiteford et al. 2009; Ledergerber and Dessimoz 2011; Zheng and Tian 2014). With differential elution from oligo(dT) columns followed by microarray analyses, one can estimate the ratio between long and short A tails (Du and Richter 2005; Beilharz and Preiss 2007; Meijer et al. 2007; Novoa et al. 2010; Cui et al. 2013). This approach allowed genome-wide estimation of poly(A) tail length, yet the resolution and accuracy of the measurements were highly limited, precluding deeper investigation of mRNA tailing. Thus, many questions remain unresolved: (1) Which mRNAs are affected by cytoplasmic polyadenylation? (2) At which developmental stage does cytoplasmic polyadenylation occur globally? (3) Is GLD-2 a main enzyme for cytoplasmic polyadenylation? (4) Does GLD-2 act specifically on a subset of mRNAs, and, if so, how is the specificity achieved?
Overcoming the technical difficulties for determining poly(A) tail length, we recently developed a method called “TAIL-seq,” which enabled us to explore the 3′ extremity of mRNA using the Illumina sequencing platform (Chang et al. 2014). In brief, total RNA is depleted of rRNAs by using affinity beads and is ligated to the 3′ adaptor that contains biotin residues (Supplemental Fig. S1A, left). Following fragmentation by RNase T1, the 3′-most fragments are purified by using streptavidin beads and are ligated to the 5′ adaptor. After reverse transcription and PCR amplification, the library is sequenced following a paired-end sequencing protocol: 51 cycles for read 1 (forward direction) to identify the transcript, and 231 cycles for read 2 (reverse direction) to examine the 3′ tail sequences. As the sequence quality of the homopolymeric T [corresponding to the poly(A) tail] is very low, TAIL-seq analyzes the raw fluorescence signals from read 2, from which a machine learning algorithm detects the transition from poly(T) to non-T sequences. Because the transition occurs at the boundary between the poly(A) tail and the 3′ UTR, one can use this information to precisely measure poly(A) tail length.
Apart from allowing high-resolution, genome-wide measurement of poly(A) tail length, TAIL-seq has some unique strengths. First, it uses the regular sequencing platform, so one can run the library in any Illumina sequencer, unlike “PAL-seq.” PAL-seq also offers a genome-wide measurement of poly(A) tail length with high resolution (Subtelny et al. 2014) but requires a special manipulation of a Genome Analyzer II (a discontinued model) and an additional imaging step. Second, TAIL-seq preserves the sequence information of the 3′ end so as to reveal 3′ end modifications such as uridylation and guanylation. TAIL-seq also detects numerous other types of 3′ ends such as endonucleolytic cleavage sites and decay intermediates.
The ability to cover the complex terminome is the strength of TAIL-seq, but, at the same time, the wide coverage of the TAIL-seq library inevitably reduces sensitivity, which limits the application of TAIL-seq. Thus, we here modified TAIL-seq to increase the sensitivity. The new method (“mTAIL-seq” [mRNA TAIL-seq]) allows us to investigate poly(A) tail regulation in Drosophila oocytes and eggs where poly(A) tail length has not been accurately measured yet. We discovered a major phase of polyadenylation during late oogenesis and a modulation of poly(A) tails upon egg activation. By comparing mTAIL-seq data with ribosome profiling data, we further found a strong coupling between poly(A) regulation and translational activation in fly eggs.
Results
mTAIL-seq: a solution for limited materials
For the original version of TAIL-seq, a large amount of total RNA (∼100 µg) was needed to achieve enough sequencing depth for mRNA (Supplemental Fig. S1A, left; Chang et al. 2014). In order to improve the sensitivity, we decided to use splint ligation, which allowed us to capture RNAs with a specific type of terminus. Note that we did not use oligo(dT) affinity purification so as to avoid a potential bias toward long poly(A) tails. Splint ligation has been used to generate various cDNA libraries, and it has been shown that splint ligation does not cause a significant bias over a wide range of poly(A) tails except for very short A tails (less than ∼8 nt) (Subtelny et al. 2014).
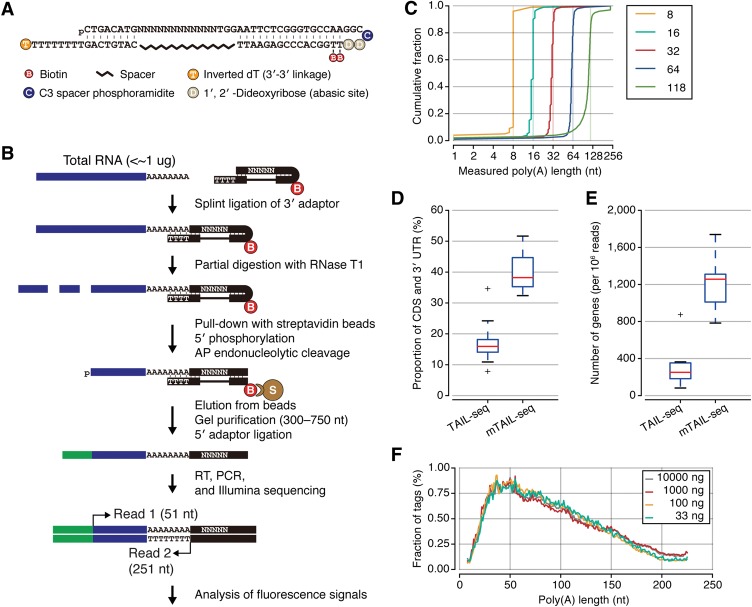
For splint ligation, stable annealing of the bridge oligo and the 3′ adaptor was a major issue because the TAIL-seq 3′ adaptor contains degenerate sequences that are used to improve sequencing performance and monitor uneven amplification (Chang et al. 2014). Initially, we used splint ligation in a conventional way that uses a bridge oligo between the 3′ adaptor and target RNA (Supplemental Fig. S1B, mTAIL-seq-1). However, ligation efficiency was poor due to weak base-pairing between the 3′ adaptor and bridge. To stably anchor the bridge to the 3′ adaptor, we designed a hairpin adaptor instead of using two single-stranded oligos (Fig. 1A; Supplemental Fig. S1B, mTAIL-seq-2). The intervening random sequences are bypassed by an ethylene glycol spacer. We confirmed that ligation was efficient and specific to A-tailed substrates (Supplemental Fig. S1B).
Figure 1.
Design and performance of mTAIL-seq. (A) Design of the 3′ hairpin adaptor. (N) Random sequence. (B) Schematic description of the experimental procedure. (Blue bars) mRNAs; (black bars) 3′ adaptors. Random sequence (N) and thymine (T) shown in 3′ adaptors are abbreviated proportional to the original length (shown in A). (B) Biotin; (S) streptavidin bead. (C) Accuracy assessment using poly(A) spike-ins. A cumulative graph of poly(A) tail length of chemically synthesized spike-ins (A8, A16, A32, A64, and A118) measured by the TAIL-seq algorithm. (D) A box plot showing the read proportion of coding sequences (CDSs) and 3′ UTRs in TAIL-seq and mTAIL-seq. For comparison, 12 libraries of TAIL-seq and 13 libraries of mTAIL-seq made from HeLa cells were used. The box indicates the first and third quartiles, and the internal bar refers to the median. Whiskers denote the lowest and highest values within 1.5 times the interquartile range of the first and third quartiles, respectively. (E) A box plot showing the number of detected genes that are normalized by 1 million reads in TAIL-seq and mTAIL-seq. Box and whisker plots are shown as in D. (F) Global distributions of poly(A) tails (8–225 nt) from four different amounts of HeLa RNA.
mTAIL-seq has several distinct features in the library construction procedure (Fig. 1B; Supplemental Fig. S1A; see the Materials and Methods). First, the 3′ hairpin adaptor specifically captures poly(A)+ RNA, so we can omit the rRNA depletion step, which is expensive and time-consuming. Second, the 3′ hairpin adaptor has two abasic sites that can easily be cut by apurinic/apyrimidinic endonuclease 1 (APE1). The cleavage helps elution of the ligated RNA from the bead and allows reverse transcription by releasing the opposite strand. Third, we changed the range of size fractionation from 500–1000 nt to 300–750 nt to increase gel elution efficiency. The remaining steps of library preparation and data analysis are similar to the previous version, with minor changes (see detailed information in the Materials and Methods). We validated the performance of the mTAIL-seq analysis pipeline by using spike-ins of known poly(A) tail length (Fig. 1C).
Compared with TAIL-seq, mTAIL-seq provided significantly more mRNA reads that are mapped to coding sequences (CDSs) and 3′ UTRs (Fig. 1D). Expectedly, the tags were derived mainly from near the annotated 3′ ends (Supplemental Fig. S1C). In terms of sensitivity, mTAIL-seq detected ∼1250 genes per million reads on average, which is approximately five times greater than the original TAIL-seq (Fig. 1E). It allowed us to analyze thousands of genes even from a small scale run on Illumina MiSeq, which reduces the cost of sequencing. It is noteworthy that mTAIL-seq detected 643 genes with at least 50 poly(A)+ tags even from 33 ng of total RNA, which corresponds to ∼1000 HeLa cells. Four experiments covering a broad dynamic range of input RNA showed reproducible results, indicating that mTAIL-seq is a robust technique (Fig. 1F; Supplemental Fig. S1D).
As expected, mTAIL-seq provides longer median lengths than TAIL-seq (Supplemental Fig. S1E) because the splint ligation used in mTAIL-seq cannot capture certain types of tails such as very short A tails (below 8 nt) or those with 3′ modifications. Uridylation is the most frequent modification of poly(A) tails and is found mainly on short tails <25 nt (Chang et al. 2014; Lim et al. 2014). Monouridylation and diuridylation are the most prevalent uridylation types. Since uridylated tails are not efficiently ligated to the adaptor with eight Ts (Supplemental Fig. S1F, mTAIL-seq-T8), we sought to capture uridylated tails by synthesizing and mixing two additional hairpin adaptors that carry one or two adenosines at the overhang (Supplemental Fig. S1F, mTAIL-seq-T7A1 and mTAIL-seq-T6A2). With the mixture of adaptors, we could detect uridylated tails, albeit at a lower frequency as compared with original TAIL-seq (Supplemental Fig. S1F). Thus, mTAIL-seq is adjustable to enrich a specific type of terminus by changing the design of adaptors with different sequences.
In conclusion, both TAIL-seq and mTAIL-seq have unique strengths suitable for particular purposes. TAIL-seq offers a comprehensive view of the 3′ terminome that covers all types of RNA termini. On the other hand, mTAIL-seq can be more practical if one is interested in a specific type of RNA terminus, such as poly(A)+ mRNAs. For its enhanced sensitivity and reduced cost and time, mTAIL-seq is useful especially when only a small amount of biological sample is available and/or when many samples need to be analyzed and compared.
Global poly(A) tail length measurement in Drosophila
Previous studies on cytoplasmic polyadenylation focused mainly on specific individual mRNAs with critical roles in developmental processes. In this study, to gain a transcriptomic landscape of cytoplasmic polyadenylation, we applied mTAIL-seq on Drosophila oocytes and embryos. Of note, we initially used the original TAIL-seq protocol for Drosophila early embryos (0–2 h after egg laying [AEL]) and S2 cells, which are relatively easy to obtain in a sufficient quantity. We found that the uridylation frequency in these samples is far lower than that in HeLa and NIH3T3 cells, which implies that uridylation may play a limited role in flies (Supplemental Fig. S2A).
Using mTAIL-seq with a T8 adaptor, we monitored poly(A) tail length at six different time points during early embryo development, ranging from 0 to 4.5 h AEL (Supplemental Fig. S2B). Because major activation of zygotic transcription occurs ∼2 h AEL, the samples up to 2 h AEL represent an early stage of development at which transcription is silenced (Tadros and Lipshitz 2009). We expected that poly(A) tail length would increase globally as described in fertilized eggs of Xenopus and zebrafish (Subtelny et al. 2014), but, surprisingly, the median length of poly(A) tails of early embryos did not significantly increase in the 0- to 2-h period, implying that poly(A) tail elongation may occur at an earlier stage prior to fertilization in flies (Supplemental Fig. S2B).
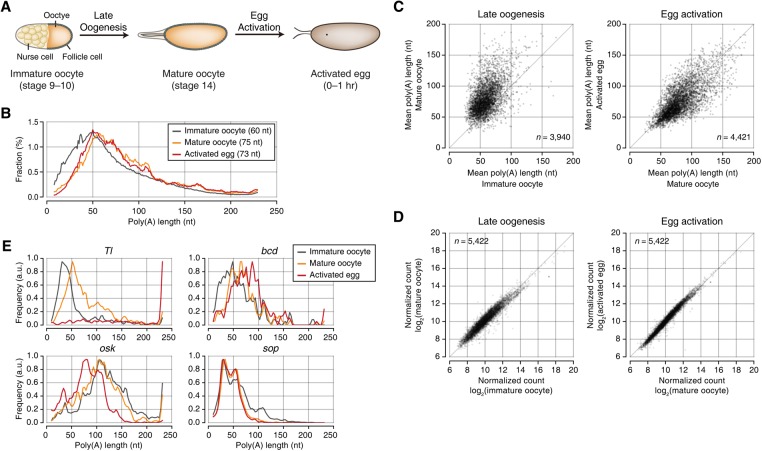
To determine the developmental stage at which cytoplasmic polyadenylation takes place, we examined three stages of female gametes: immature oocyte (stage 9–10 egg chamber), mature oocyte (stage 14 egg chamber), and activated egg (0–1 h AEL) (Fig. 2A; Bastock and St Johnston 2008; He et al. 2011). Drosophila ovarian development comprises 14 distinct stages. Two critical events, oocyte maturation and egg activation, are required for the production of functional embryos. At stage 9–10, each egg chamber contains one immature oocyte and 15 nurse cells. Nurse cells provide maternal mRNAs and proteins to the oocyte and break down shortly after this stage. At around stage 12–13, the immature oocyte undergoes maturation to yield a metaphase I-arrested mature oocyte (stage 14) (Resnick et al. 2009; Von Stetina and Orr-Weaver 2011; Laver et al. 2015). The mature oocyte is ovulated from the ovary and pass through the reproductive tract, which triggers egg activation and exit from meiosis. In Drosophila, egg activation is induced by mechanical pressure independent of fertilization (Heifetz et al. 2001; Horner and Wolfner 2008). Thus, we collected activated but unfertilized eggs for 1 h after they were laid instead of fertilized embryos. This allowed us to examine maternal mRNAs upon egg activation, avoiding the compounding effects from zygotic transcription. From small amounts of oocyte and egg samples (corresponding to <5 µg of total RNA), we performed mTAIL-seq with MiSeq and measured the poly(A) tail length of 3664 genes [with at least 50 poly(A)+ tags in all three samples]. Two biological replicates at each stage showed a high degree of reproducibility (Supplemental Fig. S2C; Supplemental Table S1).
Figure 2.
Global lengthening of poly(A) tails during Drosophila oogenesis. (A) Schematic illustration of late oogenesis and egg activation in Drosophila. Global poly(A) tail lengths are addressed at three different stages: immature oocyte, mature oocyte, and activated egg. “Stage 9–10 egg chamber,” “stage 14 egg chamber,” and “unfertilized but activated egg” are indicated as immature oocyte, mature oocyte, and activated egg, respectively. These terms are used throughout the text. (B) Global distributions of poly(A) tails at three stages. The median poly(A) tail lengths is 60 nt in immature oocytes, 75 nt in mature oocytes, and 73 nt in activated eggs. The result from biological replicates is shown in Supplemental Figure S2D. (C) Scatter plots showing the changes of poly(A) tail lengths upon late oogenesis and egg activation, respectively. The mean poly(A) tail lengths from two biological replicates were averaged. The median of mean poly(A) tail lengths is 58 nt in immature oocytes, 76 nt in mature oocytes, and 70 nt in activated eggs. (D) Changes of mRNA abundance upon late oogenesis and egg activation measured by RNA sequencing (RNA-seq). (E) Examples of individual genes. mTAIL-seq tags were plotted in 3-nt-wide bins and then smoothened with a Hanning window (width = 5). The frequency along the Y-axis was normalized by the maximum value at each stage. Note that as “read 2” runs up to only 231 cycles, longer poly(A) tails are presented as 231 nt.
Interestingly, we observed a drastic difference in poly(A) length distribution between immature oocytes and mature oocytes, while only a minor change was seen between mature oocytes and activated eggs (Fig. 2B; Supplemental Fig. S2D). The median length in the global profile increased from 60 nt in immature oocytes to 75 nt in mature oocytes (Fig. 2B). At the gene level, most mRNA species (3365 out of 3940 genes, 85.4%) were polyadenylated during late oogenesis (Fig. 2C, left). The median of mean increased from 58 nt to 76 nt. For most genes, cytoplasmic polyadenylation is initially activated at oocyte maturation in Drosophila, as reported for some genes like cycB and c-mos in Xenopus (Sheets et al. 1994). In contrast, upon egg activation, the median length did not increase at the global level (median: 73 nt; median of mean: 70 nt) (Fig. 2B,C), although there were some gene-specific modulations (Fig. 2C, right). To confirm that this dramatic change of poly(A) tails during late oogenesis was not due to transcription, we performed RNA sequencing (RNA-seq) on the same samples used for mTAIL-seq and found that individual mRNA abundance was largely unchanged during late oogenesis and egg activation (Fig. 2D; Supplemental Table S2). Additionally, there was no substantial correlation between mRNA abundance change and poly(A) length change (Supplemental Fig. S2E). Taken together, these analyses indicate that the changes of poly(A) tail length may be caused by cytoplasmic polyadenylation, not nascent transcription.
For validation, we next examined several individual genes that were previously studied (Fig. 2E; Salles et al. 1994; Benoit et al. 2005, 2008; Vardy and Orr-Weaver 2007; Vardy et al. 2009). The mTAIL-seq results were validated by high-resolution poly(A) tail (Hire-PAT) assay (Supplemental Fig. S2F; Bazzini et al. 2012). As expected, embryo patterning-related genes, such as Tl and bcd, showed a dramatic increase of poly(A) tail length during late oogenesis and egg activation, whereas sop (ribosomal protein S2) remained nearly unchanged. In the case of embryo posterior determinant osk, the poly(A) tail was relatively long in immature oocytes and mature oocytes, which differs from the previous report showing elongation at this stage (Benoit et al. 2005). However, given the earlier studies reporting the presence of Osk protein in immature oocytes (Kim-Ha et al. 1995; Yoshida et al. 2004) and the enhancement of osk translation by cytoplasmic polyadenylation at the posterior pole before late oogenesis (Castagnetti and Ephrussi 2003), it is likely that the osk mRNA indeed has a long tail and is actively translated in immature oocytes. Therefore, adenylation of some mRNAs may occur prior to stage 9–10.
In conclusion, our mTAIL-seq experiments provide an accurate profile of poly(A) length at the genomic level, revealing dynamic regulation of poly(A) tails during Drosophila oogenesis and egg activation.
Distinct patterns of poly(A) tail regulation
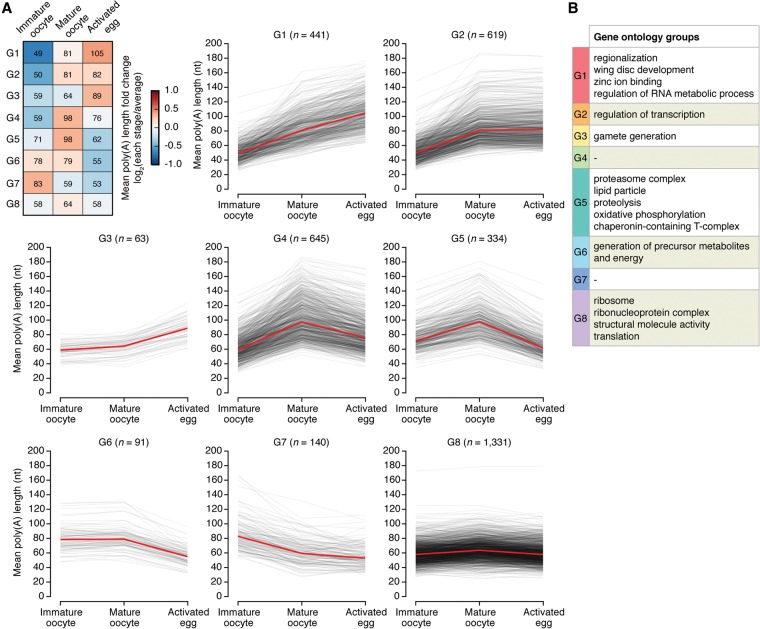
Although poly(A) tail length increases during late oogenesis and is maintained during egg activation at the global level, many individual genes show interesting temporal regulation patterns. Based on these dynamic changes, we classified 3664 genes into eight groups (Fig. 3A; Supplemental Fig. S3A,B; Supplemental Table S3; see the Materials and Methods). Groups 1, 2, and 3 show an increase throughout late oogenesis and egg activation. Transcripts in these groups have considerably shorter poly(A) tails at the immature oocyte stage than other transcripts. Specifically, group 1 contains 441 transcripts whose poly(A) tails increase continuously throughout late oogenesis and egg activation. The median length of the poly(A) tails changed from 49 nt to 105 nt. This group includes several well-known targets of cytoplasmic polyadenylation such as Tl, bcd, and cycB, which is consistent with previous studies (Salles et al. 1994; Benoit et al. 2005, 2008; Vardy and Orr-Weaver 2007; Vardy et al. 2009). Interestingly, gene ontology (GO) analysis of group 1 reveals enrichment for terms such as “regionalization,” “wing disc development,” “zinc ion binding,” and “regulation of RNA metabolic processes” (Fig. 3B; Supplemental Table S4). It is tempting to speculate that group 1 may include some unknown developmental regulators that are poised to act in the early embryo through cytoplasmic polyadenylation.
Figure 3.
Dynamic regulation of poly(A) tails. (A) Classification of transcripts according to the changes of poly(A) tail length into eight groups. (Top left) Fold changes from the average to the poly(A) length of each stage are shown in heat map. The median of the mean poly(A) length of each group is presented in the heat map. Patterns of poly(A) length changes at the individual gene level are presented in the line graphs. Red lines indicate the median. See the Materials and Methods for the classification method and see Supplemental Table S3 for the full list. (B) Functional categorization of genes in each group by GO analysis (false discovery rate [FDR] < 0.1). For overlapping terms, a representative term is selectively shown. The full list is in Supplemental Table S4.
Next, groups 4 and 5 show fluctuating patterns: lengthening during late oogenesis and shortening during egg activation. Thus, transcripts in these groups are polyadenylated specifically during late oogenesis and undergo deadenylation afterward. What stops their polyadenylation and triggers deadenylation upon egg activation is interesting but unclear at this point. Groups 4 and 5 consist of functionally diverse genes, but many of them encode proteins involved in proteolysis and oxidative phosphorylation. These proteins may need to be transiently produced in mature oocytes and silenced in early embryos immediately following egg activation.
Groups 6 and 7 show descending patterns (Fig. 3A). Transcripts in these groups have relatively long poly(A) tails in immature oocytes (78 nt and 83 nt for group 6 and group 7, respectively) as compared with those in other groups (58 nt for all detected genes). It is possible that these transcripts are cytoplasmically adenylated earlier (before and at stage 9–10) than other transcripts. Alternatively, but not mutually exclusively, the transcripts in groups 6 and 7 may retain their long tails by resisting deadenylation in immature oocytes. Group 6 is enriched with genes related to the generation of precursor metabolites and energy. Rapid deadenylation of these metabolic genes suggests that metabolic pathways may need to be reprogrammed at the onset of animal development.
Group 8 shows little changes in poly(A) tail length profile (<20 nt difference across three stages). This group is enriched with genes with constitutive functions such as ribosomal subunits and translation (Fig. 3B; Supplemental Table S4).
To understand the mechanism underlying the selectivity of cytoplasmic polyadenylation, we searched for sequence motifs enriched in each group. However, the analysis did not reveal any known motifs, such as CPEs (data not shown). While vertebrate CPEs are well known to play a central role in coordinating cytoplasmic polyadenylation, the role of Drosophila CPEs remains unclear. Although a fly homolog of CPEB, Orb, was reported to physically and genetically interact with a homolog of GLD-2, Wispy, during oogenesis (Benoit et al. 2008), CPE sequences have not been found commonly in Wispy target mRNAs (Coll et al. 2010; Cui et al. 2013). We suspect that the control of poly(A) tail length may be governed by multiple sequence motifs working in combination as opposed to one master regulatory element, such as CPE.
Cytoplasmic polyadenylation by Wispy
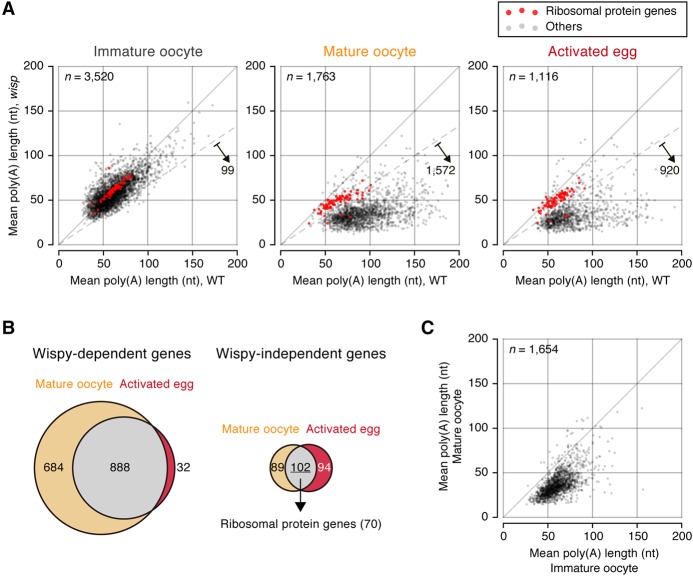
To verify the global cytoplasmic polyadenylation that we observed in late oogenesis, we carried out mTAIL-seq on wisp mutants (Supplemental Fig. S4A; Supplemental Table S5). Wispy is a noncanonical poly(A) polymerase that is expressed exclusively in maturing oocytes and early embryos (Benoit et al. 2008; Cui et al. 2008; Lee et al. 2014). Wispy acts on mRNAs and microRNAs (miRNAs). The specificity of Wispy on individual transcripts was addressed previously with a microarray approach (Cui et al. 2013) but needs to be investigated with higher resolution. In immature oocytes, poly(A) tail lengths in wisp mutants were similar to those in wild type (median of mean length: 56 nt vs. 58 nt), indicating that the activity of Wispy may be limited until this stage (Fig. 4A, left panel).
Figure 4.
Defects of cytoplasmic polyadenylation in wisp mutants. (A) Comparison of poly(A) tail lengths between wild type and the wisp mutant in each stage. The mean poly(A) tail lengths from two biological replicates are averaged. n = 2. The median of mean poly(A) tail lengths of wisp mutants is 56 nt in immature oocytes, 34 nt in mature oocytes, and 32 nt in activated eggs. The dashed line marks 1.5-fold reduction. Red dots represent ribosomal protein genes. (B) Wispy-dependent groups in mature oocytes and activated eggs are defined as in A and are presented in the Venn diagram at the left. The gray region contains genes that have dependency on both stages. Orange and red refer to stage-specific genes in mature oocytes and activated eggs, respectively. Genes independent of Wispy are depicted similarly in the Venn diagram at the right. (C) A scatter plot showing the changes of poly(A) tail length in wisp mutants upon late oogenesis.
In stark contrast, mature oocytes displayed a marked difference between wild type and wisp mutants (Fig. 4A, middle panel). The mutant had substantially shorter poly(A) tails than the wild type (median of mean length: 34 nt and 76 nt, respectively). We also observed a comparable difference in activated eggs (median of mean length: 32 nt and 70 nt in the mutant and wild type, respectively) (Fig. 4A, right panel). Gene-level analyses revealed that most mRNA species have shorter poly(A) tails in wisp mutants (98.9% and 97.8% of detected genes in mature oocytes and activated eggs, respectively). Moreover, when examining those genes that displayed changes in poly(A) length by >1.5-fold (Fig. 4A, indicated by dashed lines), 89.2% and 82.4% of genes carried shorter poly(A) tails in mutant mature oocytes and activated eggs, respectively. Thus, Wispy is responsible for most, if not all, polyadenylation events at these developmental stages. This observation is consistent with the above results (Fig. 2) showing that cytoplasmic polyadenylation takes place mainly during late oogenesis, although Wispy may act either before stage 9 or after egg activation on some select transcripts. We also noticed that mRNA abundance is modestly reduced in activated eggs of wisp mutants as compared with those of the wild type (Supplemental Fig. S4B), suggesting that Wispy may increase the stability of maternal mRNAs during embryogenesis.
This data allowed us to identify a group of genes that are refractory to Wispy (Fig. 4A, shown by red dots). This group includes 191 genes (10.8%) and 196 genes (17.6%) detected in mature oocytes and activated eggs, respectively (Fig. 4B). Notably, most of them encode ribosomal proteins (Fig. 4B). Consistently, these mRNA species belong to “group 8,” whose poly(A) tails do not change in length in wild type during both late oogenesis and egg activation (Fig. 3). These data suggest that the transcripts encoding ribosomal proteins may specifically escape from cytoplasmic polyadenylation.
Of note, it was previously reported that genes involved in mitochondrial function are independent of Wispy (Cui et al. 2013); however, we found that such genes displayed changes in poly(A) tail length in a Wispy-dependent manner (Supplemental Fig. S4C, shown as green dots). The apparent discrepancy between the studies is likely because the previous approach relied on an oligo(dT) column. The column captures mRNAs with long A tails (more than ∼40 nt) indiscriminately (Cui et al. 2013).
Our analysis also revealed that, in the absence of Wispy, poly(A) length continues to decrease instead of staying the same (Fig. 4C). The median length changed from 56 nt (at stage 9–10) to 34 nt (at stage 14) (Fig. 4C). This result implies that Wispy may be required for not only polyadenylation but also protection against deadenylation during late oogenesis.
Correlation between poly(A) tail length and translational efficiency (TE)
To understand the functional consequences of cytoplasmic polyadenylation, we compared poly(A) tail length with TE. It was shown recently that poly(A) length correlates with TE in zebrafish and frog embryos before zygotic transcription, while there is no such correlation in somatic cells (Subtelny et al. 2014). It remains unknown whether invertebrate embryos have comparable regulatory mechanism at the genomic scale.
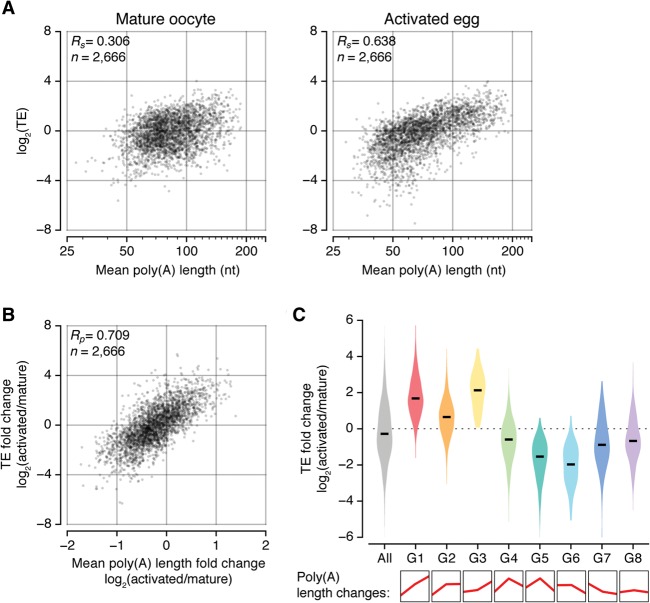
Orr-Weaver and colleagues (Kronja et al. 2014) previously measured TE by ribosome profiling in mature oocytes and activated eggs (0–2 h). To match the developmental stage, we generated a poly(A) tail profile of the 0- to 2-h activated eggs in addition to the 0- to 1-h activated eggs (Fig. 5A; Supplemental Fig. S5A). The comparison of the poly(A) profile and ribosome profile showed a clear correlation between poly(A) tail length and TE in activated eggs (Rs = 0.638) (Fig. 5A, right panel). Thus, like in vertebrates, protein synthesis is mainly and globally dictated by the poly(A) tail in early embryos of Drosophila. This result confirms earlier studies on individual genes (Benoit et al. 2008; Coll et al. 2010) and further suggests that animals have a highly conserved mechanism for the regulation of the earliest translation events.
Figure 5.
Functional association of poly(A) tail length and translation during egg activation. (A) Comparison of poly(A) tail length with TE, which was estimated by Kronja et al. (2014). TE was calculated by dividing ribosome density over RNA abundance from two biological replicates. The median of TE at each stage was adjusted to 0. “Rs” refers to Spearman correlation coefficient. (B) A scatter plot showing the correlation between mean poly(A) length changes and TE changes upon egg activation. “Rp” refers to Pearson correlation coefficient. (C) Translational controls on poly(A) tail length profiles. Violin plots showing the differences in TE changes between eight groups that were defined in Figure 3A. The poly(A) length changes of each group are simplified in the bottom panel. The black line represents the median.
A notable observation from this analysis is that the correlation is modest in mature oocytes (Rs = 0.306) (Fig. 5A, left panel). This was unexpected because the global poly(A) distribution does not change substantially during egg activation (Fig. 2B). Nevertheless, it is noteworthy that many individual genes are modulated in poly(A) tails during egg activation (Fig. 2C, right). The changes in poly(A) tail length during egg activation correlate well to the changes in TE (Fig. 5B; Supplemental Fig. S5B). These results suggest that while global elongation occurs during late oogenesis, the additional modulation of poly(A) tail length during egg activation may be important for translational control. The polysome/monosome ratio in activated eggs is fivefold higher than that in mature oocytes, indicating that translation is globally up-regulated during egg activation (Kronja et al. 2014). Thus, in flies, polyadenylation and translational activation appear to be partly separated. Polyadenylation begins during late oogenesis, while translational activation occurs later during egg activation.
We next compared TEs of different groups of transcripts that show distinct patterns of poly(A) tail length (Figs. 3A, 5C). Transcripts in groups 1, 2, and 3 whose poly(A) tails are continuously elongated showed a dramatic increase of TE upon egg activation (Fig. 5C). In contrast, genes in groups 5 and 6, which include many energy metabolism-related genes, are translationally suppressed. Poly(A) tails continue to be modified during egg activation, presumably by concurrent polyadenylation and deadenylation. For instance, transcripts encoding the vacuolar H+ ATPase subunit and cytochrome C oxidase subunit are deadenylated and translationally suppressed during egg activation. Transcripts in group 8 [those with little changes in poly(A) tails] tend to be translated at constant rates. We obtained similar results when we applied another index for translation rate, the ribosome occupancy, which is the ratio of RNA associated with polysomes over monosomes (Supplemental Fig. S5C; Kronja et al. 2014). Taken together, our analyses demonstrate that the regulation of the poly(A) tail shapes the translational landscape in early embryos.
Discussion
In conclusion, mTAIL-seq is a potent tool for investigating poly(A) tails. This method costs substantially less and takes less time than the original version (TAIL-seq), as mTAIL-seq omits the affinity-based rRNA depletion step and runs on MiSeq. Moreover, mTAIL-seq can be applied to any species, even those for which the rRNA depletion kit is unavailable. mTAIL-seq is improved markedly in sensitivity (by ∼1000 fold) so that one can now analyze small amounts of biological samples (as little as ∼33 ng of total RNA).
Our present study using mTAIL-seq allowed us to accurately document the dynamic and temporal regulation of poly(A) tail length during oogenesis and egg activation in flies. Specifically, we confirmed that Wispy-deficient oocytes fail to induce global elongation of poly(A) tails during late oogenesis, indicating that lengthening of poly(A) tails at this stage is fully dependent on cytoplasmic polyadenylation. During egg activation, we further found a modest yet significant modulation of poly(A) tail length. These changes correlate strongly with the changes in TE, implying that poly(A) tail length may function as a critical factor for translation at this developmental stage.
Based on stage-specific changes of poly(A) tail length, maternal transcripts can be classified into eight groups. Notably, we identified groups 1, 2, and 3, which include transcripts that continue to be polyadenylated through two consecutive stages and are translated efficiently in activated eggs. Known targets of cytoplasmic polyadenylation that are associated with cell cycle (cycB and cycA) and embryonic patterning (bcd and Tl) are enriched in groups 1 and 2 (Supplemental Table S3; Salles et al. 1994; Benoit et al. 2005, 2008; Vardy and Orr-Weaver 2007; Vardy et al. 2009). In addition, many genes related to RNA metabolism belong to these groups. Genes in the small RNA pathway such as AGO1, AGO2, aub, and gw (a homolog of GW182) are found in group 1. Genes associated with RNA degradation and translational repression (such as smg, pum, and brat) and the components of the deadenylase complexes (twin, [a homolog of CCR4], Not1, PAN2, and CG11486 [PAN3]) also belong to group 1 or 2. Thus, many key factors involved in mRNA destabilization are maternally inherited and may be induced post-transcriptionally by cytoplasmic polyadenylation during late oogenesis and egg activation.
Using wisp mutants, we confirmed the previous finding that global poly(A) lengthening is dependent on Wispy (Benoit et al. 2008; Cui et al. 2008, 2013). Furthermore, in wisp mutants, we found that poly(A) tail lengths are not fixed but get shortened during late oogenesis, suggesting that Wispy may protect poly(A) tails from deadenylation. In Xenopus oocytes and mouse neurons, the CPEB-associated complex contains a deadenylase, PARN, and a poly(A) polymerase, Gld2, which balance out the poly(A) tail lengths in the absence of cellular signals (Kim and Richter 2006; Udagawa et al. 2012). Upon induction of CPEB phosphorylation by meiotic maturation or synaptic stimulation, deadenylase is released from the complex, leading to poly(A) tail elongation. Likewise, the dynamic balance between Wispy and deadenylase may determine poly(A) tail length during Drosophila oogenesis and in egg activation. It is interesting that, although there is no homolog of PARN in flies, the regulatory principle may be conserved in both vertebrates and invertebrates: Polyadenylation and deadenylation work together to shape poly(A) tails. In future studies, it will be necessary to identify a deadenylase/exonuclease responsible for deadenylation in Drosophila oocytes and embryos and compare it with the vertebrate machineries.
Our data show that the dynamic adjustment of poly(A) tail length shapes the translatome in early embryos. In contrast, mature oocytes do not show a strong correlation even though the global poly(A) tail lengths are similar in those two stages. What, then, makes activated eggs so special? One possible hypothesis is the dispersion of repressive complexes such as P bodies upon egg activation. Recently, superresolution imaging unveiled that mature oocytes contain electron-dense bodies that are composed of various mRNPs, including translational repressors such as Me31B and CUP (Weil et al. 2012). Egg activation disassembles the complex and facilitates remodeling of specialized ribonucleoprotein particles, which may trigger subsequent translational activation (Krauchunas et al. 2012; Weil et al. 2012; Kronja et al. 2014). Additionally, the PAN GU (PNG) kinase complex, which relieves PUM-dependent translational repression of cycA, cycB, and smg at egg activation (Tadros et al. 2007; Vardy and Orr-Weaver 2007; Vardy et al. 2009; Kronja et al. 2014), may have a role in the global translational activation. It remains to be investigated how the poly(A) tail length is fine-tuned and mechanistically coupled to translation at this interesting developmental stage.
The global profiling of poly(A) tails by mTAIL-seq provides a comprehensive resource for the regulation of poly(A) tails in Drosophila oocyte-to-embryo development. It is noted, however, that because the experiments were performed with the whole egg chamber and embryos, the current data set cannot offer accurate information for locally regulated mRNAs. In cases of mRNAs whose protein production is spatially restricted for proper development, it will be necessary in the future to measure poly(A) tail length in specific cellular compartments.
Studies using various tissues such as neurons and cells from other species will help to reveal the mechanisms as well as the conserved features in the regulation of poly(A) tails. By virtue of high sensitivity, low cost, technical robustness, and broad accessibility, mTAIL-seq will be a potent tool to improve our understanding of mRNA tailing and translational control.
Materials and methods
Construction of the mTAIL-seq library
Total RNAs were extracted from HeLa cells or Drosophila samples by TRIzol reagent (Invitrogen, 15596-018). Total RNA (∼1–5 µg) was ligated to a 3′ hairpin adaptor using T4 RNA ligase 2 (New England Biolabs, M0239) overnight. 3′ ligated RNA was partially digested by RNase T1 (Ambion, AM2283) and subjected to streptavidin beads (Invitrogen, 11206D). 5′ phosphorylation by PNK reaction (Takara, 2021B) and endonucleolytic cleavage by APE1 reaction (New England Biolabs, M0282) were performed on beads. Subsequently, RNA was eluted by 2× RNA loading dye and gel-purified by 6% urea-PAGE gel in the range of 300–750 nt. The purified RNAs were ligated to the 5′ adaptor, subjected to reverse transcription (Invitrogen, 18080–085), and amplified by PCR using Phusion DNA polymerase (Thermo, F-530L). PCR products were purified by AMPure XP beads (Beckman, A63881). The library was sequenced on Illumina MiSeq (51 × 251 paired end run) with 50% of the PhiX control library (Illumina, FC-110-3001) and 10% of the spike-in mixture. The spike-ins were prepared and mixed as previously described (Chang et al. 2014). The 3′ hairpin adaptor sequence used for mTAIL-seq was /5Phos/CTGACATGNNNNNNNNNNNNTGGAATTCTCGGGTGCCAAGGC/iSpC3//idSp//idSp//iBiodT//iBiodT/GGCACCCGAGAATT/iSp18/CATGTCAGTTTTTTTT/3InvdT/. N refers to a random sequence, /5Phos/ refers to 5′ phosphorylation, /iSpC3/ refers to internal C3 phosphoramidite, /idSp/ refers to internal 1′,2′-dideoxyribose, /iBioT/ refers to internal biotinylated deoxythymine, /iSp18/ refers to an internal 18-atom hexaethyleneglycol spacer, and /3InvdT/ refers to 3′ inverted deoxythynime.
mTAIL-seq analysis
The detailed procedure of poly(A) length measurement was identical to that of TAIL-seq (Chang et al. 2014) except for variations in usage of the 3′ hairpin adaptor. Genes with ≥50 poly(A) tags were analyzed. A geometric mean of poly(A) lengths was used as a representative value and is referred as “mean poly(A) length” because a distribution of intragenic poly(A) lengths is a lognormal-like distribution. For replicates, the average of geometric mean lengths was used in analyses.
Drosophila stocks and oocyte/egg collection
Fly lines of w1118 and wispKG5287 were obtained from the Bloomington Drosophila Stock Center, and tud1 was from the Kyoto Stock Center. w1118 was used as a wild-type control. wispKG5287 was previously described as a null allele of wisp (Benoit et al. 2008). Immature (stage 9–10) and mature (stage 14) egg chambers were collected by hand dissection in Grace′s unsupplemented insect medium (Gibco, 11595-030) from 3- or 4-d-old female flies. Unfertilized activated eggs were produced from w1118 virgin females mated to sterile males (sons of tud1 mothers) (Boswell and Mahowald 1985). Fly eggs and embryos were collected on grape juice plates for the designated time frame at 25°C.
RNA-seq analysis
Total RNA was extracted with TRIzol (Invitrogen, 15596-018), and the quality was checked by an Agilent 2100 Bioanalyzer. rRNA was depleted from total RNA using a Ribo-Zero kit (Epicentre, MRZH11124). RNA-seq libraries were constructed by Macrogen, Inc., using Illumina TruSeq RNA sample preparation kit version 2. Sequencing reads derived from cDNA libraries described above were processed by using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit). First, the 3′ adaptor sequence was removed, and trimmed reads were filtered by Phred quality score (fastq_quality_filter -Q 33 -p 30 -q 90). The sequence reads were aligned to ERCC RNA spike-ins using STAR version 2.4.2a (Dobin et al. 2013) with options --alignIntronMin 99999 --alignEndsType EndToEnd. Reads that did not match to any spike-in were aligned to UCSC (University of California at Santa Cruz) dm6 genome assembly using RSEM version 1.2.25 with STAR (Li and Dewey 2011; Dobin et al. 2013), and splicing junction annotations were generated from the NCBI RefSeq (downloaded from UCSC Genome Browser on December 12, 2014). The reduced RefSeq transcript set for nonoverlapping representation was prepared as previously described (Chang et al. 2014). The reads mapped to ERCC spike-ins were counted by using htseq-count (Anders et al. 2015). Next, the expected counts from RSEM (Li and Dewey 2011) were normalized with spike-ins by using RUVg (k = 1) in R package RUVSeq (Supplemental Table S2; Risso et al. 2014). For analysis, transcripts with insufficient reads (<100 normalized reads in any library) were removed.
Classification and functional categorization of genes
We classified genes based on the difference of poly(A) tail lengths between consecutive developmental stages. First, genes with a <20-nt difference across three stages were regarded as an unchanged group (group 8). Next, we set a 10-nt difference between the adjacent two stages as a criteria to discriminate changes of poly(A) tail length: elongated or shortened (≥10 nt difference) or unchanged (<10 nt difference). A group showing elongation of poly(A) tails in late oogenesis and shortening in egg activation was further subdivided into two groups depending on the value of sgn (elongated length or shortened length). Additionally, three groups with shortened poly(A) tails in late oogenesis were merged, since each group had a small number of members. The gene list of each group is in Supplemental Table S3. Functional annotation was done for each gene group using DAVID bioinformatics tools (Huang da et al. 2009). For the background population, members of all groups (3664 genes) were used, and GO terms with false discovery rate (FDR) < 0.1 were selected (Supplemental Table S4).
Ribosome profiling analysis
RPF (ribosome profiling) and RNA-seq data were downloaded from a publicly available database (GSE52799) (Kronja et al. 2014). Sequencing reads were trimmed into 27-nt-long sequences and then filtered with Phred quality score. RPF and RNA-seq tags were counted and normalized by using RSEM (Li and Dewey 2011). To minimize a tendency from ribosomes accumulating near the start codon, reads with 5′ ends mapping within the first 50 nt of each ORF were disregarded (Ingolia et al. 2009; Subtelny et al. 2014). TE was calculated by the TPM (transcripts per million) ratio of RPF to RNA-seq, and the median of log2(TE) was adjusted to 0. Genes with ≥10 TPM in both RPF and RNA-seq libraries were included in the analysis.
Accession Numbers
Sequencing data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (accession no. GSE83732)
Supplementary Material
Acknowledgments
We thank the members of our laboratory for discussion and technical help. This work was supported by IBS-R008-D1 of the Institute for Basic Science from the Ministry of Science, ICT, and Future Planning of Korea (to J.L., M.L., A.S., H.C., and V.N.K.) and BK21 Research Fellowships from the Ministry of Education of Korea (J.L. and A.S.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.284802.116.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastock R, St Johnston D. 2008. Drosophila oogenesis. Curr Biol 18: R1082–R1087. [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz TH, Preiss T. 2007. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA 13: 982–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, Mitou G, Chartier A, Temme C, Zaessinger S, Wahle E, Busseau I, Simonelig M. 2005. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Dev Cell 9: 511–522. [DOI] [PubMed] [Google Scholar]
- Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M. 2008. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development 135: 1969–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell RE, Mahowald AP. 1985. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell 43: 97–104. [DOI] [PubMed] [Google Scholar]
- Castagnetti S, Ephrussi A. 2003. Orb and a long poly(A) tail are required for efficient oskar translation at the posterior pole of the Drosophila oocyte. Development 130: 835–843. [DOI] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, Kim VN. 2014. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell 53: 1044–1052. [DOI] [PubMed] [Google Scholar]
- Coll O, Villalba A, Bussotti G, Notredame C, Gebauer F. 2010. A novel, noncanonical mechanism of cytoplasmic polyadenylation operates in Drosophila embryogenesis. Genes Dev 24: 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Sackton KL, Horner VL, Kumar KE, Wolfner MF. 2008. Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation. Genetics 178: 2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Sartain CV, Pleiss JA, Wolfner MF. 2013. Cytoplasmic polyadenylation is a major mRNA regulator during oogenesis and egg activation in Drosophila. Dev Biol 383: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrogio A, Nagaoka K, Richter JD. 2013. Translational control of cell growth and malignancy by the CPEBs. Nat Rev Cancer 13: 283–290. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Richter JD. 2005. Activity-dependent polyadenylation in neurons. RNA 11: 1340–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliseeva IA, Lyabin DN, Ovchinnikov LP. 2013. Poly(A)-binding proteins: structure, domain organization, and activity regulation. Biochemistry (Mosc) 78: 1377–1391. [DOI] [PubMed] [Google Scholar]
- He L, Wang XB, Montell DJ. 2011. Shining light on Drosophila oogenesis: live imaging of egg development. Curr Opin Genet Dev 21: 612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifetz Y, Tram U, Wolfner MF. 2001. Male contributions to egg production: the role of accessory gland products and sperm in Drosophila melanogaster. Proc Biol Sci 268: 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner VL, Wolfner MF. 2008. Transitioning from egg to embryo: triggers and mechanisms of egg activation. Dev Dyn 237: 527–544. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Richter JD. 2006. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell 24: 173–183. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM. 1995. Translational regulation of oskar mRNA by Bruno, an ovarian RNA-binding protein, is essential. Cell 81: 403–412. [DOI] [PubMed] [Google Scholar]
- Krauchunas AR, Horner VL, Wolfner MF. 2012. Protein phosphorylation changes reveal new candidates in the regulation of egg activation and early embryogenesis in D. melanogaster. Dev Biol 370: 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I, Yuan B, Eichhorn SW, Dzeyk K, Krijgsveld J, Bartel DP, Orr-Weaver TL. 2014. Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell Rep 7: 1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. 2004. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci 101: 4407–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver JD, Marsolais AJ, Smibert CA, Lipshitz HD. 2015. Regulation and function of maternal gene products during the maternal-to-zygotic transition in Drosophila. Curr Top Dev Biol 113: 43–84. [DOI] [PubMed] [Google Scholar]
- Ledergerber C, Dessimoz C. 2011. Base-calling for next-generation sequencing platforms. Brief Bioinform 12: 489–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Choi Y, Kim K, Jin H, Lim J, Nguyen TA, Yang J, Jeong M, Giraldez AJ, Yang H, et al. 2014. Adenylation of maternally inherited microRNAs by Wispy. Mol Cell 56: 696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. 2014. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159: 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Bushell M, Hill K, Gant TW, Willis AE, Jones P, de Moor CH. 2007. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res 35: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Richter JD. 2001. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol 2: 521–529. [DOI] [PubMed] [Google Scholar]
- Norbury CJ. 2013. Cytoplasmic RNA: a case of the tail wagging the dog. Nat Rev Mol Cell Biol 14: 643–653. [DOI] [PubMed] [Google Scholar]
- Novoa I, Gallego J, Ferreira PG, Mendez R. 2010. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol 12: 447–456. [DOI] [PubMed] [Google Scholar]
- Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. 2008. A combinatorial code for CPE-mediated translational control. Cell 132: 434–448. [DOI] [PubMed] [Google Scholar]
- Resnick TD, Dej KJ, Xiang Y, Hawley RS, Ahn C, Orr-Weaver TL. 2009. Mutations in the chromosomal passenger complex and the condensin complex differentially affect synaptonemal complex disassembly and metaphase I configuration in Drosophila female meiosis. Genetics 181: 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD. 2007. CPEB: a life in translation. Trends Biochem Sci 32: 279–285. [DOI] [PubMed] [Google Scholar]
- Risso D, Ngai J, Speed TP, Dudoit S. 2014. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 32: 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles FJ, Lieberfarb ME, Wreden C, Gergen JP, Strickland S. 1994. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science 266: 1996–1999. [DOI] [PubMed] [Google Scholar]
- Salles FJ, Richards WG, Strickland S. 1999. Assaying the polyadenylation state of mRNAs. Methods 17: 38–45. [DOI] [PubMed] [Google Scholar]
- Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M. 1994. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev 8: 926–938. [DOI] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042. [DOI] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. 2007. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell 12: 143–155. [DOI] [PubMed] [Google Scholar]
- Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD. 2012. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell 47: 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy L, Orr-Weaver TL. 2007. The Drosophila PNG kinase complex regulates the translation of cyclin B. Dev Cell 12: 157–166. [DOI] [PubMed] [Google Scholar]
- Vardy L, Pesin JA, Orr-Weaver TL. 2009. Regulation of Cyclin A protein in meiosis and early embryogenesis. Proc Natl Acad Sci 106: 1838–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina JR, Orr-Weaver TL. 2011. Developmental control of oocyte maturation and egg activation in metazoan models. Cold Spring Harb Perspect Biol 3: a005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Keller W. 1996. The biochemistry of polyadenylation. Trends Biochem Sci 21: 247–250. [PubMed] [Google Scholar]
- Weil TT, Parton RM, Herpers B, Soetaert J, Veenendaal T, Xanthakis D, Dobbie IM, Halstead JM, Hayashi R, Rabouille C, et al. 2012. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat Cell Biol 14: 1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill L, Belloc E, Bava FA, Mendez R. 2012. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol 19: 577–585. [DOI] [PubMed] [Google Scholar]
- Whiteford N, Skelly T, Curtis C, Ritchie ME, Lohr A, Zaranek AW, Abnizova I, Brown C. 2009. Swift: primary data analysis for the Illumina Solexa sequencing platform. Bioinformatics 25: 2194–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Muller HA, Wodarz A, Ephrussi A. 2004. PKA-R1 spatially restricts Oskar expression for Drosophila embryonic patterning. Development 131: 1401–1410. [DOI] [PubMed] [Google Scholar]
- Zheng D, Tian B. 2014. Sizing up the poly(A) tail: insights from deep sequencing. Trends Biochem Sci 39: 255–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.