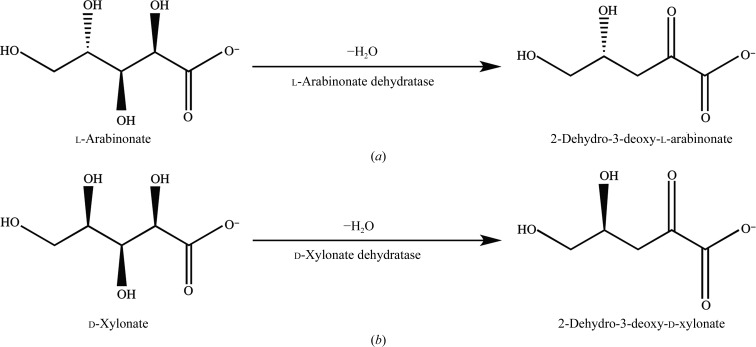
l-Arabinonate dehydratase and d-xylonate dehydratase from the IlvD/EDD family were crystallized by the vapour-diffusion method. Diffraction data sets were collected to resolutions of 2.40 and 2.66 Å from crystals of l-arabinonate dehydratase and d-xylonate dehydratase, respectively.
Keywords: l-arabinonate dehydratase, d-xylonate dehydratase, IlvD/EDD enzymes, [Fe–S] cluster, Rhizobium leguminosarum bv. trifolii, Caulobacter crescentus
Abstract
l-Arabinonate dehydratase (EC 4.2.1.25) and d-xylonate dehydratase (EC 4.2.1.82) are two enzymes that are involved in a nonphosphorylative oxidation pathway of pentose sugars. l-Arabinonate dehydratase converts l-arabinonate into 2-dehydro-3-deoxy-l-arabinonate, and d-xylonate dehydratase catalyzes the dehydration of d-xylonate to 2-dehydro-3-deoxy-d-xylonate. l-Arabinonate and d-xylonate dehydratases belong to the IlvD/EDD family, together with 6-phosphogluconate dehydratases and dihydroxyacid dehydratases. No crystal structure of any l-arabinonate or d-xylonate dehydratase is available in the PDB. In this study, recombinant l-arabinonate dehydratase from Rhizobium leguminosarum bv. trifolii (RlArDHT) and d-xylonate dehydratase from Caulobacter crescentus (CcXyDHT) were heterologously expressed in Escherichia coli and purified by the use of affinity chromatography followed by gel-filtration chromatography. The purified proteins were crystallized using the hanging-drop vapour-diffusion method at 293 K. Crystals of RlArDHT that diffracted to 2.40 Å resolution were obtained using sodium formate as a precipitating agent. They belonged to space group P21, with unit-cell parameters a = 106.07, b = 208.61, c = 147.09 Å, β = 90.43°. Eight RlArDHT molecules (two tetramers) in the asymmetric unit give a V M value of 3.2 Å3 Da−1 and a solvent content of 62%. Crystals of CcXyDHT that diffracted to 2.66 Å resolution were obtained using sodium formate and polyethylene glycol 3350. They belonged to space group C2, with unit-cell parameters a = 270.42, b = 236.13, c = 65.17 Å, β = 97.38°. Four CcXyDHT molecules (a tetramer) in the asymmetric unit give a V M value of 4.0 Å3 Da−1 and a solvent content of 69%.
1. Introduction
Lignocellulosic biomass is an important source of hexose and pentose sugars for the biorefinery industry. Apart from biofuels, the bioconversion of lignocellulosic hexose and pentose sugars can provide various platform chemicals which can be used as precursors for polymeric materials (Menon & Rao, 2012 ▸). The hemicellulose part of the lignocellulose biomass contains a significant amount of d-xylose and l-arabinose (Schädel et al., 2010 ▸). In nature, some archaea, bacteria and fungi are known to metabolize l-arabinose (Boguta et al., 2014 ▸; Brouns et al., 2006 ▸; Dien et al., 1996 ▸) and d-xylose (Okamoto et al., 2014 ▸; Gu et al., 2010 ▸) by using different nonphosphorylated oxidative pathways. In the Weimberg pathway, l-arabinose or d-xylose is converted to α-ketoglutarate, which is an intermediate metabolite in the tricarboxylic acid (TCA) cycle (Weimberg, 1961 ▸), or, in the Dahms pathway, the pentose sugars are converted to glycolaldehyde and pyruvate (Dahms, 1974 ▸). In both pathways, l-arabinonate dehydratase (EC 4.2.1.25) catalyzes the removal of one water molecule from l-arabinonate to produce 2-dehydro-3-deoxy-l-arabinonate (Fig. 1 ▸ a). Similarly, d-xylonate dehydratase (EC 4.2.1.82) catalyzes the removal of one water molecule from d-xylonate to produce 2-dehydro-3-deoxy-d-xylonate (Fig. 1 ▸ b) (Stephens et al., 2007 ▸; Dilworth et al., 1986 ▸; Weimberg, 1961 ▸). Several bacterial and yeast strains have been engineered to metabolize l-arabinose or d-xylose through the oxidative pathway and to produce biofuels, in addition to other value-added industrial chemicals (Kurosawa et al., 2015 ▸; Jin et al., 2014 ▸; Demeke et al., 2013 ▸; Xiao et al., 2012 ▸; Bettiga et al., 2009 ▸; Meijnen et al., 2009 ▸; Matsushika et al., 2009 ▸).
Figure 1.
Catalytic reactions of (a) RlArDHT and (b) CcXyDHT.
l-Arabinonate dehydratase from Rhizobium leguminosarum bv. trifolii and d-xylonate dehydratase from Caulobacter crescentus belong to the IlvD/EDD enzyme family. Enzymes in this family contain iron–sulfur [Fe–S] clusters as a prosthetic group (Andberg et al., 2016 ▸; Nunn et al., 2010 ▸; Stephens et al., 2007 ▸) and, using spectroscopic methods, active enzymes from this family have been found to contain either a [4Fe–4S] cluster (Watanabe et al., 2006 ▸; Rodriguez et al., 1996 ▸; Flint et al., 1993 ▸) or a [2Fe–2S] cluster (Flint & Emptage, 1988 ▸) in the active sites. There is only one crystal structure in the PDB from the IlvD/EDD family (PDB entry 2gp4; Southeast Collaboratory for Structural Genomics, unpublished work) and it lacks the [Fe–S] cluster. Therefore, structural studies of enzymes belonging to this family are needed in order to understand their structure–function relationship and the role of the [Fe–S] cluster. Here, we describe the overproduction, purification, crystallization and X-ray diffraction analysis of two enzymes from the IlvD/EDD family.
2. Materials and methods
2.1. Macromolecule production
The araD gene encoding R. leguminosarum l-arabinonate dehydratase (RlArDHT) and the xylD gene encoding C. crescentus d-xylonate dehydratase (CcXyDHT) were purchased as codon-optimized synthetic genes from GeneArt, Germany and a Strep-Tag II (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys) was added at the N-terminus (the deposited GenBank accession numbers for araD and xylD are KT260159 and KT260154, respectively). The genes were cloned into a pBAT4 expression vector, as described in Andberg et al. (2016 ▸). Other information on macromolecule production is given in Table 1 ▸.
Table 1. Macromolecule-production information.
| RlArDHT | CcXyDHT | |
|---|---|---|
| Source organism | R. leguminosarum bv. trifolii (GenBank accession No. KT260159) | C. crescentus (GenBank accession No. KT260154) |
| DNA source | Synthetically made | Synthetically made |
| Forward primer† | CACGTCCATGGACTGGTCTCATCCACAATTCGAGAAGAAAAAAAAAGCTGAATGGCCG | ACGTCCATGGACTGGTCTCATCCACAATTCGAGAAGTCTAATCGCACCCCGCGTC |
| Reverse primer‡ | GACGAATTCTCAATG | GTCGACGAATTCTCAATG |
| Cloning vector | pBAT4 | pBAT4 |
| Expression vector | pBAT4 | pBAT4 |
| Expression host | E. coli BL21(DE3) | E. coli BL21(DE3) |
| Complete amino-acid sequence of the construct produced§ | MDWSHPQFEKKKKAEWPRKLRSQEWYGGTSRDVIYHRGWLKNQGYPHDLFDGRPVIGILNTWSDMTPCNGHLRELAEKVKAGVWEAGGFPLEVPVFSASENTFRPTAMMYRNLAALAVEEAIRGQPMDGCVLLVGCDKTTPSLLMGAASCDLPSIVVTGGPMLNGYFRGERVGSGTHLWKFSEMVKAGEMTQAEFLEAEASMSRSSGTCNTMGTASTMASMAEALGMALSGNAAIPGVDSRRKVMAQLTGRRIVQMVKDDLKPSEIMTKQAFENAIRTNAAIGGSTNAVIHLLAIAGRVGIDLSLDDWDRCGRDVPTIVNLMPSGKYLMEEFFYAGGLPVVLKRLGEAGLLHKDALTVSGETVWDEVKDVVNWNEDVILPAEKALTSSGGIVVLRGNLAPKGAVLKPSAASPHLLVHKGRAVVFEDIDDYKAKINDDNLDIDENCIMVMKNCGPKGYPGMAEVGNMGLPPKVLKKGILDMVRISDARMSGTAYGTVVLHTSPEAAVGGPLAVVKNGDMIELDVPNRRLHLDISDEELARRLAEWQPNHDLPTSGYAFLHQQHVEGADTGADLDFLKGCRGNAVGKDSH | MDWSHPQFEKSNRTPRRFRSRDWFDNPDHIDMTALYLERFMNYGITPEELRSGKPIIGIAQTGSDISPCNRIHLDLVQRVRDGIRDAGGIPMEFPVHPIFENCRRPTAALDRNLSYLGLVETLHGYPIDAVVLTTGCDKTTPAGIMAATTVNIPAIVLSGGPMLDGWHENELVGSGTVIWRSRRKLAAGEITEEEFIDRAASSAPSAGHCNTMGTASTMNAVAEALGLSLTGCAAIPAPYRERGQMAYKTGQRIVDLAYDDVKPLDILTKQAFENAIALVAAAGGSTNAQPHIVAMARHAGVEITADDWRAAYDIPLIVNMQPAGKYLGERFHRAGGAPAVLWELLQQGRLHGDVLTVTGKTMSENLQGRETSDREVIFPYHEPLAEKAGFLVLKGNLFDFAIMKSSVIGEEFRKRYLSQPGQEGVFEARAIVFDGSDDYHKRINDPALEIDERCILVIRGAGPIGWPGSAEVVNMQPPDHLLKKGIMSLPTLGDGRQSGTADSPSILNASPESAIGGGLSWLRTGDTIRIDLNTGRCDALVDEATIAARKQDGIPAVPATMTPWQEIYRAHASQLDTGGVLEFAVKYQDLAAKLPRHNH |
The NcoI site is underlined.
The EcoRI site is underlined.
The Strep-Tag II is underlined.
For protein production, Escherichia coli BL21(DE3) expression cells were transformed with the recombinant plasmids. Large-scale protein production was performed in 2 l shake flasks containing 500 ml Luria–Bertani (LB) culture medium with 100 µg ml−1 ampicillin. The culture flasks were incubated at 303 K in a shaker incubator at 180 rev min−1. When the OD600 nm reached 0.5–0.6, protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, and cultivation was continued under identical conditions overnight (Andberg et al., 2016 ▸). Cells were harvested from the culture medium by centrifugation at 4000 rev min−1 at 277 K for 25 min. The cell pellets were suspended in an extraction buffer consisting of 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, protease inhibitor (EDTA-free, Roche) and 100 µg ml−1 lysozyme (Sigma–Aldrich, Germany). Intracellular proteins were isolated by centrifugation at 18 500 rev min−1, followed by a one-step freeze–thawing of the suspended cells at 193 K and sonication.
Crude protein samples were loaded onto a StrepTrap HP 5 ml column (GE Healthcare, Sweden) equilibrated with binding buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 5 mM MgCl2). Strep-Tag II-bound proteins were eluted with elution buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 5 mM MgCl2, 2.5 mM d-desthiobiotin). A final polishing step was conducted by gel-filtration chromatography on a Superdex 200 HR 10/30 column (GE Healthcare, Sweden) equilibrated with a buffer consisting of 50 mM Tris–HCl pH 7.5, 5 mM MgCl2. Purified samples were concentrated using an Amicon Ultra-4 centrifugal filter device with a molecular-weight cutoff of 30 kDa (Merck Millipore, Germany). The molecular weights and the purity of the enzymes were checked by SDS–PAGE with silver staining in a PhastSystem (GE Healthcare, Sweden). The enzyme activities were checked by the thiobarbituric acid (TBA) assay and the semicarbazide assay (Andberg et al., 2016 ▸).
2.2. Crystallization
Crystallization of the purified enzymes was performed by the hanging-drop vapour-diffusion method in a 24-well cell-culture plate (Greiner Bio-One, Germany) at 293 K. The following commercial crystallization kits were used: Crystal Screen, Crystal Screen 2, PEG/Ion, SaltRx 1, SaltRx 2, Index HT, PEGRx 1 and PEGRx 2 (Hampton Research). Droplets were prepared by mixing protein sample and crystallization reagent in a 1:1 ratio on a siliconized glass cover slide. The cover slides were then turned drop-side down against 500 µl crystallization reservoir reagent and sealed with grease to ensure that they were airtight. The plates were stored at 293 K. A microseeding technique (Bergfors, 2003 ▸) was applied to improve the quality of the CcXyDHT crystals. An initial condition that consisted of 3.6 M sodium formate, 5% PEG 3350 gave clusters of CcXyDHT crystals. A small cluster of crystals was removed in 4 µl crystallization reagent on a glass cover slide, crushed with a micro needle, diluted with 46 µl crystallization reagent to a final volume of 50 µl and sonicated for 3 min in a sonication bath (FinnSonic, Finland). A dog hair was dipped into the seed-stock solution and streaked through pre-equilibrated crystallization drops. Crystallization information is shown in Table 2 ▸.
Table 2. Crystallization.
| RlArDHT | CcXyDHT | |
|---|---|---|
| Method | Hanging-drop vapour diffusion | Hanging-drop vapour diffusion |
| Plate type | 24-well cell-culture plate | 24-well cell-culture plate |
| Temperature (K) | 293 | 293 |
| Protein concentration (mg ml−1) | 9.0 | 7.5 |
| Buffer composition of protein solution | 50 mM Tris–HCl pH 7.5, 5 mM MgCl2 | 50 mM Tris–HCl pH 7.5, 5 mM MgCl2 |
| Composition of reservoir solution | 4.0 M sodium formate, 0.1 M MES pH 7.0 | 3.8 M sodium formate pH 7.0, 0.1 M TES pH 7.0, 5 mM magnesium formate, 4% PEG 3350 |
| Volume and ratio of drop | 4 µl, 1:1 | 4 µl, 1:1 |
| Volume of reservoir (µl) | 500 | 500 |
2.3. Data collection and processing
RlArDHT crystals were mounted on a nylon loop and soaked in a cryoprotectant solution consisting of 4.0 M sodium formate, 0.1 M MES pH 7.0, 15% glycerol. CcXyDHT crystals were soaked with 3.9 M sodium formate pH 7.0, 0.1 M TES pH 7.0, 5 mM magnesium formate, 4% PEG 3350, 40 mM calcium d-xylonate. Data collection for RlArDHT crystals was carried out at 100 K on beamline ID14-2 at the ESRF in France at a wavelength of 0.97957 Å. Data from CcXyDHT crystals were collected remotely on beamline I04 at Diamond Light Source (DLS) in England at a wavelength of 0.97949 Å. Data processing was carried out by XDS (Kabsch, 2010 ▸). Detailed data-collection and processing statistics are shown in Table 3 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| RlArDHT | CcXyDHT | |
|---|---|---|
| Diffraction source | ID14-2, ESRF, France | I04, DLS, England |
| Wavelength (Å) | 0.97957 | 0.97949 |
| Temperature (K) | 100 | 100 |
| Detector | CCD | Pilatus 6M |
| Crystal-to-detector distance (mm) | 335.51 | 519.95 |
| Rotation range per image (°) | 0.25 | 0.5 |
| Total rotation range (°) | 180 | 180 |
| Exposure time per image (s) | 0.1 | 0.1 |
| Space group | P21 | C2 |
| a, b, c (Å) | 106.07, 208.61, 147.09 | 270.42, 236.13, 65.17 |
| α, β, γ (°) | 90, 90.43, 90 | 90, 97.38, 90 |
| Mosaicity (°) | 0.65 | 0.7 |
| Resolution range (Å) | 40.00–2.40 (2.50–2.40) | 59.69–2.66 (2.73–2.66) |
| Total No. of reflections | 779327 (88849) | 393801 (60419) |
| No. of unique reflections | 246940 (28307) | 114638 (17945) |
| Completeness (%) | 99.3 (99.0) | 99.0 (95.8) |
| Multiplicity | 3.2 (3.1) | 3.4 (3.4) |
| 〈I/σ(I)〉 | 12.3 (2.0) | 17.2 (2.1) |
| CC1/2 | 99.5 (57.8) | 99.9 (75.1) |
| R meas (%) | 10.8 (81.0) | 7.0 (82.1) |
| Overall B factor from Wilson plot (Å2) | 43 | 63 |
3. Results and discussion
Purified dehydratase enzymes contain an [Fe–S] cluster, which is necessary for enzyme activity (Andberg et al., 2016 ▸). Both the RlArDHT and the CcXyDHT protein solutions were dark brown in colour, which was a good indication that the [Fe–S] clusters had been maintained. In addition, the purified proteins were found to be active by an enzyme-activity assay as described previously (Andberg et al., 2016 ▸). Based on SDS–PAGE analysis (Supplementary Fig. S1), the molecular weights of the purified RlArDHT and CcXyDHT were approximately 64 kDa, and are in good correspondence with the theoretical molecular weights. The calculated molecular weights are 63.738 and 64.314 kDa (without the [Fe–S] cluster) for RlArDHT and CcXyDHT, respectively.
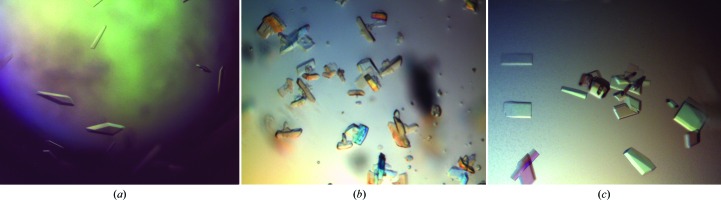
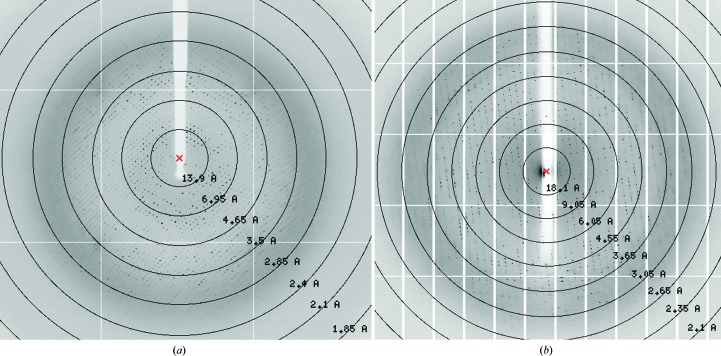
RlArDHT was crystallized in 4.0 M sodium formate, 0.1 M MES pH 7.0, which was a modification of condition No. 33 (4 M sodium formate) from Crystal Screen. Crystals appeared within two weeks (Fig. 2 ▸ a) and diffracted to 2.4 Å resolution (Fig. 3 ▸ a). The space group was P21, with unit-cell parameters a = 106.07, b = 208.61, c = 147.09 Å, β = 97.43°. Assuming eight molecules in the asymmetric unit, the calculated Matthews coefficient (V M) was 3.2 Å3 Da−1, which corresponds to a solvent content of 62% (Matthews, 1968 ▸).
Figure 2.
Crystals of (a) RlArDHT, (b) CcXyDHT before seeding and (c) CcXyDHT after seeding.
Figure 3.
Diffraction images of (a) RlArDHT and (b) CcXyDHT crystals. The black circles correspond to the resolution limit in Å.
In the initial crystallization screening of CcXyDHT, hexagonal bipyramidal crystals were obtained in various conditions. Most of the conditions contained salts, while others contained PEGs as a precipitating agent. However, the crystals showed only poor diffraction to 6 Å resolution. Additive and detergent screens were also tried, but no real improvement was observed. A modified condition that consisted of 3.6 M sodium formate pH 7.0, 5% PEG 3350 gave a cluster of bar-shaped crystals. Microseeding, together with further modification of the crystallization condition (3.8 M sodium formate pH 7.0, 0.1 M TES pH 7.0, 5 mM magnesium formate, 4% PEG 3350) gave crystals within four weeks (Fig. 2 ▸ b) which diffracted to 2.66 Å resolution (Fig. 3 ▸ b). The crystals belonged to space group C2, with unit-cell parameters a = 270.42, b = 236.13, c = 65.17 Å, β = 97.38°. Assuming four molecules in the asymmetric unit, the calculated Matthews coefficient (V M) was 4.0 Å3 Da−1, which corresponds to a solvent content of 69% (Matthews, 1968 ▸).
The structures of RlArDHT and CcXyDHT were both solved by molecular replacement using Phaser (McCoy et al., 2007 ▸). The structure of RlArDHT was solved using a modified model from PDB entry 2gp4 as a template. This resulted in a clear solution with two tetramers (Z-score = 12) in the asymmetric unit, with initial R and R free values of 37.7 and 46.7%, respectively. The structure of CcXyDHT was subsequently solved by molecular replacement using the coordinates of RlArDHT. A clear solution with a tetramer in the asymmetric unit (Z-score = 9) was found and resulted in initial R and R free values of 31.7 and 39.7%, respectively. Three-dimensional structure determination and structure refinement of both RlArDHT and CcXyDHT are ongoing.
Supplementary Material
Supplementary Figure S1.. DOI: 10.1107/S2053230X16010311/nj5256sup1.pdf
Acknowledgments
We would like to thank the ESRF and ID14-2 beamline staff for their assistance during data collection and the I04 beamline staff at DLS for the provision of remote data collection. We would also like to thank all members of the DNA Laboratory at the Biology Department of the University of Eastern Finland, Joensuu Campus for allowing us to use their protein-expression facilities. This project was supported by the Academy of Finland (decisions Nos. 256937 and 263931). The work at VTT was supported by the Finnish Centre of Excellence in the White Biotechnology–Green Chemistry programme (Academy of Finland, decision No. 118573).
References
- Andberg, M., Aro-Kärkkäinen, N., Carlson, P., Oja, M., Bozonnet, S., Toivari, M., Hakulinen, N., O’Donohue, M., Penttilä, M. & Koivula, A. (2016). Appl. Microbiol. Biotechnol., 10.1007/s00253-016-7530-8. [DOI] [PubMed]
- Bergfors, T. (2003). J. Struct. Biol. 142, 66–76. [DOI] [PubMed]
- Bettiga, M., Bengtsson, O., Hahn-Hägerdal, B. & Gorwa-Grauslund, M. F. (2009). Microb. Cell Fact. 8, 40. [DOI] [PMC free article] [PubMed]
- Boguta, A. M., Bringel, F., Martinussen, J. & Jensen, P. R. (2014). Microb. Cell Fact. 13, 97. [DOI] [PMC free article] [PubMed]
- Brouns, S. J. et al. (2006). J. Biol. Chem. 281, 27378–27388. [DOI] [PubMed]
- Dahms, A. S. (1974). Biochem. Biophys. Res. Commun. 60, 1433–1439. [DOI] [PubMed]
- Demeke, M. M., Dumortier, F., Li, Y., Broeckx, T., Foulquié-Moreno, M. R. & Thevelein, J. M. (2013). Biotechnol. Biofuels, 6, 120. [DOI] [PMC free article] [PubMed]
- Dien, B. S., Kurtzman, C. P., Saha, B. C. & Bothast, R. J. (1996). Appl. Biochem. Biotechnol. 57–58, 233–242. [PubMed]
- Dilworth, M. J., Arwas, R., McKay, I. A., Saroso, S. & Glenn, A. R. (1986). J. Gen. Microbiol. 132, 2733–2742.
- Flint, D. H. & Emptage, M. H. (1988). J. Biol. Chem. 263, 3558–3564. [PubMed]
- Flint, D. H., Emptage, M. H., Finnegan, M. G., Fu, W. & Johnson, M. K. (1993). J. Biol. Chem. 268, 14732–14742. [PubMed]
- Gu, Y., Ding, Y., Ren, C., Sun, Z., Rodionov, D. A., Zhang, W., Yang, S., Yang, C. & Jiang, W. (2010). BMC Genomics, 11, 255. [DOI] [PMC free article] [PubMed]
- Jin, L., Zhang, H., Chen, L., Yang, C., Yang, S., Jiang, W. & Gu, Y. (2014). J. Biotechnol. 173, 7–9. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kurosawa, K., Plassmeier, J., Kalinowski, J., Rückert, C. & Sinskey, A. J. (2015). Metab. Eng. 30, 89–95. [DOI] [PubMed]
- Matsushika, A., Inoue, H., Murakami, K., Takimura, O. & Sawayama, S. (2009). Bioresour. Technol. 100, 2392–2398. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Meijnen, J.-P., de Winde, J. H. & Ruijssenaars, H. J. (2009). Appl. Environ. Microbiol. 75, 2784–2791. [DOI] [PMC free article] [PubMed]
- Menon, V. & Rao, M. (2012). Prog. Energy Combust. Sci. 38, 522–550.
- Nunn, C. E., Johnsen, U., Schönheit, P., Fuhrer, T., Sauer, U., Hough, D. W. & Danson, M. J. (2010). J. Biol. Chem. 285, 33701–33709. [DOI] [PMC free article] [PubMed]
- Okamoto, K., Uchii, A., Kanawaku, R. & Yanase, H. (2014). Springerplus, 3, 121. [DOI] [PMC free article] [PubMed]
- Rodriguez, M., Wedd, A. G. & Scopes, R. K. (1996). Biochem. Mol. Biol. Int. 38, 783–789. [PubMed]
- Schädel, C., Blöchl, A., Richter, A. & Hoch, G. (2010). Plant Physiol. Biochem. 48, 1–8. [DOI] [PubMed]
- Stephens, C., Christen, B., Fuchs, T., Sundaram, V., Watanabe, K. & Jenal, U. (2007). J. Bacteriol. 189, 2181–2185. [DOI] [PMC free article] [PubMed]
- Watanabe, S., Shimada, N., Tajima, K., Kodaki, T. & Makino, K. (2006). J. Biol. Chem. 281, 33521–33536. [DOI] [PubMed]
- Weimberg, R. (1961). J. Biol. Chem. 236, 629–635. [PubMed]
- Xiao, H., Li, Z., Jiang, Y., Yang, Y., Jiang, W., Gu, Y. & Yang, S. (2012). Metab. Eng. 14, 569–578. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1.. DOI: 10.1107/S2053230X16010311/nj5256sup1.pdf