The monoclonal antibody UIC2 binds specifically to the extracellular region of the human P-glycoprotein in a conformationally sensitive manner. The crystal structure of the Fab fragment of UIC2 is reported.
Keywords: monoclonal antibodies, UIC2/Fab, multidrug resistance, human ABC-dependent transporter P-glycoprotein, multidrug resistance
Abstract
P-glycoprotein (P-gp) is a polyspecific ATP-dependent transporter linked to multidrug resistance in cancers that plays important roles in the pharmacokinetics of a large number of drugs. The drug-resistance phenotype of P-gp can be modulated by the monoclonal antibody UIC2, which specifically recognizes human P-gp in a conformation-dependent manner. Here, the purification, sequence determination and high-resolution structure of the Fab fragment of UIC2 (UIC2/Fab) are reported. Purified UIC2/Fab binds human P-gp with a 1:1 stoichiometry. Crystals of UIC2/Fab are triclinic (space group P1), with unit-cell parameters a = 40.67, b = 44.91, c = 58.09 Å, α = 97.62, β = 99.10, γ = 94.09°, and diffracted X-rays to 1.6 Å resolution. The structure was determined by molecular replacement and refined to 1.65 Å resolution. The asymmetric unit contains one molecule of UIC2/Fab, which exhibits a positively charged antigen-binding surface, suggesting that it might recognize an oppositely charged extracellular epitope of P-gp.
1. Introduction
Multidrug resistance (MDR) remains a significant problem in cancer chemotherapy (Gottesman et al., 2006 ▸) and in the treatment of microbial infections (Stavri et al., 2006 ▸). One mechanism of MDR is the overexpression of ATP-dependent efflux pumps, represented by ATP-binding cassette (ABC) transporters such as ABCB1, also known as P-glycoprotein (P-gp; Ambudkar et al., 1999 ▸). MDR conferred by the members of the family of ABC efflux transporters is achieved by pumping a wide range of structurally different drugs out of cancer cells, thus reducing their effective intracellular concentrations and cytotoxicity (Gottesman & Ling, 2006 ▸). The hope of circumventing MDR and improving the efficacy of cancer therapy has led to extensive investigations into the mechanisms of MDR and to a wide-ranging search for potential MDR-reversing agents.
One possible approach to overcome P-gp-mediated MDR is to develop monoclonal antibodies (mAb) targeting the efflux pumps directly. UIC2 (Mechetner & Roninson, 1992 ▸) is a mAb which recognizes extracellular epitopes of human P-gp in a conformation-dependent manner. Importantly, the treatment of MDR cells with UIC2 inhibits drug efflux, increases intracellular drug accumulation and enhances the cytotoxicity of anticancer drugs (Mechetner & Roninson, 1992 ▸). Furthermore, it has been observed that the relative amount of UIC2 bound to the surface of P-gp-expressing cells increases upon pre-treatment with P-gp substrates such as vinblastine, vincristine, taxol, actinomycin D, gramicidin D, cyclosporin A, reserpine and verapamil (Mechetner et al., 1997 ▸). Similarly, the reactivity of UIC2 towards P-gp also increases in the presence of ATP-depleting agents such as 2-deoxy-d-glucose, the ATP synthase inhibitor oligomycin A and the cytochrome c oxidase inhibitors sodium azide and potassium cyanide (Mechetner et al., 1997 ▸). Conversely, using permeabilized cells it has been shown that UIC2 reactivity decreases in the presence of ATP, ADP and their analogs (Druley et al., 2001 ▸).
Because of its sensitivity to the conformational states of P-gp, UIC2 has been widely used for mechanistic studies of P-gp. For example, using P-gp embedded in phospholipids in nanodiscs, it has been shown that UIC2 binding to P-gp uncouples ATP hydrolysis of P-gp from its drug efflux (Ritchie et al., 2011 ▸). Changes in the conformation of P-gp could be detected by mAb competition (Nagy et al., 2001 ▸; Goda et al., 2002 ▸) and thus can be employed for substrate classification (Nagy et al., 2004 ▸). Human P-gp is known for its substantial conformational flexibility (Chufan et al., 2015 ▸) and thus its structure determination remains difficult. In light of its conformation-dependent reactivity, UIC2 could be used to trap human P-gp in one conformation and the resulting complex may be suitable for crystallization. Towards this goal, we purified UIC2 from a hybridoma cell line, obtained its Fab fragment by papain digestion and determined the amino-acid sequence of the Fab. We show that purified UIC2/Fab is fully capable of interacting with P-gp in a concentration-dependent manner. UIC2/Fab was crystallized and its structure was determined to 1.65 Å resolution.
2. Materials and methods
2.1. Macromolecule production
2.1.1. Purification of the Fab of the UIC2 monoclonal antibody
The hybridoma (UIC2/A) producing the UIC2 monoclonal antibody was developed by Mechetner and Roninson from BALB/c mice immunized with BALB/c 3T3-1000 cells, as described previously (Mechetner & Roninson, 1992 ▸). UIC2 mAb from the medium of the hybridoma (obtained from E. B. Mechetner) was purified by Protein A antibody affinity chromatography. Fab fragments were isolated using a Fab Preparation Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to the instructions provided by the manufacturer. Briefly, the dissolved UIC2 antibody typically reached a concentration of 20–30 mg ml−1 and was transferred to digestion buffer containing cysteine using desalting columns. This solution was added to spin columns containing immobilized papain. After 4–6 h of incubation with mixing at 37°C, the digest of the UIC2 antibody containing Fab and Fc fragments was collected by centrifugation and applied onto a pre-equilibrated Protein A column with PBS. The flowthrough fraction containing the Fab was collected by centrifugation. These fractions were pooled and the Fab was concentrated to 10–15 mg ml−1.
2.1.2. Purification of human P-gp
The procedures for the purification of human P-gp from High-Five insect cells have been well documented and consist of the preparation of P-gp-enriched crude membranes (Sarkadi et al., 1992 ▸; Germann et al., 1990 ▸) followed by purification of the protein to homogeneity (Frank et al., 2016 ▸). Briefly, frozen cell pellet was thawed at room temperature and resuspended in homogenization buffer (50 mM Tris–HCl pH 7.5, 50 mM mannitol, 2 mM EGTA, 0.5% aprotinin, 2 mM DTT, 1 mM AEBSF). After swelling on ice for 30 min, the resuspended cells were homogenized by 20 strokes using a Dounce homogenizer. After a low-speed spin at 3500g for 5 min, the supernatant of the homogenate was centrifuged at 100 000g for 40 min. The pellet was resuspended in washing buffer (50 mM Tris–HCl pH 7.5, 300 mM mannitol, 1 mM EGTA, 0.5% aprotinin, 1 mM DTT, 1 mM AEBSF) using a syringe. The suspension was centrifuged again at 100 000g for a further 40 min. The pelleted membrane was resuspended in storage buffer (50 mM Tris–HCl pH 7.5, 300 mM mannitol, 1 mM EGTA, 0.5% aprotinin, 1 mM DTT, 1 mM AEBSF, 10% glycerol) and stored at −80°C. To purify human P-gp, the crude membrane was solubilized by the addition of DDM solution (10%) to a final concentration of 2% in 30 min. The solution was then centrifuged at 100 000g for 1 h and the supernatant was mixed with Ni–NTA resin pre-equilibrated in washing buffer (20 mM Tris–HCl pH 7.5, 75 mM NaCl, 15% glycerol, 2 mM β-mercaptoethanol, 0.0675% DDM, 0.04% sodium cholate, 30 mM imidazole). The resin was then washed with washing buffer (20 mM Tris–HCl pH 7.4, 75 mM NaCl, 15% glycerol, 5 mM β-mercaptoethanol, 0.0675% DDM, 0.04% sodium cholate, 30 mM imidazole) and incubated at 4°C by stirring at low speed. After 12 h incubation, the admixture of solubilized membrane and Ni–NTA resin was packed into a column and washed with the same buffer until no protein could be detected in the flowthrough. Human P-gp was eluted with washing buffer supplemented with 300 mM imidazole. The eluate was concentrated by centrifugation at 3000g to a final concentration of 15–20 mg ml−1 and centrifuged at 580 000g for 60 min. The resulting supernatant was applied onto a Superdex 200 column pre-equilibrated with buffer consisting of 20 mM Tris–HCl pH 7.5, 75 mM NaCl, 2% glycerol, 2 mM β-mercaptoethanol, 0.0675% DDM, 0.04% sodium cholate. The human P-gp protein from the pooled peak fractions was concentrated and stored at −80°C.
2.1.3. Blue native PAGE (BN-PAGE) analysis of the P-gp–Fab complex
Purified human P-gp dissolved in 20 mM Tris–HCl pH 7.5, 75 mM NaCl, 2% glycerol, 2 mM β-mercaptoethanol, 0.0675% DDM, 0.04% sodium cholate was mixed with UIC2/Fab in different molar ratios and incubated at room temperature for 30 min to allow formation of the complex. The complex was then subjected to BN-PAGE analysis following the procedure described previously (Ma & Xia, 2008 ▸).
2.1.4. Sequence confirmation of the complementarity-determining regions (CDRs) of UIC2/Fab
Total RNA from UIC2/A hybridoma cells (Mechetner & Roninson, 1992 ▸) resuspended in PBS with 1% aprotinin and 1 mM PMSF was isolated using an RNA-isolation kit from Qiagen, Valencia, California, USA. cDNA was prepared from total RNA using the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific, Waltham, Massachusetts, USA) as specified by the manufacturer’s protocol. The regions of interest in the heavy or light chain were amplified using the primers listed in Table 1 ▸. The resulting amplicons were subcloned in TOPO TA Cloning vector. The sequences of the amplicons were verified using primers 5–8 in Table 1 ▸.
Table 1. Primers used to determine the sequences of the CDRs.
| Primer | Chain | Covered region | Sequence | |
|---|---|---|---|---|
| 1 | Forward primer | H | 11–18 | 5′-TTA-GTG-AAG-CCT-GGA-AGG-GTC-CCT-3′ |
| 2 | Reverse primer | H | 115–122 | 5′-ATA-GAC-CGA-TGG-GGC-TGT-TGT-TTT-3′ |
| 3 | Forward primer | L | 2–6 | 5′-ATG-TTT-TGA-TGA-CCC-AAA-3′ |
| 4 | Reverse primer | L | 111–116 | 5′-ATG-GAT-ACA-GTT-GGT-GCA-GCA-T-3′ |
| 5 | Forward primer | M13 | 5′-GTA-AAA-CGA-CGG-CCA-G-3′ | |
| 6 | Reverse primer | M13 | 5′-CAG-GAA-ACA-GCT-ATG-AC-3′ | |
| 7 | Forward primer | T7 | 5′-ATT-AAC-CCT-CAC-TAA-AGG-GA-3′ | |
| 8 | Reverse primer | T7 | 5′-TAA-TAC-GAC-TCA-CTA-TAG-GG-3′ |
2.2. Crystallization
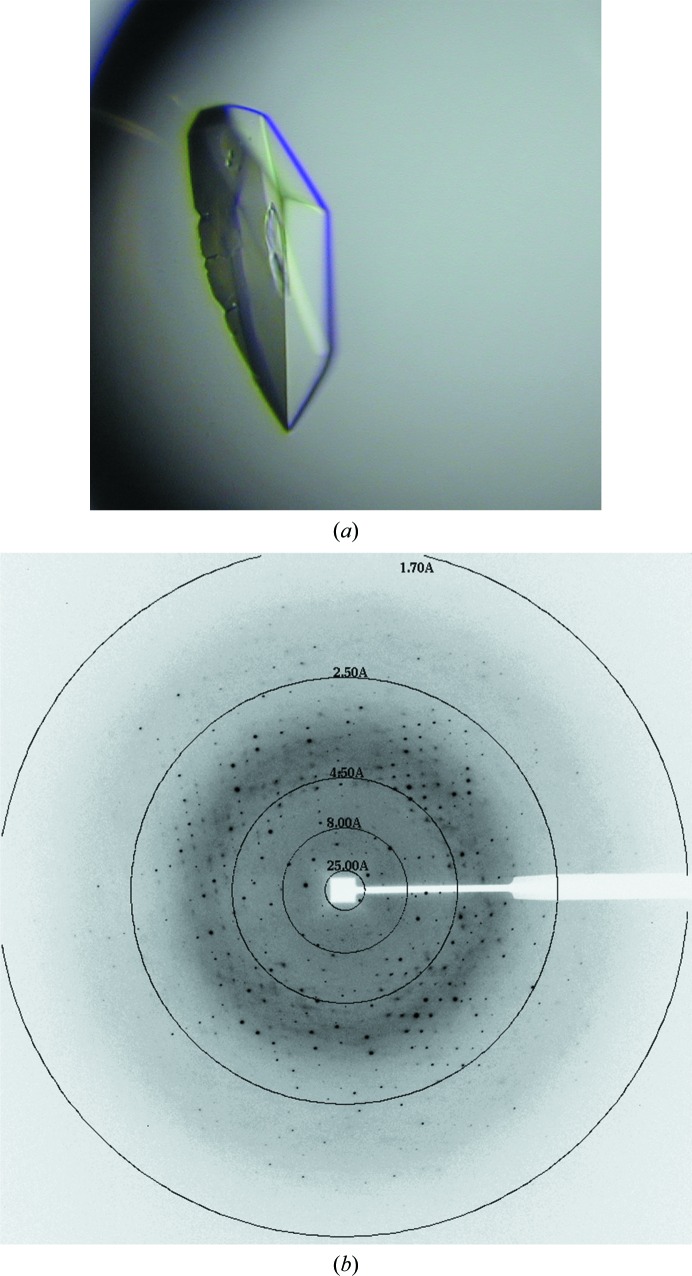
Crystallization conditions for UIC2/Fab were screened in a 96-well format using the Wizard HT (Emerald Bio, Bainbridge Island, Washington, USA) and PEGRx HT (Hampton Research, Aliso Viejo, California, USA) screens. Vapor-diffusion droplets were set up robotically by mixing 250 nl protein solution (10 mg ml−1 in PBS) and 250 nl precipitant solution using a Mosquito liquid dispenser (TTP Labtech, Melbourn, England). The Wizard screen produced crystals in drops A6 (0.1 M citrate pH 5.5, 20% PEG 3000), B9 (0.1 M HEPES pH 7.5, 20% PEG 8000) and D12 (0.1 M acetate pH 4.5, 2 M zinc acetate, 20% PEG 1000), and the PEGRx screen showed hits in drops B7 (0.1 M sodium acetate pH 4.5, 30% PEG 1500), B12 (0.1 M Tris pH 8.0, 30% PEG 2000 MME), C9 (0.1 M Tris pH 8.0, 30% PEG 2000 MME), C10 (0.1 M sodium acetate pH 4.5, 30% PEG 5000 MME) and C11 (0.1 M bis-tris pH 6.5, 8% PEG 5000 MME). The most easily reproducible condition turned out to be B12 in PEGRx. After optimization, crystals of UIC2/Fab grew under vapor-diffusion conditions at 21°C in 25% PEG 2000 MME buffered with 0.1 M Tris–HCl pH 7.0 (Fig. 1 ▸). Crystals were cryoprotected using reservoir solution augmented with 20% glycerol and were then flash-cooled in liquid propane. Crystallization information is given in Table 2 ▸.
Figure 1.
UIC2/Fab crystals and an X-ray diffraction image. (a) Crystal of UIC2/Fab of approximately 0.2 mm in size. (b) X-ray diffraction pattern of a UIC2/Fab crystal.
Table 2. Crystallization.
| Method | Vapor diffusion |
| Plate type | VDX |
| Temperature (K) | 294.15 |
| Protein concentration (mg ml−1) | 10 |
| Buffer composition of protein solution | PBS |
| Composition of reservoir solution | 100 mM Tris–HCl pH 7.0, 25% PEG 2000 MME |
| Volume and ratio of drop | 8 µl, 1:1 ratio |
| Volume of reservoir (ml) | 1 |
2.3. Data collection and processing
A cryoprotected crystal was mounted at 100 K on the goniometer of beamline 22BM (SER-CAT) at the Advanced Photon Source (APS), Argonne National Laboratory (ANL). A total of 360° of oscillation data with 0.5° oscillation per frame were collected on a MAR225 detector (Rayonix, Evanston, Illinois, USA) at a crystal-to-detector distance of 150 mm using X-rays at a wavelength of λ = 1.069 Å. Data reduction was performed in HKL-2000 (HKL X-ray, Charlottesville, Virginia, USA; Otwinowski & Minor, 1997 ▸). Molecular-replacement solutions were obtained using MOLREP (Vagin & Teplyakov, 2010 ▸) in the CCP4 package (Winn et al., 2011 ▸). Atomic positions, temperature factors and an automatically determined TLS model were refined in phenix.refine (v.2363; Adams et al., 2010 ▸). All manual modeling was performed with Coot (Emsley et al., 2010 ▸). The best crystals of UIC2/Fab diffracted X-rays to better than 1.6 Å Bragg spacing using synchrotron radiation (Fig. 1 ▸ and Table 3 ▸).
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | 22-BM, APS, ANL |
| Wavelength (Å) | 1.06882 |
| Temperature (K) | 100 |
| Detector | MAR225 CCD |
| Crystal-to-detector distance (mm) | 150.0 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 360 |
| Exposure time per image (s) | 2 |
| Space group | P1 |
| a, b, c (Å) | 40.67, 44.91, 58.09 |
| α, β, γ (°) | 97.62, 99.10, 94.09 |
| Mosaicity (°) | 0.37–0.73 |
| Resolution range (Å) | 50.0–1.65 |
| Total No. of reflections | 176673 |
| No. of unique reflections | 46314 |
| Completeness (%) | 95.1 (69.0†) |
| Multiplicity | 3.8 (2.9) |
| Anomalous completeness (28.37–4.5 Å) (%) | 98.1 |
| Anomalous CC (28.37–4.5 Å) | 0.04 |
| 〈I/σ(I)〉 | 13.1 (1.34‡) |
| R r.i.m. | 0.042 (0.663) |
| R p.i.m. | 0.024 (0.441) |
| Overall B factor from Wilson plot (Å2) | 27.73 |
The outer shell is 69% complete owing to the triclinic symmetry.
The crystal clearly diffracted to better than 1.6 Å resolution but the compromise of a cutoff at 1.65 Å was made taking the requirement of ∼70% completeness into account. At 1.71 Å resolution I/σ(I) falls below 2.22.
2.4. Structure solution and refinement
Since the structures of antibodies are well conserved, molecular-replacement phasing attempts succeeded with a number of available antibody structures from the Protein Data Bank (PDB). The best solution, however, was obtained using PDB entry 3hi5 (Zhang et al., 2009 ▸) as the search model. The structure was initially refined with all residues in the CDRs set to alanine residues (UIC2-tmp in Supplementary Fig. S1). H atoms were later included on riding positions. Water molecules were added using automated procedures and five glycerol molecules were also identified. The amino-terminal glutamine residue of the light chain showed some features that are reminiscent of pyroglutamic acid (PCA). Given the apparent low occupancy, a PCA residue was not modeled. All S atoms showed strong anomalous density, confirming that the sequence assignment was correct (Supplementary Fig. S2). It should be noted that the Cys–Cys bridges 134–194 and 140–195 in the light and heavy chains, respectively, showed multiple conformations owing to disorder or radiation damage (Supplementary Fig. S3). The final statistics for the refined structure are given in Table 3 ▸.
The light and heavy chains were numbered according to the Kabat scheme using abnum (http://www.bioinf.org.uk/abs/abnum). Initial assignments of the sequence of CDR residues were made based on the shape of the difference density and were later either confirmed or corrected according to the sequence obtained by sequencing the cDNA prepared from the UIC2-producing hybridoma (UIC2/A; Mechetner & Roninson, 1992 ▸) cells using light-chain or heavy-chain specific primers (Table 1 ▸). The resulting sequences matched the electron density for the CDRs in the structure very well (Fig. 3). However, it should be noted that in the constant region of mouse UIC2 the side chains of residues Phe82, Thr83, Phe84 and Thr108 of the heavy chain do not agree well with the weighted 2F o − F c electron-density map (Supplementary Fig. S4). Crystallography, even at 1.65 Å resolution, may not reveal the nature of residues unambigously, as is the case for Phe82 and Phe84. While Thr83 recovered towards the end of the refinement to appear as a threonine, the same cannot be said for Thr108, which still might be better interpreted as a serine as far as its surrounding electron density is concerned.
3. Results and discussion
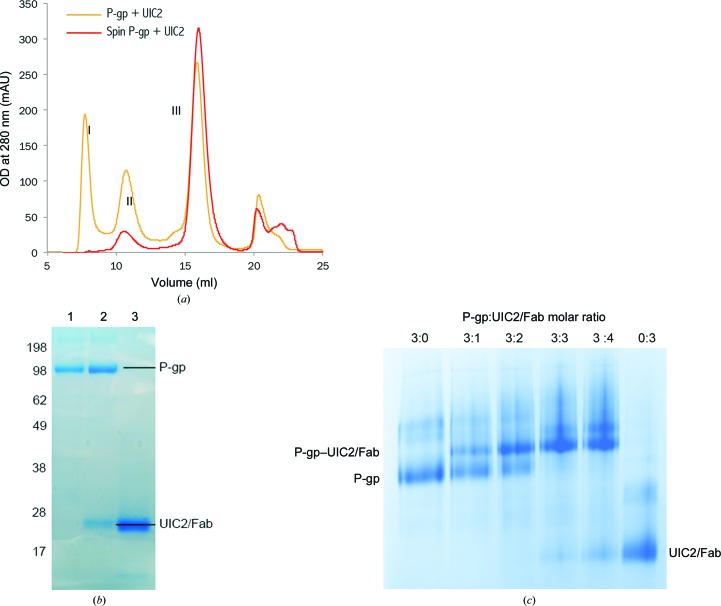
After treatment of the UIC2 mAb with immobilized papain protease, the reaction products were separated to produce the Fab fragment, with a final yield of UIC2/Fab of about 50%. The functional integrity of the Fab fragment was evaluated by its binding ability to purified human P-gp. When UIC2/Fab was incubated with isolated human P-gp in a molar ratio of 3:1 followed by size-exclusion chromatography (SEC), three peaks appeared (Fig. 2 ▸ a). The eluate under peak II is the P-gp–UIC2/Fab complex, which was confirmed by SDS–PAGE analysis (Fig. 2 ▸ b). Incubation of UIC2/Fab with P-gp in different molar ratios showed near-stoichiometric binding, as demonstrated by the dose-dependent shift of the P-gp band on blue native PAGE (Fig. 2 ▸ c), suggesting a fully functional UIC2/Fab preparation.
Figure 2.
Purification and characterization of UIC2 and its Fab fragment. (a) Size-exclusion chromatography of human P-gp in complex with UIC2/Fab. Two P-gp samples were used: one with a high-speed centrifugation (red line) and one without (orange line). Peak I is an aggregation peak, peak II is the P-gp–UIC2/Fab complex and peak III is UIC2/Fab. (b) SDS–PAGE gel showing purified human P-gp, the P-gp–UIC2/Fab complex [peak II in (a)] and UIC2/Fab alone. (c) Blue native PAGE showing the formation of human P-gp–UIC2/Fab complex at different molar ratios.
3.1. Structure of UIC2/Fab and its comparison to MRK16
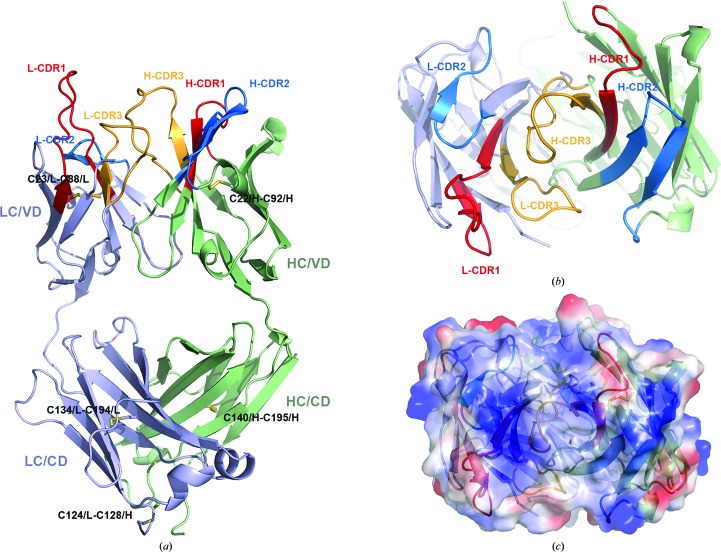
A representation of the entire structure of UIC2/Fab is shown in Figs. 3 ▸(a) and 3 ▸(b). In this structure 99% of the residues are clustered in the energetically favored regions, as demonstrated by the Ramachandran plot (Table 4 ▸). The structure of UIC2/Fab consists of a light chain (LC) and a heavy chain (HC); each chain has a variable domain (VD) and a constant domain (CD) with the typical IgG fold that is stabilized by a total of five pairs of disulfide bridges. Disulfide bridges are found in the VDs between the residues Cys23 and Cys88 of the LC, and Cys22 and Cys92 of the HC. Disulfide bonds are also found in the CDs between the residues Cys134 and Cys194 of the LC, and Cys140 and Cys195 of the HC. There is a disulfide bond crossing the CDs of the LC and HC between residues Cys124/L and Cys128/H.
Figure 3.
Structure of UIC2/Fab. (a) Cartoon representation of the structure of UIC2/Fab. The heavy chain (HC) is shown in green and the light chain (LC) in purple. The variable and constant domains are as labeled. The light and heavy chain CDR1s are colored red, CDR2s are blue and CDR3s are coral. (b) The structure of UIC2/Fab is viewed from the antigen side, showing the arrangement of CDRs. (c) Electrostatic potential surface showing the positive charge for the CDRs.
Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 28.37–1.65 (1.686–1.650) |
| Completeness (%) | 94.4 |
| No. of reflections, working set | 46002 (2124) |
| No. of reflections, test set | 2300 (112) |
| Final R cryst | 0.169 (0.297) |
| Final R free | 0.203 (0.305) |
| No. of non-H atoms | |
| Total | 3770 |
| Protein | 3430 |
| Ligand (glycerol) | 24 |
| Solvent | 316 |
| Total | 7097 |
| R.m.s. deviations | |
| Bonds (Å) | 0.01 |
| Angles (°) | 1.05 |
| Average B factors (Å2) | |
| Protein | 38.16 |
| Ligand (glycerol) | 49.53 |
| Solvent (water) | 41.12 |
| Ramachandran plot | |
| Most favored (%) | 99.55 |
| Allowed (%) | 0.45 |
The CDRs in the UIC2/Fab structure are well determined, with comparable B factors to the rest of the structure. Their rigidity is in part attributable to extensive crystal contacts. An electrostatic potential surface plot reveals that the CDRs have a positively charged surface (Fig. 3 ▸ c), with a concentration of positive potential from the CDR2s of both the LC and HC. Interestingly, another human P-gp-specific monoclonal antibody, MRK-16 (Vasudevan et al., 1998 ▸), that binds to the same general region also displays a highly positive electrostatic potential for the CDRs, suggesting that patches of negatively charged extracellular surface on P-gp may be important determinants for recognition by these monoclonal antibodies.
Supplementary Material
PDB reference: antibody fragment of UIC2, 5jue
Supporting Information.. DOI: 10.1107/S2053230X16009778/rl5121sup1.pdf
Acknowledgments
The authors wish to thank the staff of the SER-CAT beamline at APS, ANL for assistance in data collection. This study used the DNA Core of NCI and the high-performance Biowulf Linux cluster (http://hpc.nih.gov) at the NIH. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Ambudkar, S. V., Dey, S., Hrycyna, C. A., Ramachandra, M., Pastan, I. & Gottesman, M. M. (1999). Annu. Rev. Pharmacol. Toxicol. 39, 361–398. [DOI] [PubMed]
- Chufan, E. E., Sim, H.-M. & Ambudkar, S. V. (2015). Adv. Cancer Res. 125, 71–96. [DOI] [PMC free article] [PubMed]
- Druley, T. E., Stein, W. D. & Roninson, I. B. (2001). Biochemistry, 40, 4312–4322. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Frank, G. A., Shukla, S., Rao, P., Borgnia, M. J., Bartesaghi, A., Merk, A., Mobin, A., Esser, L., Earl, L. A., Gottesman, M. M., Xia, D., Ambudkar, S. V. & Subramaniam, S. (2016). Mol Pharmacol. 90, 35–41. [DOI] [PMC free article] [PubMed]
- Germann, U. A., Willingham, M. C., Pastan, I. & Gottesman, M. M. (1990). Biochemistry, 29, 2295–2303. [DOI] [PubMed]
- Goda, K., Nagy, H., Mechetner, E., Cianfriglia, M. & Szabó, G. Jr (2002). Eur. J. Biochem. 269, 2672–2677. [DOI] [PubMed]
- Gottesman, M. M. & Ling, V. (2006). FEBS Lett. 580, 998–1009. [DOI] [PubMed]
- Gottesman, M. M., Ludwig, J., Xia, D. & Szakács, G. (2006). Discov. Med. 6, 18–23. [PubMed]
- Ma, J. & Xia, D. (2008). J. Appl. Cryst. 41, 1150–1160. [DOI] [PMC free article] [PubMed]
- Mechetner, E. B. & Roninson, I. B. (1992). Proc. Natl Acad. Sci. USA, 89, 5824–5828. [DOI] [PMC free article] [PubMed]
- Mechetner, E. B., Schott, B., Morse, B. S., Stein, W. D., Druley, T., Davis, K. A., Tsuruo, T. & Roninson, I. B. (1997). Proc. Natl Acad. Sci. USA, 94, 12908–12913. [DOI] [PMC free article] [PubMed]
- Nagy, H., Goda, K., Arceci, R., Cianfriglia, M., Mechetner, E. & Szabó, G. Jr (2001). Eur. J. Biochem. 268, 2416–2420. [DOI] [PubMed]
- Nagy, H., Goda, K., Fenyvesi, F., Bacsó, Z., Szilasi, M., Kappelmayer, J., Lustyik, G., Cianfriglia, M. & Szabó, G. Jr (2004). Biochem. Biophys. Res. Commun. 315, 942–949. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Ritchie, T. K., Kwon, H. & Atkins, W. M. (2011). J. Biol. Chem. 286, 39489–39496. [DOI] [PMC free article] [PubMed]
- Sarkadi, B., Price, E. M., Boucher, R. C., Germann, U. A. & Scarborough, G. A. (1992). J. Biol. Chem. 267, 4854–4858. [PubMed]
- Stavri, M., Piddock, L. J. & Gibbons, S. (2006). J. Antimicrob. Chemother. 59, 1247–1260. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vasudevan, S., Tsuruo, T. & Rose, D. R. (1998). J. Biol. Chem. 273, 25413–25419. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zhang, H., Liu, J.-H, Yang, W., Springer, T., Shimaoka, M. & Wang, J.-H. (2009). Proc. Natl Acad. Sci. USA, 106, 18345–18350. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: antibody fragment of UIC2, 5jue
Supporting Information.. DOI: 10.1107/S2053230X16009778/rl5121sup1.pdf