Abstract
MicroRNAs (miRNAs) are small, non-coding single-stranded RNAs that suppress protein expression by binding to the 3′ untranslated regions of their target genes. Many studies have shown that miRNAs have important roles in congenital heart diseases (CHDs) by regulating gene expression and signaling pathways. We previously found that miR-30c was highly expressed in the heart tissues of aborted embryos with ventricular septal defects. Therefore, this study aimed to explore the effects of miR-30c in CHDs. miR-30c was overexpressed or knocked down in P19 cells, a myocardial cell model that is widely used to study cardiogenesis. We found that miR-30c overexpression not only increased cell proliferation by promoting cell entry into S phase but also suppressed cell apoptosis. In addition, we found that miR-30c inhibited dimethyl sulfoxide-induced differentiation of P19 cells. miR-30c knockdown, in contrast, inhibited cell proliferation and increased apoptosis and differentiation. The Sonic hedgehog (Shh) signaling pathway is essential for normal embryonic development. Western blotting and luciferase assays revealed that Gli2, a transcriptional factor that has essential roles in the Shh signaling pathway, was a potential target gene of miR-30c. Ptch1, another important player in the Shh signaling pathway and a transcriptional target of Gli2, was downregulated by miR-30c overexpression and upregulated by miR-30c knockdown. Collectively, our study revealed that miR-30c suppressed P19 cell differentiation by inhibiting the Shh signaling pathway and altered the balance between cell proliferation and apoptosis, which may result in embryonic cardiac malfunctions.
Introduction
MicroRNAs (miRNAs) are small, non-coding single-stranded RNAs that are ubiquitously expressed in plants, nematodes and human cells. miRNAs bind to the 3′ untranslated region (UTR) of their target genes, forming an RNA-induced silencing complex that mediates degradation of the target gene mRNA or inhibits translation of the target proteins.1, 2 miRNAs are involved in development, apoptosis, differentiation, hormone secretion and various physiological processes.3, 4, 5 miRNAs have critical roles in cardiovascular development, and their expression profiles change with different pathological conditions. For example, miR-29 is involved in cardiac fibrosis; miR-145 and miR-92 regulate cardiac angiogenesis; miR-30 has a role in cardiac apoptosis; and miR-26 is affected by modified ionic channel function.6 Studies have indicated that miRNAs have important roles in cardiogenesis and congenital heart diseases (CHDs).7, 8 The heart is known to be the earliest functional organ formed in the process of embryonic development. Heart development is spatiotemporally regulated, which includes accurate control of gene expression and signaling pathways, such as the Wnt signaling pathway, the Sonic hedgehog (Shh) signaling pathway, and a series of important morphological changes.9, 10 Understanding the molecular mechanisms involved in CHDs is crucial for developing new therapeutic interventions.
Previously, our microarray data showed that miR-30c was highly expressed in the heart tissues of aborted embryos with ventricular septal defects, but the role of miR-30c in heart development is not known (data not shown). miR-30c belongs to the miR-30 family, which is evolutionarily conserved in different species (Table 1). Modulating the expression of miR-30b and miR-30c may affect vascular calcification.11 Cardiomyocyte-specific miR-30c overexpression caused dilated cardiomyopathy.12 In addition, miR-30c was reported to be an independent predictor of a good response to tamoxifen therapy in advanced breast cancer patients, and the miR-30c/VIM/TWF1 signaling cascade has also been associated with clinical outcome in breast cancer patients.13, 14, 15 miR-30c negatively regulated REDD1 expression in human hematopoietic and osteoblast cells after gamma-irradiation.16 In this study, we investigated miR-30c involvement in cardiac malformations.
Table 1. Mature sequences of miR-30c from different species.
| Species | Sequences |
|---|---|
| Dre | UGUAAACAUCCUACACUCUCAG |
| Ssc | UGUAAACAUCCUACACUCUCAGC |
| Ptr | UGUAAACAUCCUACACUCUCAGC |
| Mmu | UGUAAACAUCCUACACUCUCAGC |
| Hsa | UGUAAACAUCCUACACUCUCAGC |
| Rno | UGUAAACAUCCUACACUCUCAGC |
| Gga | UGUAAACAUCCUACACUCUCAGCU |
| Lla | UGUAAACAUCCUACACUCUCAGC |
Abbreviations: Dre, Danio rerio; Gga, Gallus gallus; Hsa, Homo sapiens; Lla, Lagothrix lagotricha; Mmu, Mus musculus; Ptr, Pan troglodytes; Rno, Rattus norvegicus; Ssc, Sus scrofa.
The Shh signaling pathway is known to regulate specification, patterning and growth of multiple embryonic organs, including cardiac development.17 Shh initiates signaling by binding to its receptor Ptch (Patched), resulting in Smo (Smoothened) dissociation from Gli proteins and activation of the Gli proteins. Gli proteins are transcriptional factors that control expression of Shh target genes. The Shh pathway has a critical role during development in a time- and position-dependent manner by regulating patterning and maintenance of proliferative niches. In animals, loss of Shh leads to several cardiac abnormalities, including ventricular hypoplasia,18 septation defects19 and outflow tract shortening.20 It is a key signal in determining left–right asymmetry, including positioning the heart on the left side of the body.21 We confirmed that Gli2, one of the primary transcriptional factors in the Shh signaling pathway, is a potential target gene of miR-30c.22 Shh signaling via Gli1/2 was considered to be sufficient to initiate the expression of cardiac muscle transcription factors, leading to cardiomyogenesis in aggregated P19 cells.23 However, how miR-30c affects the cardiac lineage commitment and differentiation through Shh signaling remains elusive.
P19 cells, also called carcinoma stem cells, are able to differentiate into embryonic myocardial cells when exposed to dimethyl sulfoxide (DMSO).24 Therefore, we used this cell line to explore the possible molecular mechanisms of heart development by detecting the cardiac-specific transcription factors and upstream signaling pathways during cardiac cell differentiation.25, 26
In this study, we examined the underlying mechanisms of heart development by analyzing proliferation, apoptosis and differentiation in response to miR-30c overexpression or knockdown in P19 cells. Our findings suggest that miR-30c has an important role in the regulation of cardiac cell proliferation and/or cell survival and provide further insight into heart development.
Materials and methods
Plasmid constructs
The mature sequence of mouse mmu-miR-30c (5′-TGTAAACATCCTACACTCTCAGC-3′) was annealed and inserted into the EcoRI/AgeI sites of the pPG/miR/eGFP/blasticidin vector (GenePharma, Shanghai, China) using the following primers: sense, 5′-AATTCGTGTAAACATCCTACACTCTCAGCGTTTTGGCCACTGACTGACGCTGAGAGTAGGATGTTTACACA-3′ and antisense, 5′-CCGGTGTGTAAACATCCTACTCTCAGCGTCAGTCAGTGGCCAAAACGCTGAGAGTGTAGGATGTTTACACG-3′.
To knock down mmu-miR-30c, a miRNA sponge was used to suppress endogenous expression of miR-30c. The complementary binding sites (5′-GCTGAGAGTGTAGGATGTTTACA-3′) were inserted into the same vector.
The 3′ UTR or mutant 3′ UTR regions of Gli2 were PCR-amplified and inserted into the XhoI/NotI-digested vector psiCHECK-2 (GenePharma) containing firefly and Renilla luciferase reporter genes. The mutant sites are shown in bold in the following sequences: wild type 3′ UTR region of Gli2: 5′-GGGTCTCTCCTGGCCTGTTTACA-3′ mutant 3′ UTR region of Gli2: 5′-GGGTCTCTCCTGGCCACAAAACA-3′. To validate the efficiency of the miRNA sponge, the miR-30c sponge sequence was inserted into the psiCHECK-2 vector. To confirm that miR-30c overexpression or knockdown by the miRNA sponge was functional, the wild type or mutant 3′ UTR of twinfilin1 (Twf1), a previously identified target gene of miR-30c, was inserted into the psiCHECK-2 vector as described.27
Cell culture, miRNA transfection and establishment of stable cell lines
P19 cells obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) were cultured in modified Eagle's medium (α-MEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 mg ml−1 streptomycin, and 100 U ml−1 penicillin in a 5% CO2 incubator at 37 °C. A miR-30c mimic or inhibitor (Ambion, Foster City, CA, USA) was transiently transfected into P19 cells using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. To establish stable cell lines, the plasmids were transfected into P19 cells using Lipofectamine 2000. The cells were then selected with 60 μg ml−1 of blasticidin for 2 weeks (Invitrogen). The establishment of miR-30c-overexpressing cells was confirmed by quantitative real-time RT-PCR (qRT-PCR) (Table 2).
Table 2. TaqMan assay IDs of genes used in the TaqMan qPCR.
| Gene | TaqMan assay ID |
|---|---|
| mmu-miR-30c | 001014 |
| U6 | 001973 |
| cTnT | Mm01290256_m1 |
| Nkx2.5 | Mm01309813_s1 |
| Gata4 | Mm00484689_m1 |
| Gli2 | Mm01293111_m1 |
| Ptch | Mm00436026_m1 |
| β-actin | Mm00607939_s1 |
Luciferase assay
Reporter plasmids were transfected into the P19 cells stably overexpressing miR-30c or the P19 miR-30c knockdown cells. The Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) was used to analyze the firefly and Renilla luciferase activities according to the manufacturer's protocols.
Induction of cell differentiation
Cell differentiation was carried out as previously described.28 Briefly, cells were cultured in α-MEM medium supplemented with 10% FBS, 100 U ml−1 penicillin, 100 mg ml−1 streptomycin and 1% DMSO(Sigma, St Louis, MO, USA) for 4 days. The cells formed embryoid bodies, which were then transferred and cultured in 6 cm dishes with complete medium for an additional 8 days. Cells were harvested on differentiation days 0, 4, 8, 10 and 12. Morphological changes were photographed using a phase-contrast inverted microscope (Nikon Eclipse TE300, Tokyo, Japan) equipped with a digital camera (Nikon, E4500, Japan). RT-PCR was used to determine the expression of cardiac troponin T (cTnT), Gata4 and Nkx2.5 during differentiation.
Cell proliferation assays: CCK-8 assay, BrdU cell proliferation assay and flow cytometry
To measure cell growth, stable cell lines were cultured in 96-well plates for seven consecutive days. A Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) was used to measure cell growth at the indicated time point according to the manufacturer's instructions.
Stable cell lines were seeded in 96-well plates for 5 days. Cell proliferation was determined using a BrdU Cell Proliferation ELISA Kit (Abcam, Cambridge, MA, USA) according to the manufacturer's instructions.
To analyze the cell cycle, stable cell lines were synchronized by serum deprivation for 24 h, followed by replacement with complete medium for the indicated times (0, 8, 16, 24 and 32 h). The cells were harvested using trypsin/EDTA, washed twice with PBS, and fixed in 70% ethanol at −20 °C overnight. Then, the cells were stained with propidium iodide (PI) solution and analyzed using a BD FACS can flow cytometer and Cell Quest software (BD Biosciences, San Jose, CA, USA).
Apoptosis assays: Hoechst 33258 staining, electron microscopy, measurement of caspase-3 activity and flow cytometry
Cells were cultured in serum-deprived α-MEM for 24 h to induce apoptosis. For Hoechst staining, cells were first washed with PBS for 3 min and then stained with Hoechst 33258 (Beyotime, Nantong, China) for 5 min. Digital images were taken using a SPOT camera system (Nikon, E4500, Japan).
For electron microscopy analysis, cells were harvested using trypsin/EDTA, washed with PBS, and fixed with 2.5% glutaraldehyde for 2 h. The cells were then dehydrated with an ethanol gradient for 10 min each time and coated with gold (15 min). Images were acquired using a scanning electron microscope system (SEM; Supra 55, Carl Zeiss).
A caspase-3 colorimetric assay kit (KeyGen, Nanjing, China) was used to determine caspase-3 activity. Cells were collected, washed with PBS, lysed on ice for 1 h, vortexed for 10 s every 20 min and then centrifuged for 1 min at 10 000 g at 4 °C. Aliquots of the supernatant were mixed with 5 μl of substrate and incubated at 37 °C for 4 h in dark. The absorbance values were measured at 400–405 nm.
For analysis of apoptosis by flow cytometry, cells were harvested, washed and resuspended in 1 ml binding buffer, followed by staining with annexin V-fluorescein isothiocyanate (V-FITC) and PI at room temperature for 15 min (BioVision, CA). Flow cytometry was used to analyze the FITC and PI fluorescence signals.
Quantitative real-time RT-PCR
To determine miRNA levels, miRNAs were exacted using a miRVana PARIS kit (Ambion, Foster City, CA, USA) according to the manufacturer's protocols. RNA concentration was determined with a NanoDrop 2000 spectrophotometer (NanoDrop Tech., Rockland, DE, USA). A TaqMan MicroRNA Reverse Transcription Kit and miRNA-specific stem-loop primers (Life Technologies, Carlsbad, CA, USA) were then used to carry out reverse transcription of the total RNA into cDNA. U6 levels were used as an internal control. Real-time PCR (TaqMan) was conducted in 96-well plates on an ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). The TaqMan probe IDs of miR-30c and U6 are shown in Table 2.
To determine gene expression levels, total RNA was extracted from P19 cells using TRIzol reagent (Invitrogen). cDNA was reverse transcribed using a High Capacity cDNA Reverse Transcription Kit (Life Technologies). qPCR (TaqMan) was initiated at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. β-actin was used as an internal control. The TaqMan probe IDs of all genes are listed in Table 2.
Western blotting
Antibodies against Gli2, Ptch1 and β-actin were purchased from Abcam. Cells were lysed using a Total Protein Extraction Kit (KeyGen) on ice for 30 min. The supernatant was collected by centrifuging at 14 000 g for 15 min at 4 °C. Protein concentrations were determined using a BCA protein assay kit (KeyGen). Protein samples were subjected to western blotting and probed using antibodies against Gli2, Ptch1 or β-actin.
Immunofluorescence analysis
Cells were cultured on sterile cover slides overnight, fixed with methanol for 30 min, blocked and incubated with an antibody against Gli2, cTnT or Gata4 (Abcam) overnight. Cells were then stained with FITC-conjugated secondary antibody. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole, Sigma). Images were collected using a Zeiss Axioplan-2 epifluorescence microscope.
Statistical analysis
Experiments were performed three times. All data are shown as the mean±s.d. Statistical analysis was performed using an unpaired Student's t-test with SPSS software version 13.0 (SPSS, Inc., Chicago, IL, USA). P-values <0.05 were considered statistically significant.
Results
Establishment of stable cell lines
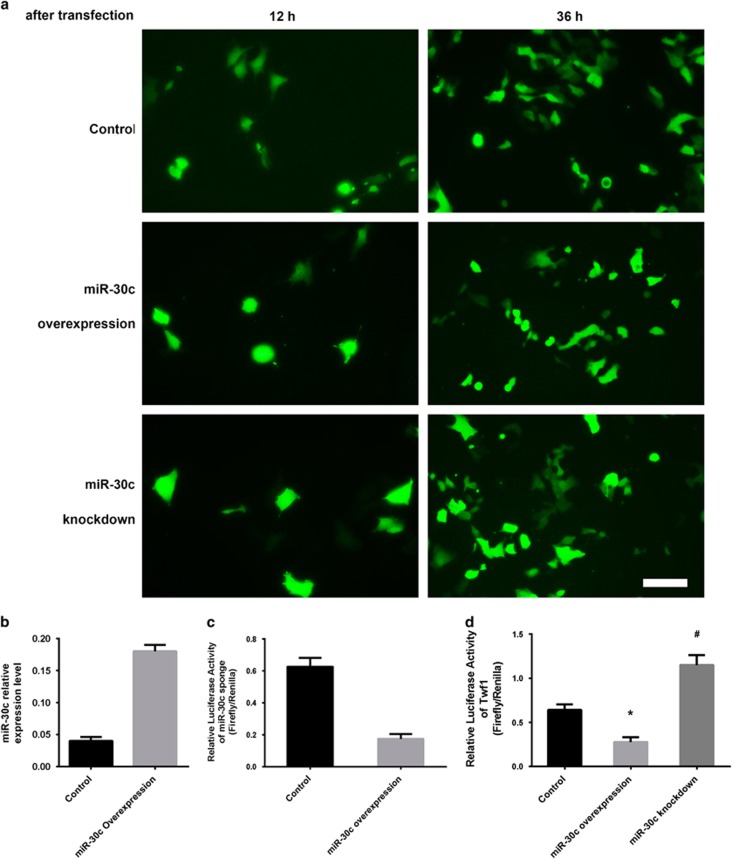
Plasmids, including the overexpression vector, the knockdown vector of mmu-miR-30c and the empty vector, were transiently transfected into P19 cells. The transfection efficiencies were similar between different groups as indicated by GFP expression (Figure 1a). The stable cell lines were selected by blasticidin. Western blotting showed that GFP was stably expressed after DMSO treatment for 10 days, indicating that the stable cell lines were successfully established (Supplementary Figure 1). The stable miR-30c-overexpressing P19 cell line was confirmed by qPCR (Figure 1b). To validate the efficiency of the miR-30c sponge, we co-transfected a luciferase reporter containing the miR-30c sponge sequence and a miR-30c-overexpressing plasmid into P19 cells. A luciferase assay showed that the luciferase activity was significantly suppressed by miR-30c, indicating that the miR-30c sponge was effective (Figure 1c). To confirm that the miR-30c overexpression or knockdown in P19 cells was functional, we transfected a luciferase reporter containing a wild type or mutant 3′ UTR of Twf1, a previously identified target gene of miR-30c,27 into stable miR-30c-overexpressing or miR-30c knockdown P19 cells. Luciferase assays showed that miR-30c overexpression significantly reduced the luciferase activity, while miR-30c knockdown increased it (Figure 1d).
Figure 1.
Establishment of stable miR-30c-overexpressing or silenced P19 cell lines. (a) GFP expression was observed by fluorescence microscopy after transfection of the miR-30c overexpression or knockdown vector or a control vector into the P19 cells at the indicated time points. Scale bar, 50 μm. (b) qRT-PCR analysis of miR-30c expression in P19 cells after blasticidin selection. The data are presented as the mean±s.d. of three experiments. (c) Luciferase assay of the miR-30c sponge efficiency. The data are presented as the mean±s.d. of three experiments. (d) Luciferase assay of miR-30c with a Twf1 3′ UTR. Luciferase reporters containing a wild type (WT) or mutant (mut) 3′ UTR of Twf1 were transfected into P19 stable cell lines. The data are presented as the mean±s.d. of three experiments. (*P<0.05 comparing miR-30c overexpression with control cells; #P<0.05 comparing miR-30c knockdown with control cells).
Effects of miR-30c on P19 cell proliferation
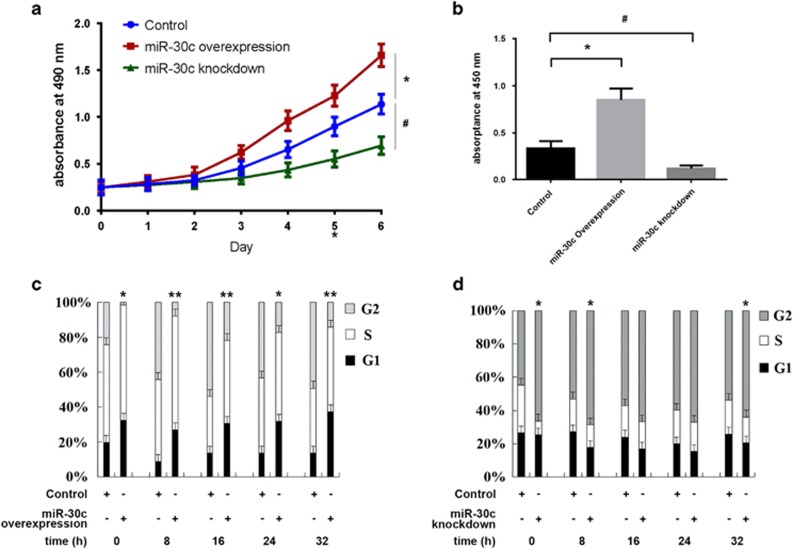
The CCK-8 assay was used to determine the effect of miR-30c overexpression or knockdown on the growth of P19 cells. As shown in Figure 2a, miR-30c-overexpressing P19 cells grew much faster than control cells, while miR-30c knockdown cells grew much more slowly (P<0.05). BrdU ELISA analysis showed that miR-30c overexpression increased P19 cell proliferation, while miR-30c knockdown decreased it (Figure 2b). In addition, miR-30c had similar effects on the cell cycle. Flow cytometry of the cell cycle distribution showed a significantly higher percentage of miR-30c-overexpressing cells in S phase (Figures 2c, *P<0.05, **P<0.001), while miR-30c knockdown cells showed a significantly lower percentage of cells in S phase (Figures 2d, *P<0.05). In addition to stably expressing miR-30c in cells, we also transiently transfected P19 cells with a miR-30c mimic or inhibitor. The transiently expressed miR-30c mimic increased, while the miR-30c inhibitor suppressed, cell growth (Supplementary Figure 2A, *,#P<0.05) and proliferation (Supplementary Figure 2B, *,#P<0.05), which was consistent with the results from the stable cell lines.
Figure 2.
Effects of miR-30c on cell proliferation. (a) Cell growth was measured for seven consecutive days using a CCK-8 assay kit. (b) BrdU ELISA analysis for measuring cell proliferation after cells were cultured for 5 days. (c, d) Flow cytometry analysis of the cell cycle of miR-30c-overexpressing (c) or miR-30c knockdown (d) cells. The data are presented as the mean±s.d. of three experiments (*P<0.05; **P<0.01 comparing miR-30c overexpression with control cells; #P<0.05 comparing miR-30c knockdown with control cells).
Effects of miR-30c on P19 cell apoptosis
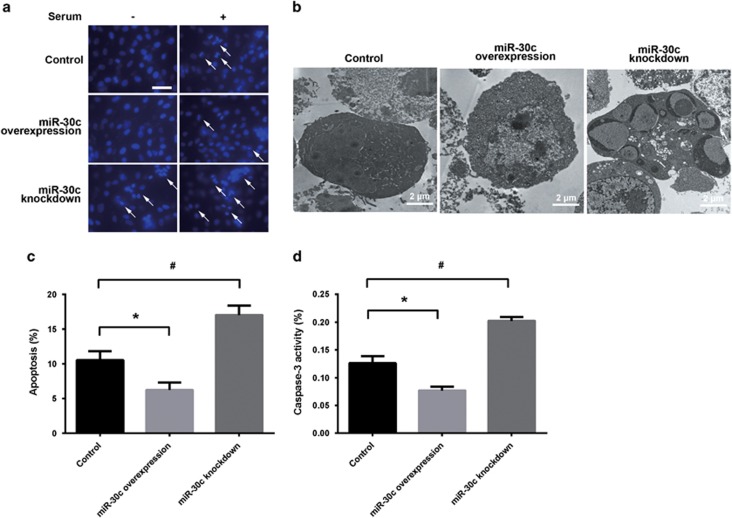
Hoechst staining is a classic, quick and easy method used to detect apoptosis. The chromatin in apoptotic cells is condensed, and the nuclei from apoptotic cells are dense or fragmented white spots under a fluorescence microscope after Hoechst staining, while the nuclei of normal cells appear blue. As shown in Figure 3a, miR-30c overexpression significantly suppressed serum starvation-induced apoptosis, while miR-30c knockdown increased cell apoptosis. Electron microscopy, which is a powerful method for morphologically observing differentiating nuclei and organelles, was used to confirm apoptosis. As shown in Figure 3b, there were few typical morphological changes in the miR-30c overexpression group, but obvious apoptotic morphological changes were observed in the miR-30c knockdown group after serum starvation for 24 h. Annexin V-FITC, which binds to phosphatidylserine, was used to detect apoptosis. Cells were induced to undergo apoptosis by serum starvation for 24 h. Overexpression of miR-30c reduced the percentage of apoptotic cells, while knockdown of miR-30c increased it in response to serum deprivation (Figures 3c, *,#P<0.05). The same results were observed in P19 cells transiently expressing either a miR-30c mimic or inhibitor (Supplementary Figure 2C, *,#P<0.05). In addition, caspase-3 activity assays showed that miR-30c overexpression suppressed cell apoptosis, while miR-30c knockdown increased the number of apoptotic cells in response to serum starvation (Figure 3d, *,#P<0.05). Collectively, these results indicated that miR-30c protected P19 cells from serum deprivation-induced apoptosis.
Figure 3.
Effects of miR-30c on cell apoptosis. (a) Hoechst staining of cell nuclei. White arrow indicates fragmented nucleus. Scale bar, 50 μm. (b) Electron microscopy analysis of apoptosis in miR-30c overexpression or knockdown cells. (c) Flow cytometry analysis of cell apoptosis. (d) Caspase-3 activity assay of cell apoptosis. The data are presented as the mean±s.d. of three experiments (*P<0.05 comparing miR-30c overexpression with control cells; #P<0.05 comparing miR-30c knockdown with control cells).
Effects of miR-30c on the differentiation of P19 cells
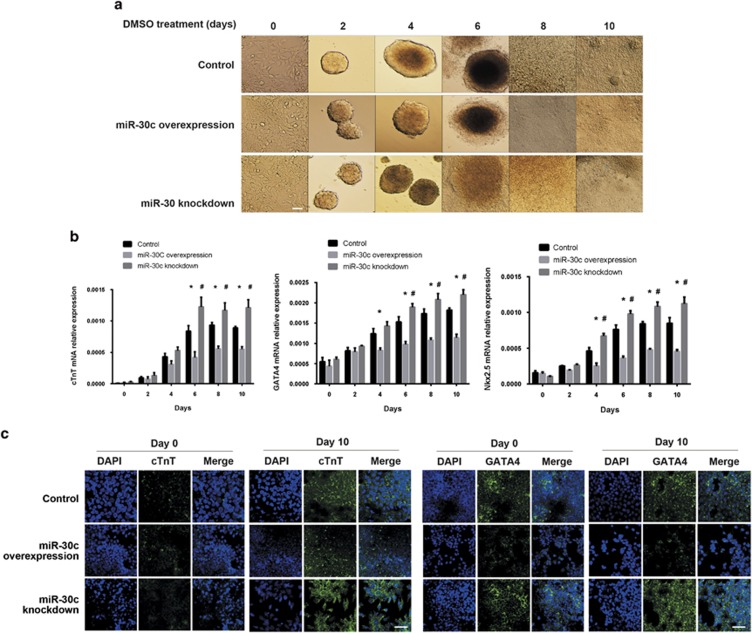
We observed morphological changes during DMSO-induced differentiation of the P19 cells into myocardial cells and quantified specific myocardial markers (cTnT, Gata4 and Nkx2.5) in the miR-30c overexpression or knockdown cell lines. Figure 4a shows the cell morphology changes from day 0 to day 10. Compared with the control groups, there were fewer beating cell clusters in the miR-30c overexpression group, while there were many more beating cell clusters in the miR-30c knockdown group. In addition, we used qRT-PCR to analyze the relative RNA expression levels of the myocardial markers cTnT, Gata4 and Nkx2.5 on days 0, 2, 4, 6, 8 and 10 after DMSO treatment. The expression of these markers is known to be upregulated during differentiation of mouse P19 cells into myocardial cells (Figure 4b, *,#P<0.05). Expression of all the marker genes gradually increased during the process of differentiation. Compared to the controls, the expression levels of these marker genes were globally much lower in miR-30c-overexpressing cells, and they were much higher in the miR-30c knockdown cells. Furthermore, an immunofluorescence assay was performed to show the expression level of cTnT and Gata4 in response to DMSO treatment. As shown in Figure 4c, the expressions of both cTnT and Gata4 were induced by DMSO treatment, and their levels were much lower in the miR-30c-overexpressing cells and much higher in the miR-30c knockdown cells compared with the control group.
Figure 4.
Effects of miR-30c on the differentiation of P19 cells. (a) miR-30c overexpressing or knockdown P19 cells were stimulated to differentiate by DMSO for 10 days. Images are shown from days 0, 2, 4, 6, 8 and 10. Scale bar, 50 μm. (b) qRT-PCR analysis of expression levels of myocardial cell markers at the indicated time points during the induction of differentiation. The data are presented as the mean±s.d. of three experiments (*P<0.05 comparing miR-30c overexpression with control cells; #P<0.05 comparing miR-30c knockdown with control cells). (c) Immunostaining of myocardial cell markers cTnT and Gata4 at the indicated time points. DAPI was used to stain the nuclei. Scale bar, 50 μm.
miR-30c targets Gli2 and regulates the Shh signaling pathway
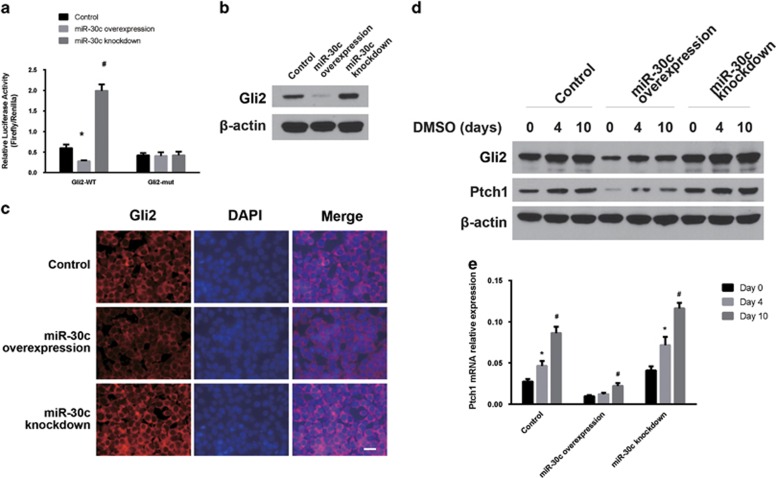
Gli2, a transcriptional factor of the Shh signaling pathway, was predicted to be a potential target gene of miR-30c by a variety of online gene analysis programs, including TargetScan 5.1. The binding of miR-30c to the target site in the Gli2 3′ UTR was detected using the Dual Luciferase Reporter system. As shown in Figure 5a, miR-30c overexpression significantly reduced the luciferase activity of the pGL3-Gli2-3′ UTR wild type (WT) reporter but not the mutated construct (mut), while miR-30c knockdown significantly rescued the luciferase activity of the pGL3-Gli2-3′ UTR reporter but not the mutated construct (*,#P<0.05). Furthermore, western blotting showed that miR-30c reduced, while miR-30c knockdown increased, the protein levels of Gli2 in P19 cells (Figure 5b). These results suggest that Gli2 is a potential target gene of miR-30c. We also conducted immunofluorescence staining of Gli2 in the cells and found that Gli2 expression was much weaker in miR-30c-overexpressing P19 cells, while it was slightly stronger in miR-30c knockdown cells compared with the control group (Figure 5c). Ptch1 is an important receptor of the Shh signaling pathway and is transactivated by Gli2. Because miR-30c targets Gli2, we therefore tested whether miR-30c regulates Ptch1 expression when the cells were induced to differentiate by DMSO. As shown in Figures 5d and e, DMSO treatment globally induced both protein and mRNA levels of Gli2 and Ptch1. miR-30c overexpression substantially inhibited Gli2 and Ptch1 expression before DMSO treatment and maintained a lower expression of the genes compared with the controls after DMSO treatment. miR-30c knockdown had the opposite effect. Furthermore, we transiently transfected a miR-30c mimic or inhibitor into P19 cells. Consistent with the results in the stable cell lines, expression levels of Gli2 and Ptch1 were reduced by the miR-30c mimic and increased by the miR-30c inhibitor in P19 cells (Supplementary Figure 3A). The Shh signaling pathway is involved in various developmental pathways. To determine if miR-30c targets the Shh signaling pathway, we employed the MC3T3-E1 cell line, a widely used murine osteoblast development model. Transient transfection of a miR-30c mimic or inhibitor in the MC3T3-E1 cells suppressed or increased Gli2 and Ptch1 expression, respectively (Supplementary Figure 3B), similar to the P19 cardiac development model.
Figure 5.
Effects of miR-30c on the Shh signaling pathway. (a, b) Identification of Gli2 as a potential target gene of miR-30c. (a) Dual luciferase reporter assay of miR-30c with a Gli2 3′ UTR reporter. The data are presented as the mean±s.d. of three experiments (*P<0.05 comparing miR-30c overexpression with control cells; #P<0.05 comparing miR-30c knockdown with control cells). (b) Western blotting of Gli2 protein levels in stable miR-30c overexpression or knockdown cell lines. (c) Immunostaining of Gli2 in stable miR-30c overexpression or knockdown cell lines. DAPI was used to stain the nuclei. Scale bar, 20 μm. (d, e) Ptch1 protein (d) and mRNA (e) levels were determined by western blotting or qRT-PCR, respectively, in miR-30c overexpression or knockdown cell line after treatment with DMSO for the indicated days. (*P<0.05 comparing Ptch1 mRNA levels in the cells on day 4 with those on day 0; #P<0.05 comparing Ptch1 mRNA levels in the cells on day 10 with those of day 0).
Discussion
CHDs, which account for ~40% of perinatal deaths, are the most common group of major birth defects. They are a serious medical problem that causes heavy social and economic burdens worldwide. CHDs have been intensively studied by the international community and much progress has been made in recent years.29 CHDs comprise numerous abnormalities in the structure and function of the heart and great vessels.30, 31 Most CHDs are correlated with gene deletions and mutations.32, 33 Thus, we explored the genetic and molecular mechanisms of CHDs. miRNAs have critical roles in cardiovascular development, and their expression profile changes correspond to different pathological conditions. miRNAs can be used as novel biomarkers and therapeutic targets of CHDs.34, 35, 36
In this study, we investigated the function of miR-30c in CHDs. We used P19 cells, which are commonly used to study cardiogenesis, as a myocardial cell model. We established stable P19 cell lines overexpressing miR-30c or knocked down miR-30c using a ‘miRNA sponge' that contains multiple binding sites in its 3′ UTR for a miRNA or miRNA seed family.28, 37, 38, 39 The stable cell lines were induced to differentiate into cardiomyocytes using DMSO. Significant changes were observed in their morphology and the levels of cardiac markers, such as cTnT, Gata4 and Nkx2.5. cTnT is a myocardial-specific protein that indicates late myocardial differentiation. Nkx2.5, one of the earliest identified myocardial markers, is predominantly expressed in the heart and cardiac progenitor cells. It has important functions in heart formation and development.40, 41, 42 Cardiogenesis is compromised in the absence of Nkx2.5,43 and multiple Nkx2.5 mutations have been found in diseased heart tissues.44, 45, 46 Gata4, which belongs to the cardiac Gata subfamily, is an important transcription factor in heart development. Gata4 interacts with Nkx2.5 to regulate myocardial cell migration and differentiation. We found that miR-30c overexpression inhibited, while miR-30c knockdown increased, the differentiation of P19 cells into cardiomyocytes, as indicated by the morphological changes and the expression of myocardial markers.
Embryonic fetal heart growth depends on the balance between cardiomyocyte proliferation and apoptosis.47 Inadequate proliferation or excess apoptosis will directly or indirectly lead to CHDs,48 which are often caused by altered proliferation and/or apoptosis in the septum, neighboring tissues or myocardium.49 miR-30c overexpression/knockdown may affect cell proliferation or apoptosis, resulting in inhibition of the differentiation of P19 cells into cardiomyocytes. miR-30c overexpression promoted cell proliferation, and miR-30c knockdown inhibited cell proliferation. Using Hoechst staining, electron microscopy, annexin V-FITC staining and caspase-3 activity assays, we found that miR-30c overexpression suppressed P19 cell apoptosis, while miR-30c knockdown increased cell apoptosis. Thus, by promoting cell growth and protecting cells from apoptosis, miR-30c inhibited the differentiation of P19 cells to cardiomyocytes.
Furthermore, we demonstrated that miR-30c regulates the Shh signaling pathway via a newly identified miR-30c target gene, Gli2. The three Gli proteins, Gli1, 2 and 3, are highly similar in amino-acid sequences.50 Gli1 knockout mice did not display organ malformations, but Gli2 knockout mice showed heart malformations, which were similar to Shh knockout mice. The phenotype of Gli3 knockout mice was different from the Shh knockout mice.51 Silencing of Gli2 inhibited Shh signaling.52 It has been reported that Shh signaling promoted the expressions of the cardiac progenitor markers Nkx2.5 and Gata4 in P19 cells,23 which were blocked by Shh inhibition.53 Ptch1, which is the receptor of Shh and a transcriptional target of Gil2, transduces the Shh signal. Our data showed that Ptch1 expression was suppressed by miR-30c and increased by miR-30c knockdown. Given that miR-30c inhibited expression of its target gene Gli2, miR-30c may suppress differentiation of P19 cells by inhibiting the Shh signaling pathway.
In summary, we demonstrated that miR-30c overexpression suppressed, while miR-30c knockdown increased, cardiomyocyte differentiation in P19 cells by regulating the Shh signaling pathway. In addition, miR-30c altered the balance between cell proliferation and apoptosis, which might result in embryonic cardiac malformations. These findings contribute to a better understanding of the mechanisms underlying cardiac abnormalities.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81370200), the National Natural Science Foundation of Jiangsu Province of China (Grant No. BK20131075), the Key Medical Personnel Foundation of Jiangsu Province (Grant No. RC2011021) and the Nanjing Medical Science and Technique Development Foundation (Grant No. QRX11107). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceived and designed the experiments: XHL, ZBY and SPH. Performed the experiments: XHL. Analyzed the data: XHL and XSH. Contributed reagents/materials/analysis tools: ZBY, SSZ, JX and MML. Wrote the paper: XHL. Study supervision: SPH and YZP.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Supplementary Material
References
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature 2004; 432: 231–235. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol 2004; 16: 223–229. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 2003; 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol 2004; 11: 214–218. [DOI] [PubMed] [Google Scholar]
- Tijsterman M, Plasterk RH. Dicers at RISC; the mechanism of RNAi. Cell 2004; 117: 1–3. [DOI] [PubMed] [Google Scholar]
- Sardu C, Marfella R, Santulli G, Paolisso G. Functional role of miRNA in cardiac resynchronization therapy. Pharmacogenomics 2014; 15: 1159–1168. [DOI] [PubMed] [Google Scholar]
- Dickinson BA, Semus HM, Montgomery RL, Stack C, Latimer PA, Lewton SM et al. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail 2013; 15: 650–659. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang X, Ren XP, Chen J, Liu H, Yang J et al. MicroRNA-494 targeting both proapoptotic and antiapoptotic proteins protects against ischemia/reperfusion-induced cardiac injury. Circulation 2010; 122: 1308–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 2001; 105: 851–862. [DOI] [PubMed] [Google Scholar]
- Harvey RP. NK-2 homeobox genes and heart development. Dev Biol 1996; 178: 203–216. [DOI] [PubMed] [Google Scholar]
- Balderman JA, Lee HY, Mahoney CE, Handy DE, White K, Annis S et al. Bone morphogenetic protein-2 decreases microRNA-30b and microRNA-30c to promote vascular smooth muscle cell calcification. J Am Heart Assoc 2012; 1: e003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen WJ, van der Made I, van den Oever S, Hiller M, de Boer BA, Picavet DI et al. Cardiomyocyte-specific miRNA-30c over-expression causes dilated cardiomyopathy. PLoS ONE 2014; 9: e96290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo K, Ogata S, Misumi Y, Takami N, Sinohara H, Ikehara Y. Cloning, structure and expression of cDNA for mouse contrapsin and a related protein. Biochem J 1991; 276(Pt 2): 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanic M, Yanowsky K, Rodriguez-Antona C, Andres R, Marquez-Rodas I, Osorio A et al. Deregulated miRNAs in hereditary breast cancer revealed a role for miR-30c in regulating KRAS oncogene. PLoS ONE 2012; 7: e38847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockhorn J, Yee K, Chang YF, Prat A, Huo D, Nwachukwu C et al. MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast Cancer Res Treat 2013; 137: 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XH, Ha CT, Fu D, Xiao M. Micro-RNA30c negatively regulates REDD1 expression in human hematopoietic and osteoblast cells after gamma-irradiation. PLoS ONE 2012; 7: e48700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev 2001; 15: 3059–3087. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 1996; 383: 407–413. [DOI] [PubMed] [Google Scholar]
- Tsukui T, Capdevila J, Tamura K, Ruiz-Lozano P, Rodriguez-Esteban C, Yonei-Tamura S et al. Multiple left-right asymmetry defects in Shh(-/-) mutant mice unveil a convergence of the shh and retinoic acid pathways in the control of Lefty-1. Proc Natl Acad Sci USA 1999; 96: 11376–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer LA, Kirby ML. Sonic hedgehog maintains proliferation in secondary heart field progenitors and is required for normal arterial pole formation. Dev Biol 2009; 330: 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M, Johnson RL, Stern CD, Kuehn M, Tabin C. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell 1995; 82: 803–814. [DOI] [PubMed] [Google Scholar]
- Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol 2006; 26: 3365–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianakopoulos PJ, Skerjanc IS. Hedgehog signaling induces cardiomyogenesis in P19 cells. J Biol Chem 2005; 280: 21022–21028. [DOI] [PubMed] [Google Scholar]
- Skerjanc IS. Cardiac and skeletal muscle development in P19 embryonal carcinoma cells. Trends Cardiovasc Med 1999; 9: 139–143. [DOI] [PubMed] [Google Scholar]
- van der Heyden MA, van Kempen MJ, Tsuji Y, Rook MB, Jongsma HJ, Opthof T. P19 embryonal carcinoma cells: a suitable model system for cardiac electrophysiological differentiation at the molecular and functional level. Cardiovasc Res 2003; 58: 410–422. [DOI] [PubMed] [Google Scholar]
- van der Heyden MA, Defize LH. Twenty one years of P19 cells: what an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc Res 2003; 58: 292–302. [DOI] [PubMed] [Google Scholar]
- Bockhorn J, Dalton R, Nwachukwu C, Huang S, Prat A, Yee K et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun 2013; 4: 1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang L, Wang H, Xu G, Zhu S, Li M et al. Effects of miR-19b knockdown on the cardiac differentiation of P19 mouse embryonic carcinoma cells. Mol Med Rep 2015; 11: 2504–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojnarska O, Grajek S, Katarzynski S, Kramer L. Predictors of mortality in adult patients with congenital heart disease. Cardiol J 2009; 16: 341–347. [PubMed] [Google Scholar]
- Allan L. Antenatal diagnosis of heart disease. Heart 2000; 83: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleftheriades M, Tsapakis E, Sotiriadis A, Manolakos E, Hassiakos D, Botsis D. Detection of congenital heart defects throughout pregnancy; impact of first trimester ultrasound screening for cardiac abnormalities. J Matern Fetal Neonatal Med 2012; 25: 2546–2550. [DOI] [PubMed] [Google Scholar]
- Bruneau BG. The developmental genetics of congenital heart disease. Nature 2008; 451: 943–948. [DOI] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science 2006; 313: 1922–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T, Catalucci D, Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc Res 2008; 79: 562–570. [DOI] [PubMed] [Google Scholar]
- Zhu S, Cao L, Zhu J, Kong L, Jin J, Qian L et al. Identification of maternal serum microRNAs as novel non-invasive biomarkers for prenatal detection of fetal congenital heart defects. Clin Chim Acta 2013; 424: 66–72. [DOI] [PubMed] [Google Scholar]
- Chang CC, Yang YJ, Li YJ, Chen ST, Lin BR, Wu TS et al. MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma. Oral Oncol 2013; 49: 923–931. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA 2010; 16: 2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 2007; 4: 721–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Hu X, Yu Z, Peng Y, Zhu J, Liu X et al. Effect of miR-20b on apoptosis, differentiation, the BMP signaling pathway and mitochondrial function in the P19 cell model of cardiac differentiation in vitro. PLoS ONE 2015; 10: e0123519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci USA 1993; 90: 8145–8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turbay D, Wechsler SB, Blanchard KM, Izumo S. Molecular cloning, chromosomal mapping, and characterization of the human cardiac-specific homeobox gene hCsx. Mol Med 1996; 2: 86–96. [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kasahara H, Bartunkova S, Schinke M, Komuro I, Inagaki H et al. Vertebrate homologs of tinman and bagpipe: roles of the homeobox genes in cardiovascular development. Dev Genet 1998; 22: 239–249. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev 1995; 9: 1654–1666. [DOI] [PubMed] [Google Scholar]
- Reamon-Buettner SM, Borlak J. Somatic NKX2-5 mutations as a novel mechanism of disease in complex congenital heart disease. J Med Genet 2004; 41: 684–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamon-Buettner SM, Hecker H, Spanel-Borowski K, Craatz S, Kuenzel E, Borlak J. Novel NKX2-5 mutations in diseased heart tissues of patients with cardiac malformations. Am J Pathol 2004; 164: 2117–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draus JM Jr, Hauck MA, Goetsch M, Austin EH 3rd, Tomita-Mitchell A, Mitchell ME. Investigation of somatic NKX2-5 mutations in congenital heart disease. J Med Genet 2009; 46: 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorina P, Corradi D, Pinelli S, Maestri R, Lagrasta C, Buscaglia M et al. Apoptotic/mytogenic pathways during human heart development. Int J Cardiol 2004; 96: 409–417. [DOI] [PubMed] [Google Scholar]
- Levy M, Maurey C, Celermajer DS, Vouhe PR, Danel C, Bonnet D et al. Impaired apoptosis of pulmonary endothelial cells is associated with intimal proliferation and irreversibility of pulmonary hypertension in congenital heart disease. J Am Coll Cardiol 2007; 49: 803–810. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Bartelings MM, Deruiter MC, Poelmann RE. Basics of cardiac development for the understanding of congenital heart malformations. Pediatr Res 2005; 57: 169–176. [DOI] [PubMed] [Google Scholar]
- Villavicencio EH, Walterhouse DO, Iannaccone PM. The sonic hedgehog-patched-gli pathway in human development and disease. Am J Hum Genet 2000; 67: 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Frias ML, Bermejo E, Frias JL. The VACTERL association: lessons from the Sonic hedgehog pathway. Clin Genet 2001; 60: 397–398. [DOI] [PubMed] [Google Scholar]
- Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J et al. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development 1998; 125: 2533–2543. [DOI] [PubMed] [Google Scholar]
- Clement CA, Kristensen SG, Mollgard K, Pazour GJ, Yoder BK, Larsen LA et al. The primary cilium coordinates early cardiogenesis and hedgehog signaling in cardiomyocyte differentiation. J Cell Sci 2009; 122: 3070–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.