Abstract
Upon interaction of the CD95 receptor with its ligand, sequential association of the adaptor molecule FADD (MORT1), pro-forms of caspases-8/10, and the caspase-8/10 regulator c-FLIP leads to the formation of a death-inducing signaling complex. Here, we identify polo-like kinase (Plk) 3 as a new interaction partner of the death receptor CD95. The enzymatic activity of Plk3 increases following interaction of the CD95 receptor with its ligand. Knockout (KO) or knockdown of caspase-8, CD95 or FADD prevents activation of Plk3 upon CD95 stimulation, suggesting a requirement of a functional DISC for Plk3 activation. Furthermore, we identify caspase-8 as a new substrate for Plk3. Phosphorylation occurs on T273 and results in stimulation of caspase-8 proapoptotic function. Stimulation of CD95 in cells expressing a non-phosphorylatable caspase-8-T273A mutant in a rescue experiment or in Plk3-KO cells generated by CRISPR/Cas9 reduces the processing of caspase-8 prominently. Low T273 phosphorylation correlates significantly with low Plk3 expression in a cohort of 95 anal tumor patients. Our data suggest a novel mechanism of kinase activation within the Plk family and propose a new model for the stimulation of the extrinsic death pathway in tumors with high Plk3 expression.
Keywords: polo-like kinase, caspase-8, CD95/Fas receptor, apoptosis, kinase regulation
Introduction
Apoptosis represents a form of programmed cell death that utilizes two major routes: the death receptor pathway (extrinsic) and the mitochondrial pathway (intrinsic)1, 2. Binding of a death ligand like the trimerized CD95L to the tumor necrosis factor (TNF) receptor superfamily members on the cell surface, e.g., CD95 (APO-1/Fas), TNF related apoptosis-inducing ligand (TRAIL) receptors, or TNF receptor 1 (TNFR1), leads to recruitment of the adaptor protein FADD (Fas-associated death domain, MORT1)3 and caspase-8 and -10 into a death-inducing signaling complex (DISC)4. FADD contains a death domain (DD) and a death-effector domain (DED). Via its DD, FADD binds to the DD of CD953. The DED of FADD recruits the DED-containing procaspase-8 into the DISC. This mechanism triggers caspase-8 activation resulting in the subsequent cleavage of downstream effector caspases5,6. Remarkably, multiple lines of evidence support a key role of CD95 in non-apoptotic signaling pathways that promote tumorigenesis7. The presence of CD95 in almost all tumor cells indicates a potential role in tumor development8. Hence, apoptotic- and non-apoptotic roles need to be considered when analyzing CD95 signaling in cancer cells.
The activation of the death receptors CD95, TRAILR1 and TRAILR2 was shown to induce cell death in a large variety of cancer cells. These observations provided the rationale for the development of TRAIL- and CD95-receptor agonists, which have demonstrated robust anticancer activity in a number of preclinical studies. For example, agonistic antibodies targeting CD95 are capable of inducing apoptosis in a wide range of cancer cells9. However, the systemic treatment of mice with CD95-agonistic antibodies resulted in hepatotoxicity10. Considering these results recombinant ligands, such as APO010, have been designed and have entered clinical trials. Similar to TNF and CD95L, TRAIL also induces apoptosis in cancer cells. Most remarkably, the systemic treatment with TRAIL in vivo killed tumor cells without causing toxicity, suggesting a cancer-selective mechanism of action. Despite this promising initial result, clinical trials revealed broad tolerability, and the therapeutic benefit of TRAIL-receptor agonists was rather limited11. To develop alternative strategies based on CD95- and TRAIL-receptor signaling-mediated apoptosis to combat cancer, research efforts are required to unravel the signal transduction machinery triggered by death receptor ligands, and how the resistance or sensitivity to death receptor-induced apoptosis is controlled.
Protein kinases represent a very attractive family of cancer targets as cancer cells have dysregulated kinase activity that enhances cell proliferation, migration and invasion, and confers apoptosis resistance12,13. In a very broad phylogenetic context, polo-like kinases (Plks) are master regulators of cell cycle progression14,15. Mammalian cells contain multiple Plk family members: Plk1, Plk2/Snk, Plk3/Fnk/Prk, Plk4/Sak and Plk516,17. A Plk consists of a conserved N-terminal serine/threonine kinase domain and a less conserved C-terminal substrate-binding domain, also known as the Polo box domain (PBD)18,19. Plks play a fundamental role in tumorigenesis, operating within a complex signaling network in cancer cells17,20,21,22. The subcellular localization, enzymatic activity and substrate interaction of Plks are controlled by their PBD23. Plk3 is an immediate early gene24,25. Plk3 has been suggested as a possible tumor suppressor because it was found to be downregulated in certain types of cancer (lung, head and neck) and its genetic locus is located in a chromosomal region that is often involved in the loss of heterozygosity in tumor cells24,26. Although genome- and kinome-wide screenings suggest a role of Plk3 in the regulation of apoptotic signaling27,28, the mechanism remains unknown.
Using immunoprecipitation (IP) experiments with Plk3-specific antibodies we searched Plk3-interacting partners that are involved in apoptotic signaling. Our analyses revealed that CD95 and other components of the DISC interact with Plk3 upon CD95 engagement, and this leads to the enzymatic activation of Plk3 via a novel kinase activation mechanism within the Plk family. We identified caspase-8 as a novel substrate of Plk3 that directly phosphorylates procaspase-8 on T273 in cells, promoting its activation, and subsequent proapoptotic function following CD95 receptor stimulation. Caspase-8 threonine phosphorylation identified in our study represents a novel mechanism that mediates extrinsic apoptosis. Remarkably, caspase-8 phosphorylation on T273 correlates with Plk3 expression in a pathological situation such as anal cancer, highlighting Plk3 as a possible enhancer of signals transmitted via the extrinsic death pathway.
Results
Identification of CD95 in immunoprecipitates of Plk3
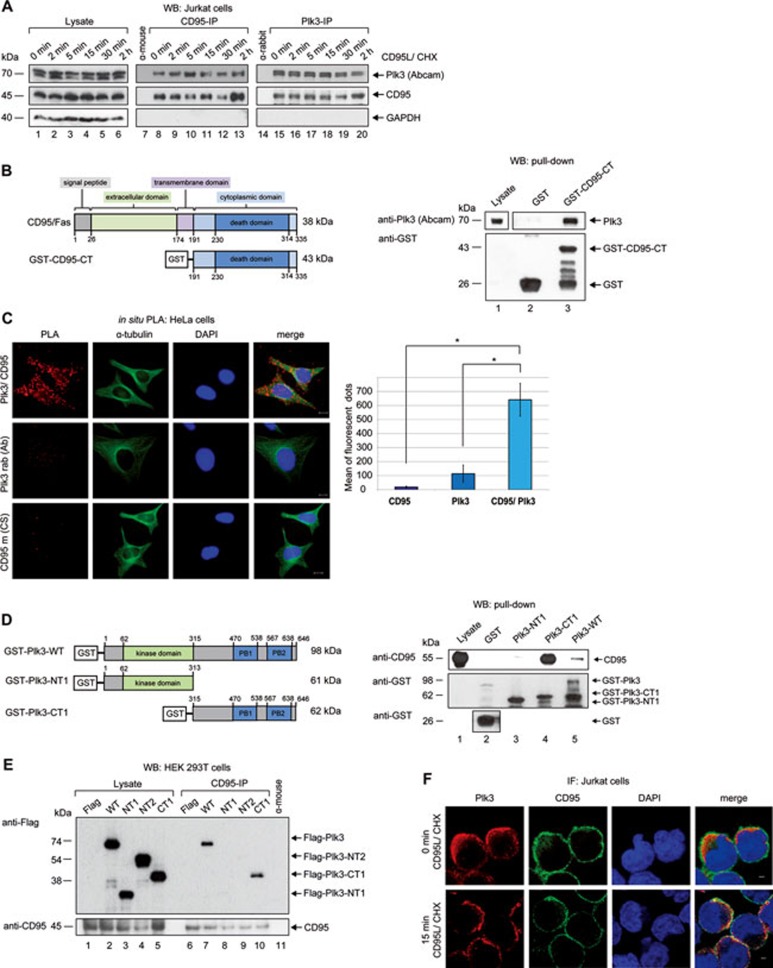
We first immunoprecipitated Plk3 from different cancer cell lines and searched for signal transducers of apoptosis in the immunoprecipitates using specific antibodies. One of the proteins co-precipitated with Plk3 was the CD95 receptor (Figure 1A, lane 15 vs 14). Following IP of extracts from HeLa and Jurkat cells that express CD95 at high levels using CD95- or Plk3-antibody, complexes containing CD95 and Plk3 were detected in both cell types (Figure 1A, lanes 8, 15; Supplementary information, Figure S1A, lane 8). Subsequently, we analyzed the association of CD95 and Plk3 in more detail to determine the cellular relevance of our novel findings. In a time course experiment we tested whether stimulation of CD95 by its ligand CD95L influences the intensity of the interaction with Plk3. The IP using CD95- or Plk3-specific antibody revealed that both proteins interact independent of stimulation of the CD95 receptor (Figure 1A, lanes 8, 15; Supplementary information, Figure S1A, lane 8). Ligand stimulation of CD95 did not further increase the interaction (Figure 1A, lanes 8-13 and lanes 15-20; Supplementary information, Figure S1A, lanes 8-13). IP of Plk3 from CD95-depleted cells showed a reduction of CD95 signals in western blot experiments compared with cells treated with a control siRNA (Supplementary information, Figure S1B). Pull-down assays with lysates of HeLa cells using the GST-fused intracellular domain of CD95 (Figure 1B, left panel) as bait confirmed the association with endogenous Plk3 (Figure 1B, right panel). The quantification of results obtained by using the proximity ligation assay (PLA) corroborated the immediate closeness of both proteins (Figure 1C). Furthermore, we performed binding studies with CD95 based on the pull-down assays and IP experiments using full-length Plk3 and various subdomains of Plk3 (Plk3-NT1, -NT2 and -CT1; Figure 1D and 1E). Once again we could confirm the interaction of both full-length proteins and demonstrated that the C-terminal moiety of Plk3 including the interdomain and the PBD of Plk3, but not the N-terminal half of Plk3, associates with CD95 (Figure 1D and 1E).
Figure 1.
Plk3 interacts with CD95. (A) Jurkat cells were treated with CD95L and CHX for different time periods. CD95 or Plk3 was immunoprecipitated from cell lysates with anti-CD95 mouse or anti-Plk3 rabbit (Abcam) antibodies. Normal mouse or rabbit IgG was used as a control. The precipitated proteins were immunoblotted for Plk3 and CD95. The Plk3 (Abcam) antibody recognized two bands corresponding to the information by the manufacturer. (B) Schematic representation of the CD95/Fas receptor and the GST-fused cytoplasmic C-terminal subdomain of CD95 (GST-CD95-CT) (left panel). Binding of GST-CD95-CT with endogenous Plk3 was analyzed in pull-down assays. GST-CD95-CT or GST alone (control) was incubated with the lysates of HeLa cells (right panel). Endogenous Plk3 was detected by immunoblotting. (C) The interaction of Plk3 and CD95 was monitored via in situ Proximity Ligation Assay (PLA). HeLa cells were labeled with anti-Plk3 (rabbit) and anti-CD95 (mouse) antibodies. Single antibody staining (Plk3/CD95) was used as a control. Scale bar: 10 μm. The quantification is shown in the right panel. Differences between single and double antibody staining were statistically significant by Student's t-test (*P ≤ 0.05). (D) Schematic representation of recombinant GST-fused full-length Plk3 (GST-Plk3-WT) and truncated forms of Plk3 (GST-Plk3-NT1 and -CT1) (left panel) used in pull-down assays. GST-Plk3-WT or Plk3 subdomains were incubated with Jurkat cell lysates for pull-down assays. Endogenous CD95 was detected by immunoblotting. (E) HEK 293T cells were transfected with Flag-Plk3-WT and its truncated forms (Supplementary information, Figure S2D) for 24 h. Endogenous CD95 was immunoprecipitated from cell lysates with anti-CD95 mouse antibody. Normal mouse IgG was used as a control. The precipitated proteins were immunoblotted for CD95 and Flag. (F) Co-localization of Plk3 and CD95 with or without CD95L treatment was studied using immunofluorescence microscopy. Cells were stained for Plk3, CD95 and DNA. Representative image acquisition was performed using a confocal laser-scanning microscope (CLSM) and rendering of confocal z stacks was performed using the LAS AF software. Scale bar: 2 μm.
Next, we investigated the subcellular localization of Plk3 and CD95 in cells via immunofluorescence microscopy. While we could confirm the localization of CD95 to the cellular membrane in Jurkat and HeLa cells, Plk3 was found predominantly at the inner membrane (Figure 1F and Supplementary information, Figure S1C and S1D). An overlap of Plk3 and CD95 as seen in yellow was found in some regions next to the plasma membrane (Figure 1F and Supplementary information, Figure S1C and S1D). To analyze Plk3's localization in more detail, we separated the cytoplasmic, membrane, and nuclear-cytoskeletal proteins from HeLa cells and evaluated first in western blots the efficacy of the separation process by monitoring the distribution of different subcellular marker proteins (Supplementary information, Figure S2A, lower part). Endogenous Plk3 was identified in different compartments depending on the antibody used: while the Abcam antibody labeled Plk3 mostly in the membrane fraction and gave a weak signal in the nuclear-cytoskeletal fraction, the BD antibody identified Plk3 predominantly in the cytoplasm and smaller amounts in the membrane fraction (Supplementary information, Figure S2A). Stimulation of HeLa cells with CD95L for 2 h did not alter the distribution of endogenous Plk3 (Supplementary information, Figure S2A). In addition, in Jurkat and HeLa cells Flag-tagged Plk3 was found at the cellular membrane and in HeLa cells also in the cytoplasm akin to the localization of endogenous Plk3 (Supplementary information, Figure S2B). While the BD antibody recognized predominantly a fast migrating form, the Abcam antibody detected slow migrating forms of endogenous Plk3 in lysates of HeLa and Jurkat cells (Supplementary information, Figures S2A and S1A; Figure 1A). The incubation of cell lysates with lambda phosphatase did not change the migration pattern of the doublet detected with the Abcam antibody indicating that the difference in migration is not due to phosphorylation of Plk3 (Supplementary information, Figure S2C). When using the BD antibody we could detect the fast migrating Plk3 isoform in the cytoplasm, and the slow migrating isoform in the membrane fraction (Supplementary information, Figure S2A). In addition, we examined the binding domains of the antibodies used in our study. Flag-tagged forms of Plk3 and its subdomains were generated for subsequent expression in HEK 293T cells (Supplementary information, Figure S2D). The polyclonal Plk3 antibody from Abcam labeled the Flag-tagged Plk3-subdomains NT1 and NT2, but not CT1, CT2, NT3 and NT4, indicating the antibody recognizes the N-terminal portion of Plk3 (Supplementary information, Figure S2E), which shows low homology compared with other Plk family members. The monoclonal antibody from BD recognized the Flag-tagged Plk3-subdomains CT1, NT2 and NT4, but not NT1, NT3 and CT2, suggesting that it recognizes an epitope located within the linker region connecting the kinase domain and the PBD (Supplementary information, Figure S2E). Thus, both tested antibodies specifically recognize different, less conserved subdomains of Plk3, consistent with the manufacturers' specifications. The BD antibody distinguished between isoforms/subpopulations of Plk3 in different subcellular compartments.
Plk3 associates with procaspase-8
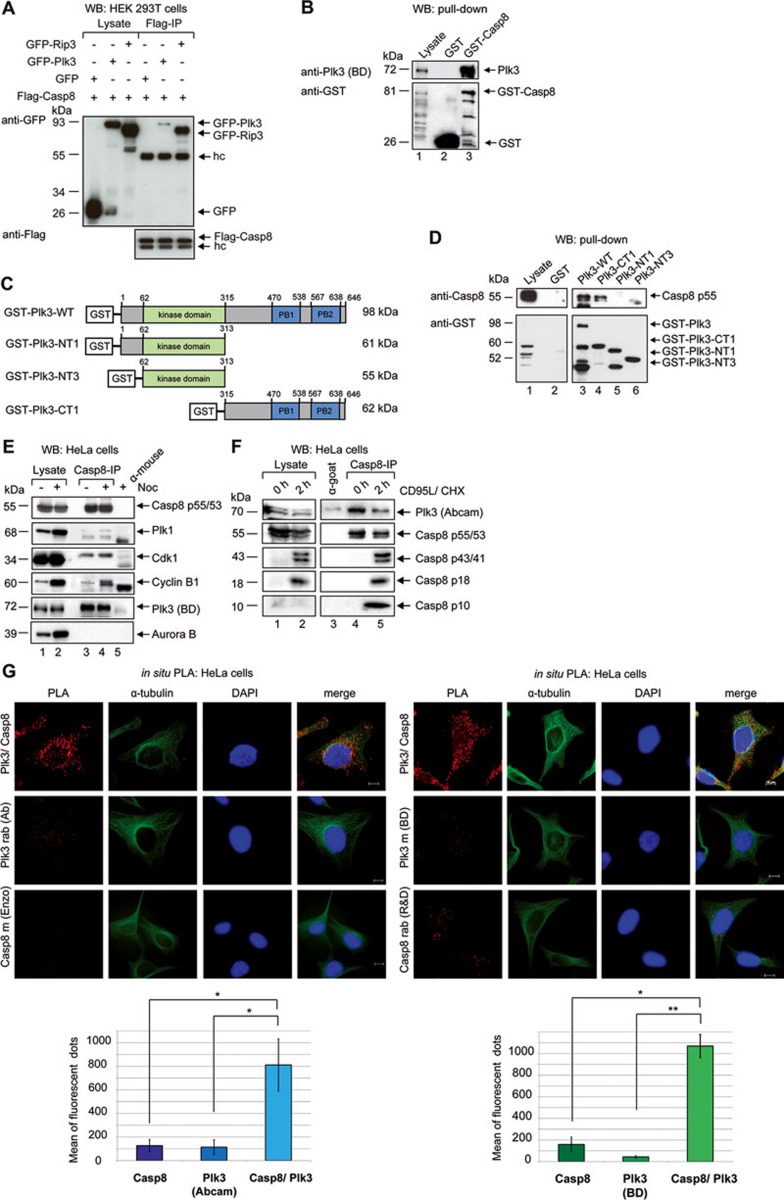
Upon interaction of the CD95 receptor with its ligand, sequential association of the adaptor molecule FADD (MORT1), pro-forms of caspases-8 and -10, and the caspase-8/10 regulator c-FLIP lead to the formation of the DISC29. We next tested whether Plk3 interacts with DISC components other than CD95. GFP-Plk3, GFP-Rip3 and Flag-tagged procaspase-8 were co-expressed in HEK 293T cells followed by Flag IP and western blot using GFP-specific antibodies (Figure 2A). This analysis confirmed the interaction of Rip3 and procaspase-830 and provided first evidence for an association of Plk3 and procaspase-8 (Figure 2A). Pull-down assays with GST-fused procaspase-8 using lysates of HeLa cells demonstrated its interaction with endogenous Plk3 (Figure 2B). To investigate the Plk3 interactome, we characterized the Plk3 complex by quantitative mass spectrometry (MS) using a SILAC (stable isotope labeling by amino acids in cell culture)-based strategy. HeLa cells expressing a Flag-tagged version of Plk3 were labeled with heavy stable isotopes of arginine (Arg+10) and lysine (Lys+8), while HeLa cells expressing the Flag-tag only were labeled with light ones (Lys+0, Arg+0). Upon lysis of the two differentially SILAC-labeled cell batches, Plk3 was immunoprecipitated via its Flag-tag in the heavy isotope-labeled lysate; proteins immunoprecipitated in the light-labeled lysate served as a negative control. The immunoprecipitated proteins were analyzed by MS. A protein with an at least threefold enrichment of heavy versus light peptides was considered as specifically co-purified with Plk3. Among the identified interaction partners of Plk3 was caspase-8, supporting the notion that Plk3 participates in signaling downstream of the DISC (Table 1 and Supplementary information, Table S1).
Figure 2.
Plk3 and procaspase-8 interact. (A) HEK 293T cells were co-transfected with GFP-Plk3 or GFP-Rip3 and Flag-procaspase-8. GFP-Rip3 was used as a positive control for the co-IP with procaspase-8. Precipitates were immunoblotted for Flag and GFP. (B) The binding ability of GST-fused procaspase-8 was analyzed in pull-down assays. GST-procaspase-8 or GST alone (as a control) was incubated with HeLa cell lysates. The pull-down was analyzed by immunoblotting for Plk3 and GST. (C) Schematic representation of recombinant GST-fused full-length Plk3 (GST-Plk3-WT) and truncated forms of Plk3 (GST-Plk3-NT1, -NT3 and -CT1) used in pull-down assays. (D) GST-Plk3-WT or Plk3 subdomains were incubated with HeLa cell lysates for pull-down assays. The immunoblot analysis was performed using caspase-8- and GST-antibodies. (E) HeLa cells were treated with 50 ng/ml nocodazole (Noc) for 16 h to enrich for mitotic cells. Endogenous caspase-8 was immunoprecipitated from cell lysates. Normal mouse IgG was used as a control. The immunoprecipitates and the lysates were immunoblotted for caspase-8, Plk1, Cdk1, Cyclin B1, Plk3 and Aurora B. (F) HeLa cells were treated with 50 ng/mL CD95L and CHX for 0 h and 2 h. Endogenous caspase-8 was immunoprecipitated from cell lysates with anti-caspase-8 p18 goat antibody. Normal goat IgG (Santa Cruz) was used as a control. The immunoprecipitates and the lysates were immunoblotted for Plk3, caspase-8, cleaved caspase-8 and p10. (G) The interaction of Plk3 and procaspase-8 (Casp8) was monitored via in situ PLA. HeLa cells were labeled with the anti-Plk3 (Abcam, rabbit) and anti-Casp8 (mouse) (left panel) or anti-Plk3 (BD, mouse) and anti-Casp8 (rabbit) (right panel) antibodies. Single antibody staining (Plk3 or Casp8) was used as a control. Scale bar: 10 μm. Each bar represents the mean value ± SD (n = 3). The differences between single and double antibody staining were statistically significant by Student's t-test (lower panels) (*P ≤ 0.05; **P ≤ 0.005).
Table 1. Interactome of exogenous Plk3 obtained from SILAC.
| Gene name | Protein | Ratio H/L |
|---|---|---|
| ACAD9 | Acyl-CoA dehydrogenase family member 9, mitochondrial | 17.8 |
| CDC73 | Parafibromin | 9.1314 |
| ITPR1 | Inositol 1,4,5-trisphosphate receptor type 1 | 7.5376 |
| IGF2BP2 | Insulin-like growth factor 2 mRNA-binding protein 2 | 5.4704 |
| PLK3 | Serine/threonine-protein kinase Plk3; Serine/threonine-protein kinase Plk | 4.125 |
| CASP8 | Caspase-8 | 3.7484 |
| FAF2 | FAS-associated factor 2 | 3.59579 |
| AURKA; AURKC | Aurora kinase A; Aurora kinase C | 3.4786 |
| SSBP1 | Single-stranded DNA-binding protein; Single-stranded DNA-binding protein, mitochondrial | 3.2498 |
| TRAF7 | E3 ubiquitin-protein ligase TRAF7 | 3.2126 |
| HUWE1 | E3 ubiquitin-protein ligase HUWE1 | 3.1139 |
With Flag-Plk3 co-immunoprecipitated proteins with a “normalized heavy/light ratio” (H/L) of > 3 were considered as high-confidence members of the Plk3 interactome; see Supplementary information, Table S1 for the original data of the complete mass spectrometric analysis.
Plks are characterized by their PBD, which mediates protein interactions and provides target specificity. Our experiments with full-length Plk3 and its subdomains (Figure 2C) demonstrated that both the full-length Plk3 and Plk3-CT1, which comprises of the PBD of Plk3, interact with procaspase-8 (Figure 2D). In an IP-analysis of endogenous protein we confirmed the association of procaspase-8 with Plk1 and Cdk1/cyclin B1 in mitotic cells31,32 and corroborated the interaction of caspase-8 and Plk3, which is independent of mitosis (Figure 2E). Stimulation of CD95 receptor induced processing of procaspase-8 with increasing levels of p18 and p10 (Figure 2F). Simultaneous IP of procaspase-8, whose levels decline upon CD95 stimulation, showed decreasing amounts of co-precipitated Plk3 (Figure 2F, right panel). The quantification of the PLA using Plk3 antibodies targeting the N-terminal (Abcam) or the interdomain of Plk3 (BD) confirmed the close proximity of both endogenous proteins in HeLa cells (Figure 2G, upper and lower panel).
Caspase-8 is phosphorylated at residue T273 by Plk3 in vivo
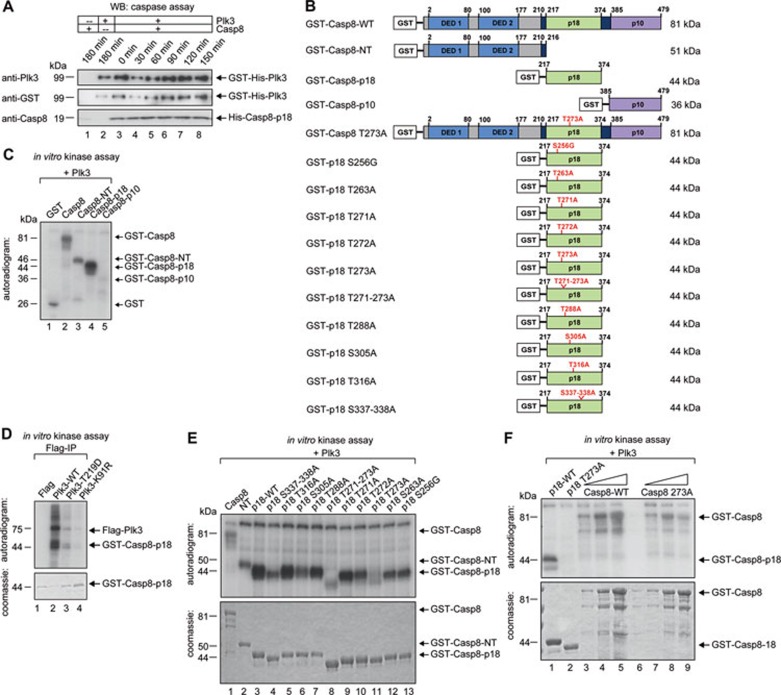
To investigate the functional role of the interaction of Plk3 and procaspase-8, we co-incubated Plk3, and procaspase-8 expressed in bacteria which undergoes spontaneous autoactivation. First we analyzed whether Plk3 could serve as a substrate for caspase-8. Prolonged incubation did not reduce the level of Plk3 significantly, suggesting that Plk3 is not proteolytically cleaved by caspase-8 (Figure 3A). To determine whether procaspase-8 might be an authentic substrate of Plk3, we incubated purified recombinant GST-fused full-length procaspase-8 and its subdomains (Figure 3B) with Plk3 purified from baculovirus-infected cells in the presence of [γ32P]ATP. While the N-terminal domain (NT) containing DED1 and DED2 was phosphorylated only to a lesser extent, the p18 subunit and procaspase-8 were labeled prominently (Figure 3C). In line with previous observations33,34, Flag-tagged, wild-type (WT) Plk3 purified from mammalian cells had the highest kinase activity compared with the phospho-mimicking Plk3 mutant T219D and the kinase-dead Plk3 (K91R) (Figure 3D). WT Plk3 phosphorylated the p18 subdomain more efficiently compared with Plk3-T219D and -K91R (Figure 3D). Next, we substituted alanine for serine or threonine at all conserved serine and threonine sites within p18 and expressed them as GST-tagged mutants of p18 in bacteria (Figure 3B). Kinase assays with Plk3 revealed that p18-T273A was not phosphorylated by Plk3, indicating that the sequence including T273 (Figure 3E) is a Plk3 phosphorylation site in vitro. This site is in agreement with the published consensus motif for Plk3 phosphorylation (www.phosphosite.org; Supplementary information, Figure S3A and S3B) and recent data obtained from an analysis of peptide libraries and mass spectrometry35 (Supplementary information, Figure S3B). The phylogenetic alignment of the p18 subunit of procaspase-8 revealed that T273 is highly conserved among different species (Supplementary information, Figure S3C). Next, we examined the phosphorylation of full-length procaspase-8. While the mutation T273A blocked the phosphorylation of p18 completely, it also strongly inhibited the phosphorylation of full-length procaspase-8 suggesting that T273 is the major phosphorylation site for Plk3 in vitro (Figure 3F).
Figure 3.
Plk3 phosphorylates procaspase-8 at T273. (A) Recombinant GST-His-fused Plk3 (0.1 μg) was incubated with recombinant active His-tagged caspase-8-p18 and -p10 (10 U) at 30 °C for the indicated time periods. The reaction mix was blotted against Plk3 (BD), GST and caspase-8. (B) Schematic representation of the recombinant GST-fused procaspase-8 (GST-Casp8-WT), N-terminal part (GST-NT), C-terminal subdomains of procaspase-8 (GST-p18 and -p10) and its mutated forms used for in vitro kinase assays. (C) Recombinant GST-fused procaspase-8 and its subdomains were incubated with Plk3 for an in vitro kinase assay and visualized by autoradiography. (D) HEK 293T cells were transfected with Flag-tagged Plk3-WT and its mutants (T219D, K91R). The proteins were immunoprecipitated using anti-Flag M2 affinity gel. Recombinant GST-fused caspase-8-p18 was incubated with immunoprecipitated Plk3 for an in vitro kinase assay, analyzed by SDS-PAGE and stained with Coomassie (lower panel), and visualized by autoradiography (upper panel). (E) GST-Casp8-WT, NT, p18 and point mutants of p18, all verified by sequencing in both directions, were incubated with recombinant GST-His-fused Plk3 (0.1 μg) for an in vitro kinase assay, analyzed by SDS-PAGE, stained with Coomassie (lower panel), and visualized by autoradiography (upper panel). (F) Recombinant GST-fused p18-WT, p18 T273A and increasing amounts of Casp8-WT and Casp8 T273A were incubated with recombinant GST-His-fused Plk3 (0.1 μg) for an in vitro kinase assay, analyzed by SDS-PAGE, stained with Coomassie (lower panel), and visualized by autoradiography (upper panel).
The database 'PHOSIDA'36 that contains 70 000 phosphorylation sites from kinase-enriched samples obtained using a quantitative MS technology provided evidence for the phosphorylation of caspase-8 T273 in vivo (Supplementary information, Figure S3D). To further determine whether procaspase-8 is a bona fide substrate of Plk3, we raised an affinity-purified antibody against a phosphopeptide containing phospho-T273. The new antibody detected caspase-8 and its subunit p18 only after Plk3 phosphorylation (Supplementary information, Figure S4A). In contrast, mutation of T273 to alanine (Supplementary information, Figure S4B) or inhibition of Plk3 by increasing concentrations of the small molecule Plk inhibitor BI2536 (Supplementary information, Figure S4C) or phosphatase treatment of caspase-8 and p18 phosphorylated by Plk3 (Supplementary information, Figure S4D) abolished the recognition indicating the high specificity of the novel pT273 antibody. In addition, we tested the new antibody for the analysis of procaspase-8 phosphorylated by Plk1 or Cdk1/cyclin B1. An antibody for pS387 recognized procaspase-8 phosphorylated by Cdk1/cyclin B1 (Supplementary information, Figure S4F), whereas the antibody pT273 did not detect procaspase-8 phosphorylated by Plk1 or Cdk1/cyclin B1 (Supplementary information, Figure S4E and S4F) supporting the specificity of the novel caspase-8 pT273 antibody.
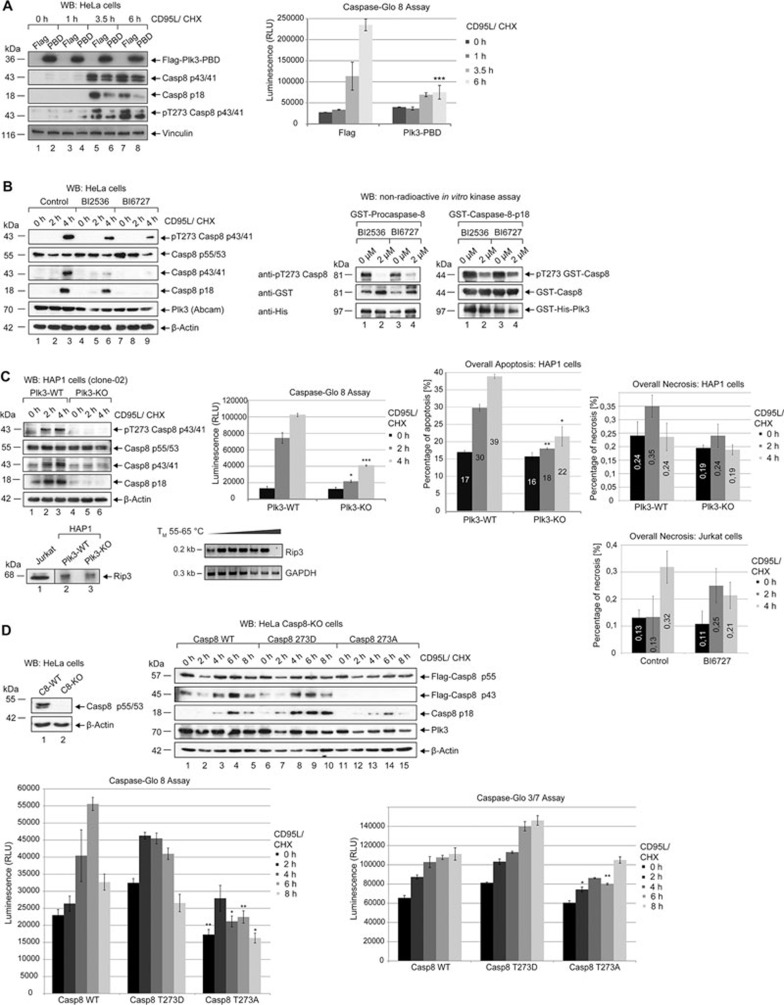
To further explore caspase-8 phosphorylation at T273 in vivo after interaction of the CD95 receptor with its ligand, we tried to knock-down the Plk3 mRNA efficiently by RNAi. However, a recent genome-wide analysis has revealed that high mRNA turnover rates are associated with reduced susceptibility to silencing by siRNA37. Within the Plk family, Plk3 was shown to have the highest turnover rate, which could explain why an efficient depletion is hard to obtain37. As an alternative strategy for the inhibition of Plk3 function, we overexpressed the Plk3-PBD, which represents the binding domain for the association with caspase-8, as dominant negative form of Plk3. This led to reduced phosphorylation at T273 predominately detectable in the proteolytic intermediate p43/41 of caspase-8 after 3.5-6 h of CD95 stimulation (Figure 4A, left panel). In the PBD-overexpressing cells, the processing of procaspase-8 was downregulated accompanied by low levels of p18 (Figure 4A, left panel) and reduced caspase-8 activity with a significance of p ≤ 0.0005 after 6 h of CD95 stimulation (Figure 4A, right panel) as compared with mock transfected cells, suggesting that the inhibition of Plk3 function might hamper CD95L-induced processing of procaspase-8. Moreover, clinical Plk inhibitors, BI2536 and BI6727, which not only inhibit Plk1, but also Plk2 and Plk3, were used to reduce Plk3 activity17,20,38,39. Both inhibitors downregulated phosphorylation of caspase-8 at T273 at least partially as demonstrated by monitoring the phosphorylation of the caspase-8 intermediate p43/41 (Figure 4B, left panel) and downregulated Plk3 activity in kinase assays (Figure 4B, right panel). In addition, we tested two Plk3 knock-out (KO) HAP1 clones that were generated using the CRISPR/Cas9 system (Figure 4C, Supplementary information, Figure S5C). Sequencing of the genomic DNA confirmed a deletion at the target site that leads to a KO of Plk3 (Supplementary information, Figure S5A, left panel). By reverse transcription we generated cDNA from mRNA isolated from WT and KO cells and analyzed the presence of the CRISPR/Cas 9-generated deletion in Plk3 mRNA. Sequencing of the Plk3 cDNA confirmed the CRISPR/Cas 9-generated deletion in the Plk3 mRNA. We performed a PCR using a 5′ primer located within the deleted region and a 3′ primer matching to the downstream Plk3 sequence. Only the WT cDNA but not the deleted form gave rise to a PCR signal (Supplementary information, Figure S5A, right panel). To monitor Plk3 protein expression, we performed immunofluorescence staining, and confirmed the KO of the Plk3 expression (Supplementary information, Figure S5B). The experiment also showed a background staining of the Abcam antibody.
Figure 4.
Inhibiting the function of Plk3 impairs the activation of procaspase-8. (A) HeLa cells were transfected with Flag-fused Plk3-PBD (Flag-Plk3-CT1) or Flag empty vector as a control. 24 h later cells were treated with 50 ng/ml CD95L and CHX for the indicated time periods. The lysates were blotted against Flag, cleaved caspase-8, pT273 caspase-8 and Vinculin (left panel). Caspase-8 activity was determined using a Caspase-Glo 8 assay (right panel). Each bar represents the mean value ± SD (n = 3). The differences between cells that were transfected with Flag or Plk3-PBD were statistically significant by Student's t-test (***P ≤ 0.0005). (B) HeLa cells were pretreated with 2 μM BI2536 or 2 μM BI6727 for 2 h. Untreated cells were used as a control (Control). Subsequently cells were treated with 50 ng/mL CD95L and CHX in the absence (Control) or in the presence of 2 μM BI2536 or BI6727 for the indicated time periods. Lysates were blotted against pT273 caspase-8, caspase-8, cleaved caspase-8, Plk3 and β-actin (left panel). Recombinant GST-fused procaspase-8 and p18 were incubated with recombinant GST-His-fused Plk3 (0.1 μg) in absence or presence of 2 μM BI2536 or BI6727 for 30 min at 37 °C for an in vitro kinase assay. Samples were immunoblotted against pT273 caspase-8, GST and His (right panel). (C) Plk3-WT and Plk3-KO (clone-02) HAP1 cells were treated with CD95L and CHX for the indicated time periods. The lysates were blotted against pT273 caspase-8, caspase-8, cleaved caspase-8 and β-actin (left panel). Caspase-8 activity was determined using a Caspase-Glo 8 assay (middle panel). The overall apoptosis and necrosis were measured by Annexin V/7 AAD FACS analysis (right upper and lower panels). Each bar represents the mean value ± SD (n = 3). The differences between Plk3-WT cells and Plk3-KO cells were statistically significant by Student's t-test (*P ≤ 0.05; **P ≤ 0.005; ***P 0.0005). The protein level of Rip3 in Hap1 and Jurkat cells was monitored by western blot (lower left panel). The mRNA from Hap1 cells was reverse transcribed and Rip3 expression was monitored by PCR with an annealing temperature of 55-65 °C (lower middle panel). GAPDH served as a control. Jurkat cells were treated with 100 ng/mL CD95L and CHX in the absence (Control) or in the presence of 2 μM BI6727 for the indicated time periods. The overall necrosis was measured by Annexin V/7 AAD FACS analysis (lower right panel). Each bar represents the mean value ± SD (n = 3). (D) Lysates of HeLa Casp8-WT and Casp8-KO cells were immunoblotted against caspase-8 and β-actin (left upper panel). In Casp8-KO cells Flag-caspase-8 WT, T273A or T273D mutant was expressed followed by treatment with CD95L and CHX for the indicated time periods. Lysates were blotted against Flag, cleaved caspase-8, Plk3 and β-actin (right upper panel). Caspase-8 and -3/7 activities were determined using Caspase-Glo 8 and 3/7 assays (lower panels). Each bar represents the mean value ± SD (n = 3). The differences between caspase-8 WT and T273A were statistically significant by Student's t-test (*P ≤ 0.05; **P ≤ 0.005).
In different Plk3-KO clones stimulated with CD95L neither caspase-8 phosphorylation at T273 nor caspase-8 processing was detectable (Figure 4C, left panel and Supplementary information, Figure S5C, left panel). The analysis of the enzymatic activity supported the inhibition of caspase-8 activation in Plk3-KO cells upon CD95 stimulation (Figure 4C, middle panel and Supplementary information, Figure S5C, right panel), suggesting that Plk3 is a critical stimulator for procaspase-8 processing and activity. Moreover, the levels of overall apoptosis in Plk3-KO cells upon CD95 stimulation were significantly reduced as compared with the Plk3-WT cells (Figure 4C, right panel) supporting the role of Plk3 in apoptosis induction. The level of overall necrosis in HAP1 cells expressing a very low but detectable level of RIP3 (Figure 4C, lower left panel) was not significantly increased upon Plk3 deficiency after CD95 stimulation (Figure 4C, right panel). Moreover, in Jurkat cells expressing RIP3 and MLKL, BI6727 treatment, which inhibits Plk3's enzymatic activity, did not cause an increase of necrosis upon CD95L stimulation (Figure 4C, lower right panel).
In order to stably KO the expression of procaspase-8 in the HeLa cells, a 20 nucleotide target sequence located within its exon 7 was selected, and inserted into the CRISPR vector for subsequent transfection of HeLa cells. Single clones were selected and checked for the expression of procaspase-8 (Figure 4D, upper left panel). For a rescue experiment we used procaspase-8 KO cells and replaced caspase-8 by similar amounts of transfected procaspase-8 (WT) or its mutants (T273A, T273D; Figure 4D, upper right panel). The replacement with the non-phosphorylatable T273A mutant followed by CD95-stimulation led to very little caspase-8 processing as monitored by the levels of p18 (Figure 4D, upper right panel). In contrast, the T273D mutant or procaspase-8 (WT), which is phosphorylated at T273 in HeLa cells upon receptor stimulation, showed higher levels of activation (Figure 4D, upper right panel). Moreover, we performed Caspase-Glo 8 assays to analyze the activity of both phospho-mimic (T273D) and dephospho-mimic (T273A) mutants of procaspase-8 that were expressed in caspase-8-KO cells. Upon CD95 stimulation the T273D mutant showed a significant increase of enzymatic activity of caspase-8 and -3/7 compared with T273A-expressing cells (Figure 4D, lower panels).
Interaction of the CD95 receptor with its ligand stimulates Plk3 activity
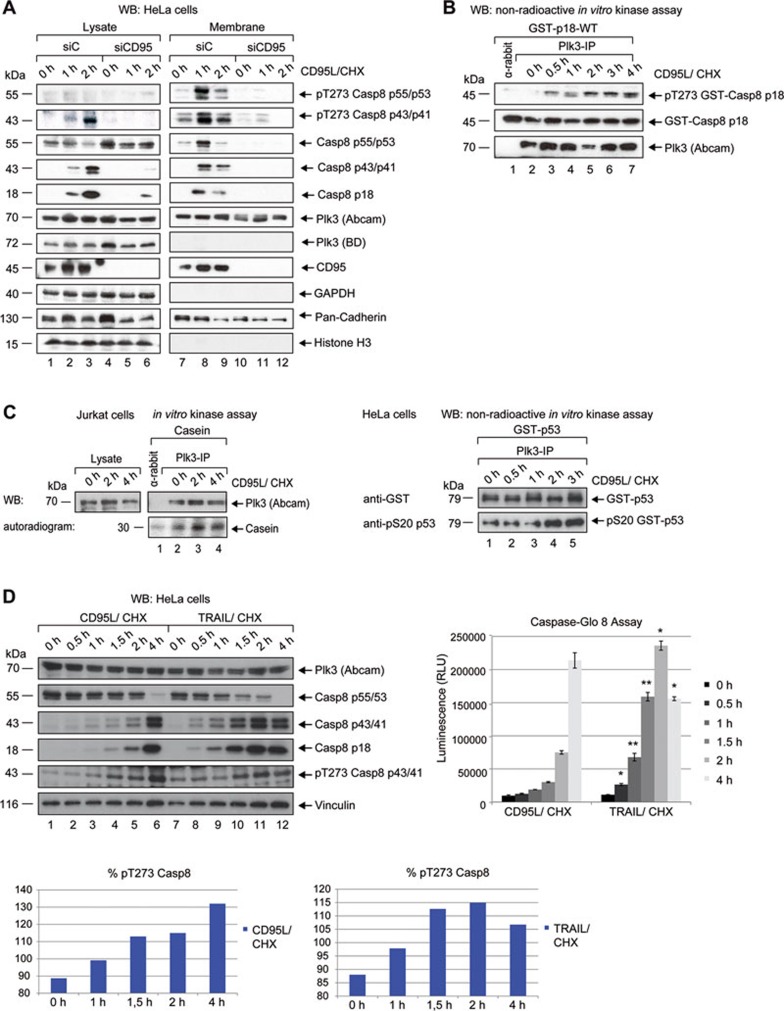
Our experiments have shown an increase of T273 phosphorylation in the proteolytic intermediate p43/41 of procaspase-8 in whole HeLa cell lysates 3.5-4 h after addition of CD95L (Figure 4A and 4B). We also tested the colorectal cancer cell line HCT 116 and observed an increase of the T273 phospho-signal and caspase-8 processing 2 h following stimulation (Supplementary information, Figure S5D, upper panel). To study the T273 phosphorylation at the membrane, where the association of CD95 and Plk3 occurs, the membrane fraction was subjected to further analysis. HeLa cells and CD95-depleted HeLa cells were both treated with CD95L: in the membrane fractions the phosphorylation at T273 of procaspase-8 and p43/41 was weak without CD95L stimulation, but 1 h later a strong phospho-signal intensity was visible, which was clearly reduced at 2 h of stimulation, likely due to further procaspase-8 processing (Figure 5A). Depletion of CD95 almost completely blocked phosphorylation of caspase-8 (Figure 5A). In parallel, immunoprecipitated Plk3 from CD95-stimulated Jurkat (Figure 5B) or HeLa (Supplementary information, Figure S5E) cells was subjected to in vitro kinase assays showing that Plk3 activity using caspase-8 or p18 as substrates was detectable after 30 min in Jurkat cells or 1 h in HeLa cells and increased with time (Figure 5B and Supplementary information, Figure S5E, lower panel). In addition, to verify the activation of Plk3, we tested the artificial substrate casein and the authentic substrate p53. An increasing phosphorylation of both proteins could be detected upon ligand stimulation of CD95 (Figure 5C and Supplementary information, Figure S5D, lower panel) supporting the model of Plk3 activation upon CD95 stimulation. To investigate this mechanism in another caspase-8-dependent pathway, cells were treated with the death ligand TRAIL (Figure 5D). Upon stimulation with TRAIL, procaspase-8 processing, enzymatic activation and phosphorylation at T273 took place in HeLa (Figure 5D) and A549 cells (Supplementary information, Figure S5F, left and right panels) showing the stimulation with both ligands, CD95L and TRAIL, leads to an increase of caspase-8 phosphorylation at T273 (Figure 5D, lower panel). Simultaneous treatment of cells with the Plk inhibitor BI6727 blocked the phosphorylation of caspase-8 completely (Supplementary information, Figure S5G) indicating that Plk3 could be activated by two stimuli, CD95L and TRAIL.
Figure 5.
CD95 stimulation induces Plk3 activation and phosphorylation of procaspase-8. (A) HeLa cells were transfected with CD95 siRNA (siCD95) or a scrambled siRNA (siC) as control. 48 h later, cells were treated with CD95L and CHX for the indicated time periods. The whole cell lysate and the membrane fraction were immunoblotted against pT273 caspase-8, caspase-8, cleaved caspase-8, Plk3, CD95 and different subcellular markers (GAPDH, cytoplasm; histone H3, nucleus and pan-cadherin, plasma membrane). (B) Jurkat cells were treated with CD95L and CHX for the indicated time periods. Endogenous Plk3 was immunoprecipitated with rabbit anti-Plk3 (Abcam) and incubated with recombinant GST-fused p18-WT for 30 min at 37 °C. Samples from the in vitro kinase assays were immunoblotted against pT273 caspase-8, GST and Plk3. Normal rabbit IgG was used as a control. (C) Endogenous Plk3 was immunoprecipitated from Jurkat (left panel) and HeLa (right panel) cells and analyzed by western blot or was incubated with casein (left panel) or GST-fused p53 (right panel) for an in vitro kinase assay and visualized by autoradiography (lower left panel) or by western blot (upper left and right panels). Samples were blotted against p53-pS20 and GST (right panel). (D) HeLa cells were treated with CD95L or TRAIL and CHX for the indicated time periods. The lysates were immunoblotted against Plk3, caspase-8, cleaved caspase-8, pT273 caspase-8 and Vinculin (left upper panel). Caspase-8 activity was determined by Caspase-Glo 8-Assay (right upper panel). Each bar represents the mean value ± SD (n = 3). The differences between CD95L and TRAIL treatment were statistically significant by Student's t-test (*P ≤ 0.05; **P ≤ 0.005). Phospho-signal of pT273 caspase-8 was quantified by ImageJ and standardized to the Vinculin signal (lower panels).
Recently, it was demonstrated that the binding of the PBD of Plk1 to a target protein/substrate at the proper subcellular location can disrupt the intramolecular interaction between the PBD and kinase domain (KD) and relieve the inhibition of Plk1's kinase activity by the PBD40. To find out whether procaspase-8 as PBD-binding protein might play a similar role for the activation of Plk3, we expressed increasing amounts of procaspase-8 in cells expressing the myc-tagged N-terminal (including the KD) and the V5-tagged C-terminal portion (including the PBD) of Plk3 (Supplementary information, Figure S6A). IPs with myc- or V5-specific antibody revealed in the absence of procaspase-8 an interaction of the tagged KD- and the PBD-containing domains (Supplementary information, Figure S6A and S6B). Simultaneous expression of both domains with low amounts of procaspase-8 seemed to stabilize the interaction, but upon a certain threshold of procaspase-8 expression the interaction between two Plk3 domains decreased (Supplementary information, Figure S6B, right panel). In addition, we stimulated cells expressing the myc-tagged N-terminal and the V5-tagged C-terminal portion of Plk3 with CD95L (Supplementary information, Figure S6C). The anti-myc IP revealed directly after stimulation an increased binding of the KD- and PBD-containing domains followed by their partial dissociation suggesting that Plk3 is regulated at the DISC through the modulation of the interaction of the KD- and PBD-containing domains. Upon dissociation of both domains coprecipitated procaspase-8 was increasingly phosphorylated at T273 (Supplementary information, Figure S6C, right panel).
DISC formation is required for Plk3 activation
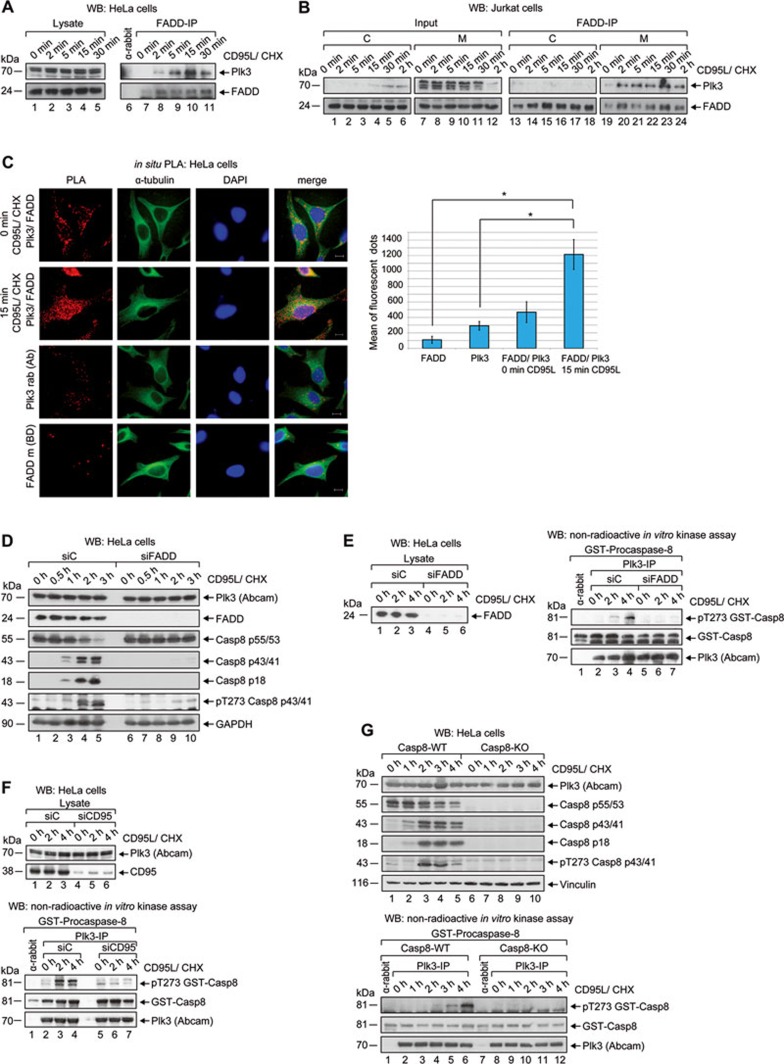
Although our previous experiments suggest that Plk3 associates with CD95 and procaspase-8, the role of DISC formation for Plk3 activation remains elusive. Engagement of CD95 by CD95L results in receptor aggregation and recruitment of the DD-containing adaptor protein FADD/MORT1 to the intracellular domain of CD95 through homophilic DD/DD interaction. Recruitment of the procaspase-8, which has two DED motifs within its prodomain41,42, occurs through a DED/DED interaction with FADD leading to DISC formation3. Since we could show that stimulation of the extrinsic pathway with CD95L or TRAIL is associated with an increase of Plk3 activity, we investigated whether DISC formation plays a role for Plk3 activation. Following CD95L treatment, FADD started to associate with Plk3 within 2 min (Figure 6A). Fractionation of cytoplasm and membrane revealed that the interaction of FADD and Plk3 took place at the membrane (Figure 6B). PLA demonstrated that the association of FADD and Plk3 increased upon CD95 stimulation (Figure 6C, left and right panels). Depletion of FADD prevented processing of procaspase-8 upon CD95 stimulation (Figure 6D), phosphorylation of procaspase-8 at T273 (Figure 6D) and activation of Plk3 as demonstrated in an in vitro kinase assay using procaspase-8 as substrate (Figure 6E, right panel). The depletion of CD95 also prevented activation of Plk3 (Figure 6F, lower panel). In caspase-8 KO cells the processing of procaspase-8 could not take place upon CD95 stimulation (Figure 6G, upper panel), and CD95 stimulation failed to activate Plk3 in an in vitro kinase assay (Figure 6G, lower panel). Taken together, depletion of FADD, CD95 or KO of caspase-8 prevented Plk3 activation upon CD95 stimulation, suggesting that the DISC formation is required for the activation of Plk3.
Figure 6.
Plk3 interacts with FADD and DISC formation is required for Plk3 activation. (A) HeLa cells were treated with CD95L and CHX for the indicated time periods. Endogenous FADD was immunoprecipitated with rabbit anti-FADD and immunoblotted against Plk3 and FADD. (B) Jurkat cells were treated with CD95L and CHX for the indicated time periods. Cytoplasmic and membrane fractions were separated using hypotonic buffer. Endogenous FADD was immunoprecipitated with rabbit anti-FADD and immunoblotted for Plk3 and FADD. (C) The potential interaction between Plk3 and FADD was monitored via in situ PLA. HeLa cells were treated with CD95L and CHX for the indicated time periods and labeled with the anti-Plk3 (rabbit) and anti-FADD (mouse) antibodies. Single antibody staining (Plk3 or FADD) was used as a control. Scale bar: 10 μm. The quantification is shown on the right. Each bar represents the mean value ± SD (n = 3). The differences between single and double antibody staining were statistically significant by Student's t-test (*P ≤ 0.05). (D) HeLa cells were transfected with FADD siRNA (siFADD) or a control siRNA (siC) for 48 h followed by the treatment with CD95L and CHX for the indicated time periods. The lysates were immunoblotted against Plk3, FADD, caspase-8, cleaved caspase-8, pT273 caspase-8 and GAPDH. (E) HeLa cells were transfected with FADD siRNA (siFADD) or a control siRNA (siC). Cells were treated with CD95L and CHX for the indicated time periods. The lysates were immunoblotted against FADD (left panel). Immunoprecipitated endogenous Plk3 was incubated with recombinant GST-fused procaspase-8 for an in vitro kinase assay. Samples were immunoblotted against pT273 caspase-8, GST and Plk3 (right panel). (F) HeLa cells were transfected with CD95 siRNA (siCD95) or a control siRNA (siC) and then treated with CD95L and CHX for the indicated time periods. Lysates were immunoblotted against Plk3 and CD95 (upper panel). Immunoprecipitated Plk3 was incubated with GST-fused procaspase-8 for an in vitro kinase assay. Samples were immunoblotted against pT273 caspase-8, GST and Plk3 (lower panel). (G) HeLa Casp8-WT and Casp8-KO cells were treated with CD95L and CHX for the indicated time periods. Lysates were immunoblotted against Plk3, caspase-8, cleaved caspase-8, pT273 caspase-8 and vinculin (upper panel). Immunoprecipitated Plk3 was incubated with GST-fused procaspase-8 for an in vitro kinase assay. Samples were immunoblotted against pT273 caspase-8, GST and Plk3 (lower panel).
Caspase-8 is phosphorylated on T273 in primary anal cancer
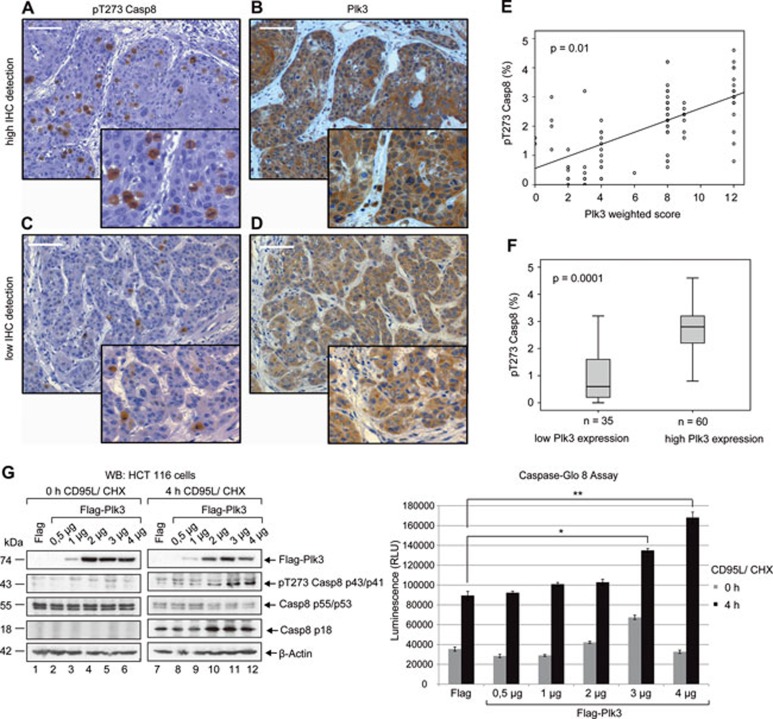
We have demonstrated that the activation of Plk3 in a physiological situation such as CD95L treatment triggers caspase-8 phosphorylation. We therefore asked whether caspase-8 T273 phosphorylation occurs in pathological situations including anal cancer where Plk3 shows different levels of expression. We speculated that T273 phosphorylation of caspase-8 may represent a new mechanism through which Plk3 activity in transformed cells determines the signaling intensity of the extrinsic death pathway. Considering the functional and clinical importance of cytotoxic T lymphocytes bearing CD95L in gastrointestinal tumors43, we analyzed a cohort of 95 anal cancer patients using antibodies against Plk3 and pT273 caspase-8 (Figure 7A-7D). Using our novel phospho-specific antibody we were able to show that caspase-8 is phosphorylated on T273 in primary anal cancer tissues (Figure 7A and 7C). The median pT273 caspase-8 index for all 95 patients was 2.2% with a range of 0% - 4.6%. The weighted score (WS) of Plk3 expression (intensity [1-3] × percentage [0-4]) showed a significant correlation with pT273 caspase-8 signals in corresponding tumor tissues (p = 0.01; Figure 7E). Because of the limited number of patients, the weighted score for Plk3 expression was arbitrarily dichotomized: a Plk3 score of 6 or below was classified as “low Plk3 expression”, and a score above 6 as “high Plk3 expression”. The correlation of this dichotomized variable with pT273 caspase-8 signal intensity was determined (Figure 7F). The median pT273 caspase-8 signal was significantly reduced (p = 0.001) in tumors with a low Plk3 expression.
Figure 7.
Plk3 protein expression correlates with phosphorylation of procaspase-8 at T273 in primary anal tissues. (A-D) Examples of anal cancer biopsies with high (A and B) and low (C and D) immunohistochemical detection of pT273 Casp8 and Plk3 in tumor cells. Original magnification × 100, × 400 inlets, scale bar: 100 μm. (E) Association of the immunohistochemical detection of pT273 Casp8 and the Plk3-weighted score (intensity of staining × percent of positive tumor cells) in pretreatment biopsies of 95 patients with anal carcinoma. (F) Association of the immunohistochemical detection of pT273 Casp8 with high (score: 6-12) and low (score: 0-4) Plk3 expression. The tick line is the median value, the solid box is the interquartile range and the whiskers are the 5th and 95th percentiles. n = number of patients. (G) HCT-116 cells were transfected with increasing amount of vector DNA expressing Flag-Plk3 or Flag alone as a control followed by the treatment with CD95L and CHX for the indicated time periods. The lysates were immunoblotted against Plk3, pT273 caspase-8, caspase-8 and β-actin (left panel). Caspase-8 activity was determined by Caspase-Glo 8 Assay (right panel). Each bar represents the mean value ± SD (n = 3). The differences between Flag- and Plk3-trasfected cells were statistically significant by Student's t-test (*P ≤ 0.05; **P ≤ 0.005).
The 5-year cumulative incidence of local failure (i.e., non-complete response at re-staging after chemoradiation or locoregional relapse after initial complete response) and distant metastases were significantly reduced in patients with a high pT273 caspase-8 expression (8% vs 39.6%, P = 0.001) and (9.2% vs 20.9%, P = 0.033), while a high Plk3 immunoreactivity was significantly related to a reduced local (13.7% vs 37.1%, P = 0.011) but not to distant failure (11% vs 22.2%, P = 0.13). Moreover, 5-year disease-free (DSF) and overall survival (OS) was superior for tumors showing a high pT273 caspase-8 signal (DSF: 91% vs 74%, P = 0.02 and OS: 87% vs 60%, P = 0.002) and high Plk3 expression (DSF: 90% vs 69%, P = 0.02 and OS: 79% vs 83%, P = 0.03) compared with patients with low pT273 caspase-8 signals and low Plk3-expressing carcinomas. Taken together, our study reveals a correlation between the level of Plk3 protein expression and the phosphorylation of procaspase-8 at T273 in primary cancer tissues. Moreover, our data demonstrate the prognostic significance of pT273 caspase-8 staining for anal cancer patients.
To investigate whether caspase-8 phosphorylation at T273 could modulate caspase-8 activity in gastrointestinal cancer cells, we used plasmid titration as an efficient method to vary the expression rate of Plk3 (Figure 7G, left panel). Immunoblot with our phospho-specific antibody revealed that the upregulation of Plk3 expression correlates with increasing levels of caspase-8 phosphorylation at T273 following CD95 stimulation. Remarkably, strong Plk3 expression sensitized cells to CD95-mediated apoptosis as indicated by increasing levels of p18 and caspase-8 activity upon CD95 stimulation (Figure 7G, left and right panels) supporting the idea that phosphorylation of T273 enhances caspase-8 activity in gastrointestinal cancer.
Discussion
The activation of the DISC within the CD95/CD95L system occurs through an “induced proximity” mechanism involving dimerization of procaspase-8 molecules, which facilitates activation through subsequent self-processing5,44. However, our knowledge about the complex mechanisms that regulate this apoptotic pathway in the context of cancer development is still incomplete. Various reports revealed that the overexpression of exogenous Plk3 induces apoptosis45,46. Moreover, a genome-wide screen revealed Plk3 as modulator of caspase activity27. A kinome-wide screening for modulators of cisplatin response in ovarian cancers found that the inhibition of Plk3 sensitized SKOV3 cells to cisplatin28. Despite these indications that Plk3 regulates apoptosis the mechanistic details remain completely elusive.
Here, we report the constitutive association of Plk3 to CD95 in different types of cancer cells. Plk3 binds with its C-terminal moiety, which includes the interdomain and the PBD, to CD95 (Figure 1D and 1E). This is the first example for an interaction of a cell surface receptor with a member of the Plk family. Various studies have described the association of transmembrane receptors to downstream protein kinases, but this interaction requires the stimulation of the receptor by their corresponding ligands47,48,49. An example is that of TRANCE, a TNF family member, and its receptor, TRANCE-R, which regulate dendritic cell and osteoclast function. TRANCE activates the antiapoptotic serine/threonine kinase Akt/PKB through a signaling complex including c-Src and TRAF6, which is recruited to the TRANCE-R upon receptor engagement50. As a consequence of this association, TRAF-6 increases the kinase activity of c-src leading to phosphorylation of downstream substrates. However, the constitutive association of CD95 with protein kinases that is independent of extracellular stimuli as we describe here for CD95 and Plk3 has not been reported yet.
Upon CD95 stimulation with its ligand and the DISC formation the kinase activity of Plk3 was found to be upregulated (Supplementary information, Figure S7A). Depletion or KO of individual DISC components blocked processing of procaspase-8 and enzymatic activation of Plk3 (Supplementary information, Figure S7A). Previous reports have documented that Polo kinases are regulated by phosphorylation and proteolysis51. Human Plk1 has been shown to be activated in mitosis by phosphorylation at S137 and T210 by Aurora kinases as well as by binding of a phosphorylated peptide to the PBD40,51. In mammalian cells, this activation is supported by Bora, which may help to open the closed conformation of Plk1 and thereby expose the T-loop to upstream activating kinases. Remarkably, while a phospho-mimicking mutation at T210E increases the kinase activity of Plk1 markedly52, the corresponding mutation in Plk3 (T219D) blocked kinase activity (Figure 3D). Functional studies on Plk3 have not provided evidence yet that a phosphorylation of the T-loop is required for the activation of its kinase activity. While the constitutive association with the CD95 receptor is not sufficient for the enzymatic activation of Plk3, the CD95L induced assembly of the DISC was found to be required for the activation of Plk3 (Supplementary information, Figure S6D). In the case of Plk1 the binding of the PBD to phosphorylated targets at the proper subcellular location facilitates the dissociation of the PBD from kinase domain thereby relieving the inhibition of the PBD. The interaction of the PBD of Plk3 to DISC components could have a similar function for relieving its inhibitory impact on the kinase domain. Our data indicate that in the presence of sufficient procaspase-8 that is recruited to the DISC upon stimulation of the CD95 receptor the association of the KD with the PBD-containing domain of Plk3 is impaired by binding of procaspase-8 to the Plk3-PBD (Supplementary information, Figure S6D). This is accompanied at least with a partial activation of Plk3. In summary, the mechanisms that activate Plk3 seem to have common traits compared with Plk1. Novel insights on the regulation of Plk3 by phosphorylation and structural information on Plk3 with DISC components like procaspase-8 is required to improve our understanding of the activation of Plk3 at the DISC.
Moreover, we could demonstrate that Plk3 phosphorylates procaspase-8 at T273 in vitro and in vivo. The association of both proteins is mediated by the PBD of Plk3. Previously, it was shown that phospho-Ser/Thr-containing motifs bind the PBD from Plk153. However, the PBD is also capable of binding proteins that are unphosphorylated as has been shown for microtubule-associated protein Map20554. Another example of a noncanonical interaction was shown for the Plk1 homologue, Cdc5 in yeast, which interacts with the yeast protein Dbf455. Dbf4 associated with the Cdc5's PBD through a noncanonical R-S-I-E-G-A protein motif in Dbf4, which is not phosphorylated. Despite mutations in the PBD pincer it still interacts strongly to Dbf4 indicating that regions other than the classical pincer domain could mediate the interaction. Although we do not know yet whether the interaction of Plk3 with caspase-8 is phospho-dependent, the phosphorylation of caspase-8 by Plk3 is the first example for a pro-apoptotic phosphorylation of procaspase-8. Previous studies described phosphorylation events at Y380 by Src, S305 by Plk1, S364 by p38-MAPK, S387 by Erk1/2 and Cdk1/cyclin B1 within procaspase-8 that inhibit its processing32,56,57,58. Our study demonstrates the phosphorylation of the catalytic subunit p18 at T273 by Plk3. Using embedded-atom potential-based molecular-dynamics simulations, we found that a phosphorylation at this site, which is located in the second helix of the p18 subunit, is oriented exactly toward the second catalytic subunit, p10 (Supplementary information, Figure S7B). By interacting with R430, C426 or the backbone of p10 the phosphorylation of T273 could increase the affinity or stability of the catalytically active heterodimer thereby increasing the overall activity of caspase-8. Interestingly, the ribosomal S6 kinase 2 (RSK2), a member of the p90 ribosomal S6 kinase (p90RSK) family was reported to phosphorylate caspase-8 at T263 thereby mediating caspase-8 ubiquitination and degradation through the proteasome pathway59. Despite the close vicinity of both phosphorylation sites we could not detect reduced stability of procaspase-8 phosphorylated at T273 compared with the non-phosphorylated counterpart. Remarkably, S305 in procaspase-8 is phosphorylated by Plk1 during mitosis31,32. By phosphorylating S387 in procaspase-8, Cdk1/cyclin B1 generates a phospho-epitope for the binding of the PBD of Plk131. As overexpression of Plk1 and Plk3 were previously shown to have opposing effects on the survival of mammalian cells: promoting carcinogenesis versus induction of apoptosis45,46,60, their roles (anti-apoptotic versus pro-apoptotic) in the regulation of caspase-8 is in line with previous studies.
The expression of Plk3 in human cancer was found to be reduced in head and neck, lung and liver61. We analyzed the profile of caspase-8 in primary anal cancer and studied whether caspase-8 may be less phosphorylated at T273 in the pathological situations where Plk3 is downregulated. High Plk3 expression correlated with phosphorylation of caspase-8 at T273. High levels of Plk3 in cells of gastrointestinal origin showed high sensitivity toward CD95 stimulation compared with low Plk3 expressing cells. We could demonstrate for the first time that high Plk3 protein expression and high pT273 levels provide a survival benefit for cancer patients, highlighting the tumor suppressor function of Plk3 in human cancer. The observed correlation of Plk3 expression and T273 phosphorylation indicates that Plk3 expression in cancer tissues modulates the sensitivity of the extrinsic death pathway in primary tissues. Notably, a reduced local tumor control and distant failure significantly correlated with the pT273 signal, while only local failure correlated with high Plk3 immunoreactivity. This could indicate the presence of additional kinases that may activate caspase-8 by phosphorylation on T273 especially during metastatic progression or the presence of yet unknown factors which may increase the activity of Plk3 within the local tumor tissue. Among these factors, local accumulation of CD8+ tumor infiltrating lymphocytes carrying the CD95L may contribute to elevated levels of Plk3 activity. Supporting this assumption, recent findings indicate favorable survival rates in patients with anal cancer presenting a high density of intratumoral CD8+ cells62. Whether the presence of infiltrating CD8+ cells influences the activity of Plk3 and the level of pT273 caspase-8 in cancer cells is currently under investigation in our laboratory.
Materials and Methods
Antibodies
The following antibodies were used according to the respective manufacturers' recommendations — (1) Abcam: Caspase 8-p10 (catalog number ab2553/clone number C502S), pan-Cadherin (ab16505), GAPDH (ab9485), Nucleolin (ab22758), Plk3/PRK (ab33119); (2) BD Biosciences: FADD (556402/A66-2), Fas/ CD95 (610198/13/Fas); (3) BD Pharmingen: Calnexin (610524/37/Calnexin), Plk3 (556518/B37-2); (4) Cell Signaling Technology: Aurora B (3094S), phospho-Chk2 (pT68, 2661), cleaved Caspase-8 (D391/ 18C8), Cyclin B1 (4138S), Fas (8023/4C3), Phospho-p53 (pS20, 9496); (5) Enzo Life Sciences: Caspase-8 (ALX-804-429-C050/ C15); (6) MBL: Lamin B1 (JM-3046-100/ ZL-5); (7) Merck Millipore: Histone H3 (05-499/6.6.2), Plk1 (05-844/35-206), Plk3 (ABS89), Vimentin (MAB3400/V9); (8) R&D Systems: Caspase-8 (AF1650); (9) Santa Cruz Biotechnology: Caspase-8 p18 (sc-6136/C-20), Cdc2 p34 (Cdk1) (sc-54/17), FADD (sc-5559/H-181), GFP (sc-9996/B-2), GST (sc138/B-15); (10) Sigma-Aldrich: β-Actin (A5441/AC-15), Flag tag (FlagM2, F1804), Vinculin (V4505/VIN-11-5). To generate a polyclonal antibody against caspase-8 phosphorylated on T273, rabbits were immunized using the caspase-8-peptide containing phospho-T273 (C-GALTT(pT)FEELHFE-amide). The antibodies were affinity purified. Phospho-Caspase-8 (pS387) antibodies were generated and tested in our laboratory31. Secondary antibodies against rabbit (NA934V) and mouse (NXA931) primary antibodies were obtained from GE Healthcare.
Cell culture and transfection
Cell lines were purchased from ATCC and cultured according to their guidelines. Parental Plk3-WT and Plk3-KO (HZGHC003037c002 and HZGHC003037c011) HAP1 cells were purchased from Horizon Genomics. DNA and siRNA transfections were performed using TurboFect™ in vitro Transfection Reagent (Fermentas) and LipofectamineRNAiMAX Transfection Reagent (Invitrogen), respectively according to the manufacturer's instructions. Cell lysis was performed in RIPA buffer as described63. The control siRNA (siC) was from Qiagen, siCD95/Fas (sc-29311) and siFADD (sc-35352) from Santa Cruz Biotechnology.
Generation of plasmids and mutagenesis
Primer sequences used here will be provided upon request. Deletion fragments of CD95, Plk3 and procaspase-8 were generated by standard techniques. Site-directed mutagenesis was performed using the QuikChange protocol and PfuUltra II Fusion HS DNA polymerase (Stratagene). Procaspase-8-WT, its subfragments (NT, p18, p10) and its mutants were inserted into the HindIII/XbaI sites of the pGEX 5X-3 vector (GE Healthcare). 3 × Flag-tagged procaspase-8-WT and its mutants (T273A, -T273D) were inserted into the BamHI/XbaI sites of the pcDNA3.1-V5 vector (Invitrogen). Plk3-WT and its subfragments were inserted into the BamHI and EcoRI sites of the Myc-, V5- or 3xFlag-tagged pcDNA3.1-Hygro+ vector (Invitrogen) and the pGEX 5X-3 vector. All constructs were confirmed by sequencing.
Generation of Casp8- and Plk3-KO cell lines using the CRISPR/Cas9 system
To stably KO the expression of procaspase-8 in HeLa cells, selected sequences were individually inserted into the CRISPR vector pXH1 within the BamHI/BbsI restriction sites. pXH1 was generated by exchanging the U6 promoter of the pX260 vector with a human H1-promoter for high frequency expression of sgRNA. Sequences for sgRNAs used in the present study will be provided upon request. Cells were transfected with the CRISPR vector and selected for 72 h in medium containing Puromycin (0.75 g/ml). Subsequently single clones were selected through serial dilution. Clones were verified by PCR and sequencing.
Parental Plk3-WT and Plk3-KO (HZGHC003037c002 and HZGHC003037c011) HAP1 cells were purchased from Horizon Genomics. HAP1 Plk3-KO clones were engineered using CRISPR/Cas9. Exon 2 (NM_004073) was selected for guide RNA design (GCTGATGCGGCTTGGCGACG). Both HAP1 clones contain a 13 bp deletion in exon 2, causing a frameshift in Plk3 gene.
Separation of subcellular compartments
Cytoplasmic, nuclear, membrane and cytoskeletal proteins were extracted using a Compartmental Protein Extraction Kit (Millipore). Cytosolic and membrane separation from Jurkat cells was performed using hypotonic buffer.
Immunoprecipitation and GST pull-down
Cell lysates were incubated with Protein G Sepharose beads (GE Healthcare) and the specific antibody overnight at 4 °C. Immunoprecipitates were washed 3× with ice-cold buffer (20 mM Tris-HCl pH 8.2, 150 mM NaCl, 1% (v/v) TritonX-100). GST pull-down assays were performed as previously described31.
Apoptosis induction and analysis
To induce apoptosis, cells were stimulated with SuperFasLigand (100 ng/ml) or SuperKillerTRAIL (50 ng/ml) (Enzo Life Sciences) in the presence of 1 μg/ml of cycloheximide (CHX, Sigma-Aldrich). Overall apoptosis was measured by staining the cells with Annexin V (556422, BD Biosciences)/7 AAD (559925, BD Biosciences) according to the manufacturer's protocol followed by their analysis using a FACScan (BD Biosciences). The early necrotic cells were considered to be Annexin V negative but 7 AAD positive, the apoptotic cells were considered to be positive for both Annexin V and 7 AAD64. The data were analyzed using the BD Cell Quest pro software (version 5.2.1, BD). Overall apoptosis was presented as the mean value ± SD (n = 3). The significance of differences between populations of data was assessed according to the Student's two-tailed test (*P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005).
In vitro kinase assay
This assay using recombinant purified Plk3, Plk1 and Cdk1 (ProQinase), respectively was performed as described previously31.
Caspase-8 and -3/7 assay
Caspase-Glo 8 and -Glo 3/7 assay kits (Promega) were used according to the manufacturer's instructions. The measured luminescence (RLU) was presented as the mean value ± SD (n = 3). The significance of differences between populations of data was assessed according to the Student's two-tailed test (*P ≤ 0.05; **P 0.005; ***P ≤ 0.0005).
Immunofluorescence assays
Cells were fixed and permeabilized with methanol (−20 °C). Mouse monoclonal anti-α-Tubulin-FITC antibody (Sigma-Aldrich) was used for staining of cytoskeleton. FITC goat anti-mouse (Alexa Fluor 488), Cy3 goat anti-rabbit (Alexa Fluor 594), goat anti-rat (Alexa Fluor 647) (Invitrogen) were used as secondary antibodies. DNA was stained using DAPI (4′,6-diamidino-2-phenylindol-dihydrochlorid) (Roche). Slides were examined using an Axio Imager 7.1 microscope (Zeiss), images were taken at room temperature using an inverted confocal laser-scanning microscope (CLSM) (Leica CTR 6500). Z stacks with optical sections of 1-μm intervals were collected using a 40×, 1.3 NA, and 60× oil, 1.4 NA, objective lenses and the LAS AF acquisition and imaging software (Leica). 3D rendering of confocal z stacks was performed using the LAS AF software (Leica).
Proximity ligation assay (PLA)
The assay was performed according to the manufacturer's (Olink Biosciences) protocol. DNA was stained with DAPI, cytoskeleton with a FITC-labeled α-tubulin antibody. The slides were analyzed using an Axio Observer microscope (Carl Zeiss), imaged and analyzed using the Axio Vision software (version 4.8.2.0, Carl Zeiss). The mean number of fluorescent dots was determined by counting of dots in three different fields, each field having approximately 15-20 cells (means ± SD). The significance of differences between single and double antibody staining was assessed according to the two-tailed Student's t-test (*P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005).
Mass spectrometry and data acquisition
Metabolic labeling of HeLa cells via SILAC was performed as described65. The proteins purified from the light and heavy-labeled cell batches were mixed in equimolar amounts and were analyzed by mass spectrometry as described66. Proteins with a “normalized heavy/light ratio” of > 3 were considered as high-confidence members of the Plk3 interactome. The full list of identified/quantified proteins is given in Supplementary information, Table S1.
Patients' characteristics
After institutional review board approval, 95 patients, treated homogeneously with curative intent 5-fluorouracil (5-FU)/mitomycin C-based chemoradiotherapy (CRT) for anal squamous cell carcinoma at the Departments of Radiotherapy of the University Hospital (Frankfurt) and of the University Medical Center (Göttingen) who provided informed consent, were included in this study67.
Scoring for Plk3 expression and phosphorylation of Caspase- 8-T273
Pretreatment formalin-fixed, paraffin-embedded (FFPE) biopsy tissue was subject to an standardized horseradish peroxidase technique (DAKO Envision Flex) with primary anti-Plk3 (Abcam) and pT273 caspase-8 antibodies applied at a 1:100 dilution. Plk3 immunoreactivity was analyzed considering both the percentage of positive cells (1 (0%-25%), 2 (26%-50%), 3 (51%-75%) and 4 (> 75%)) and the intensity of staining scored as 1+ (weak), 2+ (moderate) and 3+ (intense). These parameters were multiplied to produce an individual weighted score (WS). A WS ≤ 6 was defined as “low” and a WS of > 6 as “high” Plk3 expression. Phosphorylation of caspase-8-T273 was assessed by dividing the number of positive tumor cells by the total number of tumor cells, multiplied by 100 and a correlation to the Plk3-WS was analyzed according to a Spearman's rank test. Disease-free (DFS) and overall survival (OS) were defined as the time of start of CRT to death by anal cancer or any reasons or the day of the last follow-up. Survival parameters were determined according to the Kaplan-Meier method and significances were calculated by the log-rang test using SPSS 21 software.
Statistical methods
All experiments were performed at least in triplicate. Standardization and statistics were determined as described68,69. For paired t-tests, all experimental groups were compared with their respective groups. Significant differences (*P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005) are indicated in the figures with asterisks.
Author Contributions
CH, HU, CR and KS designed the study, interpreted the data and wrote the manuscript; CH, MR, FR, YM, TO, RM, and MS performed the experiments and analyzed the data.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
This work was supported by grants from the German Cancer Consortium (DKTK) (Heidelberg), Wilhelm Sander-Stiftung, Deutsche Krebshilfe, Carls-Stiftung, and BANSS Stiftung. We thank Denis Kudlinzki for the computer-based analysis of the caspase-8 conformation.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Interaction of Plk3 and CD95.
Analysis of the subcellular localization of Plk3.
The Plk3 consensus site and phylogenetic alignment of caspase-8-p18.
In vitro validation of the new phospho-caspase-8 antibody (pT273).
Phosphorylation of caspase-8 by Plk3.
Analysis of intramolecular interaction of Plk3 subdomains.
Proposed model for the function of Plk3 in the extrinsic apoptotic signaling and influence of the phosphorylation of procaspase-8 at T273 on its 3D structure.
Mass spectrometric analysis of the interactome of exogenous Plk3 obtained from SILAC.
References
- Galluzzi L, Kepp O, Kroemer G. Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol 2012; 13:780–788. [DOI] [PubMed] [Google Scholar]
- Oberst A, Green DR. It cuts both ways: reconciling the dual roles of caspase 8 in cell death and survival. Nat Rev Mol Cell Biol 2011; 12:757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischkel FC, Hellbardt S, Behrmann I, et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J 1995; 14:5579–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischkel FC, Lawrence DA, Tinel A, et al. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J Biol Chem 2001; 276:46639–46646. [DOI] [PubMed] [Google Scholar]
- Medema JP, Scaffidi C, Kischkel FC, et al. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC). EMBO J 1997; 16:2794–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi C, Fulda S, Srinivasan A, et al. Two CD95 (APO-1/Fas) signaling pathways. EMBO J 1998; 17:1675–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Villalba A, Llorens-Bobadilla E, Wollny D. CD95 in cancer: tool or target? Trends Mol Med 2013; 19:329–335. [DOI] [PubMed] [Google Scholar]
- Peter ME, Budd RC, Desbarats J, et al. The CD95 receptor: apoptosis revisited. Cell 2007; 129:447–450. [DOI] [PubMed] [Google Scholar]
- Trauth BC, Klas C, Peters AM, et al. Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science 1989; 245:301–305. [DOI] [PubMed] [Google Scholar]
- Ogasawara J, Watanabe-Fukunaga R, Adachi M, et al. Lethal effect of the anti-Fas antibody in mice. Nature 1993; 364:806–809. [DOI] [PubMed] [Google Scholar]
- Lemke J, von Karstedt S, Zinngrebe J, Walczak H. Getting TRAIL back on track for cancer therapy. Cell Death Differ 2014; 21:1350–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Targeting Polo-like kinases: a promising therapeutic approach for cancer treatment. Transl Oncol 2015; 8:185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 2009; 9:28–39. [DOI] [PubMed] [Google Scholar]
- Glover DM. Polo kinase and progression through M phase in Drosophila: a perspective from the spindle poles. Oncogene 2005; 24:230–237. [DOI] [PubMed] [Google Scholar]
- Lee KS, Park JE, Asano S, Park CJ. Yeast polo-like kinases: functionally conserved multitask mitotic regulators. Oncogene 2005; 24:217–229. [DOI] [PubMed] [Google Scholar]
- Eckerdt F, Yuan J, Strebhardt K. Polo-like kinases and oncogenesis. Oncogene 2005; 24:267–276. [DOI] [PubMed] [Google Scholar]
- Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov 2010; 9:643–660. [DOI] [PubMed] [Google Scholar]
- Holtrich U, Wolf G, Brauninger A, et al. Induction and down-regulation of PLK, a human serine/threonine kinase expressed in proliferating cells and tumors. Proc Natl Acad Sci USA 1994; 91:1736–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Grenfell TZ, Yarm FR, Erikson RL. Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. Proc Natl Acad Sci USA 1998; 95:9301–9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Kappel S, Kramer A, et al. Toxicity modelling of Plk1-targeted therapies in genetically engineered mice and cultured primary mammalian cells. Nat Commun 2011; 2:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R, Strebhardt K. Plk1: unexpected roles in DNA replication. Cell Res 2013; 23:1251–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BT, Strebhardt K. Polo-like kinase 1: target and regulator of transcriptional control. Cell Cycle 2006; 5:2881–2885. [DOI] [PubMed] [Google Scholar]
- Yuan J, Kramer A, Eckerdt F, Kaufmann M, Strebhardt K. Efficient internalization of the polo-box of polo-like kinase 1 fused to an Antennapedia peptide results in inhibition of cancer cell proliferation. Cancer Res 2002; 62:4186–4190. [PubMed] [Google Scholar]
- Li B, Ouyang B, Pan H, et al. Prk, a cytokine-inducible human protein serine/threonine kinase whose expression appears to be down-regulated in lung carcinomas. J Biol Chem 1996; 271:19402–19408. [DOI] [PubMed] [Google Scholar]
- Holtrich U, Wolf G, Yuan J, et al. Adhesion induced expression of the serine/threonine kinase Fnk in human macrophages. Oncogene 2000; 19:4832–4839. [DOI] [PubMed] [Google Scholar]
- Dai W, Li Y, Ouyang B, et al. PRK, a cell cycle gene localized to 8p21, is downregulated in head and neck cancer. Genes Chromosomes Cancer 2000; 27:332–336. [DOI] [PubMed] [Google Scholar]
- Yi CH, Sogah DK, Boyce M, Degterev A, Christofferson DE, Yuan J. A genome-wide RNAi screen reveals multiple regulators of caspase activation. J Cell Biol 2007; 179:619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S, Bisanz KM, Peralta LA, et al. RNAi screening of the kinome identifies modulators of cisplatin response in ovarian cancer cells. Gynecol Oncol 2010; 118:220–227. [DOI] [PubMed] [Google Scholar]
- Peter ME, Krammer PH. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ 2003; 10:26–35. [DOI] [PubMed] [Google Scholar]
- Mandal P, Berger SB, Pillay S, et al. RIP3 induces apoptosis independent of pronecrotic kinase activity. Mol Cell 2014; 56:481–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthess Y, Raab M, Sanhaji M, Lavrik IN, Strebhardt K. Cdk1/cyclin B1 controls Fas-mediated apoptosis by regulating caspase-8 activity. Mol Cell Biol 2010; 30:5726–5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthess Y, Raab M, Knecht R, Becker S, Strebhardt K. Sequential Cdk1 and Plk1 phosphorylation of caspase-8 triggers apoptotic cell death during mitosis. Mol Oncol 2014. [DOI] [PMC free article] [PubMed]
- Jiang N, Wang X, Jhanwar-Uniyal M, Darzynkiewicz Z, Dai W. Polo box domain of Plk3 functions as a centrosome localization signal, overexpression of which causes mitotic arrest, cytokinesis defects, and apoptosis. J Biol Chem 2006; 281:10577–10582. [DOI] [PubMed] [Google Scholar]
- Xie S, Wu H, Wang Q, et al. Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway. J Biol Chem 2001; 276:43305–43312. [DOI] [PubMed] [Google Scholar]
- Salvi M, Trashi E, Cozza G, Franchin C, Arrigoni G, Pinna LA. Investigation on PLK2 and PLK3 substrate recognition. Biochim Biophys Acta 2012; 1824:1366–1373. [DOI] [PubMed] [Google Scholar]
- Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res 2011; 39(Database issue):D253–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Sander C, Marks D. mRNA turnover rate limits siRNA and microRNA efficacy. Mol Syst Biol 2010; 6:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer 2006; 6:321–330. [DOI] [PubMed] [Google Scholar]
- Raab M, Pachl F, Kramer A, et al. Quantitative chemical proteomics reveals a Plk1 inhibitor-compromised cell death pathway in human cells. Cell Res 2014; 24:1141–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Shen C, Wang T, Quan J. Structural basis for the inhibition of Polo-like kinase 1. Nat Struct Mol Biol 2013; 20:1047–1053. [DOI] [PubMed] [Google Scholar]
- Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 1996; 85:803–815. [DOI] [PubMed] [Google Scholar]
- Muzio M, Chinnaiyan AM, Kischkel FC, et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 1996; 85:817–827. [DOI] [PubMed] [Google Scholar]
- Reissfelder C, Stamova S, Gossmann C, et al. Tumor-specific cytotoxic T lymphocyte activity determines colorectal cancer patient prognosis. J Clin Invest 2015; 125:739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. An induced proximity model for caspase-8 activation. J Biol Chem 1998; 273:2926–2930. [DOI] [PubMed] [Google Scholar]
- Wang Q, Xie S, Chen J, et al. Cell cycle arrest and apoptosis induced by human Polo-like kinase 3 is mediated through perturbation of microtubule integrity. Mol Cell Biol 2002; 22:3450–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn CW, Hennigan RF, Dai W, Sanchez Y, Stambrook PJ. Incomplete cytokinesis and induction of apoptosis by overexpression of the mammalian polo-like kinase, Plk3. Cancer Res 2000; 60:6826–6831. [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell 2010; 141:1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd CE. Lymphocyte signaling: adapting new adaptors. Curr Biol 1998; 8:R805–808. [DOI] [PubMed] [Google Scholar]
- Kleber S, Sancho-Martinez I, Wiestler B, et al. Yes and PI3K bind CD95 to signal invasion of glioblastoma. Cancer Cell 2008; 13:235–248. [DOI] [PubMed] [Google Scholar]
- Wong BR, Besser D, Kim N, et al. TRANCE, a TNF family member, activates Akt/PKB through a signaling complex involving TRAF6 and c-Src. Mol Cell 1999; 4:1041–1049. [DOI] [PubMed] [Google Scholar]
- Zitouni S, Nabais C, Jana SC, Guerrero A, Bettencourt-Dias M. Polo-like kinases: structural variations lead to multiple functions. Nat Rev Mol Cell Biol 2014; 15:433–452. [DOI] [PubMed] [Google Scholar]
- Jang YJ, Ma S, Terada Y, Erikson RL. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J Biol Chem 2002; 277:44115–44120. [DOI] [PubMed] [Google Scholar]
- Elia AE, Rellos P, Haire LF, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 2003; 115:83–95. [DOI] [PubMed] [Google Scholar]
- Archambault V, D'Avino PP, Deery MJ, Lilley KS, Glover DM. Sequestration of Polo kinase to microtubules by phosphopriming-independent binding to Map205 is relieved by phosphorylation at a CDK site in mitosis. Genes Dev 2008; 22:2707–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Weinreich M. Dbf4 regulates the Cdc5 Polo-like kinase through a distinct non-canonical binding interaction. J Biol Chem 2010; 285:41244–41254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R, Raab M, Matthess Y, Becker S, Knecht R, Strebhardt K. pERK 1/2 inhibit Caspase-8 induced apoptosis in cancer cells by phosphorylating it in a cell cycle specific manner. Mol Oncol 2014; 8:232–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cursi S, Rufini A, Stagni V, et al. Src kinase phosphorylates Caspase-8 on Tyr380: a novel mechanism of apoptosis suppression. EMBO J 2006; 25:1895–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado-Kristensson M, Melander F, Leandersson K, Ronnstrand L, Wernstedt C, Andersson T. p38-MAPK signals survival by phosphorylation of caspase-8 and caspase-3 in human neutrophils. J Exp Med 2004; 199:449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller D, Crosetto N, Blagoev B, et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol 2006; 8:163–169. [DOI] [PubMed] [Google Scholar]
- Smith MR, Wilson ML, Hamanaka R, et al. Malignant transformation of mammalian cells initiated by constitutive expression of the polo-like kinase. Biochem Biophys Res Commun 1997; 234:397–405. [DOI] [PubMed] [Google Scholar]
- Helmke C, Becker S, Strebhardt K. The role of Plk3 in oncogenesis. Oncogene 2016; 35:135–147. [DOI] [PubMed] [Google Scholar]
- Hu WH, Miyai K, Cajas-Monson LC, Luo L, Liu L, Ramamoorthy SL. Tumor-infiltrating CD8+ T lymphocytes associated with clinical outcome in anal squamous cell carcinoma. J Surg Oncol 2015; 112:421–426. [DOI] [PubMed] [Google Scholar]
- Yuan J, Kramer A, Matthess Y, et al. Stable gene silencing of cyclin B1 in tumor cells increases susceptibility to taxol and leads to growth arrest in vivo. Oncogene 2006; 25:1753–1762. [DOI] [PubMed] [Google Scholar]
- Prieto A, Diaz D, Barcenilla H, et al. Apoptotic rate: a new indicator for the quantification of the incidence of apoptosis in cell cultures. Cytometry 2002; 48:185–193. [DOI] [PubMed] [Google Scholar]
- Oellerich T, Gronborg M, Neumann K, Hsiao HH, Urlaub H, Wienands J. SLP-65 phosphorylation dynamics reveals a functional basis for signal integration by receptor-proximal adaptor proteins. Mol Cell Proteomics 2009; 8:1738–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej S, Kuvardina ON, Oellerich T, et al. PADI4 acts as a coactivator of Tal1 by counteracting repressive histone arginine methylation. Nat Commun 2014; 5:3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodel F, Wieland U, Fraunholz I, et al. Human papillomavirus DNA load and p16INK4a expression predict for local control in patients with anal squamous cell carcinoma treated with chemoradiotherapy. Int J Cancer 2015; 136:278–288. [DOI] [PubMed] [Google Scholar]
- Spankuch-Schmitt B, Bereiter-Hahn J, Kaufmann M, Strebhardt K. Effect of RNA silencing of polo-like kinase-1 (PLK1) on apoptosis and spindle formation in human cancer cells. J Natl Cancer Inst 2002; 94:1863–1877. [DOI] [PubMed] [Google Scholar]
- Spankuch-Schmitt B, Wolf G, Solbach C, et al. Downregulation of human polo-like kinase activity by antisense oligonucleotides induces growth inhibition in cancer cells. Oncogene 2002; 21:3162–3171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Interaction of Plk3 and CD95.
Analysis of the subcellular localization of Plk3.
The Plk3 consensus site and phylogenetic alignment of caspase-8-p18.
In vitro validation of the new phospho-caspase-8 antibody (pT273).
Phosphorylation of caspase-8 by Plk3.
Analysis of intramolecular interaction of Plk3 subdomains.
Proposed model for the function of Plk3 in the extrinsic apoptotic signaling and influence of the phosphorylation of procaspase-8 at T273 on its 3D structure.
Mass spectrometric analysis of the interactome of exogenous Plk3 obtained from SILAC.