Abstract
Proteins involved in DNA double-strand break (DSB) repair localize within the promyelocytic leukemia nuclear bodies (PML-NBs), whose disruption is at the root of the acute promyelocytic leukemia (APL) pathogenesis. All-trans-retinoic acid (RA) treatment induces PML-RARα degradation, restores PML-NB functions, and causes terminal cell differentiation of APL blasts. However, the precise role of the APL-associated PML-RARα oncoprotein and PML-NB integrity in the DSB response in APL leukemogenesis and tumor suppression is still lacking. Primary leukemia blasts isolated from APL patients showed high phosphorylation levels of H2AX (γ-H2AX), an initial DSBs sensor. By addressing the consequences of ionizing radiation (IR)-induced DSB response in primary APL blasts and RA-responsive and -resistant myeloid cell lines carrying endogenous or ectopically expressed PML-RARα, before and after treatment with RA, we found that the disruption of PML-NBs is associated with delayed DSB response, as revealed by the impaired kinetic of disappearance of γ-H2AX and 53BP1 foci and activation of ATM and of its substrates H2AX, NBN, and CHK2. The disruption of PML-NB integrity by PML-RARα also affects the IR-induced DSB response in a preleukemic mouse model of APL in vivo. We propose the oncoprotein-dependent PML-NB disruption and DDR impairment as relevant early events in APL tumorigenesis.
The DNA damage response (DDR) includes cell cycle arrest and transcriptional and post-translational activation of genes involved in DNA repair and triggering of apoptosis. Deficiencies in DDR are fundamental to the etiology of most human cancers.1
The DNA double-strand breaks (DSBs) arise from endogenous and exogenous sources, including replication errors, chemical mutagens, and ionizing radiation (IR).2 The sensing phase of the DSB response includes their recognition by the MRE11/RAD50/NBN complex, ATM protein activation, histone H2AX phosphorylation at Ser139 (γ-H2AX), MDC1, and 53BP1 recruitment. γ-H2AX is an initial DSB sensor for subsequent accumulation and post-translational modification (PTM) of signaling and repair proteins to form the so-called IR-induced foci (IRIF).3, 4, 5, 6, 7, 8 Finally, DSBs are removed through either the non-homologous end-joining (NHEJ) or the homologous recombination repair (HRR) pathways.9
Many proteins involved in the DDR localize within the promyelocytic leukemia (PML)-nuclear bodies (PML-NBs).10, 11, 12, 13, 14, 15, 16 PML-NBs are nuclear organelles implicated in cellular processes relevant to tumor suppression (e.g., PTM, DDR, transcriptional regulation, induction of apoptosis, and senescence).16, 17, 18, 19, 20, 21 PML-NBs increase in number and change their subnuclear distribution in response to DNA damage and represent structures where protein complexes are assembled, anchored, and/or post-translationally modified.11, 13, 15, 16, 22 PML also undergoes PTMs, SUMOylation representing one of the main modification necessary for the proper NB biogenesis and regulation of the cellular response to the DNA damage.23
Aberrant levels of the PML protein and loss of PML-NBS integrity were reported in acute promyelocytic leukemia (APL) and in other tumors.24 PML gene was originally cloned in APL leukemic cells carrying the t(15;17) detectable in over 95% of APLs. This translocation involves the PML and the retinoic acid receptor-α (RARα) genes, and generates the PML-RARα fusion gene.25, 26, 27 PML-RARα acts as a transcriptional repressor antagonizing myeloid differentiation and promoting the self-renewal capacity of APL-initiating cells.28, 29 PML-RARα also competitively inhibits the oligomerization of wild-type (WT) PML proteins, leading to the disruption of PML-NBs into nuclear ‘microspeckles'.30 In APL cells, all-trans retinoic acid (RA) treatment restores NB integrity, reverses PML-RARα-mediated transcriptional inhibition, and induces PML-RARα degradation and terminal cell differentiation.29, 31, 32
To date, it is yet unclear to what extent the DSBS repair is dependent upon PML and PML-NBS function. Here, the relationship existing between PML-NBS integrity and IR-induced DSBS sensing, signaling, and repair has been investigated in leukemic cells derived from APL individuals, myeloid cell lines expressing or not the PML-RARα and in a PML-RARα preleukemic mouse model in vivo. Data obtained show that the expression of PML-RARα in myeloid cells causes basal damage and a defective DSBS response, highlighting the pivotal role of PML-NBs in coordinating and regulating the early and late events of DDR in APL. Overall, our results suggest that PML-RARα-dependent PML-NBS disruption and DDR impairment are relevant early events in APL tumorigenesis.
Results
The integrity of PML-NBs is required for a proper DSBs rejoining
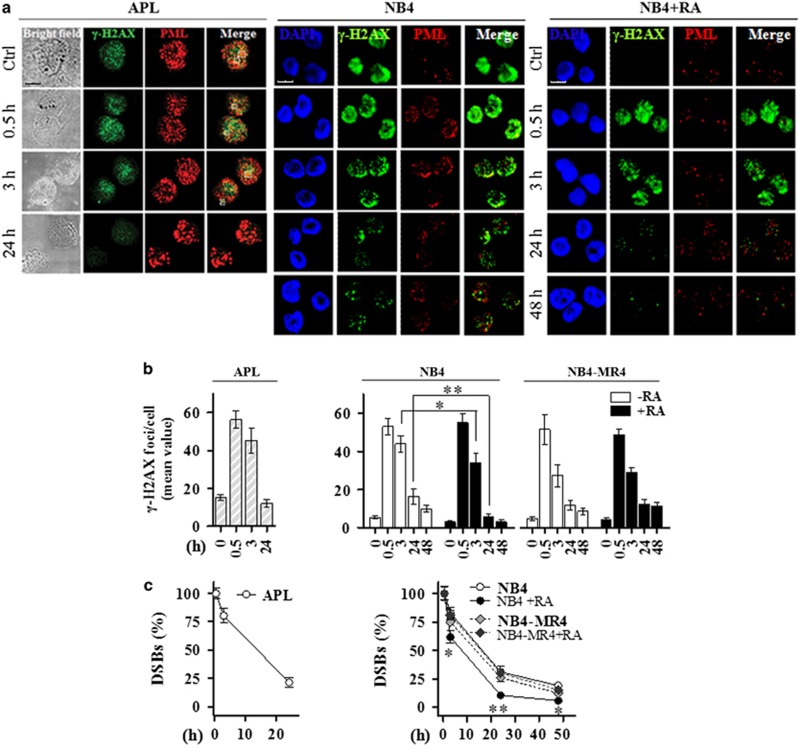
Double immunofluorescence staining with anti-PML and anti-γ-H2AX antibodies allowed the detection of disassembled PML-NBs and high number of γ-H2AX foci in primary blasts isolated at diagnosis from three APL patients, harboring the chromosomal translocation t(15;17) resulting in the PML-RARα fusion product (also confirmed by RT-PCR, Figures 1a and b). Biological and clinical features of these APL cases are reported in Supplementary Table S1. Similar results were observed in the APL-derived NB4 cell line and in its RA-resistant derived subclone NB4-MR4 (Figure 1a and Supplementary Figure S1A).
Figure 1.
PML-NB integrity and γ-H2AX dephosphorylation kinetics. (a) Representative images of the double immunofluorescence analysis of γ-H2AX (Alexa Fluor 488, green fluorophore) and PML (Alexa Fluor 610, red fluorophore) foci in APL blasts untreated (Ctrl) and exposed to 1 Gy of X-rays and fixed after 0.5, 3, and 24 h (cell image: bright field; confocal microscopy images, magnification × 63), and NB4 cells treated or not with 1 μM RA for 72 h, and then exposed to IR and fixed after 0.5, 3, 24, and 48 h (counterstain: DAPI; confocal microscopy images, magnification × 63). (b) Quantification of the mean number of γ-H2AX foci/cell and (c) analysis of the rejoining capability measured as percentage of DSBs in untreated and irradiated APL blasts derived from 3 different individuals, and NB4 and NB4-MR4 cells untreated or treated with 1 μM RA for 72 h and then irradiated with 1 Gy. For NB4 and NB4-MR4 DSB rejoining graphs; * and ** indicate significant differences of RA-treated NB4 cells with respect to RA-untreated NB4 cells and NB4-MR4 cells. Mean values were derived from the analysis of 100 cells in 3 independent experiments±S.D. For DSBs, the mean number of γ-H2AX foci/cell measurable at 0.5 h after IR was taken as 100%. *P<0.05; **P<0.01. Confocal analysis was performed using the LCS Leica confocal microscope (Leica Microsystems, Heidelberg, Germany)
The DSBs repair capability after 1 Gy of X-rays was evaluated as mean value of γ-H2AX foci/cell and as percentage of residual DSBs at each time point. In primary APL blasts, the number of γ-H2AX foci reached a peak after 0.5 h from IR, and slowly decreased after 3 h (∼45 foci/cell and 80% of unrepaired DSBs). A high number of unrepaired DSBs persisted after 24 h from IR, highlighting a delay in the DSBS repair in APL blasts (∼12 foci/cell and 24% of unrepaired DSBs) (Figures 1b and c).
Confocal microscopy also revealed a high number of γ-H2AX foci/cell in the untreated NB4 and RA-resistant NB4-MR4 cells (Figures 1 and b and Supplementary Figure S1). The peak in the mean value of γ-H2AX foci/cell was reached at 0.5 h from IR in both RA-untreated and RA-treated cells. A significant difference in the mean value of γ-H2AX foci/cell and in the percentage of residual DSBs was observed after 3 h from IR (~80% of unrepaired DSBs in NB4, NB4-MR4, and RA-treated NB4-MR4 cells; ~60% of unrepaired DSBs in RA-treated NB4 cells; P<0.05; Figures 1b and c). Similar to APL blasts, the DSB rejoining profile in NB4 cells showed that at 24–48 h from IR the number of persisting DSBs was significantly higher in NB4 cells and in NB4-MR4 cells, than in RA-treated NB4 cells (Figures 1b and c). In these cells PML-NBs were restored following PML-RARα degradation and granulopoiesis was induced in a time-dependent and IR-independent manner, as revealed by an increased expression of he myeloid differentiation marker CD11b (Supplementary Figures S1B and C).
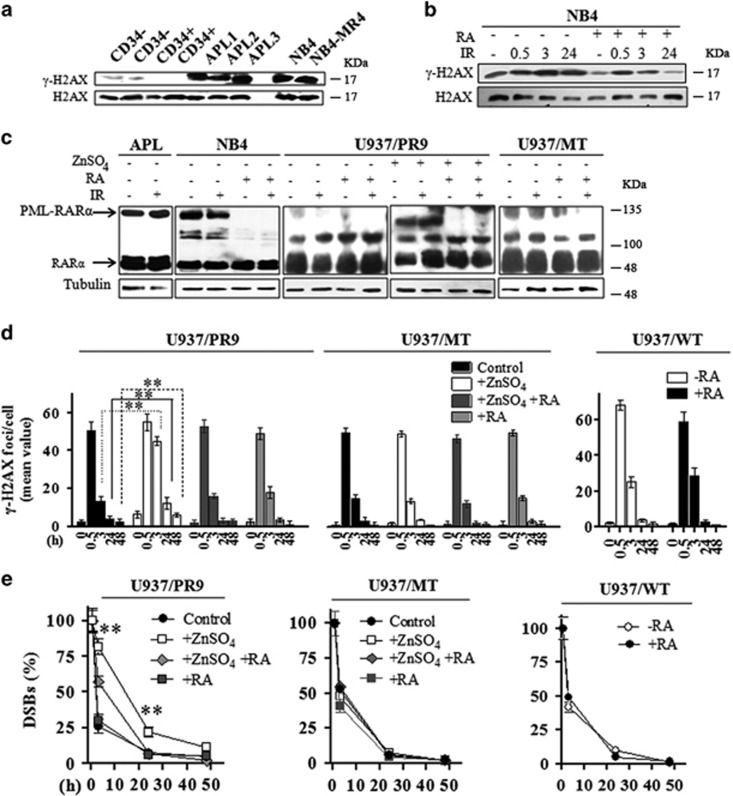
By immunoblot analysis we further observed that γ-H2AX was undetectable or barely detectable in human CD34+ hematopoietic progenitor cells (HPCs) and CD34− mononuclear cells (MNCs), respectively, whereas high levels of γ-H2AX were observed in PML-RARα-expressing samples from three APL patients, as well as in NB4 and NB4-MR4 cells (Figure 2a). RA treatment in NB4 cells decreased the levels of γ-H2AX when compared with untreated cells. Following IR, in NB4 cells the highest levels of γ-H2AX were detectable after 3 h, persisting until 24 h, whereas in RA-treated cells γ-H2AX levels reached a peak at 0.5 h and gradually decreased until 24 h (Figure 2b). IR treatment did not exert any effect on RARα or PML-RARα expression levels in primary APL blasts and NB4 cells (Figure 2c).
Figure 2.
(a) Representative immunoblot analysis of H2AX and H2AX phosphorylation at the Ser139 residue in untreated human CD34− and CD34+ cells isolated from the peripheral blood of normal donors, in three APL patients, in NB4 and NB4-MR4 cells. (b) Representative immunoblot analysis of H2AX phosphorylation in NB4 cells treated or not with 1 μM RA for 72 h and then irradiated with 1 Gy of X-rays and lysed after 0.5, 3, and 24 h. (c) Representative immunoblot analysis of RARα and PML-RARα expression levels in APL blasts, NB4, U937/PR9, and U937/MT cells exposed to IR and lysed after 0.5 h; before irradiation, NB4 cells were treated or not with 1 μM RA for 72 h, whereas U937/PR9 cells were treated or not with 100 μM ZnSO4 for 8 h and then with 1 μM RA for 72 h, as indicated; filters were probed with anti-RARα antibody, and tubulin was used as loading control. (d) Quantification of the mean number of γ-H2AX foci/cell and (e) analysis of the rejoining capability measured as percentage of DSBs in U937/PR9, U937/MT, and U937/WT cells treated or not with 100 μM ZnSO4 for 8 h and/or with 1 μM RA for 72 h, as indicated; cells were then exposed to IR and fixed at the indicated times. Mean values were derived from the analysis of 100 cells in three independent experiments±S.D. For DSBs, the mean number of γ-H2AX foci/cell measurable at 0.5 h after IR was taken as 100%. **P<0.01. For U937/PR9 DSB rejoining graph; **indicates significant differences of ZnSO4-treated U937/PR9 cells with respect to U937/PR9 control cells. Confocal analysis was performed using the LCS Leica confocal microscope (Leica Microsystems)
IR-treated U937/PR9 cells induced to express the PML-RARα oncoprotein by ZnSO4 provided results similar to those observed in APL blasts and NB4 cells. U937/PR9+ZnSO4 cells displayed ~80%, 20%, and 10% of persisting DSBs after 3, 24, and 48 h from IR, respectively (Figures 2d and e). After RA treatment in U937/PR9+ZnSO4 cells, resulting in PML/RARα degradation and PML-NBS reformation, the percentage of persisting DSBs was 60%, 10%, and 2% after 3, 24, and 48 h, respectively. Interestingly, similar γ-H2AX foci/cell mean values and DSBS rejoining profiles were measurable, before or after treatment with ZnSO4 and/or RA, in myeloid cell lines expressing the WT PML protein such as U937/MT, U937/WT, HL60, and HL60-R, a RA-resistant subclone of HL60 (Figures 2d and e and Supplementary Figures S2A–C). Of note, the IR treatment did not exert any effect on RARα or PML-RARα expression levels in U937/PR9 and U937/MT cells (Figure 2c). This further suggested a role for the PML-NB disruption following PML-RARα expression in the DSB rejoining proficiency of myeloid cells.
The integrity of PML-NBs is required for the recruitment of 53BP1 to the DSBs
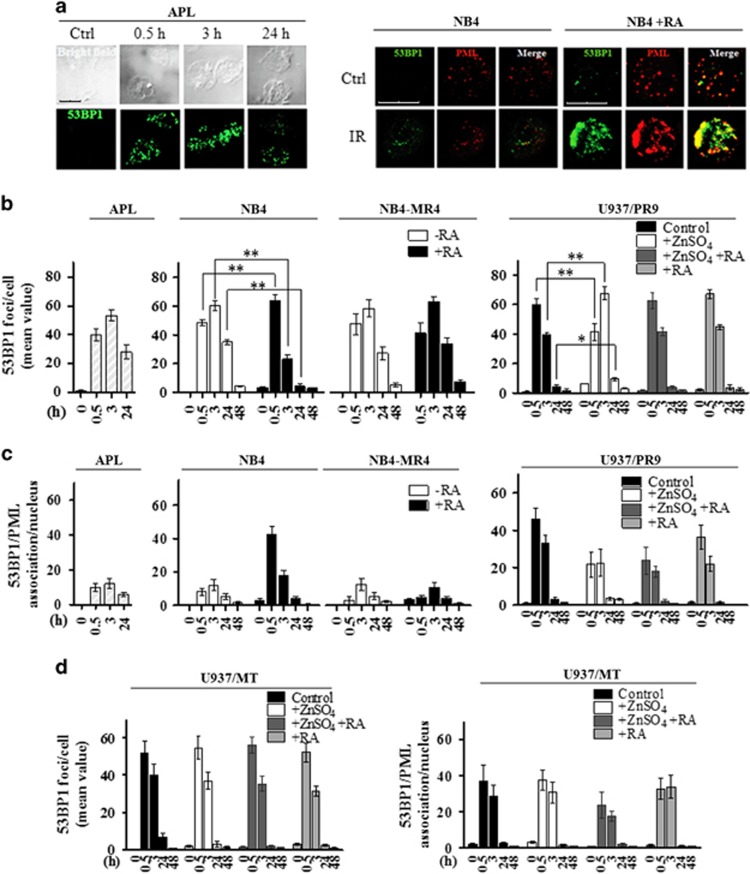
53BP1 accumulates within the PML-NBs and is recruited into IRIF after DSBS induction, promoting the activation of the repair signaling.33 Therefore, we studied the DSB kinetics by counting the number of 53BP1 foci in primary APL cells and NB4 and NB4-MR4 cells after 0.5, 3, and 24 h from irradiation with 1 Gy. We found that PML-NBS integrity is required for 53BP1 localization into the nuclei and for 53BP1 foci formation after DSBS induction. In fact, 53BP1 was barely detectable in non-irradiated APL blasts and NB4 cells, probably because of a weak basal expression of 53BP1 or of its pan-nuclear dispersion into the disassembled PML-NBs. On the contrary, 53BP1 colocalized with PML within the restored PML-NBs following RA treatment of NB4 cells (Figure 3a). After IR-induced damage, the 53BP1 foci number and colocalization with PML was significantly lower in RA-untreated APL blasts and NB4 and NB4-MR4 cells compared with RA-treated NB4 cells (Figures 3a–c). Thus, restoration of the 53BP1 foci within the reformed PML-NBs may occur as a consequence of the RA-induced PML-RARα degradation.
Figure 3.
PML-NB integrity and 53BP1 recruitment to the DSBs. (a) Representative images of 53BP1 foci disappearance in APL blasts untreated (Ctrl) and exposed to 1 Gy and fixed after 0.5, 3, and 24 h, and in RA-untreated (NB4) and RA-treated (NB4+RA) cells non-irradiated (Ctrl) and exposed to IR and fixed after 0.5 h (cell image: bright field; confocal microscopy images, magnification × 63; 53BP1 (Alexa Fluor 488, green fluorophore); PML (Alexa Fluor 610, red fluorophore)). (b) Quantification of 53BP1 foci/cell, reported as the mean value of 53BP1 foci in APL blasts, NB4 and NB4-MR4 cells untreated or treated with 1 μM RA for 72 h, and U937/PR9 cells either untreated or exposed for 8 h to 100 μM ZnSO4 and then treated or not with 1 μM RA for 72 h. After ZnSO4 and/or RA treatment, cells were irradiated with 1 Gy and fixed at the indicated time points. (c) Quantification of 53BP1 foci association events with PML per nucleus in APL blasts, NB4, NB4-MR4, and U937/PR9 cells. (d) Quantification of 53BP1 foci/cell and 53BP1 foci association events with PML per nucleus in U937/MT cells. Before irradiation, U937/MT cells were either untreated or exposed for 8 h to 100 μM ZnSO4 and then treated or not with 1 μM RA for 72 h. Mean values were derived from the analysis of 100 cells in three independent experiments±S.D. *P<0.05; **P<0.01. Confocal analysis was performed using the LCS Leica confocal microscope (Leica Microsystems)
We evaluated the DSBs rejoining efficiency in cells expressing the PML-RARα oncoprotein and in cells expressing the WT PML by counting the number of 53BP1 foci/cell in untreated and irradiated cells (Figures 3b and d and Supplementary Figure S2D). After 0.5 h from IR, APL blasts and NB4 cells showed mean numbers of 53BP1 foci/cell lower than those calculated in cells expressing WT PML or in cells where PML-RARα was degraded by RA. After 3 h from IR, APL blasts and NB4 cells showed a number of 53BP1 foci significantly higher than that measurable in cells expressing WT PML or where the PML-RARα was degraded by RA. Finally, after 24 and 48 h from IR, the residual mean value of 53BP1 foci/cell scored in cells expressing the PML-RARα fusion protein was significantly higher compared with those expressing WT PML (Figures 3b and d and Supplementary Figure S2D).
PML-RARα does not increase chromosomal in stability, S phase, and cell death
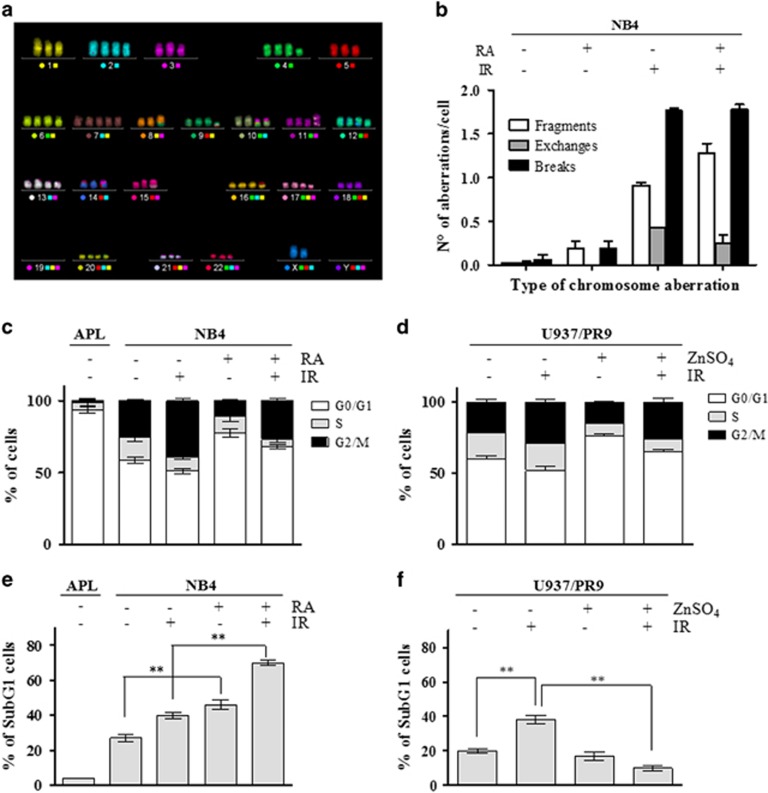
To evaluate whether the residual γ-H2AX and 53BP1 foci/cell observed after 24 and 48 h from IR in APL blasts and PML-RARα-expressing cells may correlate with chromosome instability, the multicolor FISH (mFISH) analysis was performed in NB4 cells, either untreated or exposed to RA, in combination with exposure to 1 Gy. The karyotype of NB4 was established considering conserved translocations that appear in >90% of the cells analyzed in controls. Karyotype was hypotetraploid and can be summarized as follows: 80 XX, −X, −X, −1, −3, −5, +7, −8, −14, −15, −18, −19, −19, −19, −19, −21, −22, t(8′–9), (9′–8), t(10–19), t(10–19), t(14–19), t(15′–17), t(16–5) t(17′–15), t(17′–15) (Figures 4a and b). The analysis of chromosomal exchanges in each sample was performed ignoring basal translocations observed in control cells. Results obtained indicated that RA alone did not exert any effect on the frequency of chromosome aberrations over control values and did not modify IR-induced break frequency (Figure 4b). Although break frequency remain unchanged, RA pretreatment reduced the frequency of IR-induced chromosome exchanges (from 2 to 0.9 exchanges/cell in NB4 control and RA-treated irradiated samples, respectively), whereas it increased the frequency of chromosomal excess fragments (from 1.8 to 3.9 fragments/cells in NB4 control and RA-treated irradiated samples, respectively) (Figure 4b). The only difference observed in RA-treated versus untreated NB4 samples was in terms of quality of the IR-induced chromosome damage rather than in terms of quantity (total breaks). Unfortunately, cytogenetic analysis could not be performed on primary APL samples because of the very slow proliferation rate of blasts isolated from the peripheral blood that are blocked in the G0/G1 phase of the cell cycle (Figure 4c).
Figure 4.
Chromosomal damage, cell cycle distribution, and apoptosis in PML-RARα-expressing cells. (a) Representative image of an mFISH-stained NB4 karyotype. Karyotype was established considering conserved translocations that appear in >90% of the cells analyzed in controls. Metaphases were captured with the Axio Imager M1 microscope (Carl Zeiss, Oberkochen, Germany). Karyotyping and cytogenetic analysis of each single chromosome was performed by the ISIS software. (b) Frequency per cell of chromosomal aberrations (i.e., fragments, exchanges, and breaks) after irradiation with 1 Gy in NB4 cells untreated or exposed to 1 μM RA for 72 h. (c) Cell cycle distribution of APL blasts and of NB4 cells untreated or treated with 1 μM RA for 72 h and then irradiated and fixed after 24 h. (d) Cell cycle distribution of U937/PR9 cells untreated or treated with 100 μM ZnSO4 for 8 h to induce PML-RARα expression and then irradiated and fixed after 24 h. (e) Sub-G1 population analyzed by flow cytometry in APL blasts and in NB4 cells untreated or treated with 1 μM RA for 72 h, and then irradiated and fixed after 24 h. (f) Sub- G1 population analyzed by flow cytometry in U937/PR9 cells untreated or treated with 100 μM ZnSO4 for 8 h, and then irradiated and fixed after 24 h. Mean values were derived from three independent experiments±S.D. **P<0.01
As H2AX is phosphorylated either during the S phase of the cell cycle or as a consequence of cell death induction,34 we analyzed the cell cycle profile and the sub-G1 population of APL blasts and myeloid cell lines. In APL blasts, >90% of cells were in G1, 2% in S and 8% in G2/M phases of the cell cycle (Figure 4c), whereas the sub-G1 population was ~4% (Figure 4e). Cell cycle analysis in NB4 and U937/PR9 cells treated or not with RA and ZnSO4, respectively, untreated or irradiated with 1 Gy, showed that the high number of γ-H2AX foci/cell scored in PML-RARα-expressing cells did not correlate with the percentage of cells in the S phase (Figures 4c and d). Moreover, in NB4 and U937/PR9 cells, no association was observed between the number of γ-H2AX foci/cell and cell death, both in basal conditions and after 24 h from IR exposure (Figures 4e and f). Overall, data obtained indicated that the high basal levels of γ-H2AX foci observed in APL blasts and PML-RARα-expressing cells are not associated with the cell cycle S-phase or with cell death.
The integrity of PML-NBs is required for ATM activation
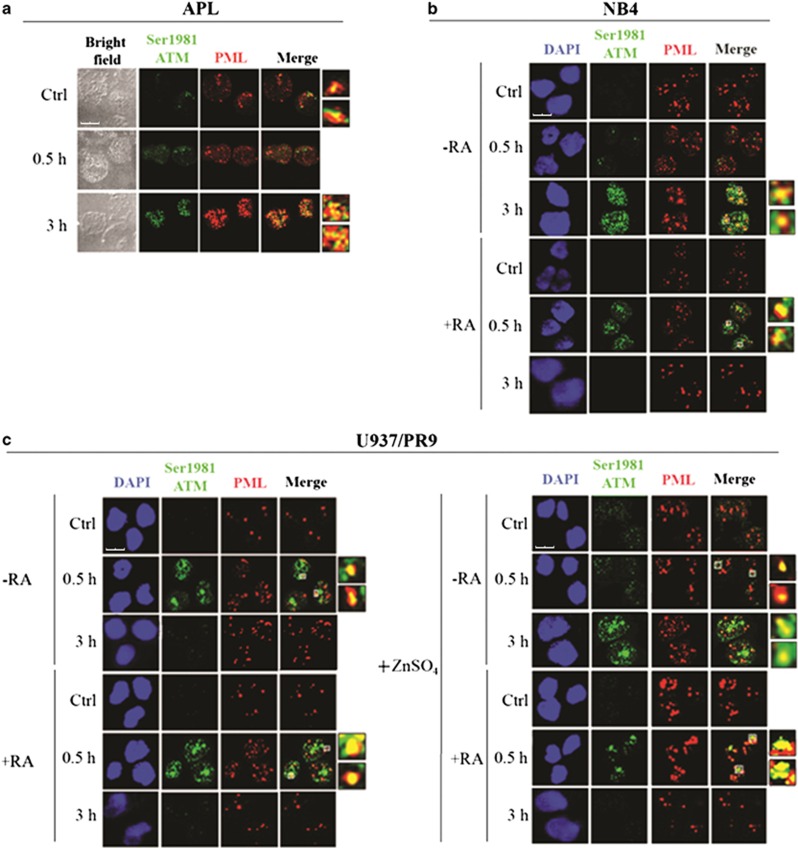
ATM kinase, a critical regulator of the DDR, is rapidly phosphorylated at the Ser1981 residue in response to IR, and its active form in turn phosphorylates H2AX at Ser139, as well as several other component of the DDR (e.g., NBN at Ser 343 and CHK2 at Thr68).2 We evaluated the role of PML-NBS integrity in ATM phosphorylation at Ser1981 and activation in response to IR. ATM resulted poorly activated in non-irradiated PML-RARα-expressing APL blasts, its Ser1981-phosphorylated form colocalizing with PML at 3 h from the irradiation with 1 Gy (Figure 5a). Similar to primary APL blasts, pSer1981-ATM foci were barely detectable in non-irradiated NB4 and U937/PR9+ZnSO4 cells (Figures 5b and c). In PML-RARα-expressing cells, ATM activation and colocalization with PML was well detectable after 3 h from IR, whereas in cells expressing the WT PML (i.e., U937/WT and U937/PR9) an accumulation of pSer1981-ATM in PML foci was already visible after 0.5 h from IR (Figure 5c and Supplementary Figure S3A). Interestingly, RA treatment in both NB4 cells and Zn-induced U937/PR9 restored the pSer1981-ATM/PML colocalization pattern into PML-NBs after 0.5 h from IR (Figures 5b and c).
Figure 5.
PML-NB integrity and ATM activation. Double immunofluorescence of pSer1981-ATM (Alexa Fluor 488, green fluorophore) and PML (Alexa Fluor 610, red fluorophore) foci performed in cells untreated or irradiated with 1 Gy and fixed after 0.5 and 3 h. Representative images of the analysis performed in (a) APL blasts (cell image: bright field), (b) NB4 cells untreated or treated with 1 μM RA for 72 h previous to IR (counterstain: DAPI), and (c) U937/PR9 cells either untreated or exposed for 8 h to 100 μM ZnSO4 and then untreated or treated with 1 μM RA for 72 h previous to IR (counterstain: DAPI). Some colocalization signals have been highlighted and marked within the cell by a white square. Confocal microscopy images, magnification × 63; LCS Leica confocal microscope (Leica Microsystems)
The integrity of PML-NBs is required for DSBS signal transduction
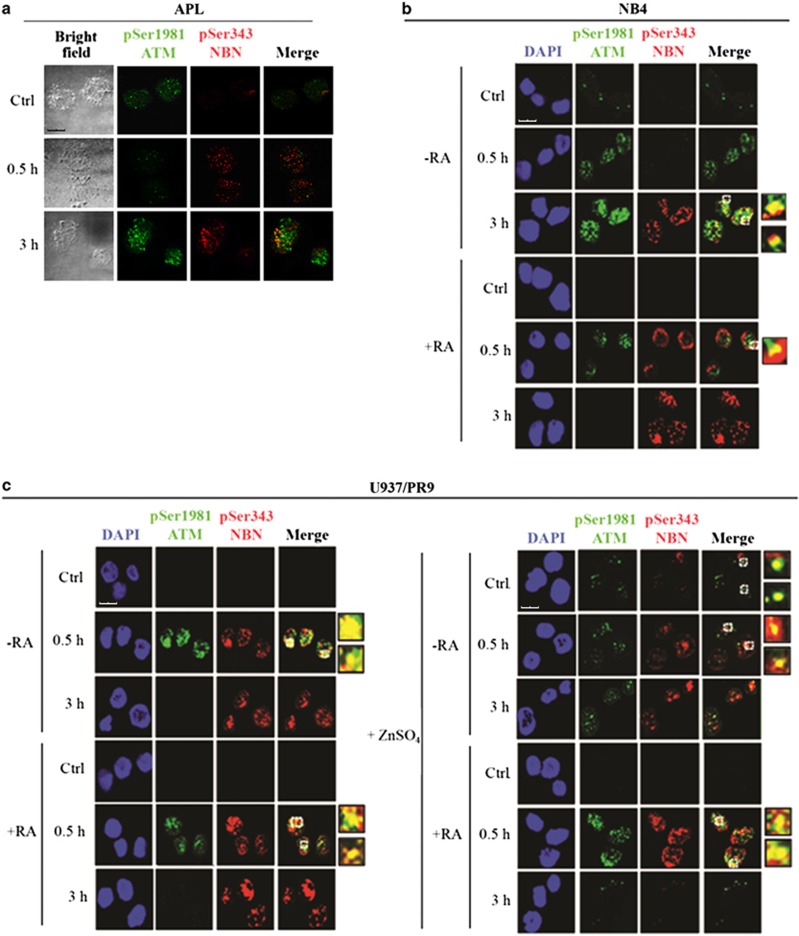
Co-immunofluorescence experiments with anti-pSer1981-ATM and anti-pSer343-NBN antibodies indicated that ATM and NBN phosphorylation were significantly induced after 3 h from irradiation with 1 Gy in PML-RARα-expressing cells (Figure 6). In PML-RARα cells treated with RA (NB4+RA and Zn-induced U937/PR9+RA) and in cells expressing the WT PML (U937/WT and U937/PR9 cells), a high number of pSer1981-ATM foci, colocalizing with pSer343-NBN, was visible at 0.5 h from IR; at 3 h from IR, pSer1981-ATM foci disappeared whereas NBN foci persisted (Figures 6b and c and Supplementary Figure S3B). These findings are in agreement with the proposed function of NBN in several phases of the DDR.35, 36
Figure 6.
PML-NB integrity and ATM-dependent NBN phosphorylation. Double immunofluorescence of pSer1981-ATM (Alexa Fluor 488, green fluorophore) and pSer343-NBN (Alexa Fluor 610, red fluorophore) foci performed in cells untreated or irradiated with 1 Gy and fixed after 0.5 and 3 h. Representative images of the analysis performed in (a) APL blasts (cell image: bright field), (b) NB4 cells untreated or treated with 1 μM RA for 72 h previous to IR (counterstain: DAPI), and (c) U937/PR9 cells either untreated or exposed for 8 h to 100 μM ZnSO4 and then treated or not with 1 μM RA for 72 h previous to IR (counterstain: DAPI). Some colocalization signals have been highlighted and marked within the cell by a white square. Confocal microscopy images, magnification × 63; LCS Leica confocal microscope (Leica Microsystems)
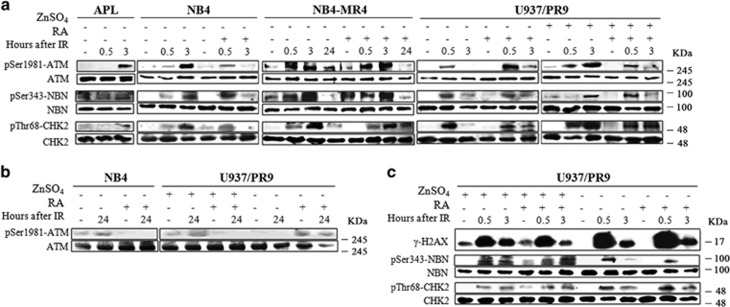
Immunoblot analysis further showed that in PML-RARα-expressing cells, ATM, NBN, and CHK2 proteins were phosphorylated in response to IR although with a delayed kinetics (Figure 7a). Indeed, the phosphorylation signal of ATM, NBN, and CHK2 reached a peak after 3 h from IR, ATM phosphorylation being visible up to 24 h from irradiation (Figures 7a and b), whereas in cells expressing WT PML, the ATM, NBN, and CHK2 proteins were phosphorylated after 0.5 h from IR, the phosphorylation signal being strongly reduced after 3 and 24 h (Figures 7a and b and Supplementary Figure S4A). These data indicate that PML-NBS integrity is necessary for a correct ATM–CHK2 axis activation.22 Furthermore, immunoblot analysis showed that H2AX, NBN, and CHK2 expression and phosphorylation levels were not affected in U937/PR9 cells treated or not with ZnSO4, then exposed to cycloheximide and finally irradiated, thus suggesting that de novo protein synthesis is not involved in the DDR signaling defects detected in APL cells (Figure 7c).
Figure 7.
Phosphorylation of ATM kinase and its substrates in PML-RARα-expressing cells. (a) Immunoblot analysis of pSer1981-ATM, pSer343-NBN, and pThr68-CHK2 phosphorylation in APL blasts, NB4, NB4-MR4, and U937/PR9 cells. Previous to IR, NB4 and NB4-MR4 cells were either untreated or exposed to 1 μM RA for 72 h, whereas U937/PR9 cells were either untreated or exposed for 8 h to 100 μM ZnSO4, and then treated or not with 1 μM RA for 72 h. Cells were then exposed to 1 Gy of X-rays and lysed after 0.5, 3, and 24 h. (b) Immunoblot analysis of ATM phosphorylation at the Ser1981 residue after 24 h from IR in NB4 and U937/PR9 cells. (c) U937/PR9 cells were first treated with 100 μM of ZnSO4, then exposed to 10 μg/ml cycloheximide, and finally irradiated with 1 Gy and lysed after 0.5 h. Immunoblots were performed using anti-γ-H2AX, anti-pSer343-NBN, anti-NBN, anti-pThr68-CHK2, and anti-CHK2 antibodies. Blots presented are exemplificative of the results obtained from two independent experiments
Defective DSBS sensing and repair in a preleukemic mouse model of APL
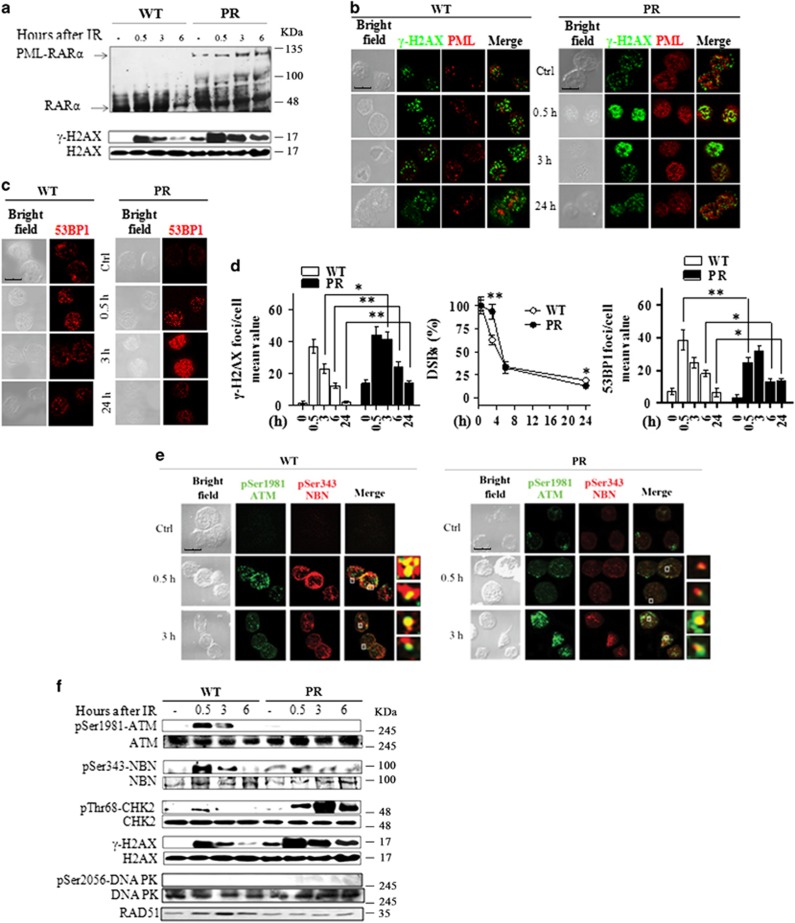
To evaluate the role of PML-RARα expression in the DSBS sensing and signaling in vivo, we used the preleukemic model of APL (i.e., PR mice).37 In this model, the disease develops after a long latency, suggesting that additional alterations cooperate with PML-RARα for leukemia development. We confirmed that in Lin− HPCs from these mice the expression levels of RARα and PML-RARα were not affected by IR. Of note, the levels of γ-H2AX were higher in untreated PR as compared with WT mice (Figures 8a and b).
Figure 8.
In vivo validation of the DDR in the preleukemic mouse model of APL. WT and preleukemic mice knock-in for PML-RARα (PR) were irradiated with 5.5 Gy of X-rays and sacrificied after 0.5, 3, 6, and 24 h. Lin− cells were isolated from the bone marrow of three pooled mice. (a) Immunoblot analysis of RARα and PML-RARα expression, and of H2AX phosphorylation at Ser139 residue, in untreated and irradiated WT and PR mice. (b) Representative images of the double immunofluorescence analysis of γ-H2AX (Alexa Fluor 488, green fluorophore) and PML (Alexa Fluor 610, red fluorophore) foci in untreated and irradiated WT and PR mice. (c) Representative images of the 53BP1 foci in untreated and irradiated WT and PR mice. (d) The DSBs rejoining analysis was reported as the mean value of γ-H2AX foci/cell and as the percentage of residual DSBs in untreated and irradiated WT and PR mice. The DSBS repair was also analyzed by counting the number of 53BP1 foci/cell in untreated and irradiated WT and PR mice. Mean values were derived from the analysis of 100 cells from three independent experiments±S.D. *P<0.05, **P<0.01. (e) Representative images of the double immunofluorescence analysis of pSer1981-ATM (Alexa Fluor 488, green fluorophore) and pSer343-NBN (Alexa Fluor 610, red fluorophore) foci in WT and PR mice. (f) Immunoblot analysis of ATM phosphorylation at the Ser1981 residue, NBN phosphorylation at Ser343, CHK2 phosphorylation at Thr68, DNA-PK phosphorylation at Ser2056, and RAD51 expression. Cell image: bright field; confocal microscopy images, magnification × 63; LCS Leica confocal microscope (Leica Microsystems)
PR mice HPCs showed a microsplecked nuclear pattern of PML-NBs whose number increased after irradiation with 5.5 Gy (Figure 8b). Kinetics of both γ-H2AX dephosphorylation and 53BP1 recruitment at the DSBs were delayed in preleukemic PR mice as compared with WT animals, with the persistence of significantly higher levels of unrepaired damage after 24 h from IR (P<0.05; Figures 8b–d). In addition, the ATM activation, as well as the phosphorylation of its substrates NBN and CHK2, was delayed in PR mice as compared with WT mice (Figures 8e and f). To analyze the DSBS repair pathway active in WT and PR mice, we looked at the status of the DNA-PK and RAD51 that play key roles in the NHEJ repair and HRR, respectively.9 Results showed that DNA-PK was not phosphorylated following IR either in WT or PR mice, whereas RAD51 was induced at 3 h in irradiated WT mice (Figure 8f). These results suggested that PML-RARα expression impairs the IR-induced activation of HRR in vivo.
Discussion
Disruption of PML-NBs by PML-RARα is a hallmark of APL, a model disease to understand leukemogenic pathways directed by an oncoprotein.24, 25, 26, 27, 28, 29, 30, 31, 32 Here, we report that PML-NBs play a key role in regulating early and late steps of IR-induced DSBs sensing, signaling, and repair in myeloid cells in vitro and in vivo. These events may depend on the cellular context as PML depletion/overexpression and PML-RARα ectopic expression in non-hematopoietic cells and solid tumors only impairs the late phases of HRR.16
Results obtained in a PML-RARα preleukemic knock-in mouse model strongly suggest that the PML-RARα-induced PML-NBs disruption is associated with DDR impairment and represents an early event in APL pathogenesis. Evidence obtained in myeloid cells expressing or not the PML-RARα and in RA-resistant myeloid cells lines suggested that RA-induced reformation of PML-NBs and oncoprotein degradation, two events contributing to the therapeutic effect of RA in APL, is tightly correlated with the restoration of DDR in APL cells. Moreover, our results point to a direct involvement of RA binding to the endogenous PML-RARα in the restoration of PML-NB integrity and DDR.
In agreement with previous findings obtained in APL cell lines and in hematopoietic precursors from mice expressing PML-RARα,14, 38 we showed that primary APL blasts and Lin− HPCs from PR mice were characterized by higher levels of γ-H2AX than those detectable in normal human and murine HPCs. Interestingly, high levels of γ-H2AX foci were found in human tumors and cultured cells,39 during the S phase of the cell cycle, and during apoptosis.34 The cell cycle analysis in APL blasts and in PML-RARα-expressing cells seems to exclude the possibility that H2AX phosphorylation depends on the percentage of cells in the S phase or on apoptotic events. Interestingly, PML-RARα expression increases the capability of histone deacetylase inhibitors (HDACi) to induce DNA damage and apoptosis by affecting the expression of genes involved in DNA repair mechanism.38 This may explain HDACi selectivity in causing cancer cell death in certain transformed cells.40
As γ-H2AX represents a biomarker of the DSB, but not a functional component of the DDR,2, 3, 4 we further analyzed DSBS sensing using 53BP1, a protein that accumulates within PML-NBs and is involved in DSBs repair.41, 42 The time-course analysis of 53BP1 recruitment at the DSBs in in vitro and in vivo models of APL indicated that the disruption of the PML-NBs slows its recruitment at the DSBs after IR. 53BP1 was nonrandomly associated with PML-NBs and its distribution within the nucleus was dependent on PML-NBs integrity. Furthermore, we observed that in myeloid cells the number of 53BP1 and PML associations follows the DSBs rejoining profile, indicating an association of the two proteins at the DSB foci in vivo. Indeed, a defective recruitment of 53BP1 to the damaged sites was observed in PML-RARα-expressing cells, the proper localization being re-established by RA treatment.
Overall, our findings suggest that the expression of the PML-RARα fusion protein, by destroying the PML-NBS integrity, slows down the DSBS rejoining kinetics in human APL blasts and cell lines, and in hematopoietic progenitors from the APL mouse model. This may contribute to the persistence of the higher number of γ-H2AX and 53BP1 foci at longer times from IR in APL cells (when compared with myeloid progenitors expressing normal PML protein and PML-NBs), resembling the deficiency in the repair of specific subset of DSBs described in DDR-defective cells.43 As PML-NBs are defined as baskets that contain proteins involved in the DSBS response, their disruption may cause a pan-nuclear dispersion of DDR proteins and consequently a defect in the DSBS tethering.10, 11, 13, 15, 16, 44
The disruption of the PML-NBs by PML-RARα expression does not increase the chromosomal instability in NB4 cells treated or not with RA and/or IR, although a compromised ability to maintain a genomic stability has been suggested as part of APL pathogenesis.16 Most of the somatic events in APL genomes appear to be random background mutations in the hematopoietic cells that acquired the PML-RARα initiating event.45, 46 A broad spectrum of acquired and recurrent chromosomal abnormalities, which may act as secondary events and could explain APL leukemogenesis, were reported by high-resolution single-nucleotide polymorphism array (SNP-A) analysis in ~50% matched diagnosis and remission samples from 48 APL cases. These genetic abnormalities were undetectable by conventional cytogenetics.47 Almost 90% of these lesions were not exclusive of APL, but recurrent in non-APL acute myeloid leukemias and other hematologic neoplasms.47, 48, 49, 50 Notably, mutations present in human leukemias were also identified by the sequencing of an APL mouse genome.51 Therefore, the PML-RARα initiating event may be necessary, but not sufficient, to cause APL; additional hits, largely unknown and possibly associated with the HPCs impaired capability to properly repair the DNA lesions,52 may occur and ultimately lead to the clonal expansion of leukemic blasts.
PTMs regulate multiple biological functions of PML and of PML-NBS disassembly and rebuilding during the DDR,21 and in turn PML-NBs regulate the PTMs of nuclear proteins dependent upon the ATM and ATR kinases.11, 12, 13, 53 In this context, our results suggest the existence of a feedback mechanism between PML-NBs and the ATM–NBN–CHK2 axis in the DSBS response. Beside a direct role of these proteins in modulating PML-NB roles in the IR-induced DSBS response,11 PML-NBS integrity may be necessary to rapidly activate the DSBS sensing, signaling, and repair. Our results highlight that PML-NBS disruption by PML-RARα strongly affects ATM activation, as well as CHK2 and NBN phosphorylation. This suggests that the PML-NBs integrity is necessary to properly sense DSBs in order to activate ATM and to allow ATM-dependent phosphorylation of proteins localized within PML-NBs.11, 12, 52 The delayed recruitment of 53BP1 at the DSBs in PML-RARα-expressing cells, harboring disrupted PML-NBs, together with the impaired increase of RAD51 levels in PR mice, suggest that the HRR mechanism is facilitated by intact PML-NBs in vivo.
Data here obtained in human primary APL blasts and in myeloid cells, further confirmed in a preleukemic mouse model of APL, highlighted the relevant role of PML-NBs in coordinating and regulating the DDR. These results further shed light on the events occurring at the onset of APL, suggesting that the expression of PML-RARα disrupts PML-NBs, causes DNA damage, and is responsible for a defect in the early and late steps of the DDR that in turn play a role in the pathogenesis and progression of APL.
Materials and Methods
Reagents and antibodies
The complete list of reagents and antibodies is reported in Supplementary Materials and Methods.
Human samples, cell lines, and cultures
Leukemic cells were isolated from the peripheral blood of three consenting, newly diagnosed APL individuals presenting >80% circulating blasts and classified as M3 or M3v by morphological criteria according to the French–American–British (FAB) classification. The main biologic and clinical features of patients are summarized in Supplementary Table S1. Analysis of the PML-RARα fusion gene was performed by RT-PCR as reported elsewhere.54 CD34+ HPCs and mononuclear CD34+ cell fractions were isolated from the peripheral blood of informed healthy donors according to institutional guidelines as previously reported.55 Flow cytometric analysis confirmed that >90% of the cells were CD34+.
The human myeloid leukemia cell lines were: the APL-derived NB4 cell line containing the t(15;17)56 and the RA-resistant NB4-MR4 subclone;57 the human promonocytic U937 cell line either WT (U937/WT) or carrying the PML-RARα sequence under the control of the zinc-inducible MT-I promoter (U937/PR9) or an empty MT1 vector (U937/MT);58 the human myeloblastic leukemia cell line HL60 and the RA-resistant HL60-R subclone.59 NB4-MR4 and HL60-R resistant cell lines carry mutations abrogating RA-binding capacities of PML-RARα and RARα, respectively. Cell culture conditions and treatments are reported in Supplementary Materials and Methods.
APL murine model
Experiments were performed using a previously described APL knock-in mouse model37 in accordance with the national and international laws and policies. Experimental details are described in Supplementary Materials and Methods.
Analysis of cell cycle, differentiation, and apoptosis
Cell cycle, cell differentiation, and apoptosis were analyzed with a Dako Galaxy Flow Cytometer (Dako Denmark A/S, Glostrup, Denmark) equipped with a 488 nm laser source and with the FloMax software (version 2.4e; Partec GmbH, Munster, Germany). Hypodiploid peak analysis was performed as previously described.60 Cell differentiation was evaluated by direct immunofluorescence staining using fluorochrome-conjugated anti-CD11b antibody.14, 32 Details are in Supplementary Materials and Methods.
Immunofluorescence analysis
Cells were seeded on a glass coverslip using the Shandon Cytospin III (Thermo Fisher Scientific Inc., Waltham, MA, USA), and then fixed, stained, and analyzed as described in Supplementary Materials and Methods.
Multicolor FISH
The mFISH was performed by hybridizing chromosome spreads with the 21 × mFISH Probe Kit (MetaSystems, Altlussheim, Germany) as described elsewhere.41 Karyotyping and cytogenetic analysis of each single chromosome was performed by the ISIS software (MetaSystems, Altlussheim, Germany). Detailed procedures are in Supplementary Materials and Methods.
Immunoblotting
Protein extraction and western blot analysis were performed by standard procedures reported in Supplementary Materials and Methods.
Statistical analysis
Results are shown as the mean±S.D. derived minimally from three independent experiments. Statistical significance between means was assessed by Student's t-test (GraphPad Software Inc., San Diego, CA, USA). Statistical significance was considered as P-values of <0.05.
Acknowledgments
This work was partially supported by research funding from Roma Tre University (CLA 2014 and CLA 2015 to AdM), the University of Rome ‘La Sapienza', and Italian Association for Cancer Research (AIRC IG-11949) to CN.
Author contributions
AdM designed and performed the research, analyzed data and wrote the paper. DC, AT, RP, and NIG performed experiments. FB performed cytogenetic experiments and analyzed data. IP performed in vivo experiments. SL performed experiments and analyzed data. AA contributed with reagents and analyzed data. FL-C provided primary normal and leukemic samples. PA analyzed data. SM designed the experiments in the APL mouse model. CN contributed to research design and wrote the paper.
Glossary
- APL
acute promyelocytic leukemia
- DDR
DNA damage response
- DSB
DNA double-strand break
- IR
ionizing radiation
- γ-H2AX
H2AX phosphorylation at Ser139
- HDACi
histone deacetylase inhibitor
- HPC
hematopoietic progenitors
- HRR
homologous recombination repair
- IRIF
ionizing radiation-induced foci
- mFISH
multicolor FISH
- NHEJ
non-homologous end-joining
- PML
promyelocytic leukemia
- PML-NB
PML nuclear body
- PTM
post-translational modification
- RA
all-trans retinoic acid
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Raschellà
Supplementary Material
References
- Madhusudan S, Middleton MR. The emerging role of DNA repair proteins as predictive, prognostic and therapeutic targets in cancer. Cancer Treat Rev 2005; 31: 603–617. [DOI] [PubMed] [Google Scholar]
- Pardo B, Gomez-Gonzalez B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci 2009; 66: 1039–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273: 5858–5868. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 1999; 146: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Masi A, Viganotti M, Polticelli F, Ascenzi P, Tanzarella C, Antoccia A. The R215W mutation in NBS1 impairs gamma-H2AX binding and affects DNA repair: molecular bases for the severe phenotype of 657del5/R215W Nijmegen breakage syndrome patients. Biochem Biophys Res Commun 2008; 369: 835–840. [DOI] [PubMed] [Google Scholar]
- Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci USA 2003; 100: 5057–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez G, Cilli D, Berardinelli F, Viganotti M, Ascenzi P, Tanzarella C et al. Cleavage of the BRCT tandem domains of nibrin by the 657del5 mutation affects the DNA damage response less than the Arg215Trp mutation. IUBMB Life 2012; 64: 853–861. [DOI] [PubMed] [Google Scholar]
- Scully R, Xie A. Double strand break repair functions of histone H2AX. Mutat Res 2013; 50: 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 2012; 47: 497–510. [DOI] [PubMed] [Google Scholar]
- Carbone R, Pearson M, Minucci S, Pelicci PG. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene 2002; 21: 1633–1640. [DOI] [PubMed] [Google Scholar]
- Dellaire G, Ching RW, Ahmed K, Jalali F, Tse KC, Bristow RG et al. Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J Cell Biol 2006; 175: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Jeong JH, Brown AL, Lee CH, Pandolfi PP, Chung JH et al. Promyelocytic leukemia activates Chk2 by mediating Chk2 autophosphorylation. J Biol Chem 2006; 281: 26645–26654. [DOI] [PubMed] [Google Scholar]
- Dellaire G, Bazett-Jones DP. Beyond repair foci: subnuclear domains and the cellular response to DNA damage. Cell Cycle 2007; 6: 1864–1872. [DOI] [PubMed] [Google Scholar]
- Viale A, De Franco F, Orleth A, Cambiaghi V, Giuliani V, Bossi D et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature 2009; 457: 51–56. [DOI] [PubMed] [Google Scholar]
- Boichuk S, Hu L, Makielski K, Pandolfi PP, Gjoerup OV. Functional connection between Rad51 and PML in homology-directed repair. PLoS One 2011; 6: e25814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung PL, Denissova NG, Nasello C, Hakhverdyan Z, Chen JD, Brenneman MA. Promyelocytic leukemia nuclear bodies support a late step in DNA double-strand break repair by homologous recombination. J Cell Biochem 2012; 113: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 2007; 8: 1006–1016. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de Thé H. PML nuclear bodies. Cold Spring Harb Perspect Biol 2010; 2: a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Song Y, Qian J, Liu B, Dong Y, Tian B et al. Promyelocytic leukemia protein interacts with werner syndrome helicase and regulates double-strand break repair in γ-irradiation-induced DNA damage responses. Biochemistry (Mosc) 2011; 76: 550–554. [DOI] [PubMed] [Google Scholar]
- Münch S, Weidtkamp-Peters S, Klement K, Grigaravicius P, Monajembashi S, Salomoni P et al. The tumor suppressor PML specifically accumulates at RPA/Rad51-containing DNA damage repair foci but is nonessential for DNA damage-induced fibroblast senescence. Mol Cell Biol 2014; 34: 1733–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz ML, Grishina I. Regulation of the tumor suppressor PML by sequential post-translational modifications. Front Oncol 2012; 2: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaire G, Ching RW, Dehghani H, Ren Y, Bazett-Jones DP. The number of PML nuclear bodies increases in early S phase by a fission mechanism. J Cell Sci 2006; 119: 1026–1033. [DOI] [PubMed] [Google Scholar]
- Ivanschitz L, De Thé H, Le Bras M. PML, SUMOylation, and senescence. Front Oncol 2013; 3: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurrieri C, Nafa K, Merghoub T, Bernardi R, Capodieci P, Biondi A et al. Mutations of the PML tumor suppressor gene in acute promyelocytic leukemia. Blood 2004; 103: 2358–2362. [DOI] [PubMed] [Google Scholar]
- de Thé H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 1991; 66: 675–684. [DOI] [PubMed] [Google Scholar]
- Kakizuka A, Miller WH Jr, Umesono K, Warrell RP Jr, Frankel SR, Murty VV et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell 1991; 66: 663–674. [DOI] [PubMed] [Google Scholar]
- Pandolfi PP, Alcalay M, Longo L, Fagioli M, Zangrilli D, Grignani F et al. Molecular genetics of the t(15;17) of acute promyelocytic leukemia (APPL). Leukemia 1992; 6: 120S–122S. [PubMed] [Google Scholar]
- de Thé H, Chen Z. Acute promyelocytic leukaemia: novel insights into the mechanisms of cure. Nat Rev Cancer 2010; 10: 775–783. [DOI] [PubMed] [Google Scholar]
- di Masi A, Leboffe L, De Marinis E, Pagano F, Cicconi L, Rochette-Egly C et al. Retinoic acid receptors: from molecular mechanisms to cancer therapy. Mol Aspects Med 2015; 41C: 1–115. [DOI] [PubMed] [Google Scholar]
- Dyck JA, Maul GG, Miller WH Jr, Chen JD, Kakizuka A, Evans RM. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell 1994; 76: 333–343. [DOI] [PubMed] [Google Scholar]
- Raelson JV, Nervi C, Rosenauer A, Benedetti L, Monczak Y, Pearson M et al. The PML/RAR alpha oncoprotein is a direct molecular target of retinoic acid in acute promyelocytic leukemia cells. Blood 1996; 88: 2826–2832. [PubMed] [Google Scholar]
- Nervi C, Ferrara FF, Fanelli M, Rippo MR, Tomassini B, Ferrucci PF et al. Caspases mediate retinoic acid-induced degradation of the acute promyelocytic leukemia PML/RARα fusion protein. Blood 1998; 92: 2244–2251. [PubMed] [Google Scholar]
- Foltankova V, Matula P, Sorokin D, Kozubek S, Bartova E. Hybrid detectors improved time-lapse confocal microscopy of PML and 53BP1 nuclear body colocalization in DNA lesions. Microsc Microanal 2013; 19: 360–369. [DOI] [PubMed] [Google Scholar]
- de Feraudy S, Revet I, Bezrookove V, Feeney L, Cleaver JE. A minority of foci or pan-nuclear apoptotic staining of gammaH2AX in the S phase after UV damage contain DNA double-strand breaks. Proc Natl Acad Sci USA 2010; 107: 6870–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digweed M, Sperling K. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair (Amst) 2004; 3: 1207–1217. [DOI] [PubMed] [Google Scholar]
- Cilli D, Mirasole C, Pennisi R, Pallotta V, D'Alessandro A, Antoccia A et al. Identification of the interactors of human nibrin (NBN) and of its 26 kDa and 70 kDa fragments arising from the NBN 657del5 founder mutation. PLoS One 2014; 9: e114651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westervelt P, Lane AA, Pollock JL, Oldfather K, Holt MS, Zimonjic DB et al. High-penetrance mouse model of acute promyelocytic leukemia with very low levels of PML-RARalpha expression. Blood 2003; 102: 1857–1865. [DOI] [PubMed] [Google Scholar]
- Petruccelli LA, Pettersson F, Del Rincon SV, Guilbert C, Licht JD, Miller WH Jr. Expression of leukemia-associated fusion proteins increases sensitivity to histone deacetylase inhibitor-induced DNA damage and apoptosis. Mol Cancer Ther 2013; 12: 1591–1604. [DOI] [PubMed] [Google Scholar]
- Nakamura AJ, Redon CE, Sedelnikova OA. Where did they come from? The origin of endogenous gamma-H2AX foci in tumor cells. Cell Cycle 2009; 8: 2324. [DOI] [PubMed] [Google Scholar]
- Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc Natl Acad Sci USA 2010; 107: 14639–14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieri D, Berardinelli F, Antoccia A, Tanzarella C, Sgura A. Comparison between two FISH techniques in the in vitro study of cytogenetic markers for low-dose X-ray exposure in human primary fibroblasts. Front Genet 2013; 4: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SM, Bristow RG. Discordance between phosphorylation and recruitment of 53BP1 in response to DNA double-strand breaks. Cell Cycle 2012; 11: 1432–1444. [DOI] [PubMed] [Google Scholar]
- Porcedda P, Turinetto V, Lantelme E, Fontanella E, Chrzanowska K, Ragona R et al. Impaired elimination of DNA double-strand break-containing lymphocytes in ataxia telangiectasia and Nijmegen breakage syndrome. DNA Repair (Amst) 2006; 5: 904–913. [DOI] [PubMed] [Google Scholar]
- Xu ZX, Timanova-Atanasova A, Zhao RX, Chang KS. PML colocalizes with and stabilizes the DNA damage response protein TopBP1. Mol Cell Biol 2003; 23: 4247–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012; 150: 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia [published erratum appears in N Engl J Med 2013; 369: 98]. N Engl J Med 2013; 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Seguí I, Sánchez-Izquierdo D, Barragán E, Such E, Luna I, López-Pavía M et al. Single-nucleotide polymorphism array-based karyotyping of acute promyelocytic leukemia. PLoS One 2014; 9: e100245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiu RV, Gondek LP, O'Keefe CL, Huh J, Sekeres MA, Elson P et al. New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia. J Clin Oncol 2009; 27: 5219–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin B, Erba H, Ouillette P, Roulston D, Purkayastha A, Karp J et al. Acquired genomic copy number aberrations and survival in adult acute myelogenous leukemia. Blood 2010; 116: 4958–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer CD, Coombes KR, Majewski T, Barron LL, Lerner S, Sargent RL et al. Genomic variation by whole-genome SNP mapping arrays predicts time-to-event outcome in patients with chronic lymphocytic leukemia: a comparison of CLL and HapMap genotypes. J Mol Diagn 2013; 15: 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartman LD, Larson DE, Xiang Z, Ding L, Chen K, Lin L et al. Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. J Clin Invest 2011; 12: 1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 2010; 7: 174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima K, Ohara M, Seki R, Tauchi H. Dancing on damaged chromatin: functions of ATM and the RAD50/MRE11/NBS1 complex in cellular responses to DNA damage. J Radiat Res 2008; 49: 451–464. [DOI] [PubMed] [Google Scholar]
- Lo-Coco F, Diverio D, Pandolfi PP, Biondi A, Rossi V, Avvisati G et al. Molecular evaluation of residual disease as a predictor of relapse in acute promyelocytic leukemia. Lancet 1992; 340: 1437–1438. [DOI] [PubMed] [Google Scholar]
- De Felice L, Tatarelli C, Mascolo MG, Gregorj C, Agostini F, Fiorini R et al. Histone deacetylase inhibitor valproic acid enhances the cytokine-induced expansion of human hematopoietic stem cells. Cancer Res 2005; 65: 1505–1513. [DOI] [PubMed] [Google Scholar]
- Lanotte M, Martin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3). Blood 1991; 77: 1080–1086. [PubMed] [Google Scholar]
- Shao W, Benedetti L, Lamph WW, Nervi C, Miller WH Jr. A retinoid-resistant acute promyelocytic leukemia subclone expresses a dominant negative PML-RAR alpha mutation. Blood 1997; 89: 4282–4289. [PubMed] [Google Scholar]
- Grignani F, Ferrucci PF, Testa U, Talamo G, Fagioli M, Alcalay M et al. The acute promyelocytic leukemia-specific PML-RAR alpha fusion protein inhibits differentiation and promotes survival of myeloid precursor cells. Cell 1993; 74: 423–431. [DOI] [PubMed] [Google Scholar]
- Robertson KA, Emami B, Collins SJ. Retinoic acid resistant HL-60R cells harbor a point mutation in the retinoic acid receptor ligand-binding domain that confers dominant negative activity. Blood 1992; 80: 1885–1889. [PubMed] [Google Scholar]
- Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Protoc 2006; 1: 1458–1461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.