Abstract
Synthetic biology has enabled the production of many value-added chemicals via microbial fermentation. However, the problem of low product titers from recombinant pathways has limited the utility of this approach. Methods to increase metabolic flux are therefore critical to the success of metabolic engineering. Here we demonstrate that vitamin E-derived designer micelles, originally developed for use in synthetic chemistry, are biocompatible and accelerate flux through a styrene production pathway in Escherichia coli. We show that these micelles associate non-covalently with the bacterial outer-membrane and that this interaction increases membrane permeability. In addition, these micelles also accommodate both heterogeneous and organic-soluble transition metal catalysts and accelerate biocompatible cyclopropanation in vivo. Overall, this work demonstrates that these surfactants hold great promise for further application in the field of synthetic biotechnology, and for expanding the types of molecules that can be readily accessed from renewable resources via the combination of microbial fermentation and biocompatible chemistry.
Keywords: Micelles, Metabolism, Catalysis, Synthetic Biology, Sustainable Chemistry
Micelles and Microbes
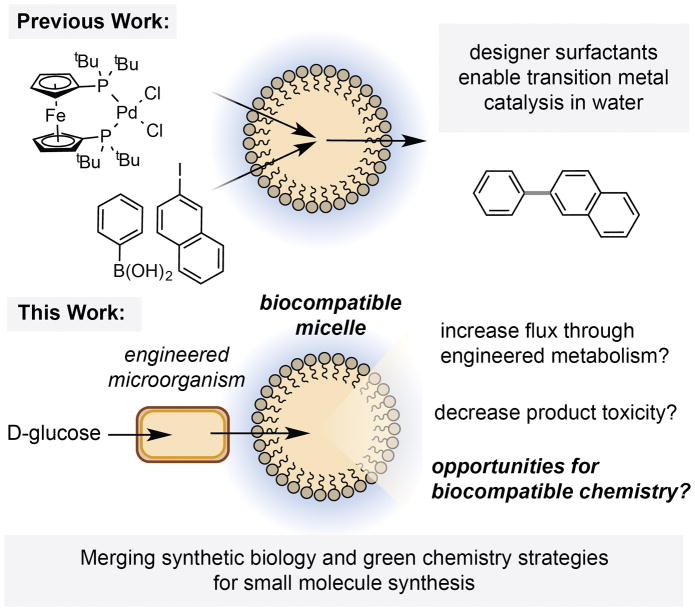
Here we disclose that vitamin E-derived synthetic micelles, originally developed for use as nano-reactors for synthetic chemistry in water, are biocompatible and accelerate flux through an engineered styrene production pathway in the microorganism E. coli NST74. We also demonstrate that these microbe-associated micelles can accommodate both heterogeneous and organic-soluble transition metal catalysts, and accelerate biocompatible cyclopropanation in vivo.
The production of small molecules of societal importance from renewable, non-petroleum derived feedstocks is arguably one of the foremost challenges facing the field of chemical synthesis.[1] Alongside advances in “green” synthetic methodologies, the use of metabolically engineered microorganisms has emerged as a viable and elegant solution to this challenge.[2] However, maximizing titers of desired products from engineered metabolic pathways remains a central challenge in synthetic biology. This is due, in part, to the regulatory elements and metabolic enzymes that have evolved to counteract metabolite accumulation, as well as the toxicity of many of these metabolites at high concentrations. These problems are typically addressed by: (i) knocking-out genes involved in negative feedback regulation or product catabolism and/or (ii) increasing metabolite flux and reducing product toxicity via in situ product removal (ISPR).[3] Genetic methods for ISPR include the heterologous expression of transmembrane efflux pumps from solvent-tolerant microbes.[4] Chemical methods for ISPR involve sequestering the desired (typically hydrophobic) metabolite into a separate organic phase.[5] This most often involves the use of an organic solvent overlay (n-dodecane), but can also employ the use of a solvent-impregnated resin (Amberlite® XAD4) or surfactant micelles (Triton X-100).[6]
In the field of green chemistry, designer surfactants have recently been shown to facilitate transition metal catalysis in water by sequestering reagents into the hydrophobic interior of a micelle.[7] However, the application of these compounds in microbial fermentations has not been explored. Here we report that the vitamin E-derived designer surfactants PTS, TPGS-750-Mand TPGS-1000 are biocompatible and accelerate flux through a styrene production pathway in E. coli, dramatically increasing production titers above the threshold of toxicity. We demonstrate that these designer micelles also accommodate both heterogeneous and organic-soluble iron catalysts for biocompatible cyclopropanation, and that co-localization of the reaction components in the micelle interior results in increased product titers and enhanced reactivity in vivo. To the best of our knowledge, this is the first example of micellar catalysis in the presence of a living microorganism.
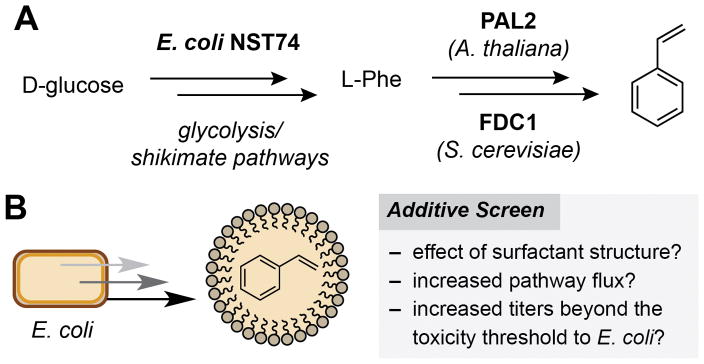
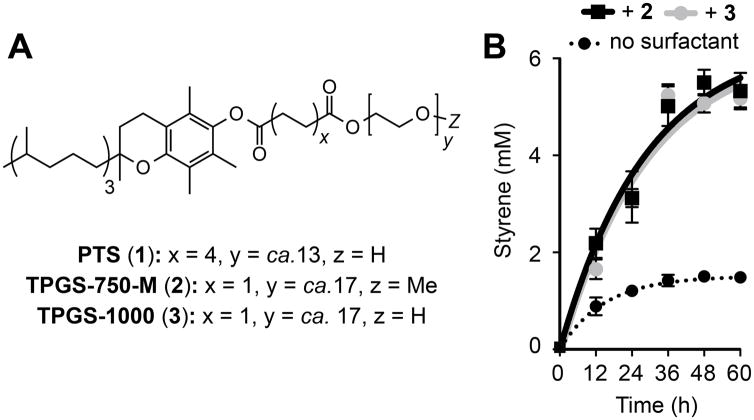
Our studies began by investigating whether micelles could increase flux through a styrene production pathway in E. coli NST74 (a feedback-deregulated L-phenylalanine overproducer). This engineered pathway consists of aphenylalanine ammonia lyase (PAL2) from Arabidopsis thalianaand a cinnamic acid decarboxylase (FDC1) from Saccharomyces cerevisiae, which together convert phenylalanine to styrene (Figure 2A).[8] The utility of this pathway is limited by the fact that styrene is toxic to E. coli at low concentrations (ca. 2.5 mM) via a mechanism that is thought to involve accumulation of the planar arene in the cell membrane (log Pow= 3.0).[9] Maximizing production from this pathway has therefore relied on genetic (overexpression of the AcrAB-TolC efflux pump) and chemical (solvent overlay and gas stripping) ISPR approaches.[4b,6g] We began by screening a range of surfactants and supramolecules that are known to bind hydrophobic compounds. We found that the addition of α- and β-(H/OMe) cyclodextrins marginally increased styrene concentrations (Table 1, Entry 2 and Table S1). Cationic and anionic surfactants generally inhibited styrene production (Table 1, Entries 4 and 5, and Table S1).[6d,10] The nonionic surfactants Triton X-100 and Tween-80 had no significant effect on this pathway (Table 1, Entries 6 and 7). Most notably, addition of the vitamin E-derived surfactants PTS (1), TPGS-750-M (2) and TPGS-1000 (3) enhanced styrene production by nearly two-fold (Table 1, Entries 8–10 and Figure 3A). Use of 5% wt/v surfactant further increased styrene production to a maximum of 5.5 mM for the O-methylated succinic acid derivative 2 (Table 1, Entry 17 and Figure 3B). This is twice the concentration that is toxic to E. coli and a four-fold improvement in overall production. This increase is also comparable to that achieved by a 20% wt/v solvent overlay of n-dodecane (Table 1, Entry 19).[6g] The addition of vitamin E O-acetate had no effect on styrene production, indicating that self-assembly of the amphiphiles in the culture medium is a prerequisite for ISPR.[11] In the presence of the third-generation designer surfactant SPGS-550-M (nok), styrene reached a maximum concentration of 0.4 mM after 24 h and then decreased over time. The reason for this is unclear, however this data suggests that the β-sitosterol moiety of the surfactant plays a role in styrene depletion in vivo (Table 1, Entry 11 and Figure S3).
Figure 2.
A) The styrene production pathway. B) The use of micelles as a strategy for ISPR from engineered E. coli.
Table 1.
Additive screen in styrene-producing cultures of engineered E. coli.
| Entry | Culture Additive | Styrene (mM) |
|
|
|---|---|---|---|---|
| 1 | None | 1.49 | 1 | |
| 2 | Me-β-Cyclodextrin | 1.86 | 1.24 | |
| 4 | 16-BAC | 0.02 | <1 | |
| 5 | SDS | 1.29 | <1 | |
| 6 | Triton X-100 | 1.70 | 1.10 | |
| 7 | Tween-80 | 1.86 | 1.33 | |
| 8 | PTS (1) | 2.70 | 1.81 | |
| 9 | TPGS-750-M (2) | 2.51 | 1.68 | |
| 10 | TPGS-1000 (3) | 2.64 | 1.78 | |
| 11 | SPGS-550-M (nok) | 0.36[e]/0.03[f] | – | |
| 12 | Vitamin E O-Ac | 1.38 | <1 | |
| 13 | PTS (1)[a] | 4.07 | 2.95 | |
| 14 | TPGS-1000 (3)[a] | 4.36 | 2.78 | |
| 15 | TPGS-750-M (2)[a] | 4.62 | 2.98 | |
| 16 | TPGS-1000 (3)[b] | 5.24 | 3.27 | |
| 17 | TPGS-750-M (2)[b] | 5.52 | 3.28 | |
| 18 | TPGS-750-M (2)[c] | 3.95 | 2.56 | |
| 19 | n-dodecane[d] | 5.50 | – |
Cells were grown aerobically in 100 mL of MM1 media in sealed 500 mL Erlenmeyer flasks at 32 °C with shaking at 220 rpm. Additives were added at 3.2 mM at the point-of-induction of the styrene-producing pathway (OD600=0.6–0.8). Styrene concentrations were determined inorganic extracts of culture aliquots by GC relative to an internal standard of 1,3,5-trimethoxybenzene using a standard curve. All data is shown as an average of three independent experiments. Detailed experimental procedures and a complete list of all surfactants tested can be found in Sections S3, S4 and Table S1.
2% wt/v,
5% wt/v,
10% wt/v,
20% wt/v,
after 24 h,
after 48 h.
Figure 3.
A) Vitamin E-derived designer surfactants 1–3. B) The effect of 5% wt/v 2 or 3 on styrene production from engineered E. coli. Experiments were performed as described in Table 1 and Section S3. Error bars represent the standard deviation of values from three independent experiments.
In order to assess whether these micelles accelerate flux through styrene biosynthesis we calculated the first derivative of the production time-course curves 12 h after surfactant addition. Comparing these values to those from control cultures containing no surfactant allowed us to express the relative rate-of-change of styrene production with respect to time ( , Table 1, Table S1). For 5% wt/v 2 and 3 we calculated a 3.3 fold increase in the relative rate-of-change of styrene production, which then decreased over time as production levels plateaued (Figure S2). Therefore as well as increasing overall styrene production levels, these surfactants also accelerated flux through this pathway. Due to the success of surfactants 2 and 3 up to this point, and their commercial availability as solids, we focused the remainder of our study on these molecules.
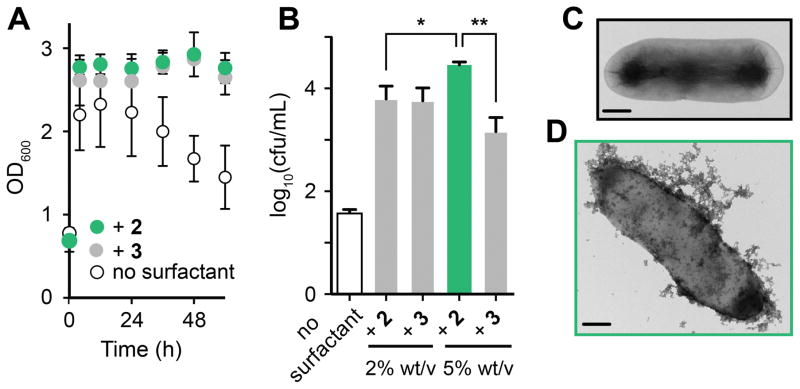
Having discovered that vitamin E surfactants increased styrene production, we next studied their biocompatibility and interaction with E. coli. We found that the growth of wild-type E. coli NST74 in media containing styrene (4 mM) was dependent on the presence of 2 or 3, confirming that styrene is toxic to E. coli at this concentration and that the presence of micelles alleviates this toxicity in vivo (Figure S5). The addition of 2 or 3 to the styrene-producing strain resulted in a prolonged stationary growth phase and an up to 200-fold increase in the number of viable cells (cfu/mL) after 60 h (Figures 4A/B and Table S2). Unexpectedly, centrifugation of cultures containing 5% w/v 2 produced a milky cell-containing supernatant and a transparent gel-like pellet, which is a hallmark of bacterial slime (Figure S7). E. coli is known to secrete biofilm-like exo-polysaccharide “slimes” to protect the cell from external environmental stress.[12] We confirmed the organism’s ability to secrete polysaccharide slime by observing the growth of black colonies on BHI-sucrose-Congo Red agar plates (Figure S8).[10b,13] The reason for slime formation occurring only in response to 5% wt/v 2 is unclear, however the less polar O-methylated micelle exterior of 2 (Z=Me, Figure 3A) could favour binding to lipids in the bacterial membrane causing enhanced disruption.
Figure 4.
Investigating the effect of 2 and 3 on styrene-producing E. coli. A) The addition of 2 or 3 alleviates the toxic effects of styrene on cell growth. B) Serial dilution and plate count assays after 60 h show an increased number of viable cells in cultures containing 2 or 3, and slime production by E. coli that may protect the cell from the effects of 2 (see Section S10 for additional data). C) and D) Cryo-TEM images of styrene-producing E. coli (C) + 2 (D). Scale bars = 0.5 μm. Data from slime-producing cultures is coloured green. Cells were grown as described in Table 1 and Section S3. Surfactants were added at 5% wt/v unless stated otherwise. Error bars represent the standard deviation of values from three independent experiments; *P < 0.05 **P < 0.005 (Welch’s T-test).
To further explore this hypothesis we imaged the cells by transmission electron microscopy (cryo-TEM). Whole-cell images showed that 2 and 3 coat the cell surface in an amorphous matrix of nanomicelles (Figure 4D, Figure S10/11). Cells grown in the presence of 2 are more densely covered than cells treated with 3, supporting our hypothesis that slime production occurs in response to an increased micelle presence at the cell membrane. Plastic-mounted cell slice images after multiple washes showed no observable micelles on either side of the cell membrane, indicating that micelles are not internalized, and that the extracellular interaction is non-covalent (Figure S12–S14).
To investigate whether the association of these micelles weakens the cell membrane we conducted a plasmid stability test. This assay uses the loss of plasmid DNA (and associated antibiotic resistance) by a microorganism when grown on non-selective media as a proxy for membrane impairment.[14] When styrene-producing cultures treated with 5% wt/v 3 were grown overnight on non-selective LB-agar only 9% regrowth on selective LB-agar was observed, indicating near-complete loss of the pTrc99 A plasmid. However, cells from slime-producing cultures containing 5% wt/v 2 retained 96% regrowth under these conditions, and cells treated with 2% wt/v 2 or 3 exhibited 52% and 44% recovery, respectively (Table S3). The interaction between vitamin E surfactants and E. coli therefore weakens the cell membrane, and slime production appears to protect the cell from this effect.
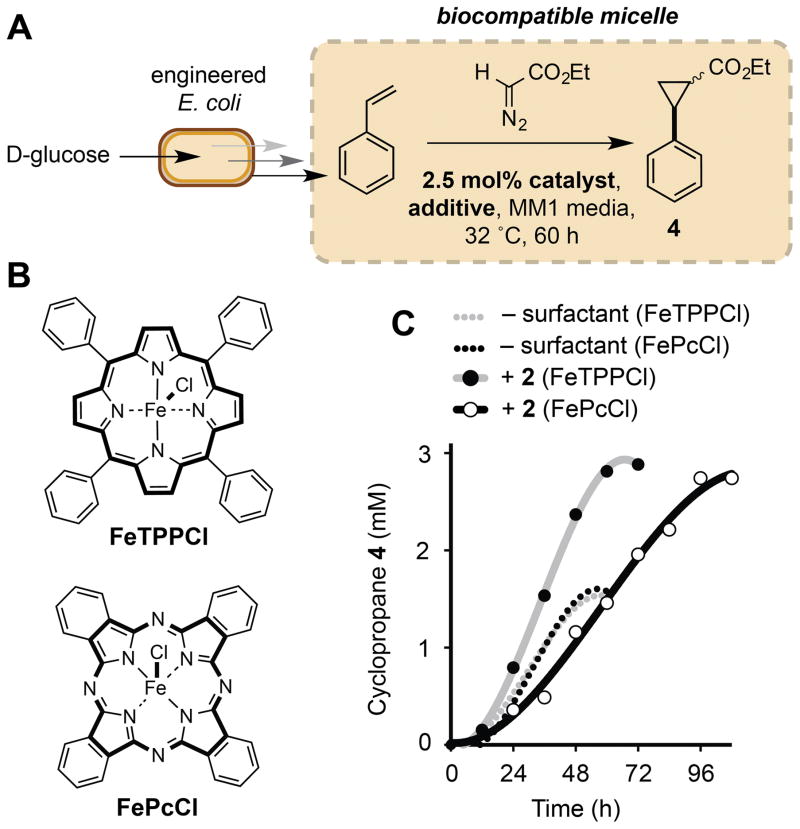
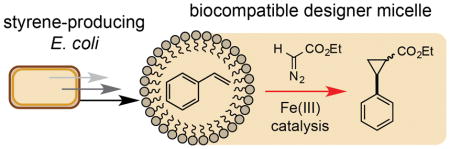
We next investigated the effect of micelles on non-enzymatic catalysis in vivo by examining the effect of 2 on our previously reported biocompatible cyclopropanation reaction (Figure 5).[15] Under our optimized conditions (3 equivethyldiazoacetate (EDA), 2.5 mol% iron (III) phthalocyanine (FePcCl)) and using 2% wt/v 2, cyclopropane production was increased by two-fold (560 mgL−1, 96% yield, Table 2, Entry 2). In contrast, using a 20% wt/v overlay of n-dodecane the titre of 4 was reduced to 49 mgL−1 and the yield of the reaction decreased to 12% (Table 2, Entry 3). This is likely a three-phase effect manifested by the heterogeneous nature of the phthalocyanine catalyst in the culture medium. Reactivity was restored to this system by using the organic-soluble catalyst tetraphenylporphyriniron(III) chloride (FeTPPCl), however the yield of 4 was reduced due to its diminished activity relative to FePcCl (Table 2, Entry 4). In contrast, both homo- and heterogeneous catalysts were highly active in cultures with TPGS-750-M micelles. The reactivity of FeTPPCl was also enhanced in the presence of 2, which reduced the reaction time by 36 h, an observation that is best rationalized by the solubility of the catalyst in the micelle interior (Figure 5C, Table 2, Entry 5). Vitamin E-derived designer micelles therefore support both homo- and heterogeneous catalysis in vivo, and, in addition to their pronounced effect on metabolite flux at the cell membrane, also increase the overall productivity and rate of biocompatible reactions outside of the cell via localization of the reaction components.
Figure 5.
Biocompatible cyclopropanation in E. coli-associated micelles. A) Biocompatible cyclopropanation. B) Organic-soluble FeTPPCl and heterogeneous FePcCl catalysts. C) Metabolite production time-course for Fe-catalyzed cyclopropanation reactions in the presence and absence of 5% wt/v 2. Reactions were conducted aerobically in 100 mL of MM1 media in sealed 500 mL Erlenmeyer flasks at 32 °C with shaking at 220 rpm. The reaction components and 2 were added at the point-of-induction of the styrene-producing pathway (OD600=0.6–0.8). Cyclopropane concentrations were determined inorganic extracts of culture aliquots by GC relative to an internal standard of 1,3,5-trimethoxybenzene using a standard curve. Data points are shown as mean values from three independent experiments. Errors bars have been removed for clarity. The iron catalyst used in each experiment is shown in parenthesis. Full experimental conditions and additional data can be found in Section S11 and Figures S18–22.
Table 2.
End-point production levels of cyclopropane 4 in the presence and absence of TPGS-750-M or n-dodecane.
| Entry | Catalyst | Additive | % Yield | Titer (mg/L) |
|---|---|---|---|---|
| 1 | FePcCl | – | 97 | 282 |
| 2 | FePcCl | 2% TPGS-750-M | 96[a] | 560 |
| 3 | FePcCl | 20% n-dodecane | 12[a] | 49 |
| 4 | FeTPPCl | 20% n-dodecane | 84 | 401 |
| 5 | FeTPPCl | 2% TPGS-750-M | 95 | 553 |
Experiments and analyses were performed as described in Figure 5. Yields refer to the relative concentrations of styrene and cyclopropane relative to an internal standard.
96 h.
The development of non-enzymatic, biocompatible reactions for use in microbial fermentations is a promising approach to expanding the types of molecules that can be accessed from renewable, non-petroleum derived feedstocks, particularly molecules of industrial importance that are not accessible using known enzymatic chemistry.[15,16] In vivo micellar catalysis represents a new way of interfacing non-enzymatic transformations with the products of microbial metabolism. In particular, it could enable the use of reactions/reagents in fermentations that would be otherwise toxic to the producing microorganism. This approach resembles strategies used by microbes that generate reactive or toxic metabolites (e.g. micro compartments, and membrane-bound organelles such as the annamoxosome).[17] These naturally compartmentalized pathways could provide inspiration for the types of intermediates and modes of reactivity that can be accommodated and utilized within a microbe-associated micelle.
Metabolite sequestration into micelles offers many benefits compared to the use of an organic solvent overlay. In the case of styrene production, significantly more solvent overlay was required for comparable ISPR from E. coli (157 mmol dodecane vs. 4 mmol 2). Sequestering styrene into dodecane also restricted the use of heterogeneous catalysts for cyclopropanation due to partitioning of the reaction components into three phases, highlighting an important limitation of using solvent overlays and non-enzymatic catalysis simultaneously.
In summary, this study establishes that (i) vitamin E designer surfactants previously developed and used exclusively for synthetic chemistry purposes are biocompatible and accelerate flux through engineered styrene biosynthesis in E. coli, outperforming surfactants previously used in the field of synthetic biotechnology; and (ii) nanomicelles formed by these molecules associate with the outer membrane of E. coli and can accelerate non-enzymatic catalysis in vivo. This represents the first example of micellar catalysis in the presence of a living microorganism, a strategy that will enable biocompatible reaction development. Future studies will focus on expanding the use of these micelles in vivo to encompass reaction manifolds that would be otherwise unproductive in fermentation media and/or toxic to E. coli.
Supplementary Material
Figure 1.
Combining engineered microorganisms and biocompatible synthetic micelles for small molecule synthesis.
Footnotes
This work was supported by the National Institutes of Health (DP2 GM105434), the Searle Scholars Program, and the European Commission (Marie Curie IOF Fellowship to S.W). We acknowledge Prof. David Nielsen (Arizona State University) for generously providing plasmids, and Dr. Maria Ericsson (Harvard Medical School) for running cryo-TEM experiments.
Supporting information for this article is given via a link at the end of the document.
References
- 1.Li C, Trost BM. Proc Natl Acad Sci U S A. 2008;105:13197–13202. doi: 10.1073/pnas.0804348105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Keasling J. Science. 2010;330:1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]; b) Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY. Nat Chem Biol. 2012;8:536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]; c) Becker J, Wittermann C. Angew Chem Int Ed. 2015;54:3328–3350. doi: 10.1002/anie.201409033. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2015;127:3383–3407. [Google Scholar]
- 3.a) Mukhopadhyay A. Trends Microbiol. 2015;23:498–508. doi: 10.1016/j.tim.2015.04.008. [DOI] [PubMed] [Google Scholar]; b) Peabody GL, Winkler VJ, Kao KC. Curr Opin Chem Eng. 2014;6:9–17. [Google Scholar]
- 4.a) Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A. Mol Syst Biol. 2011;7:487. doi: 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mingardon F, Clement C, Hirano K, Nhan M, Luning EG, Chanal A, Mukhopadhyay A. Biotechnol Bioeng. 2015;112:879–888. doi: 10.1002/bit.25511. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Jones CM, Hernández Lozada NJ, Pfleger BF. Appl Microbiol Biotechnol. 2015;99:9381–9393. doi: 10.1007/s00253-015-6963-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Volmer J, Neumann C, Bühler B, Schmid A. Appl Environ Microbiol. 2014;80:6539–6548. doi: 10.1128/AEM.01940-14. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Dunlop MJ. Biotechnol Biofuels. 2014;80:6539–6548. [Google Scholar]; f) Chen B, Ling H, Chang MW. Biotechnol Biofuels. 2013;6:21. doi: 10.1186/1754-6834-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fisher MA, Boyarskiy S, Yamada MR, Kong N, Bauer S, Tullman-Ercek D. ACS Synth Biol. 2013;3:30–40. doi: 10.1021/sb400065q. [DOI] [PubMed] [Google Scholar]
- 5.Todd DB. In: Fermentation and Biochemical Engineering Handbook. 3. Todaro CC, Vogel HC, editors. Elsevier; Oxford, UK: 2014. pp. 225–238. [Google Scholar]
- 6.a) Dhamole PB, Wang Z, Liu Y, Wang B, Feng H. Biomass Bioenergy. 2012;40:112–119. [Google Scholar]; b) Kirby J, Nishimoto M, Chow RWN, Pasumarthi VN, Chan R, Chan LJG, Petzold CJ, Keasling JD. Appl Environ Microbiol. 2014;80:6685–6693. doi: 10.1128/AEM.02155-14. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chen M, Chen X, Wan F, Zhang B, Chen J, Xiong Y. Microb Cell Fact. 2015;14:119. doi: 10.1186/s12934-015-0310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Duperray F, Jezequel D, Ghazi A, Letellier L, Shechter E. Biochim Biophys Acta. 1992;1103:250–258. doi: 10.1016/0005-2736(92)90094-3. [DOI] [PubMed] [Google Scholar]; e) Yoon S, Lee Y, Kim J, Lee S, Lee J, Kim J, Jung K, Shin Y, Keasling JD, Kim S. Biotechnol Bioeng. 2006;94:1025–1032. doi: 10.1002/bit.20912. [DOI] [PubMed] [Google Scholar]; f) Luo W, Huang J, Cai J, Xu Z. Food Sci Biotechnol. 2013;22:207–214. [Google Scholar]; g) McKenna R, Moya L, McDaniel M, Nielsen DR. Bioprocess Biosyst Eng. 2015;38:165–174. doi: 10.1007/s00449-014-1255-9. [DOI] [PubMed] [Google Scholar]; h) Staggs KW, Nielsen DR. Process Biochem. 2015;50:1487–1498. [Google Scholar]; i) Kang B, Zhang X, Wu Z, Qi H, Wang Z. Microb Biotechnol. 2013;6:540–550. doi: 10.1111/1751-7915.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Hu Z, Zhang X, Wu Z, Qi H, Wang Z. J Biotechnol. 2012;162:202–209. doi: 10.1016/j.jbiotec.2012.10.004. [DOI] [PubMed] [Google Scholar]; k) Kabay N, Cortina JL, Trochimczuk A, Streat M. React Funct Polym. 2010;70:484–496. [Google Scholar]; l) van den Berg C, Roelands M, Bussmann P, Goetheer E, Verdoes D, van der Wielen L. Ind Eng Chem Res. 2008;47:10071–10075. [Google Scholar]; m) van den Berg C, Wierckx N, Vente J, Bussmann P, de Bont J, van der Wielen L. Biotechnol Bioeng. 2008;100:466–472. doi: 10.1002/bit.21790. [DOI] [PubMed] [Google Scholar]; n) Xue Y, Qian C, Wang Z, Xu JH, Yang R, Qi H. Appl Microbiol Biotechnol. 2010;85:517–524. doi: 10.1007/s00253-009-2139-9. [DOI] [PubMed] [Google Scholar]
- 7.a) Dwars T, Paetzold E, Oehme G. Angew Chem Int Ed. 2005;44:7174–7199. doi: 10.1002/anie.200501365. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2005;117:7338–7364. [Google Scholar]; b) Handa S, Slack ED, Lipshutz BH. Angew Chem Int Ed. 2015;54:11994–11998. doi: 10.1002/anie.201505136. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2015;127:12162–12166. [Google Scholar]; c) Handa S, Wang Y, Gallou F, Lipshutz BH. Science. 2015;349:1087–1091. doi: 10.1126/science.aac6936. [DOI] [PubMed] [Google Scholar]; d) Slack ED, Gabriel CM, Lipshutz BH. Angew Chem Int Ed. 2014;53:14051–14054. doi: 10.1002/anie.201407723. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2014;126:14275–14276. [Google Scholar]; e) Lipshutz BH, Ghorai S, Abela AR, Moser R, Nishikata T, Duplais C, Krasovskiy A. J Org Chem. 2011;76:4379–4391. doi: 10.1021/jo101974u. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Lipshutz BH, Taft BR. Org Lett. 2008;10:1329–1332. doi: 10.1021/ol702755g. [DOI] [PubMed] [Google Scholar]; g) Klumphu P, Lipshutz BH. J Org Chem. 2014;79:888–900. doi: 10.1021/jo401744b. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Lipshutz BH, Peterson TB, Abela AR. Org Lett. 2008;10:1333–1336. doi: 10.1021/ol702714y. [DOI] [PubMed] [Google Scholar]; i) Lipshutz BH, Aguinaldo GT, Ghorai S, Voigtritter K. Org Lett. 2008;10:1325–1328. doi: 10.1021/ol800028x. [DOI] [PubMed] [Google Scholar]; j) Isley NA, Linstadt RTH, Kelly SM, Gallou F, Lipshutz BH. Org Lett. 2015;17:4734–4737. doi: 10.1021/acs.orglett.5b02240. [DOI] [PubMed] [Google Scholar]; k) Handa S, Fennewald JC, Lipshutz BH. Angew Chem Int Ed. 2014;53:3432–3435. doi: 10.1002/anie.201310634. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2014;126:3500–3503. [Google Scholar]; l) Tasca E, La Sorella G, Sperni L, Strukul G, Scarso A. Green Chem. 2015;17:1414–1422. [Google Scholar]; m) Gallou F, Isley NA, Ganic A, Onken U, Parmentier M. Green Chem. 2016;18:14–19. [Google Scholar]; n) Gabriel CM, Keener M, Gallou F, Lipshutz BH. Org Lett. 2015;17:3968–3971. doi: 10.1021/acs.orglett.5b01812. [DOI] [PubMed] [Google Scholar]; o) Isley NA, Hageman MS, Lipshutz BH. Green Chem. 2015;17:893–897. doi: 10.1039/C4GC01733A. [DOI] [PMC free article] [PubMed] [Google Scholar]; p) Rühling A, Galla H-J, Glorius F. Chem Eur J. 2015;21:12291–12294. doi: 10.1002/chem.201502542. [DOI] [PubMed] [Google Scholar]; q) La Sorella G, Strukul G, Scarso A. Green Chem. 2015;17:644–683. [Google Scholar]; r) Fennewald JC, Lipshutz BH. Green Chem. 2014;16:1097–1100. doi: 10.1039/C3GC42119H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKenna R, Nielsen DR. Metab Eng. 2011;13:544–554. doi: 10.1016/j.ymben.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 9.a) Seok KY, Min J, Hong HN, Park JH, Park KS, Gu MB. J Microbiol Biotechnol. 2007;17:1390–1393. [PubMed] [Google Scholar]; Kieboom J, du Bont JA. In: Bacterial Stress Responses. Storz G, Hengee-Aronis R, editors. ASM Press; Washington DC, USA: 2000. pp. 393–402. [Google Scholar]; c) Osborne SJ, Leaver J, Turner MK, Dunnill P. Enzyme Microb Technol. 1990;12:281–291. doi: 10.1016/0141-0229(90)90100-5. [DOI] [PubMed] [Google Scholar]
- 10.a) Nakata K, Tsuchido T, Matsumura Y. J Appl Micrbiol. 2010;110:568–579. doi: 10.1111/j.1365-2672.2010.04912.x. [DOI] [PubMed] [Google Scholar]; b) Pollack VA, Anderson DA. Appl Environ Microbiol. 1970;20:727–733. doi: 10.1128/am.20.5.727-733.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.In order to confirm the presence of micelles in our experiments we determined the critical micelle concentration (CMC) of 2 and 3 in MM1 media. Using a standard absorbance assay involving the water-insoluble dye 1-(2-pyridylazo)-2-napthol (PAN) we determined the CMC of 2 and 3 to be 0.007% and 0.008% wt/v, respectively (Section S10). The amount of 2 and 3 used in our in vivo experiments are above this concentration, thus confirming the presence of micelles.
- 12.a) Duguid JP. J Pathol. 1951;63:673–685. doi: 10.1002/path.1700630413. [DOI] [PubMed] [Google Scholar]; b) Ionescu M, Belkin S. Appl Environ Microbiol. 2009;75:483–492. doi: 10.1128/AEM.01616-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Arciola CR, Campoccia D, Gamberini S, Cervellati M, Donati E, Montanaro L. Biomaterials. 2002;23:4223–4239. doi: 10.1016/s0142-9612(02)00171-0. [DOI] [PubMed] [Google Scholar]; b) Freeman DJ, Falkiner FR, Keane CT. J Clin Pathol. 1989;42:872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo W, Huang J, Zhu X, Huang L, Cai J, Xu Z. Food Sci Biotechnol. 2013;22:207–214. [Google Scholar]
- 15.Wallace S, Balskus EP. Angew Chem Int Ed. 2015;54:7106–7109. doi: 10.1002/anie.201502185. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2015;127:7212–7215. [Google Scholar]
- 16.a) Wallace S, Balskus EP. Curr Opin Biotechnol. 2014;30:1–8. doi: 10.1016/j.copbio.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wallace S, Shultz EE, Balskus EP. Curr Opin Chem Biol. 2015;25:71–79. doi: 10.1016/j.cbpa.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Anbarasan P, Baer ZC, Sreekumar S, Gross E, Binder JB, Blanch HW, Clark DS, Toste DF. Nature. 2012;491:235–239. doi: 10.1038/nature11594. [DOI] [PubMed] [Google Scholar]; d) Domaille DW, Hafenstine GR, Greer MA, Goodwin AP, Cha JN. ACS Sustainable Chem Eng. 2016;4:671–675. doi: 10.1021/acssuschemeng.5b01590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Kartal B, Maalcke WJ, de Almeida NM, Cirpus I, Gloerich J, Geerts W, Op den Camp HJM, Harhangi HR, Janssen-Megens EM, Francoijs K, et al. Nature. 2011;479:127–130. doi: 10.1038/nature10453. [DOI] [PubMed] [Google Scholar]; b) SinningheDamsté JS, Strrous M, Rijpstra WIC, Hopmans EC, Geenevasen JAJ, van Duin ACT, van Niftrik LA, Jetten MSM. Nature. 2002;419:708–712. doi: 10.1038/nature01128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.