Abstract
Heat Shock Transcription Factor (HSF) family members function in stress protection and in human disease including proteopathies, neurodegeneration and cancer. The mechanisms that drive distinct post-translational modifications, co-factor recruitment and target gene activation for specific HSF paralogs are unknown. We present high-resolution crystal structures of the human HSF2 DNA-binding domain (DBD) bound to DNA, revealing an unprecedented view of HSFs that provides insights into their unique biology. The HSF2 DBD structures resolve a novel carboxyl-terminal helix that directs the coiled-coil domain to wrap around DNA, exposing paralog-specific sequences of the DBD surface, for differential post-translational modifications and co-factor interactions. We further demonstrate a direct interaction between HSF1 and HSF2 through their coiled-coil domains. Together, these features provide a new model for HSF structure as the basis for differential and combinatorial regulation to influence the transcriptional response to cellular stress.
Introduction
The Heat Shock Transcription Factors (HSFs) are found in eukaryotes from fungi to humans and impact diverse aspects of cell biology including stress adaptation, protein folding and quality control, development, and disease1–3. The remarkable diversity of HSF target genes contributes to the influence of HSF in a wide range of cellular processes4,5. Despite the recognition that HSFs play a prominent role in cellular adaptation to stress and in disease, our understanding of the mechanisms by which distinct HSF paralogs bind to genomic loci and engage in unique interactions with regulatory factors is limited. A greater understanding of HSF structure, interactions and function will benefit the development of therapeutic strategies that modulate HSFs for the treatment of human disease6,7.
The human HSF family is comprised of three members, HSF1, HSF2, and HSF43. HSFs are multi-domain transcription factors containing an amino-terminal winged helix-turn-helix DNA-binding domain, an adjacent extended coiled-coil multimerization domain, a central regulatory domain, a carboxyl-terminal coiled-coil domain and a transcriptional activation domain8–12. When activated, HSFs bind to a conserved DNA sequence known as a Heat Shock Element (HSE) consisting of inverted repeats of 5′-nGAAn-3′, which exhibits variations in sequence and geometry in target genes across the human genome5,13,14. Moreover, HSF family members interpret a diverse array of regulatory inputs that enable sophisticated tuning of the transcriptional response to stressful stimuli. Understanding the specific regulatory modalities of HSF family members could empower the ability to target specific arms of the HSF-mediated transcriptional response.
HSF1 is the most studied of the human HSF family for its role in the inducible transcription of genes encoding protein chaperones and the chaperonin TRiC, components of the ubiquitin-proteasome and autophagy pathway for misfolded protein degradation, anti-apoptotic proteins and stress adaptation factors following proteotoxic stress15,16. Under normal conditions, HSF1 is maintained in a repressed monomeric state through functional interactions with the Hsp90 chaperone and direct interactions with the chaperonin TRiC3,15. In response to proteotoxic stress, HSF1 multimerizes through the extended coiled-coil domain, accumulates in the nucleus and promotes target gene transcription. In addition to activating the protein quality control machinery in response to proteotoxic stress, HSF1 activates a large constellation of genes that are influenced by different cellular contexts. For example, in cancer cells, HSF1 promotes the transcription of a subset of pro-survival genes that only partially overlap genes that are activated in response to heat stress5,17. Moreover, the genomic binding fingerprint of HSF1 in striatal neuronal cells expressing a pathological poly-glutamine-expanded Huntingtin protein is distinct from that in cells expressing a non-pathogenic variant of Huntingtin18. These observations highlight the importance of understanding the mechanistic features of HSF1 target gene recognition and activation, in a context-dependent manner.
Like HSF1, HSF2 participates in the transcriptional regulation of genes in response to stress and is similar in overall domain structure, but exhibits genomic binding site occupancy and regulatory interactions that are distinct from HSF113,19. One of the most striking contrasts in HSF1 and HSF2 regulation is their relative stability, where HSF1 is much longer lived than HSF2 following proteotoxic stimuli20,21. HSF2 is recognized as a critical mediator of brain development and plays a role in fetal alcohol syndrome22. In addition, both HSF1 and HSF2 contribute to spermatogenesis and specific mutations in HSF2 have been associated with idiopathic azoospermia23–25. Several studies have identified a functional interaction between HSF1 and HSF2 and have observed that HSF1 and HSF2 are found in a complex in vivo22,26,27. However, the nature of the HSF1 and HSF2 interactions and their functional consequences are not well understood. Furthermore, the repertoire of HSF1 and HSF2 target genes under different cellular contexts has only recently been explored. A ChIP-seq study identified the spectrum of HSF1 and HSF2 gene targets in human K562 cells and found that despite binding similar HSE sequence motifs, HSF1 and HSF2 exhibited only a partially overlapping pattern of genomic occupancy13. This observation presents the interesting question of whether HSF1 and HSF2 target gene selection is driven by intrinsic biochemical differences in HSF1 and HSF2, or is mediated by extrinsic factors such as protein-protein interactions and post-translational modifications.
Our current knowledge of HSF structure is very limited. Since elucidation of the partial structure of the yeast K. lactis HSF DNA-binding domain, little HSF structural information has been reported8. To gain structural insights with respect to HSF2 DNA binding, we solved two high-resolution crystal structures of the human HSF2 DNA-binding domain (DBD) bound to two distinct HSEs at 1.73 and 2.10 Å. These structures revealed novel structural features that effectively invert current models of HSF DNA binding. Moreover, we demonstrate that the unique structural features identified in this study impact human HSF regulation in vitro and in vivo. Taken together, these critical insights on the architecture of HSFs while bound to DNA lay mechanistic ground for understanding how HSF structure drives combinatorial regulation that can enable precise control of target gene transcription at specific loci and in specific physiological and pathophysiological contexts.
Results
Structure of the HSF2 DBD bound to a “2-site” HSE
We solved the crystal structure of the human HSF2 DBD bound to a 2-site HSE (5′-GGTTCTAGAACC-3′), at 1.73 Å (Table 1, Supplementary Fig. 1a). The structure shows two HSF2 monomers bound to the 2-site HSE creating a dimer interface parallel to the axis of DNA (Fig. 1a). The sequence-specific interaction is largely mediated through a hydrogen bonding interaction between Arg63 and the guanine of the nGAAn HSE motif (Fig. 1b,c, Supplementary Fig. 1b). This arginine, which is positionally conserved and is critical for HSF1 DNA binding8, is located within a recognition helix containing the conserved sequence “SFVRQ” and is inserted into the major groove of DNA (Fig. 1a). In addition, numerous direct and indirect (water-mediated) contacts are made to the DNA phosphate backbone (Fig. 1b).
Table 1.
Data collection and refinement statistics (molecular replacement)
| HSF2DBD-2siteHSE | HSF2DBD-3siteHSE | |
|---|---|---|
| Data collection | ||
| Space group | C 1 2 1 | P 1 |
| Cell dimensions | ||
| a, b, c (Å) | 84.88, 39.63, 39.93 | 40.17, 47.14, 109.59 |
| α, β, γ(°) | 90.00, 91.43, 90.00 | 82.82, 90.03, 64.75 |
| Resolution (Å) | 50 – 1.73 | 50–2.1 |
| Rmerge | 0.082 (0.659)a | 0.116 (0.832)a |
| I /σI | 22.8 (2.13) | 11.9 (1.28) |
| Completeness (%) | 94.3 (89.7) | 93.9 (89.7) |
| Redundancy | 5.1 (4.1) | 2.5 (2.3) |
| Refinement | ||
| Resolution (Å) | 21.21–1.73 | 24.41–2.069 |
| No. reflections | 13023 | 38940 |
| Rwork / Rfree | 0.1479/0.1991 | 0.2171/0.2730 |
| No. atoms | 2353 | 9293 |
| Protein | 1867 | 6827 |
| Ligand (DNA) | 380 | 2164 |
| Water | 106 | 302 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.011 | 0.015 |
| Bond angles (°) | 1.396 | 1.486 |
1 Crystal was used for each structure (Supplementary Fig. 1a, 4a).
Values in parentheses are for highest-resolution shell.
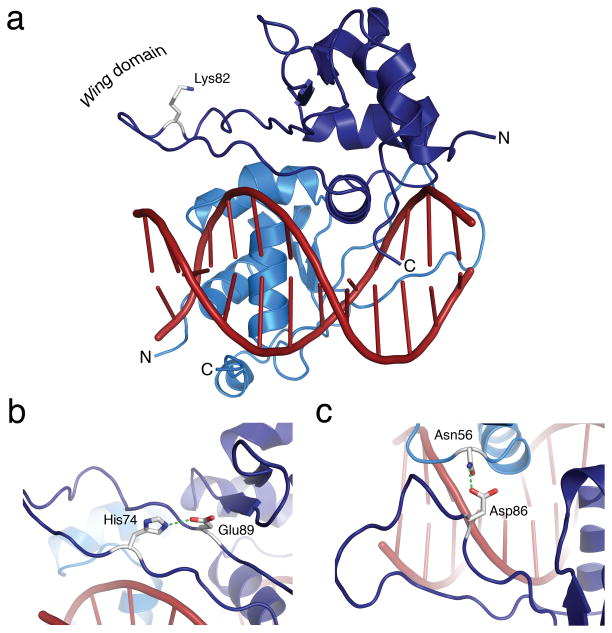
Figure 1. Structure of the HSF2 DBD bound to a 2-site HSE.
a) Overview of the HSF2 DBD dimer. (top) Individual monomers (light and dark blue) form a dimer interface parallel to the axis of HSE DNA (red). Viewing down the axis of DNA (bottom) reveals the recognition helix of the “winged helix-turn-helix” motif for each monomer inserted into the major groove of the DNA. b) Ladder diagram of the direct and indirect contacts formed between the HSF2 DBD and HSE DNA, with side chain, peptide backbone and water molecule-mediated contacts indicated. c) Arg63 of the HSF2 DBD engages in a direct contact to the guanine of the HSE nGAAn motif. d) Distance difference matrix of human HSF2 DBD and K. lactis HSF DBD depicted as a putty diagram. Distances between paired atoms ranging from 0.348 Å (thin, blue) to 3.837 Å (thick, red) indicate structural deviations between HSF2 DBD and K. lactis HSF DBD. Shown in purple are novel HSF2 structural features that are not present in the K. lactis HSF DBD structure.
A particularly interesting non-sequence specific DNA contact involves Lys72 with the phosphate backbone (Fig. 1b, Supplementary Fig. 1d). This lysine corresponds to Lys80 in HSF1, previously identified as being subject to acetylation-mediated inhibition of DNA binding28. By analogy, the HSF2 DBD structure provides empirical evidence for this residue directly contacting the DNA phosphate backbone (Supplementary Fig. 1d). While site-specific analysis of HSF2 acetylation is lacking, these results suggest that HSF2 DNA binding could be regulated in a similar fashion to that of HSF1.
A direct comparison of the HSF2 DBD structure with the K. lactis DBD structure revealed insights into the novelty of the HSF2 structures. Figure 1d depicts a difference distance matrix calculated between pairs of like atoms in the human HSF2 and K. lactis DBD (Supplementary Fig. 1c). While the DNA binding helix, encompassing Arg63, exhibits little structural deviation (thin, blue putty), structural features distal to the DNA binding surface are strikingly distinct (thick, red putty). Moreover, the HSF2 structures presented here resolve two entirely new components of the DBD, which include the wing domain and a carboxyl-terminal helix (Figure 1d, purple).
HSF2 “wing” domain topology
The wing domain of other winged helix-turn-helix transcription factors functions in direct DNA binding8,28,29. While previous structural studies of the K. lactis HSF DBD did not resolve the wing domain, the human HSF2 wing domain showed clear electron density for the entirety of the peptide backbone (Supplementary Fig. 2), with no direct or indirect contacts to DNA (Figs. 1a, 2a). The HSF2 wing domain is formed, in part, by a hydrogen bond between His74 and Glu89 that acts as a wedge between β sheets to “pry open” the wing domain for solvent exposure (Fig. 2b). Additionally, Asp86 engages in a hydrogen bond to Asn56 of the adjacently bound monomer, establishing an HSF2 dimer interface also observed in the K. lactis structure, that was proposed to be the primary function of the wing domain8 (Fig. 2c). Moreover, the side chain of HSF2 DBD Lys82, identified by in vivo proteomic studies to be covalently conjugated by SUMO2 30–32, is exposed to solvent (Fig. 2a). The importance of the wing domain is underscored by a report demonstrating that Replication Protein A subunit 70 kDa (RPA70) binds to the HSF1 wing domain, but not to HSF233. By analogy with the HSF2 DBD structure presented here, the location of the RPA70 interaction with HSF1 is predicted to occur at the most distal portion of the wing domain where little steric hindrance would be observed. The structural findings described here, together with earlier published functional studies, suggest that the HSF wing domain plays a critical regulatory role and provides a specificity component for distinct HSF family members. These regulatory events are likely enabled by the unique structural features of the HSF “wing” domain that are strikingly different from other members of the winged helix-turn-helix DBD family which utilize the wing domain to contact DNA28,29.
Figure 2. The HSF2 wing domain is solvent exposed and does not contact DNA.
a) Side view of the HSF2 DBD with the wing domain indicated on the left and the recognition helix inserted into the major groove of the HSE DNA site. Lys82, depicted as a stick model, is directed away from the DNA and exposed for modification. b) His74 and Glu89 form a hydrogen bond that acts as a wedge to pry open the wing domain. c) Asp86 engages Asn56 from the adjacent monomer to form a hydrogen bond that is predicted to strengthen the dimer interface and anchor the wing domain.
HSF2 DBD directs the coiled-coil to wrap around DNA
Current models for the topology of HSFs bound to DNA posit that the coiled-coil multimerization domain sits atop the DNA binding domain, poising the trans-activation region for interactions with the transcriptional machinery1,3,34. We observe a carboxyl-terminal helix present on the outer region of the HSF2 DBD (Fig. 3a) and Phe96, preceding this helix, is inserted into a deep hydrophobic pocket in the DBD formed by Leu14, Trp15, Trp29, and Phe35 (Fig. 3b). Gly99 enables a hairpin turn into the helix that is formed by the amphipathic sequence “LLENI” directed toward the “bottom” half of the DNA. The hydrophobic surface of this sequence fits into a hydrophobic pocket in the core of the DBD, directing the helix “down” to the opposite side of DNA (Fig. 3a,b). In addition, two conserved residues following the helix, Arg109 and Lys110, contact the DNA phosphate backbone, further directionally stabilizing the region preceding the coiled-coil multimerization domain on the opposite side of the DNA (Fig. 3a, Supplementary Fig. 3a,b). Key residues involved in the architecture of this carboxyl-terminal helix are conserved throughout HSF species, suggesting conservation of the directionality of the coiled-coil multimerization domain (Supplementary Fig. 3c).
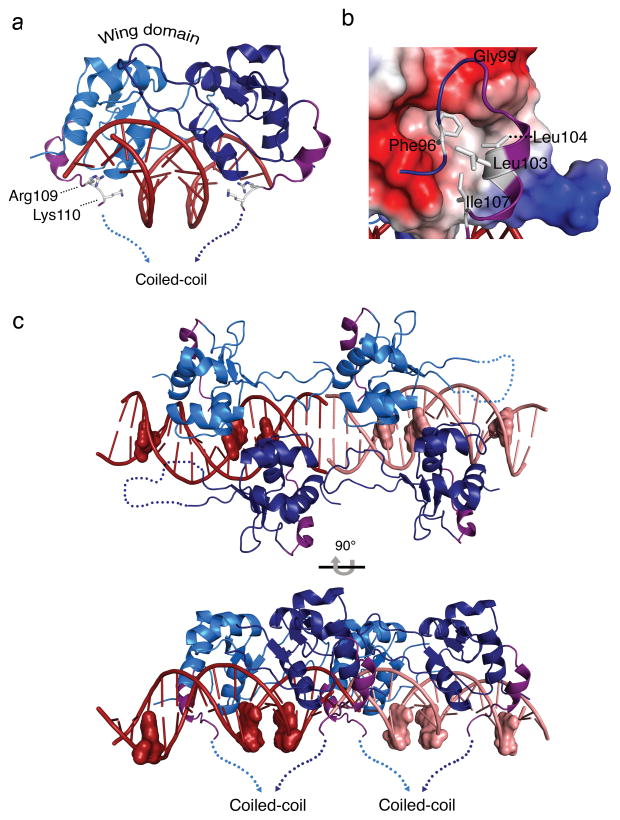
Figure 3. An HSF2 DBD carboxyl-terminal helix directs the coiled-coil multimerization domain to wrap around HSE DNA.
a) Side view of the HSF2-DBD depicting a newly resolved carboxyl-terminal helix (purple) orients the polypeptide to the underside of HSE binding site DNA, where Arg109 and Lys110 engage in direct and water-mediated DNA phosphate backbone contacts in the major groove (see Fig. S3). b) Electrostatic surface representation of the HSF2 DBD and the carboxyl-terminal helix. Red and blue represent negative and positively charged regions, respectively, and white depicts hydrophobic surfaces. The potential contours are shown on a scale from 63.41 (blue) to −63.41 kbT e−1 (red) with white representing no charge. The carboxyl-terminal helix (purple) contains two leucines (Leu103, Leu104) and an isoleucine (Ile107) that together with Phe96, pack against the hydrophobic groove presented by the core of the DBD. c) Representation of the asymmetric unit of the HSF2 DBD 3-site HSE structure. HSF2 crystallized as a dimer of dimers bound to adjacent HSE sequences shown in dark red and pink. The guanine of each nGAAn motif is represented as a surface model to show the last guanine in each HSE as unoccupied. The carboxyl-terminal helices (purple) of all four monomers direct the coiled-coil domain to the underside of the DNA, exposing the top portion of the DBD.
HSFs are thought to exist as trimeric proteins based on the tripartite nature of a canonical HSE (nnGAAnnTTCnnGAAnn), their apparent electrophoretic mobility in heat-shocked cell extracts, and the presence of an extended coiled-coil immediately downstream of the DBD10,35,36. To gain insights into the structural features of the DBD topology in an HSF trimer, we solved the crystal structure of the HSF2 DBD bound to a canonical 3-site HSE (Table 1, Supplementary Fig. 3d). Interestingly, the HSF2 DBD crystallized in the P1 space group with 2 dimers bound to two-3 site HSEs (Fig. 3c), demonstrating that even when presented with a tripartite HSE, the HSF2 DBD crystallized as a dimer, with the third nGAAn of the HSE unoccupied. The independently bound dimers pack against each other to form pseudo continuous DNA and form a “dimer of dimers”. Identical direct and indirect DNA contacts are made when compared to the HSF2 DBD 2-site HSE structure (Supplementary Fig. 3e). Furthermore, all carboxyl-terminal helices of the HSF2 DBD are directed to the opposite side of the DNA, similar to the 2-site HSE structure (Fig. 3c). While it is possible that the dimer structure observed here occurs as a function of crystal packing, HSF oligomeric states may exist outside of the canonical trimer, as has been suggested for HSF2 dimers in K562 cells37. The HSF2 coiled-coil multimerization domain is located carboxyl-terminal to the DBD, separated by a short linker sequence of 14 amino acids. Based on both the HSF2 DBD 2-site and 3-site structures, we propose that the conserved carboxyl-terminal amphipathic helix directs the extended coiled-coil opposite the DNA, allowing HSFs to “wrap-around” the DNA upon binding. This topology would permit the upper surface of the HSF DBD to be accessible for regulatory interactions that would otherwise be sterically occluded by the multimerization domain as depicted in current models3,7,34,38,39. This topology presents surfaces for geographically distinct interactions with regulatory proteins via both the DBD and the coiled-coil domain in proximity to DNA. Support for this model is rooted in reports that have identified key interactions and post-translational modifications that occur within both the DNA-binding and coiled-coil domains of transcription factors including HSF133,40–42. Thus, embracing of DNA by HSF family members would enable geographically separated regulatory interactions at both the DBD and coiled-coil domain that provide multiple input signals for HSF-mediated regulation. Given this new structural information, we propose a new model for HSF geometry, when bound to DNA, that is inverted with respect to previous models1,43.
HSFs exhibit divergent surface characteristics distal to DNA
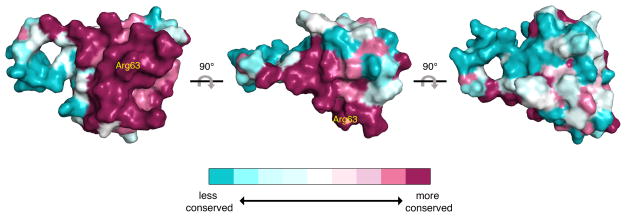
A comparison of the structures of the HSF2 DBD bound to DNA, with the sequence of the HSF1 and HSF4 DBDs, suggests that these proteins interact with DNA in a highly similar manner. To explore the nature of HSF family DBD surface features, we calculated the relative conservation of individual amino acids between HSF paralogs using the ConSurf Server44. On the HSF2 surface that contacts DNA, it is apparent that amino acid residues such as Arg63 and other residues making direct and indirect DNA contacts are conserved (Figure 4). This suggests that HSF family members would have little, if any, paralog-specific sequence preferences for DNA and provides support for extrinsic mechanisms driving differential target gene binding and activation in vivo. Examination of the HSF DBD surface distal to DNA contact sites shows little conservation of sequence or biochemical character (Figure 4), suggesting that HSFs have evolved distinct surfaces to accommodate divergent regulatory inputs within the DBD, without altering DNA-binding mechanisms. Furthermore, the wrapping of HSF coiled-coils around HSE DNA would expose the wing and other unique surfaces of the HSF1, HSF2, and HSF4 DBDs, generating a template for additional paralog-specific interactions with regulatory proteins.
Figure 4. Human HSF family members share similar DNA proximal surfaces but have distinct distal surfaces.
Surface of the HSF2 DBD with a color gradient representing relative conservation between HSF1, HSF2, and HSF4 calculated by ConSurf. Dark red surfaces represent highly conserved regions between HSF family members, whereas light blue surfaces indicate divergent regions. Arg63 (yellow) indicates the location of the HSF-DBD interface.
HSF1 and HSF2 are differentially SUMOylated in vitro
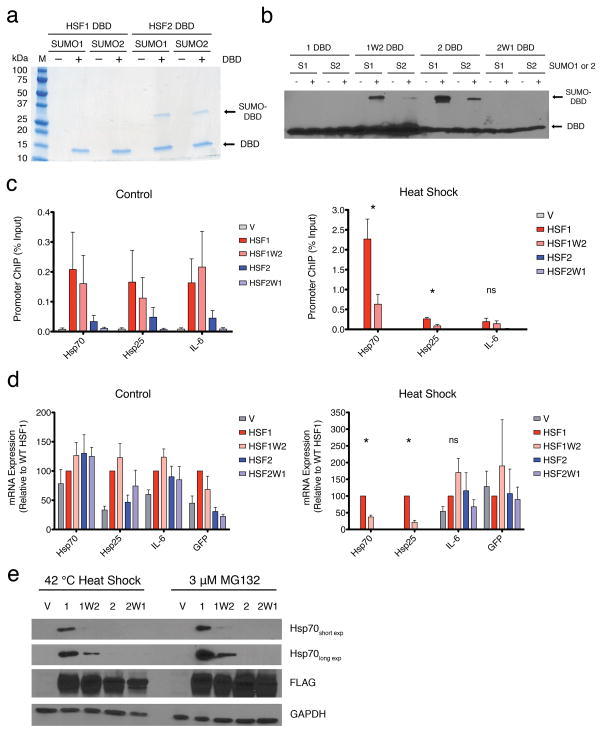
The HSF1 DBD, but not HSF2, was previously shown to interact with RPA70 in vitro and in vivo and this interaction contributes to HSF1-mediated target gene expression35. We postulated that a distinct protein-protein interaction or post-translational modification could be specific for the HSF2 DBD. Reports describing the mammalian SUMOylated proteome found that Lys82 of HSF2 is conjugated by SUMO2 but the corresponding Lys91 of HSF1 was not identified30–32. As HSF2 Lys82 localizes within the exposed wing domain (Fig. 2a), we ascertained whether there is intrinsic SUMOylation specificity between the HSF1 and HSF2 DBDs. SUMOylation is analogous to ubiquitination in that both processes utilize an E1, E2, E3 enzymatic cascade for covalent modification of Lys on substrate proteins31,45. In addition, mammals utilize three distinct SUMO paralogs, SUMO1, SUMO2, and SUMO3, where SUMO2 and SUMO3 are similar and impart different functional consequences than SUMO130. Using recombinant SUMO E1 (SAE1 and SAE2), SUMO E2 (Ubc9) and SUMO E3 (RANBPΔFG) enzymes, we examined whether SUMO1 or SUMO2 could be covalently attached to the HSF1 or HSF2 DBD in vitro. As shown in Fig. 5a, the HSF1 DBD showed little to no modification by SUMO1 or SUMO2, whereas the HSF2 DBD was efficiently modified by both SUMO paralogs. This result was recapitulated when the SUMO reactions were performed with the SUMO E3 ligase PIAS1, or in the absence of E3 ligase (Supplementary Fig. 4a). Since SUMO proteomics studies identified Lys82 in the HSF2 wing domain as a SUMO substrate, wing domain chimeras were constructed where the HSF1 DBD contains the HSF2 wing domain (HSF1W2) and vice versa (HSF2W1) (Fig. 5b, Supplementary Fig. 4b). The HSF1W2 and HSF2W1 chimeras displayed opposite SUMOylation phenotypes compared with their WT counterparts, demonstrating that the HSF1 and HSF2 wing domains harbor distinct and transportable biochemical features that drive differential SUMOylation in vitro and in vivo. The HSF4 DBD does not contain a lysine in the wing domain (Supplementary Fig. 4b), rendering this wing domain incompetent for SUMO modification. The distinct SUMOylation profiles of HSF1, HSF2 and HSF4, and the HSF1-specific interaction with RPA70 within the wing domain could, in part, underlie regulatory differences that impart target gene selectivity in vivo46–49.
Figure 5. The HSF1 and HSF2 wing domains are biochemically and functionally distinct.
a) Coomassie staining of 6xHis-tagged HSF1 and HSF2 DBDs following in vitro SUMOylation and NiNTA purification. Molecular weight markers are indicated to the left. (−) or (+) indicate the addition, or not, of the purified HSF1 or HSF2 DBD b) Anti-6xHis tag immunoblot of HSF DBD wing domain chimeras following in vitro SUMOylation. (−) or (+) indicate the inclusion or not of ATP c) ChIP analysis of HSF wing domain chimeras expressed in hsf1−/−hsf2−/− MEFs by transfection with 3xFLAG-tagged derivatives of HSF1 and HSF2 or empty vector (V). Binding to Hsp70, Hsp25, and IL-6 promoters under control (left), and heat shock (right) conditions is shown. Specificity is demonstrated by lack of signal in vector transfected cells (gray). d) mRNA expression analysis of Hsp70, Hsp25, IL-6, and GFP in hsf1−/−hsf2−/− MEFs transfected with 3xFLAG-tagged derivatives of HSF1 and HSF2 or empty vector (V). Amplification of GFP is used as a transfection and HSF mRNA expression control. Values for each gene and HSF mutant are normalized to WT HSF1 (red) as 100%. e) Immunoblot analysis of Hsp70 induction in hsf1−/−hsf2−/− MEFs transfected with 3xFLAG-tagged HSF1 and HSF2 wing domain chimera proteins following heat shock or proteasome inhibition with MG132, with GAPDH as loading control. Histograms depict mean + SEM; * : p < 0.05, ns (not significant) : p > 0.05. n=3 for ChIP experiments, n=4 for qRT-PCR and each n represents biological replicates from separate cell cultures.
The wing domain regulates HSF activity in vivo
To explore a potential role for the wing domain in differential HSF function in vivo, 3xFLAG-tagged derivatives of WT HSF1, HSF1W2, HSF2, or HSF2W1 proteins were expressed in mouse embryonic fibroblasts doubly knocked out for HSF1 and HSF2 (Fig. 5c-e). Under control conditions, in vivo DNA binding activity of WT HSF1 and HSF1W2 were indistinguishable at the Hsp70, Hsp25, and IL-6 promoters (Fig. 5c). In contrast, WT HSF2 but not HSF2W1 exhibited basal binding to the Hsp70, Hsp25, and IL-6 promoter suggesting that the endogenous HSF2 wing domain is essential for basal HSF2 DNA binding (Fig. 5c). Interestingly, under heat shock conditions, HSF1W2 was compromised in binding to the Hsp70 promoter (~3 fold reduction) and Hsp25 promoter (~5 fold reduction), whereas DNA binding is unaffected at the IL-6 promoter (Fig. 5c). This suggests that HSF wing domains have genomic locus specific effects on DNA binding. Importantly, differences in HSF DBD wing domain chimeras DNA binding in vivo is not explained by differences in DNA binding to an HSE in vitro (Supplementary Fig. 4c).
Analogous to HSF wing domain chimera DNA binding, expression of Hsp70, Hsp25, and IL-6 mRNA, is unchanged in cells expressing WT HSF1 compared to HSF1W2 under control conditions (Fig. 5d). Upon heat shock, HSF1W2 is compromised in the ability to induce Hsp70 (~37% of WT) and Hsp25 (~21% of WT) expression, but not IL-6, suggesting that the HSF wing domain has genomic locus specific effects on target gene expression (Fig. 5d). Expression of both WT HSF2 and HSF2W1 did not increase expression of Hsp70, Hsp25, or IL-6 mRNA over vector transfected cells, raising the possibility that HSF2 does not exhibit activity at these loci in the absence of HSF1. The DNA binding and gene expression analysis is further supported by immunoblotting, where cells expressing HSF1W2 were strongly compromised for activation of Hsp70 expression compared to WT HSF1 following proteotoxic conditions induced by heat shock or the proteasome inhibitor MG132 (Fig. 5e). Moreover, neither wild type nor chimeric HSF2 activated Hsp70 expression following heat shock or proteasome inhibition, despite protein levels similar to the HSF1 variants (Fig. 5e). Taken together, these results suggest a locus-specific biological function for the HSF wing domain, not involving direct DNA contacts, and lay the foundation for mechanistic studies into critical regulatory events that occur within the HSF wing domains.
HSF1 and HSF2 directly interact to form hetero-complexes
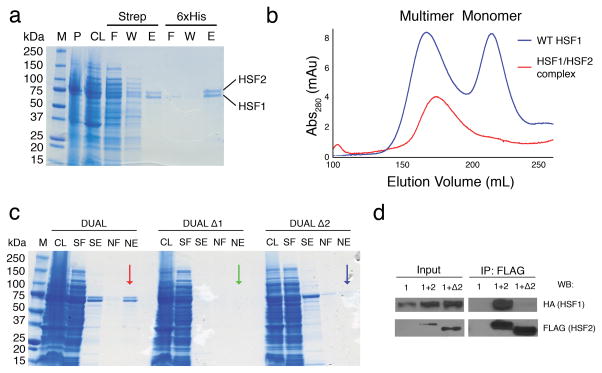
Several reports document an interaction between HSF1 and HSF2 in vivo through co-immunoprecipitation and electrophoretic mobility supershift assays, approaches that do not distinguish between direct or indirect interactions22,26,27. To ascertain if HSF1 and HSF2 directly interact, StrepII-tagged human HSF1 and 6xHis-tagged human HSF2 were co-expressed in E. coli from a single plasmid using a bi-directional promoter (Supplementary Fig. 5a). Tandem differential affinity purification resulted in co-elution of HSF1 and HSF2, suggesting that the two proteins directly interact (Fig. 6a). The HSF1-HSF2 hetero-complexes were fractionated by gel filtration, which resolves monomeric and multimeric HSF14, where the HSF1-HSF2 complex co-purified at an elution volume similar to the HSF1 multimer, demonstrating that HSF1 and HSF2 form hetero-multimeric complexes that are stable through three tandem purification steps (Fig. 6b, Supplementary Fig. 5b). Crosslinking and mass spectrometry confirmed that HSF1 and HSF2 are the only non-keratin proteins detectable in cross-linked complexes (Supplementary Fig. 5c, Supplementary Table 1).
Figure 6. HSF1 and HSF2 directly interact, enabling combinatorial regulation within the DBD and coiled-coil domains.
a) Coomassie staining of the tandem affinity purification of StrepII-HSF1 and 6xHis-HSF2. M-molecular weight marker, P - pellet, CL - cleared lysate, F - flow through, W - wash, E - elution. b) Sephacryl s400 size exclusion chromatograms of the HSF1-HSF2 hetero-complex (red), which elutes at a similar volume to a WT HSF1 multimer (blue). c) Coomassie staining of the tandem affinity purification of co-expressed WT HSF1 and WT HSF2 (DUAL; red arrow), HSF1ΔLZ1-3 and WT HSF2 (DUAL Δ1; green arrow), and WT HSF1 and HSF2ΔLZ1-3 (DUAL Δ2; blue arrow). M – molecular weight marker CL - cleared lysate, SF - strep flow through, SE - strep elution, NF - NiNTA flow through, NE - NiNTA elution. d) Co-Immunoprecipitation of HSF1 and HSF2 in MEFs. hsf1−/−hsf2−/− MEFs were transfected with HSF1-HA alone (1) or HSF1-HA + WT HSF2-3xFLAG (1+2) or HSF1 + HSF2ΔLZ1-3-3xFLAG (1+Δ2), immunoprecipitated with anti-FLAG beads prior to immunoblotting with anti-HA antibody or anti-FLAG antibody. Input= 1/100 of the IP sample
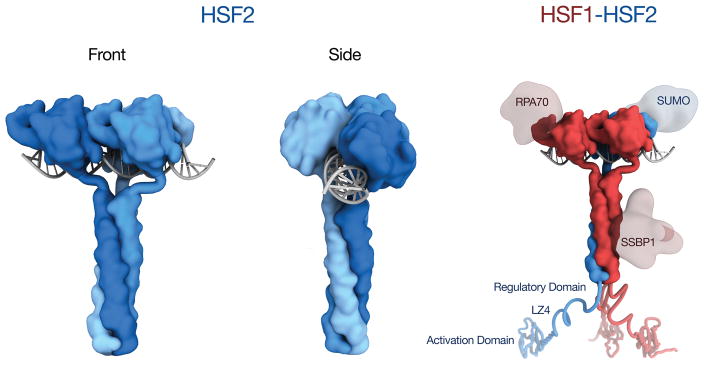
We evaluated the possibility that the HSF1-HSF2 interaction is mediated by the extended coiled-coil domain (LZ1-3), contained in both HSF1 and HSF2. Plasmids co-expressing HSF1ΔLZ1-3 and WT HSF2 (DUAL Δ1) and the reciprocal HSF1 and HSF2ΔLZ1-3 (DUAL Δ2) proteins were expressed and purified as in Fig. 6a. As shown in Fig. 7c, WT HSF1 and HSF2 co-purify from cell lysates (DUAL), but WT HSF2 fails to co-purify with HSF1 ΔLZ1-3 (DUAL Δ1) and WT HSF1 does not co-purify with HSF2 ΔLZ1-3 (DUAL Δ2). These results demonstrate that HSF1 and HSF2 form hetero-complexes directly through their respective extended coiled-coil multimerization domains in a fashion similar to other coiled-coil transcription factor multimers such as c-Fos and c-Jun40. To validate the direct interaction between HSF1 and HSF2, we performed co-immunoprecipitation experiments in hsf1−/− hsf2−/− MEFs expressing either HSF1-HA alone, or with WT HSF2 or HSF2 ΔLZ1-3. HSF1 specifically co-precipitates with WT HSF2, but not HSF2 ΔLZ1-3, demonstrating that the HSF2 coiled-coil domain is indispensable for the HSF1 interaction in vitro and in vivo (Figure 6d). Taken together, these observations suggest that in the context of the HSF coiled-coil wrapping around DNA, HSF1-HSF2 interactions would provide a template for additional differential and combinatorial regulatory events that would not exist in HSF1 or HSF2 homo-oligomeric complexes. In Fig. 7 we present a new model for HSF DNA binding, using a generic coiled-coil domain50, where front and side views of homomultimeric HSF2 depict the coiled-coil domain embracing DNA. Shown on the right is a similar model for an HSF1-2 heteromultimer bound to DNA, enabling combinatorial interactions between HSF1 complexes (RPA70, and SSBP1) and HSF2 (SUMO) regulatory modules33,41,42. The direct interaction between HSF1 and HSF2 also enables combinatorial regulation at other domains including the Regulatory Domain, LZ4, and Activation domain.
Figure 7. A new model for Heat Shock Factor DNA binding.
HSF2 homotrimers are shown on the left embracing DNA, thereby exposing both the DBD and coiled-coil for regulatory interactions. Shown on the right is an HSF1-HSF2 heterotrimer with HSF1 specific (RPA70, SSBP1) and HSF2 specific (SUMO) regulatory modules engaged with the DBD and coiled-coil.
Discussion
The HSF2 DBD structures presented in this study lay the foundation for a greater understanding of how HSFs enable complex and dynamic regulation at diverse genomic loci in distinct cellular contexts. Distinct biochemical features of HSF DBDs distal to the DNA binding surface drive differential post-translational modifications and protein-protein interactions. Such differential regulation will be enabled by HSFs wrapping around DNA, which presents divergent and geographically separated surfaces, including both the DBDs and coiled-coil domains, for regulatory inputs. Further exploration into the divergent surfaces of HSF family members may reveal additional paralog-specific regulatory events that enable complex transcriptional regulation that would not be achieved via subtle variations in genomic HSE sequences.
The evolution of transcriptional regulatory networks has been proposed to occur through variation in cis-regulatory elements upstream of target genes51, as mutations in transcriptional regulators themselves would have the potential for widespread consequences in comparison to single mutations in the regulatory sequences of specific genes. However, a growing body of evidence suggests that variation in transcription factors themselves can drive complex regulation of gene expression networks and can be enabled through gene duplication events52,53. We suggest that the human HSF family represents a quintessential example of this type of transcriptional network evolution.
The conserved and unique surfaces of human HSF paralogs resemble the evolution of MADS-box proteins in yeast, in which gene duplication enabled the neo-functionalization of MADS-box paralogs in S. cerevisiae when compared to yeast harboring a single MADS-box protein54. For example, the S. cerevisiae Mcm1 and Arg80 MADS-box paralogs maintain similar DNA-binding preferences, but exhibit diversity in co-factor binding pockets and interactions between Mcm1 and Arg80 facilitate combinatorial regulation54. A similar phenomenon could be envisioned with HSFs, where HSF1 and HSF2 could exhibit paralog interactions that influence the activity of each individual paralog.
The direct physical and functional interactions between HSF1 and HSF2 add an additional layer of complexity to the response to stress, but may also provide cells with a rheostat for more precise transcriptional output at specific HSF target genes or other chromosomal loci. HSF1 is known to function in protein misfolding disease, and to drive a unique gene expression program in cancer cells, whereas HSF2 has recently been proposed to act as a tumor suppressor, highlighting the importance of understanding differential HSF regulation55. A better understanding of such regulatory features could enable the design and development of selective molecular probes that target specific arms of the HSF-mediated transcriptional program. The transcriptional outcome of the direct interaction between HSF1 and HSF2, and the consequential regulatory interactions, presents an intricate mechanism that could impact a diverse array of biological processes in normal cells as compared to pathophysiological states. Further exploration of surface features of the HSFs, and other transcription factors, could reveal new layers of transcriptional outputs that influence cellular processes, and new ways to modulate these complex regulatory interactions.
Online Methods
Expression and purification of HSF DNA-Binding Domains
A codon-optimized DNA sequence encoding the HSF2 DNA-Binding Domain (amino acids 8–115) was cloned into the pET15b expression plasmid containing an amino-terminal 6xHis tag using InFusion (Clontech) and the plasmid was transformed into BL21(DE3) E. coli. Overnight LB cultures were back diluted 1:100 into 4 L flasks containing 2 L of LB + 100 ug/mL ampicillin, grown to an OD600 ~ 0.5 and induced with 1 mM IPTG for 5 hours. Cell pellets were lysed in 20 mL NiNTA Lysis Buffer (50 mM HEPES pH 7.5, 300 mM NaCl, 20 mM Imadazole HCl) using a sonic dismembrator for 3 × 30 sec bursts. Lysates were then cleared by centrifugation at 20,000 g for 30 minutes. Cleared lysates were incubated with 5 mL bed volume of NiNTA Agarose (Qiagen) and rotated at 4 °C for 2 hours. Following addition to a gravity filtration column, the NiNTA resin was washed with 2 × 50 mL NiNTA wash buffer (50 mM HEPES pH 7.5, 300 mM NaCl, 40 mM Imidazole HCl) and eluted with 10 mL NiNTA elution buffer (50 mM HEPES pH 7.5, 300 mM NaCl, 250 mM Imidazole HCl). For 6xHis tag cleavage, the purified HSF2 DBD was buffer exchanged using an Amicon Ultra Centricon (MWCO 3kDa) until the final Imidazole concentration was 20 mM in 10 mL. This procedure resulted in 2 mg protein yield. The buffer exchanged HSF2 DBD was then incubated with 3 units of biotinylated-thrombin (Novagen) overnight at 15 °C. Following streptavidin removal of thrombin and NiNTA removal of uncleaved HSF2DBD, the protein was concentrated to 7 mg/ml prior to crystallization trials. Expression and purification of HSF1 DBD, HSF1W2 DBD, and HSF2W1 DBD were performed as described for HSF2 DBD but without removal of the 6xHis tag. HSF1W2 DBD and HSF2W1 DBD were generated by swapping amino acids 85–97 (HSF1) and 77–89 (HSF2) using Infusion cloning.
Crystallization of the HSF2 DBD
7 mg/ml HSF2 DBD (537 μM) was premixed with 322.2 μM 2-site HSE (GGTTCTAGAACC) or 214.8 μM 3-site HSE (GGTTCTAGAATATTCCG) double stranded oligonucleotide for a final ratio of 1:1.2 protein:DNA-binding site. The resulting complex was then incubated at 25 °C for 5 minutes prior to depositing into sitting drop vapor diffusion crystallization trays (Intelliplate 3, Robbins). The HSF2 DBD:2-site HSE complex crystallized at 25 °C against 100 mM NaCl, 100 mM Bicine pH 9, and 30% PEG 550 MME. Parallelogram shaped crystals grew overnight to approximately 200 μm × 50 μm × 50 μm. Crystals were cryo-protected with 100 mM NaCl, 100 mM Bicine pH 9, and 30% PEG 400. HSF2DBD:3-site HSE complex crystallized against 170 mM ammonium-acetate, 85 mM sodium acetate pH 4.6, 25.5% PEG 4000, and 15% Glycerol. Parallelogram shaped crystals grew in 3–4 days to 50 μm × 50 μm × 25 μm.
Data collection and refinement
Diffraction data were collected on a Rigaku-007HF X-ray generator outfitted with a RaxisIV++ image plate detector, with an incident beam 1.54 Å in wavelength. Data sets were processed in HKL-200056. Matthews analysis suggested one molecule of protein and ssDNA in the asymmetric unit of the structure for the 2-site structure and four molecules of protein and four molecules of ssDNA for the 3-site structure57. The structures were phased by molecular replacement in PHENIX58 using a modified version of the 3HTS structure containing only base pairs 1–4 of the HSE. Rebuilding and real-space refinements were done in Coot59 with reciprocal space refinements in PHENIX60. Structure validation was performed with the Molprobity server. Ramachandran statistics for the 2-site HSE structure were 97% favored, 3% allowed, and 0% outliers. Ramachandran statistics of the 3-site HSE structure were 94% favored, 4% allowed, and 2% outliers. The structures described here have been submitted to the RCSB protein data bank with accession codes 5D8K (2-site HSE) and 5D8L (3-site HSE).
Calculation of the Distance Difference Matrix between the HSF2 and K. lactis DBD 5D8K and 3HTS were aligned using LSQ superposition in Coot with HSE DNA residues 1–12 as an anchor. The x, y, and z coordinates of the alignment files were then copied into Excel. The peptide backbone (C, O, Cα, N) atoms were paired based on BLAST alignment files and the distance between paired atoms was calculated using the following equation: distance = sqrt((x5D8K-x3HTS)2+(y5D8K-y3HTS)2+(z5D8K-z3HTS)2). Distances were then copied into the B-factor column of the 5D8K.pdb file prior to import into PyMOL. The putty diagram was then generated using the b-factor putty preset in PyMOL.
In vitro SUMOylation assays
SUMO E1 (SAE1/SAE2), SUMO E2 (Ubc9), SUMO1, and SUMO2 were purchased from Boston Biochem. SUMO E3 ligases RANBPΔFG and PIAS1 were purchased from Enzo Life Sciences. Reactions were performed in SUMOylation Assay Buffer (SAB; 20 mM HEPES pH 7.3, 110 mM KOAc, 2 mM Mg(Oac)2, 1 mM EGTA, 2% Tween 20, 1 μg/mL Leupeptin/Pepstatin/Aprotinin, 1 mM DTT 10 mM ATP). 20 μL reactions containing 150 nM E1, 150 nM E2, 150 nM E3, 10 μM SUMO1/2, and 1 μM HSF DBD in SAB were incubated at 30 °C for 1 hour. Reactions were terminated with 1X SDS loading buffer prior to SDS-PAGE analysis followed by immunoblotting. For colloidal blue stained samples, 100 μL reactions were incubated with 20 μL NiNTA agarose beads for 1 hour at 4 °C, washed 2× with 200 μL NiNTA wash buffer, and eluted with 50 μL NiNTA elution buffer prior to SDS-PAGE. SUMO-DBD immunoblots were performed with anti-His-tag antibody (6AT18, Abgent) (Supplementary Data Set 3).
Cell culture, transfection, Chromatin Immunoprecipitation, and qRT-PCR
HSF1/HSF2 double knock out mouse embryonic fibroblasts (hsf1−/−hsf2−/− MEFS) were a kind gift from Dr. Ivor Benjamin. Cells were maintained in DMEM (Gibco) supplemented with 10% heat inactivated FBS (Gibco), 0.1 mM non-essential amino acids (Gibco), 1X MycoZap (Lonza), and 55 μM beta-mercaptoethanol. For transfection, 4 × 106 cells were transfected with 2 μg of either empty pcDNA3.1-IRES-GFP, or pcDNA3.1-IRES-GFP with human WT HSF1, HSF1W2, HSF2, or HSF2W1 containing a carboxyl terminal 3xFLAG tag using a 4D-Nucleofector (Lonza) according to manufacturer’s instructions. Transfected cells were split into 10 cm plates for ChIP, or 6 well plates for qRT-PCR or immunoblotting. For ChIP analysis, at 36 hours post transfection cells were maintained at 37 °C or heat shocked for 30 min at 42 °C prior to crosslinking with 1% formaldehyde. After quenching with 125 mM glycine, proteins were extracted with 2 mL SDS lysis buffer (20 mM HEPES pH 7.4, 5 mM MgCl2, 1 mM EDTA, 100 mM KCl, 1% Triton-X100, 0.05% SDS, 1x halt protease inhibitor cocktail (Pierce), 1x phosphatase inhibitor cocktail (Pierce)) and sonicated 3 × 30 sec bursts. Lysates were cleared with centrifugation at 14,000 g for 15 min. FLAG tagged HSF proteins were then immunoprecipitated with Magnetic M2 FLAG Affinity resin (Sigma) for 6 hours at 4 °C. Beads were then washed 2x with lysis buffer, 2x with lysis buffer + 0.5 M NaCl, 2x with TE buffer, and eluted with 200 μL TE + 1% SDS. Crosslinks were reversed overnight at 65 °C prior to treatment with Proteinase K for 1.5 hr at 37 °C. Immunoprecipitated genomic DNA was then purified using a PCR cleanup kit (Qiagen) prior to qRT-PCR analysis. For RNA extraction, 36 hours post-transfection, cells were maintained at 37 °C or heat shocked for 1 hour at 42 °C with a 1 hour recovery. RNA was extracted with RNeasy mini-prep kits (Qiagen) according to manufacturer instructions and DNase treated with Turbo DNase (Ambion). cDNA synthesis was carried out with SuperScript III first strand kits (Invitrogen) using oligo dT as primer, and then RNAse H treated for 45 min at 37 °C prior to qRT-PCR analysis. qRT-PCR was performed with iQ SYBR Green Supermix (Bio-Rad) in 384 wells plates with a CFX-384 qRT-PCR machine (Bio-Rad). For immunoblotting analysis, transfected cells were maintained at 37 °C or heat shocked for 1 hour at 42 °C with a 6-hour recovery at 37 °C, or treated with 3 μM MG132 for 7 hours. Cell lysates were harvested using SDS lysis buffer. Lysates were cleared by centrifugation at 14,000 × g for 15 minutes prior to quantification with the BCA Assay (Thermo) followed by fractionation by SDS-PAGE and transfer to 0.2 μM nitrocellulose membrane. Anti-FLAG (Sigma), Anti-Hsp72 (Enzo), Anti-GAPDH (Santa Cruz) antibodies were used at a dilution of 1:1,000 prior to incubation with anti-rabbit or anti-mouse antibody (GE Health Sciences) at a dilution of 1:5,000 (Supplementary Data Set 3).
Statistical Analysis
For statistical analysis of ChIP and qRT-PCR data, Two-Way Analysis of Variance was performed using GraphPad Prism 5. An interaction between HSF mutant and gene was observed enabling further analysis by t-test for individual mutant and gene pairs, i.e. WT HSF1 v HSF1W2 for Hsp70.
In vitro DNA Binding
Analysis of HSF DBD binding to HSEs was carried out with fluorescence polarization as previously described4. 1 nM fluorescein labeled HSE was added to 25 mM HEPES and 150 mM NaCl. HSF DBDs were titrated into the binding reaction and relative polarization measured after each addition of DBD. Binding curves were generated in GraphPad Prism 5 using One site total binding non-linear regression curve fitting.
Purification of HSF1-HSF2 hetero-complexes
Human HSF1 with an amino-terminal StrepII Tag and human HSF2 with an amino-terminal 6xHis tag were codon optimized for expression in E. coli and cloned into the pET15b expression vector using two divergent lac-operon promoters (Supplementary Fig. S6). Overnight cultures of E. coli were backdiluted 1:100 into 2 L of LB containing 100 μg/mL ampicillin, grown to an OD600 = 0.5 and induced with IPTG for 5 hours. Cell pellets were lysed in 20 mL of Strep Binding Buffer (50 mM HEPES pH 7.5, 300 mM NaCl) and cleared by centrifugation at 20,000 × g for 30 minutes at 4°C. Cleared lysates were then applied to a StrepTrap column (GE Heath Sciences) using an Akta Pure FPLC (GE Health Sciences) at a flow rate of 0.5 mL/min. The column was washed with Strep Binding Buffer until the Abs280 reading reached zero at a flow rate of 2 mL/min. The Strep Trap column was then directly attached to a HisTrap column (GE Health Sciences) and reconnected to the FPLC. Bound proteins were eluted with Strep Binding Buffer supplemented with 5 mM desthiobiotin directly onto the HisTrap column at a flow rate of 1 mL/min for 50 mL. The StrepTrap column was then removed from the assembly and the FPLC connected directly to the HisTrap column. The HisTrap was then washed with 50 mL NiNTA wash buffer, 20 mL NiNTA wash buffer supplemented with 20 mM MgCl2 and 5 mM ATP, and an additional 50 mL NiNTA wash buffer at a flow rate of 2 mL/min. Bound proteins were eluted from the HisTrap column with NiNTA elution buffer at a flow rate of 1 mL/min. The eluate fractions containing HSF1 and HSF2 were analyzed by SDS-PAGE and Colloidal Blue staining prior to pooling, concentrating to a volume of 10 mL and loading onto a Sephacryl s400 Gel Filtration column using the Akta FPLC. The HSF1-HSF2 hetero-complexes were eluted from the s400 column using 25 mM HEPES pH 7.5 and 150 mM NaCl and fractions analyzed by SDS-PAGE and colloidal blue staining. HSF1-2 crosslinking was performed with disuccinimidyl subterate (DSS, Thermo). 2 μg of HSF1-HSF2 hetero-complexes in 25 mM HEPES pH 7.5, 150 mM NaCl were incubated with increasing concentrations of DSS for 30 minutes at room temperature. Reactions were then quenched with 25 mM Tris for 15 minutes at room temperature prior to SDS-PAGE and mass spectrometry analysis.
HSF1-HSF2 Co-immunoprecipitation
hsf1−/−hsf2−/− MEFS were transfected with HSF1-HA alone, or with WT-HSF2-3xFLAG or HSF2ΔLZ1-3-3xFLAG for as described above. Cells were crosslinked with 1.5 mM DSP (Pierce) for 30 min at room temperature according to manufacturer’s protocols. Cells were lysed in SDS lysis buffer prior to sonication 1 × 30 sec to disrupt chromatin associated HSF complexes. 1.5 mg of total protein was then immunoprecipitated using M2 FLAG affinity beads (Sigma) overnight at 4°C. Beads were then washed 3× in lysis buffer prior to elution with TE buffer + 1% SDS. HSF1-HA protein was detected with anti-HA antibody (Y-11, Santa Cruz) (Supplementary Data Set 3).
Supplementary Material
Acknowledgments
We thank M. Schumacher (Duke University) and N. Tonthat (Duke University) for valuable reagents and insights on protein expression and purification, B. Wacker for experimental assistance, J. Joutsen for valuable SUMOylation insights, A. Masoudi for assistance in calculating distance difference matrices, N. Nicely and the Duke University Crystallography Core Facility, the Duke University Proteomics Core Facility and I. Benjamin (University of Wisconsin at Madison) for the hsf1−/−hsf2−/− MEFs. This work was funded by the Duke University Core Facility Voucher Program, United States National Institutes of Health grant T32 GM007105 and R01 NS065890 (DJT), a Senior Visiting Professorship from the Sigrid Jusélius Foundation, Helsinki, Finland (DJT), The Academy of Finland and The Finnish Cancer Organization (LS).
Footnotes
Accession Codes:
5D8K (HSF2 DBD bound to 2-site HSE)
5D8L (HSF2 DBD bound to 3-site HSE)
Author Contributions
AMJ conceived and performed experiments, analyzed data, and wrote the manuscript. CWP assisted in crystallographic data analysis and presentation. LS and DJT conceived experiments, analyzed data, and wrote the manuscript.
References
- 1.Jiang S, et al. Multifaceted roles of HSF1 in cancer. Tumour Biol. 2015 doi: 10.1007/s13277-015-3674-x. [DOI] [PubMed] [Google Scholar]
- 2.Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 2010;8:e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–55. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaeger AM, Makley LN, Gestwicki JE, Thiele DJ. Genomic heat shock element sequences drive cooperative human heat shock factor 1 DNA binding and selectivity. J Biol Chem. 2014;289:30459–69. doi: 10.1074/jbc.M114.591578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendillo ML, et al. HSF1 Drives a Transcriptional Program Distinct from Heat Shock to Support Highly Malignant Human Cancers. Cell. 2012;150:549–62. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitesell L, Lindquist S. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin Ther Targets. 2009;13:469–78. doi: 10.1517/14728220902832697. [DOI] [PubMed] [Google Scholar]
- 7.Neef DW, Jaeger AM, Thiele DJ. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat Rev Drug Discov. 2011;10:930–44. doi: 10.1038/nrd3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Littlefield O, Nelson HC. A new use for the ‘wing’ of the ‘winged’ helix-turn-helix motif in the HSF-DNA cocrystal. Nat Struct Biol. 1999;6:464–70. doi: 10.1038/8269. [DOI] [PubMed] [Google Scholar]
- 9.Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science. 1993;259:230–4. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- 10.Sorger PK, Nelson HC. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell. 1989;59:807–13. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- 11.Farkas T, Kutskova YA, Zimarino V. Intramolecular repression of mouse heat shock factor 1. Mol Cell Biol. 1998;18:906–18. doi: 10.1128/mcb.18.2.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuo J, Baler R, Dahl G, Voellmy R. Activation of the DNA-binding ability of human heat shock transcription factor 1 may involve the transition from an intramolecular to an intermolecular triple-stranded coiled-coil structure. Mol Cell Biol. 1994;14:7557–68. doi: 10.1128/mcb.14.11.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vihervaara A, et al. Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc Natl Acad Sci U S A. 2013;110:E3388–97. doi: 10.1073/pnas.1305275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guertin MJ, Martins AL, Siepel A, Lis JT. Accurate prediction of inducible transcription factor binding intensities in vivo. PLoS Genet. 2012;8:e1002610. doi: 10.1371/journal.pgen.1002610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neef DW, et al. A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell Rep. 2014;9:955–66. doi: 10.1016/j.celrep.2014.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn JS, Neef DW, Thiele DJ. A stress regulatory network for coordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol Microbiol. 2006;60:240–51. doi: 10.1111/j.1365-2958.2006.05097.x. [DOI] [PubMed] [Google Scholar]
- 17.Scherz-Shouval R, et al. The reprogramming of tumor stroma by HSF1 is a potent enabler of malignancy. Cell. 2014;158:564–78. doi: 10.1016/j.cell.2014.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riva L, et al. Poly-glutamine expanded huntingtin dramatically alters the genome wide binding of HSF1. J Huntingtons Dis. 2012;1:33–45. [PMC free article] [PubMed] [Google Scholar]
- 19.Shinkawa T, et al. Heat shock factor 2 is required for maintaining proteostasis against febrile-range thermal stress and polyglutamine aggregation. Mol Biol Cell. 2011;22:3571–83. doi: 10.1091/mbc.E11-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elsing AN, et al. Expression of HSF2 decreases in mitosis to enable stress-inducible transcription and cell survival. J Cell Biol. 2014;206:735–49. doi: 10.1083/jcb.201402002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahlskog JK, et al. Anaphase-promoting complex/cyclosome participates in the acute response to protein-damaging stress. Mol Cell Biol. 2010;30:5608–20. doi: 10.1128/MCB.01506-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Fatimy R, et al. Heat shock factor 2 is a stress-responsive mediator of neuronal migration defects in models of fetal alcohol syndrome. EMBO Mol Med. 2014;6:1043–61. doi: 10.15252/emmm.201303311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mou L, et al. A dominant-negative mutation of HSF2 associated with idiopathic azoospermia. Hum Genet. 2013;132:159–65. doi: 10.1007/s00439-012-1234-7. [DOI] [PubMed] [Google Scholar]
- 24.Akerfelt M, et al. Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2. Proc Natl Acad Sci U S A. 2008;105:11224–9. doi: 10.1073/pnas.0800620105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilkerson DC, Murphy LA, Sarge KD. Interaction of HSF1 and HSF2 with the Hspa1b promoter in mouse epididymal spermatozoa. Biol Reprod. 2008;79:283–8. doi: 10.1095/biolreprod.107.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossi A, et al. The proteasome inhibitor bortezomib is a potent inducer of zinc finger AN1-type domain 2a gene expression: role of heat shock factor 1 (HSF1)-heat shock factor 2 (HSF2) heterocomplexes. J Biol Chem. 2014;289:12705–15. doi: 10.1074/jbc.M113.513242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandqvist A, et al. Heterotrimerization of heat-shock factors 1 and 2 provides a transcriptional switch in response to distinct stimuli. Mol Biol Cell. 2009;20:1340–7. doi: 10.1091/mbc.E08-08-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, et al. Structural basis of DNA recognition by PCG2 reveals a novel DNA binding mode for winged helix-turn-helix domains. Nucleic Acids Res. 2015;43:1231–40. doi: 10.1093/nar/gku1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 2010;18:177–87. doi: 10.1016/j.str.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Tammsalu T, et al. Proteome-wide identification of SUMO2 modification sites. Sci Signal. 2014;7:rs2. doi: 10.1126/scisignal.2005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendriks IA, et al. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol. 2014;21:927–36. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker J, et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat Struct Mol Biol. 2013;20:525–31. doi: 10.1038/nsmb.2526. [DOI] [PubMed] [Google Scholar]
- 33.Fujimoto M, et al. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol Cell. 2012;48:182–94. doi: 10.1016/j.molcel.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 34.Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 35.Westwood JT, Wu C. Activation of Drosophila heat shock factor: conformational change associated with a monomer-to-trimer transition. Mol Cell Biol. 1993;13:3481–6. doi: 10.1128/mcb.13.6.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu XD, Liu PC, Santoro N, Thiele DJ. Conservation of a stress response: human heat shock transcription factors functionally substitute for yeast HSF. EMBO J. 1997;16:6466–77. doi: 10.1093/emboj/16.21.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sistonen L, Sarge KD, Morimoto RI. Human heat shock factors 1 and 2 are differentially activated and can synergistically induce hsp70 gene transcription. Mol Cell Biol. 1994;14:2087–99. doi: 10.1128/mcb.14.3.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujimoto M, Nakai A. The heat shock factor family and adaptation to proteotoxic stress. FEBS J. 2010;277:4112–25. doi: 10.1111/j.1742-4658.2010.07827.x. [DOI] [PubMed] [Google Scholar]
- 39.Calderwood SK, Murshid A, Prince T. The shock of aging: molecular chaperones and the heat shock response in longevity and aging--a mini-review. Gerontology. 2009;55:550–8. doi: 10.1159/000225957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–8. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 41.Tan K, et al. Mitochondrial SSBP1 protects cells from proteotoxic stresses by potentiating stress-induced HSF1 transcriptional activity. Nat Commun. 2015;6:6580. doi: 10.1038/ncomms7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zelin E, Freeman BC. Lysine deacetylases regulate the heat shock response including the age-associated impairment of HSF1. J Mol Biol. 2015;427:1644–54. doi: 10.1016/j.jmb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalmar B, Greensmith L. Activation of the heat shock response in a primary cellular model of motoneuron neurodegeneration-evidence for neuroprotective and neurotoxic effects. Cell Mol Biol Lett. 2009;14:319–35. doi: 10.2478/s11658-009-0002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–33. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–56. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 46.Anckar J, et al. Inhibition of DNA binding by differential sumoylation of heat shock factors. Mol Cell Biol. 2006;26:955–64. doi: 10.1128/MCB.26.3.955-964.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tateishi Y, et al. Molecular basis for SUMOylation-dependent regulation of DNA binding activity of heat shock factor 2. J Biol Chem. 2009;284:2435–47. doi: 10.1074/jbc.M806392200. [DOI] [PubMed] [Google Scholar]
- 48.Niskanen EA, et al. Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol. 2015;16:153. doi: 10.1186/s13059-015-0717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seifert A, Schofield P, Barton GJ, Hay RT. Proteotoxic stress reprograms the chromatin landscape of SUMO modification. Sci Signal. 2015;8:rs7. doi: 10.1126/scisignal.aaa2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shu W, Liu J, Ji H, Lu M. Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 A resolution. J Mol Biol. 2000;299:1101–12. doi: 10.1006/jmbi.2000.3776. [DOI] [PubMed] [Google Scholar]
- 51.Sorrells TR, Booth LN, Tuch BB, Johnson AD. Intersecting transcription networks constrain gene regulatory evolution. Nature. 2015;523:361–5. doi: 10.1038/nature14613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 53.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–45. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baker CR, Hanson-Smith V, Johnson AD. Following gene duplication, paralog interference constrains transcriptional circuit evolution. Science. 2013;342:104–8. doi: 10.1126/science.1240810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bjork JK, et al. Heat-shock factor 2 is a suppressor of prostate cancer invasion. Oncogene. 2015 doi: 10.1038/onc.2015.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 57.Matthews BW. Solvent Content of Protein Crystals. Journal of Molecular Biology. 1968;33:491. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 58.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallographica Section D-Biological Crystallography. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D-Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.