Abstract
Bean production is affected by a wide diversity of fungal pathogens, among them Rhizoctonia solani is one of the most important. A strategy to control bean infectious diseases, mainly those caused by fungi, is based on the use of biocontrol agents (BCAs) that can reduce the negative effects of plant pathogens and also can promote positive responses in the plant. Trichoderma is a fungal genus that is able to induce the expression of genes involved in plant defense response and also to promote plant growth, root development and nutrient uptake. In this article, a strategy that combines in silico analysis and real time PCR to detect additional bean defense-related genes, regulated by the presence of Trichoderma velutinum and/or R. solani has been applied. Based in this strategy, from the 48 bean genes initially analyzed, 14 were selected, and only WRKY33, CH5b and hGS showed an up-regulatory response in the presence of T. velutinum. The other genes were or not affected (OSM34) or down-regulated by the presence of this fungus. R. solani infection resulted in a down-regulation of most of the genes analyzed, except PR1, OSM34 and CNGC2 that were not affected, and the presence of both, T. velutinum and R. solani, up-regulates hGS and down-regulates all the other genes analyzed, except CH5b which was not significantly affected. As conclusion, the strategy described in the present work has been shown to be effective to detect genes involved in plant defense, which respond to the presence of a BCA or to a pathogen and also to the presence of both. The selected genes show significant homology with previously described plant defense genes and they are expressed in bean leaves of plants treated with T. velutinum and/or infected with R. solani.
Keywords: biotic stress, systemic acquired resistance, induced systemic resistance, hypersensitive response, defense genes, biocontrol agent, Phaseolus vulgaris
Introduction
The common bean (Phaseolus vulgaris L.) is the most important food legume crop worldwide. Bean production is often affected by biotic and abiotic factors (Guerrero-González et al., 2011) by microorganisms, humidity, temperature… that are detected as signals for the activation of plant response mechanisms. This crop is affected by a wide diversity of fungal pathogens (Sclerotinia spp., Fusarium spp., Phytium spp., Botrytis spp.,...) among them Rhizoctonia solani JG Kühn [Teleomorph: Thanatephorus cucumeris (AB Frank) Donk] has a remarkable importance as responsible of important economic losses in this crop (Valenciano et al., 2006). R. solani is a necrotrophic pathogen responsible for the root and hypocotyl diseases. Plant infection occurs through wounds or by the direct action of the fungal mycelium, which tears the cuticle and penetrates the epidermis (Guerrero-González et al., 2011).
As a strategy to control bean infectious diseases, mainly those caused by fungi, the use of biocontrol agents (BCA) can reduce the negative effects of plant pathogens and they also can promote positive responses in the plant (Shoresh et al., 2010). The genera Trichoderma, Gliocladium, Rhizobium, Pseudomonas, are beneficial organisms that have shown good efficiency as BCAs against pathogenic microorganisms. Trichoderma (Teleomorph: Hypocrea) is a fungal genus that is found in the soil, and it is a secondary fast growing opportunistic invasive. In addition, Trichoderma biocontrol strains are able to induce the expression of genes involved in defense response and also to promote plant growth, root development, and nutrient uptake (Hermosa et al., 2012).
The relationships established between plant and micro-organisms are very diverse. When a plant is exposed to a pathogenic microorganism, the production of molecules associated to salicylic acid is increased, being this a systemic acquired resistance (SAR) response. The response of plants against non-pathogenic microorganisms is different, resulting in activation of signaling cascades that are dependent on jasmonic acid and ethylene, such as hydroperoxide lyase, peroxidase, and phenylalanine ammonia lyase, all of which belong to an induced systemic resistance (ISR) response (Druzhinina et al., 2011). Other responses result in a rapid cell death in infected tissues, then plants activate the hypersensitive response that involves the accumulation of salicylic acid, reactive oxygen species and an increased the influx of Ca2+ (Guerrero-González et al., 2011).
In the tripartite interaction of bean plants with the pathogen R. solani and a biocontrol Trichoderma species, several changes are produced in the plant, such as the increase in phenolic acid and lignin, accumulation of phytoalexins (Guerrero-González et al., 2011), and down- or up-regulation of defense-related genes expression (Mayo et al., 2015). Different categories of defense-related genes whose expression is modulated by biotic stresses have been described in bean plant interacting with pathogen and non-pathogenic microorganisms (Mayo et al., 2015).
Our hypothesis is that the combination of real time PCR with “in silico” analysis is a valid strategy to identify bean defense-related genes regulated by BCAs and/or plant pathogens. The aim is develop a systematic strategy to detect bean defense-related genes regulated by the presence of Trichoderma velutinum and/or R. solani. Finally, the procedure has been validated by the analysis of expression of the selected genes in the presence or absence of these two fungi.
Materials and Methods
Trichoderma and Rhizoctonia solani Isolates and Culture Collections
Trichoderma velutinum T028, was collected from the bean traditional production area (Protected Geographical Indication, PGI), called “Alubia La Bañeza - León” (EC Reg. n.256/2010 published on March 26th, 2010, OJEU L880/17), from a High Quality variety of beans (Figure 1) without any genetic manipulation. It was isolated from soil plot bean in the Astorga region (León, Spain). This isolate gave percentages of inhibition greater that 60% in membrane assays and 40% in direct confrontation assays with R. solani, and that was able to sporulate on potato-dextrose-agar (PDA) medium.
FIGURE 1.

Bean seeds of “Canela” variety of the Protected Geographical Indication “Alubia La Bañeza – León” (Spain).
Rhizoctonia solani R43 was isolated from bean plants of the same PGI and selected based on its high virulence. The isolated strains were stored in the collection “Pathogens and Antagonists of the Laboratory Diagnosis of Pests and Diseases” (PALDPD, University of León, León, Spain).
Isolates were inoculated on PDA (Becton Dickinson, Germany) medium and grown at 25°C in the dark for 1 week. After this incubation time T. velutinum T028 was exposed to light in order to induce the spore’s formation.
Plant Materials and Growth Conditions
Sixty bean seeds (Canela landrace, PGI “Alubia de la Bañeza –León,” Figure 1) per treatment were germinated and cultured in presence or absence of the fungi in four conditions according to the procedure previously described by Mayo et al. (2015): (i) T. velutinum (T028) isolate plus R. solani (R43) (RT028); (ii) T. velutinum isolate (T028) without pathogen (C = control) (CT028); (iii) control (without T. velutinum) with R. solani (RC) and (iv) control without fungi (CC). The culture was carried out in climatic chamber and growth conditions were performed as previously described (Mayo et al., 2015). Six bean leaves from 45 day-old plants of each treatment were randomly collected and stored at -80°C until use.
RNA Extraction and Purification
The procedures for RNA extraction were performed as described previously (Reid et al., 2006). Bean leaves were lyophilized and were ground to a fine powder in liquid nitrogen using a mortar and pestle. The powder was mixed with 20 ml of extraction buffer/g of sample (extraction buffer: 0.1% SDS, 100 mM LiCl, 10 mM EDTA, 100 mM Tris-HCl, pH9) pre-warmed at 65°C, and 20 ml/g of phenol-chloroform-isoamyl alcohol 25:24:1 (Sigma–Aldrich, St. Louis, MO, USA). Then, the mixtures, in eppendorf tubes, were centrifuged at 13,000 rpm for 10 min at 4°C. The aqueous layer was transferred to a new tube. This step was repeated twice. Nucleic acids were precipitated with 1 volume of LiCl 4 M, mixed and kept overnight at 4°C. Tubes were then centrifuged at 13,000 rpm for 30 min at 4°C, and the resulting pellets were washed with ice cold ethanol 70%-DEPC, centrifuged again at 13,000 rpm for 10 min at 4°C and air dried. Finally, the pellets were dissolved in 50–200 μl H2O-DEPC and stored at -20°C until use.
RNA concentrations and its purity were estimated from the A260/280 absorbance ratio with a NanoDrop (Thermo Scientific, Wilmington, DE, USA), considering the ideal absorbance ratio (1.8 ≤ A260/280 ≤ 2.0) and 1% agarose gel was run to visualize the integrity of the RNA.
cDNA Synthesis
Approximately 5 μg of RNA were treated with DNase using the TURBO DNAfreeTM Kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer instructions. cDNA was synthesized using High-Capacity cDNA Reverse Transcription kit (Applied-Biosystems, Foster City, CA, USA) according to the manufacture’s manual.
qPCR Conditions and Analysis
qPCR reactions were performed with 7300 System (Applied Biosystems, Foster City, CA, USA) using SYBR® Green. Each reaction was performed in 20 μl containing 10 μl of 2 X Power SYBR Green PCR Master Mix (Life Technologies), 0.2–0.3 μM primers and cDNA samples diluted 1:20. Each qPCR reaction was performed in triplicate. Reactions were run using the cycling parameter described previously (Reid et al., 2006) and the qPCR data were analyzed by the 2-ΔΔCt method (Pfaffl, 2001). In order to analyze the qPCR data, Act11 gene was used as housekeeping to determine the relative expression level of the other genes analyzed in this work (Borges et al., 2012). T. velutinum T028 strain was selected as reference strain in this study based on its positive effects on bean phenotype with and without R. solani infection (data no published). For the determination of qPCR efficiency of each primer pairs, a standard curve was performed using the following cDNA dilutions: 1:4, 1:16, 1:64; 1:256 and 1:1024. Every measurement was made in triplicate. The corresponding qPCR efficiencies (E) were calculated for every primer pair with the software 7300 System SDS software (Applied Biosystems, Foster City, CA, USA) according to the equation E = (10-1/slope - 1) × 100 (Rutledge and Stewart, 2008).
The significance of the differences between the gene expressions levels were compared by the Student’s t-test using SAS (SAS Institute Inc., 2004, Cary, NC, USA).
Results
Selection of Putative Bean Defense-Related Genes
Following an exhaustive and systematic analysis, summarized in the Figure 2, several bean genes were selected for their expression analysis in leaves from bean plants grown in interaction with T. velutinum and infected or not with Rhizoctonia solani. Thus, as result of the search in the literature, 48 genes were firstly found, showing stress and/or defense response (Table 1). Only those genes that resulted to be expressed in P. vulgaris leaves, based on transcriptomic data reported in the Phytozome database1, were considered for qPCR expression analysis in leaves. The genes for which we confirmed expression in leaves were considered for further analyses.
FIGURE 2.
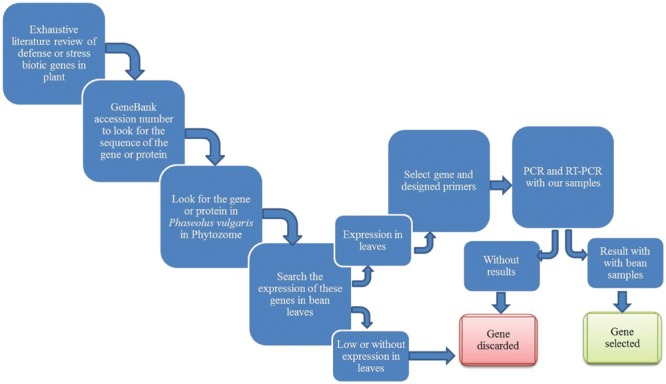
Schematic representation of the work flow set up in the present work to select bean genes involved in plant defense.
Table 1.
Genes selected for stress and/or defense response and their empirical expression in Phaseolus vulgaris leaves.
| Id | Gene | Accession number | Functional annotation | NCBI Phytozome |
|---|---|---|---|---|
| Pereira et al., 2014 | ||||
| 1 | Chit | AY357300.2 | Chitanase | |
| 2 | Glu1 | DQ093563.1 | β-1,3-glucanase | |
| 3 | Pod3 | AF485265.1 | Peroxidase | |
| 4 | PR3 | TC18606 | Chitinase class I | Phvul.009G116600 |
| 5 | Lox1 | U76687.2 | Lipoxygenase | |
| Upchurch and Ramirez, 2010 | ||||
| 6 | PPO | EF158428 | Polyphenol oxidase | |
| 7 | PR10 | AJ289155 | Stress-induced ribonuclease-like protein | |
| 8 | PR12 | BU964598 | Defensin precursor | |
| 9 | MMP2 | AY057902 | Matrix metalloproteinase 2 | |
| 10 | CHS | X53958 | Chalcone synthase | |
| 11 | AOS | DQ288260 | Allene oxide synthase | |
| 12 | HPL | AW733791 | Hydroperoxide lyase | Phvul.005G116800 |
| 13 | LOX2 | D13949 | Lipoxygenase 2 | Phvul.005G156700 |
| 14 | LOX7 | U36191 | Lipoxygenase 2 | Phvul.005G156900 |
| 15 | IPER | AF007211 | Basic peroxidase | Phvul.009G215000 |
| Borges et al., 2012 | ||||
| 16 | PR16a | CB540239 | Germin-like protein 8 | Phvul.010G129900 |
| 17 | PGIa | CB542106 | Polygalacturonase-inhibitor-like protein | |
| 18 | MAPKK | CB543156 | MEK map kinase kinase | |
| 19 | PROF | CB543496 | Profilin | |
| 20 | CNGC2 | CB542582 | Cyclin nucleotide-gated ion channel 2 | Phvul.008G036200 |
| Guerrero-González et al., 2011 | ||||
| 21 | PR1 | HO864272 | Pathogenesis related protein 1 | Phvul.003G109100 |
| 22 | PR2 | HO864270 | Pathogenesis related protein 2 | Phvul.003G109200 |
| 23 | PR4 | HO864354 | Pathogenesis related protein 4 | Phvul.006G102300 |
| 24 | PR10 | HO864271 | Pathogenesis related protein 10) | |
| 25 | LTP2 | HO864366 | Lipid-transfer protein 2 | |
| 26 | SIP | HO864290 | Syringolide-induced protein B13-1-9 | |
| 27 | DAAP | HO864358 | Defense associated acid phosphatase | |
| 28 | CHI | HO864289 | Chalcone isomerase | |
| 29 | hGS | HO864377 | Homoglutathione synthetase | Phvul.006G094500 |
| 30 | aDO1 | HO864351 | Alpha- dioxygenase 1 | |
| 31 | CPRD14 | HO864341 | CPRD14 protein | |
| 32 | OPR5 | HO864304 | 12-oxophytodienoic acid 10, 11-reductase | |
| 33 | GST22 | HO864275 | Glutathione S-transferase 22 | |
| 34 | CPRD8 | HO864396 | CPRD8 protein | |
| 35 | UDPGT | HO864301 | UDP-glucosyl transferase 72E1 | |
| 36 | ERD15 | HO864375 | ERD15 protein | |
| 37 | GTSa | HO864392 | 2,4-D inducible glutathione S-transferase | Phvul.002G241400 |
| 38 | GST15 | HO864369 | Glutathione S-transferase 15 | |
| Gallou et al., 2009 | ||||
| 39 | GST1 | J03679 | Gluthatione-S-transferase 1 | |
| Lehtonen et al., 2008 | ||||
| 40 | TSI-1 | BQ121547 | TSI-1 protein | |
| 41 | Lip | BQ112158 | Lipase-like protein | |
| 42 | Amintransf2 | BQ517030 | Aminotransferase 2 | Phvul.006G029100 |
| Bakshi and Oelmüller, 2014 | ||||
| 43 | WRKY33 | NM129404.3 | WRKY transcription factors | Phvul.008G090300 |
| Vellicce et al., 2006 | ||||
| 44 | CH5b | FE897014.1 | Endochitinase precursor | Phvul.009G116500 |
| Lorenzo et al., 2003 | ||||
| 45 | ERF1 | AF076277 | Ethylene-Responsive Transcription Factor 1 | Phvul.007G127800 |
| Moffat et al., 2012 | ||||
| 46 | ERF5 | At5g47230 | Ethylene-Responsive Transcription Factor 5 | Phvul.002G055700 |
| Kim and Hwang, 2014 | ||||
| 47 | PAL1 | KF279696 | Phenylalanine and histidine ammonia-lyase | Phvul.001G177800 |
| Sharma et al., 2013 | ||||
| 48 | OSM34 | At4g11650 | Osmotin-like protein | Phvul.002G155500 |
As result, from the 48 genes selected for their involvement in bean stress and/or defense responses, only 19 were selected which showed a detectable level of expression in bean leaves.
The selected genes can be included in nine different groups (Table 2): (i) involved in the regulation of the balance between necrotrophic and biotrophic pathogen responses: WRKY33 (WRKY transcription factor) (NM129404.3) (Bakshi and Oelmüller, 2014); (ii) pathogenesis related genes: PR1 (pathogenesis related 1) (HO864272) (Guerrero-González et al., 2011), PR2 (β 1-3 endoglucanase) (HO864270) (Guerrero-González et al., 2011), PR3 (chitinase class I) (TC18606) (Pereira et al., 2014), PR4 (pathogenesis related 4) (HO864354) (Guerrero-González et al., 2011), PR16a (germin.like protein 8) (CB540239) (Borges et al., 2012), IPER (basic peroxidase) (AF007211) (Upchurch and Ramirez, 2010), PPO (polyphenol oxidase) (EF158428) (Upchurch and Ramirez, 2010); (iii) related with the ethylene signaling pathway: ERF1 (ethylene-responsive transcription factor 1) (AF076277) (Lorenzo et al., 2003), ERF5 (ethylene-responsive transcription factor 5) (At5g47230) (Moffat et al., 2012), and CH5b (endochitinase precursor) (FE897014.1) (Vellicce et al., 2006); (iv) involved in phytoalexin biosynthesis: PAL1 (phenylalanine and histidine ammonia-lyase) (KF279696) (Kim and Hwang, 2014); (v) related in osmotin biosynthesis: OSM34 (osmitin-like protein) (At4g11650) (Sharma et al., 2013); (vi) involved in Ca2+ signaling: CNGC2 (cyclic nucleotide-gated ion channel 2) (CB542582) (Borges et al., 2012); (vii) needed for antimicrobials and oxylipins (defense signaling molecules): HPL (hydroperoxide lyase) (AW733791) (Upchurch and Ramirez, 2010), Lox2 (lipoxygenase 2) (D13949) (Upchurch and Ramirez, 2010), Lox7 (lipoxygenase 2) (Upchurch and Ramirez, 2010); (viii) GSTa (2,4-D inducible glutathione S-transferase) (HO864392) (Guerrero-González et al., 2011); and (ix) hGS (homoglutathione synthetase) (HO864377) both related with oxidative stress (Guerrero-González et al., 2011).
Table 2.
Common bean sequences used for primer design for RT-PCR analysis.
| Gene | Functional annotation | NCBI Phytozome | Forward/Reverse | Efficiency Reference |
|---|---|---|---|---|
| Reference genes | ||||
| Act11 | Actin-11 | Phvul.008G011000 | TGCATACGTTGGTGATGAGG | 1.084 |
| AGCCTTGGGGTTAAGAGGAG | ||||
| Ukn1 | Unknown | Phvul.011G023200 | ATTCCCATCATGCAGCAAAG | 0.937 |
| AGATCCCTCCAGGTCAATCC | ||||
| Balance between necrotrophic and biotrophic pathogen responses | ||||
| WRKY33 | WRKY transcription factors | Phvul.008G090300 | TTTCACAGGACAGGTTCCAGC | 0.938 |
| CCTTTGACAGAAATGACTGAAGGA | ||||
| Pathogenesis related genes | ||||
| PR1 | Pathogenesis Related 1 | Phvul.003G109100 | TGGTCCTAACGGAGGATCAC | 1.094 |
| TGGCTTTTCCAGCTTTGAGT | Mayo et al., 2015 | |||
| PR2 | Beta 1-3 Endoglucanase | Phvul.003G109200 | GTGAAGGACGCCGATAACAT | 1.048 |
| ACTGAGTTTGGGGTCGATTG | Mayo et al., 2015 | |||
| PR3 | Chitinase class I | Phvul.009G116600 | TGGAGTTGGTTATGGCAACAA | 1.034 |
| ATTCTGATGGGATGGCAGTGT | ||||
| PR4 | Pathogenesis-related 4 | Phvul.006G102300 | CGCAGTGAGTGCATATTGCT | 0.922 |
| TGTTTGTCACCCTCAAGCAC | Mayo et al., 2015 | |||
| PR16a | Germin-like protein 8 | Phvul.010G129900 | GGCAGTCTCATGGTTATGGTTT | – |
| GCATGCTCAAGTCTCAACACAT | ||||
| IPER | Peroxidase precursor | Phvul.009G215000 | GGCAAGCATTATATGGTTGAAA | – |
| GATGGCAACATCCATCACTTTA | ||||
| PPO | Polyphenol oxidase | Phvul.008G073200 | GAAGACGATGATTTGCTGGTTA | – |
| AAGAAACATTTTCCTTTGTGAAA | ||||
| Ethylene signaling pathway | ||||
| ERF1 | Ethylene-Responsive Transcription Factor 1 | Phvul.007G127800 | CGCTCTCAAGAGGAAACACTCC | 0.937 |
| TGAATCAGAAGGAGGAGGGAAT | ||||
| ERF5 | Ethylene-Responsive Transcription Factor 5 | Phvul.002G055700 | GGCTCCAAGTGGATTGAGAAC | 0.932 |
| TCAGAATCAGATAACTACAAAGCACAA | ||||
| CH5b | Endochitinase precursor | Phvul.009G116500 | CAGCCAAAGGCTTCTACACC | 0.883 |
| TTGTTTCGTGAGACGTTTGC | Mayo et al., 2015 | |||
| Phytoalexins biosynthesis | ||||
| PAL1 | Phenylalanine and histidine ammonia-lyase | Phvul.001G177800 | TGAGAGAGGAGTTGGGCACT | 1.034 |
| TTCCACTCTCCAAGGCATTC | ||||
| Osmotin biosynthesis | ||||
| OSM34 | Osmotin-like protein | Phvul.002G155500 | GAACGGAGGGTGTCACAAAATC | 0.927 |
| CGTAGTGGGTCCACAAGTTCCT | ||||
| Involved in Ca2+ signaling | ||||
| CNGC2 | Cyclic nucleotide-gated ion channel 2 | Phvul.008G036200 | ATTCAATTTGCTTGGAGACGTT | 0.98 |
| ACAGTTTTATTGAAGGCCAGGA | ||||
| Antimicrobials and oxylipins (defense signaling molecules) | ||||
| HPL | Hydroperoxide lyase | Phvul.005G116800 | TCAAGGCTACATTTGTATTTCCA | 0.984 |
| TGGTGCACATTTCTTAGTAGCAA | ||||
| Lox2 | Lipoxygenase 2 | Phvul.005G156700 | ATGCAAGGCTAAAGAGATCCAA | – |
| ATGGTGACAGGAGCTAAACACA | ||||
| Lox7 | Lipoxygenase 2 | Phvul.005G156900 | GAAGGCTTGACTTTCAGAGGAA | – |
| AACACACGAGAAGATTCAACCA | ||||
| Oxidative stress | ||||
| GSTa | 2.4-D inducible glutathione S-transferase | Phvul.002G241400 | AGGGAGTCACACTGGCTATGTT | 1.013 |
| ATGTGCCATTTGCATTTTAGTG | ||||
| hGS | Homoglutathione synthetase | Phvul.006G094500 | GTGGCTATATGGTGCGTACAAA | 1.023 |
| GAAACAAGAATGCATCTCCTCA | ||||
| Amintransf2 | Aminotransferase 2 | Phvul.006G029100 | TTCTTCCTTTTCTGCTCTTTCAA | – |
| AGATGACAAGATGCAATGATTTTT | ||||
(–) Genes that empirically showing expression but showing negative qPCR results.
However, only 14 genes were selected to the study of the expression genes because PR16a, IPER, PPO, Lox2, and Lox7, showing negative qPCR results, were finally discarded.
Selection of a Trichoderma Strain to Validate the Gene Selection Strategy
Trichoderma velutinum T028 was the selected isolate, based on its positive effect on bean growth. Thus, plants inoculated with this strain showed a significant increase in dry weight of both aerial parts and root system, including when R. solani was present in the substrate (Figure 3). Thus, when bean plants were treated with T. velutinum T028 they increased respect to control plants (CC) 4.75% their diameter of hypocotyl, 10.75% their length of root system, 4.27 and 5.51% in dry weight of aerial parts and root system, respectively. When plants were infected with R. solani, the action of T. velutinum T028 caused an increased respect to the control plant with the pathogen (RC) of the diameter of hypocotyl in 8.76, 21.15% in the length of root system, and 11.05 and 3.43% in dry weight of aerial parts and root system respectively.
FIGURE 3.
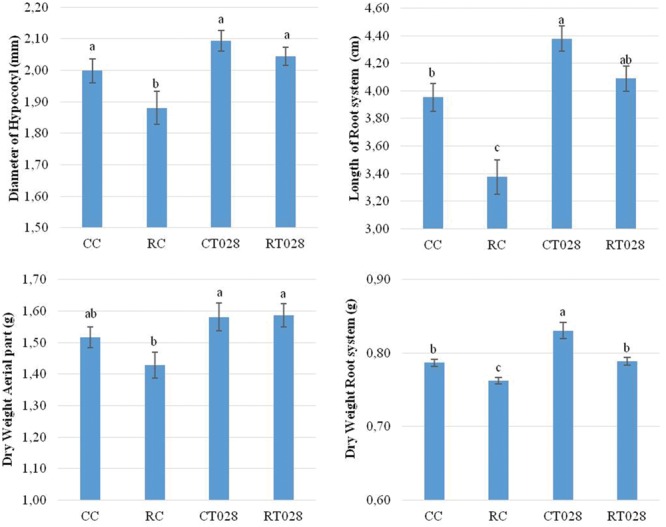
Evaluation of the diameter of the hypocotyl (above left), length of root system (above right) dry weight of the aerial part (below left) and root system (below right) of bean plants grown during 45 days after sowing. [Trichoderma velutinum T028 without pathogen (CT028), T. velutinum T028 with Rhizoctonia solani (RT028), R. solani control (RC) and control without fungus (CC)]. Differences statistically significant respect to control plants (p < 0.05) are indicated with different letters.
Based on these results, this isolate was used for further studies. In addition, this is the first report in which the effects of this strain on bean phenotype and plant gene regulation are studied.
Effect of R. solani Infection on Expression of the Selected Genes. Validation of the Procedure Used to Select Bean Genes Involved in Defense Responses (Strategy Validation I)
A significant down-regulation of expression of PR2, PR3, PR4, ERF1, ERF5, PAL1, HPL, and GTSa genes with ratios of expression ranging from 0.149 fold for PAL1 and 0.763 fold for PR3 was observed in bean plants grown in the presence of R. solani (RC) compared to control plants (CC). Conversely, expression of PR1, OSM34, CNGC2, and hGS genes was up-regulated, but with non-statistically significant differences with a ratios between 1.289 and 1.193 for PR1 and hGS, respectively (Figure 4).
FIGURE 4.
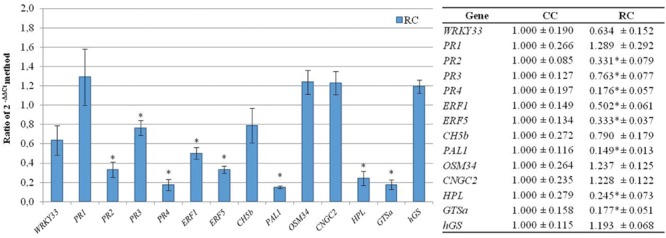
Analysis of relative expression levels of the bean defense genes selected in the present work in bean plants infected with R. solani versus their levels of expression in control plants. The data were analyzed by the 2-ΔΔCt method. The differences statistically significant respect to control plants (p < 0.05) are indicated with an asterisk.
Effect of Trichoderma on Expression of the Selected Genes (Strategy Validation II)
Trichoderma treatment also down-regulates expression of most of the bean defense-related genes, but at a lower level than R. solani. Thus, when T. velutinum T028 was in the substrate (CT028), PR2, PR3, PR4, ERF1, ERF5, PAL1, CNGC2, HPL, and GSTa were significantly down-regulated with expression ratios ranging from 0.168 for PR4 to 0.754 for ERF1. However, WRKY 33, CH5b, and hGS were up-regulated when compared with the levels of expression in control plants, with relative expression levels between 2.462 for CH5b and 1.576 for hGS (Figure 5). OSM34 was slightly but not significantly up-regulated.
FIGURE 5.
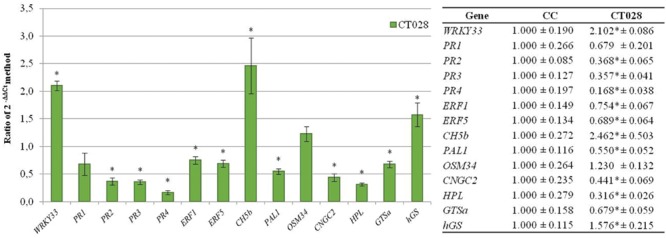
Analysis of relative expression levels of the bean defense genes selected in the present work in bean plants treated with T. velutinum versus their levels of expression in control plants. The data were analyzed as indicated in the legend to the Figure 4.
Effect of Interaction of T. velutinum and R. solani on Expression of the Selected Genes (Strategy Validation III)
When T. velutinum T028 and R. solani (RT028) were in the substrate, the genes WRKY33, PR2, PR3, PR4, ERF1, ERF5, PAL1, OSM34, HPL and GSTa were significantly down-regulated with values between 0.179 for PAL1 and 0.631 for WRKY33. In the case of PR1 and CNGC2, they were also down-regulated but not significantly respect to control plant (C). Conversely, hGS was up-regulated with a significant ratio of 1.589 respect to control plants, while CH5b was not significantly up-regulated with a ratio of 1.613 (Figure 6).
FIGURE 6.
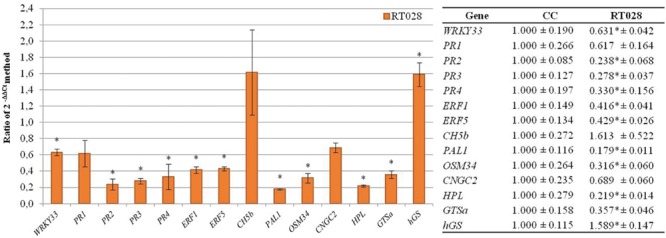
Analysis of relative expression levels of the bean defense genes selected in the present work in bean plants infected with R. solani and treated with T. velutinum versus their levels of expression in control plants. The data were analyzed as indicated in the legend to the Figure 4.
Discussion
Plants have developed some defensive strategies to perceive pathogen attack and to translate this perception into an appropriate adaptive response. During attack, plants are able to enhance their resistance (induced, acquired, hypersensitive) (Lodha and Basak, 2012). Contact with pathogenic and non-pathogenic microorganisms triggers two mechanisms: (i) SAR that is usually triggered by local infections, it provides long-term systemic resistance to pathogen attack and requires the involvement of the signal molecule salicylic acid (Durrant and Dong, 2004), and (ii) ISR that is known to result from colonization of roots by certain non-pathogenic microorganisms and is dependent on components of the jasmonic acid and ethylene signaling pathways (Shoresh et al., 2010). Then, the combination of both types of induced resistance response can protect the plant against pathogens and can even result in additive level of induced protection against pathogens through both the jasmonic acid/ethylene and salicylic acid pathways (Verhagen et al., 2006).
In the present work we developed a strategy to select genes involved in bean defense response, which would belong to those pathways, but also genes that can contribute to plant defense by other mechanisms. In this sense several previous works have described genes involved in bean defense response (Guerrero-González et al., 2011; Mayo et al., 2015). However, in the present work, by a systematic approach, 48 genes were initially considered, and 14 finally selected, which match with the criteria set up in this work: (i) they showed significant homology with previously described plant defense genes, and (ii) were expressed in bean leaves of plants treated with Trichoderma and/or infected with R. solani.
The expression of P. vulgaris defense-related genes was analyzed in leaves, although the interaction with Trichoderma and/or R. solani is initially produced at the root level, to determine if the signals generated in roots as result of this interaction are able to systematically stimulate the bean defense along long distance from roots to the leaves. The isolate T. velutinum T028 was selected following a similar strategy to that previously described (Mayo et al., 2015), and based on its positive effect on bean growth. In this work, to select a Trichoderma isolate, the results of the in vitro membrane assays and direct confrontation assays against R. solani were analyzed. Isolate Trichoderma T019 was then selected, showing a percentage of inhibition higher than 40% in the membrane assays, and/or 20% in the direct confrontation assays. This isolate also showed the best positive effects on plant phenotype among all the analyzed isolates.
WRKY transcription factors have been involved in the regulation of plant defense gene expression (Rushton and Somssich, 1998; Singh et al., 2002). Thus, WRKY33 has a role in biotic stress defense, where it regulates the balance between necrotrophic and biotrophic pathogen responses (Lippok et al., 2007; Pandey and Somssich, 2009; Birkenbihl et al., 2012). Previous studies have pointed out the involvement of Arabidopsis WRKY transcription factors in regulating the expression of PR genes by direct binding (Chen et al., 2002; Kim et al., 2006). A rapid pathogen-induced WKRY33 expression did not require salicylic acid signaling but a downregulation of this gene involved a direct activation of jasmonic acid (Bakshi and Oelmüller, 2014). In the present case, when bean plants were in contact T. velutinum T028 without pathogen, the WRKY33 gene expression was significantly up-regulated while the PR genes expression (PR2, PR3 and PR4) was significantly down-regulated compared to expression levels in plants without Trichoderma treatment. In the present work, when R. solani was added to the substrate, expression of WRKY33 was significantly down-regulated in plants with Trichoderma inoculation, while PR2, PR3 and PR4 were down-regulated. In the study by Mayo et al. (2015), the expression of PR1, PR2, PR3, and PR4 was down-regulated when beans were inoculated with R. solani.
WRKY33 is also involved in the regulation of the expression of genes modulated by components of the ethylene signaling pathway. In this work, expression of the ERF1 and ERF5 reached similar significant values either with or without Trichoderma and or R. solani in the substrate. This result contrasts with previous reports showing that ERF5 was up-regulated and WRKY33 was down-regulated in Arabidopsis infected with Alternaria brassicicola (Son et al., 2012). WRKY33 would act as a represor of ERF1 and ERF5 expression. Thus, when the expression of WRKY33 is increased, expression of ERF1 and ERF5 is down-regulated.
CH5b encodes an endochitinase precursor and it is also related with the ethylene signaling pathway. In previous works, it has been shown that, when this gene was over-expressed the R. solani symptoms were reduced in crops like Nicotiana tabacum and Brassica napus (Broglie et al., 1991). However, in this study, when bean plants were in contact with R. solani, the expression of this gene was down-regulated but not significantly, while treatment of these infected plants with T. velutinum resulted in its significant up-regulation. These results are in agreement with previous data, showing that the pathogen represses its expression, and the presence of Trichoderma induced it (Mayo et al., 2015).
PAL plays an important role in plant defense; it is involved in the biosynthesis of salicylic acid, which is related to plant systemic resistance (Nugroho et al., 2002; Chaman et al., 2003). PAL gene expression is also regulated in response to pathogen infection. In this work, the presence of T. velutinum and R. solani in the soil resulted in a significant down-regulation of this gene compared with control plants.
Osmotins have plant protective effects against pathogen infection (Narasimhan et al., 2009). In this study, when T. velutinum or R. solani were present in the soil, the expression of OSM34 was not significantly up-regulated respect to control plants, but when both fungi were in the soil at the same time, OSM34 was slightly but significantly down-regulated.
The CNGC genes can be related to early plant defense responses due to changes in ion flux, including H+ and Ca2+ influx and K+ and Cl- efflux (Atkinson et al., 1996). The up-regulation of CNGC2 can confirm the importance of ion channels for the plant resistance response (Borges et al., 2012). In this work, this gene was up-regulated when R. solani was present in the soil not significant. Conversely, CNGC2 was down-regulated in plants treated with T. velutinum. Then, the pathogen would induce an activation of hypersensitive defense mechanisms.
Hydroperoxide lyase (HPL) is involved in the production of antimicrobial and defense signaling oxylipins (Noordermeer et al., 2001; Huang et al., 2010). In this study, the presence of T. velutinum and R. solani, resulted in a down-regulation of this gene expression respect to control plants. In previous works, when tomato plants were in contact with Botrytis cinerea, HPL expression increased 24 h after gray mold infection, but after that time the expression of this gene decreased gradually (Wan et al., 2013). In the present case, after 45 days in contact with the fungus T. velutinum and/or R. solani, its expression was down-regulated, indicating that the plant identifies Trichoderma and Rhizoctonia as two invader organisms, and some of the mechanisms activated against the presence of both are similar, independently of the final response specifically activated in the plant by each one.
GSTa (2,4-D inducible glutathione S-transferase) expression also responds to pathogen attack (Mauch and Dudler, 1993) and can be induced by molecules such as salicylic acid, methyl jasmonate, abscisic acid and H2O2 (Dixon et al., 2002; Moons, 2005). In Gossypium arboretum, GST provides resistance to fungal pathogens and oxidative stress (Barthelson et al., 2010). GST expression was up-regulated during fungal infection in barley, Arabidopsis, and cotton (Dowd et al., 2004; Durrant and Dong, 2004; Lu et al., 2005). However, in banana GST was down-regulated following Fusarium oxysporum f specialis (f. sp.) cubense infection (Wang et al., 2013), which is in agreement with the present case, where the expression of GSTa was down-regulated when T. velutinum and/or R. solani were present in the soil.
hGS encodes a homoglutathione synthetase that is involved in response to oxidative stress. There is not much information about the behavior of this gene in the plant. In the present study, when bean plants were in contact with T. velutinum and/or R. solani, expression of this gene was significantly up-regulated compared to control plants. In other studies, treatment of Medicago truncatula plants with compounds that release nitric oxide, a key signaling molecule in plants, induced expression of GST but not hGS in roots (Innocenti et al., 2007). Similarly, common bean plants treated with H2O2 showed up-regulation of hGS in nodules, whereas treatments with cadmium, sodium chloride, or jasmonic acid had no effect (Loscos et al., 2008).
Conclusion
From 48 genes initially analyzed, 14 bean genes were selected in the present work and only WRKY33, CH5b and hGS showed an up-regulatory response in the presence of T. velutinum, the other genes were or not affected (OSM34) or down-regulated by the presence of this fungus. R. solani infection resulted in a down-regulation of most of the genes analyzed, except PR1, OSM34 and CNGC2 that were not affected, and the presence of both, T. velutinum and R. solani, up-regulates hGS and down-regulates all the other genes analyzed, except CH5b which was not significantly affected.
As conclusion, the strategy described in the present work has been shown to be effective to detect genes involved in plant defense, which respond to the presence of a BCA or to a pathogen and also to the presence of both. The selected genes showed significant homology with described plant defense genes and they are expressed in bean leaves of plants treated with T. velutinum and/or infected with R. solani. The proposed strategy will be very useful in studies about the interaction of bean with pathogens and biocontrol fungi.
Author Contributions
PC and SG conceived the research. SM, OG-L, and AR-G designed the research. SM, OG-L, and AR-G conducted the experiments. SM, PC, and SG analyzed data. SM, PC, SG, EC, and FS interpreted the data. SM, PC, SG, EC, and FS wrote the manuscript. All authors were agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors critically revised the manuscript. All authors approved the final version to be published.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer EVG declared a past co-authorship with one of the authors SG to the handling Editor, who ensured that the process met the standards of a fair and objective review.
Acknowledgments
The grant awarded to Sara Mayo Prieto (FPU12/00677) by the Ministry of Education, Culture and Sport (Spain) according the Resolution of April 25, 2012 (BOE of 10 May), as amended by Resolution of May 18, 2012 (BOE of 22 May), and by Resolution of October 31, 2012 (BOE of 12 November) and granted in the Resolution of December 20, 2012. Ministry of Economy and Competitiveness for National project “Farnesol as self-regulatory molecule in Trichoderma. Tyrosol and farnesol signaling in Trichoderma-bean interaction” (AGL2012-40041-C02-02). Junta de Castilla y León, Consejería de Educación for the project “Effect of terpenes and physiologically related compounds produced by Trichoderma parareesei in the development of common bean (Phaseolus vulgaris L.) and in defensive responses of this plant” (LE228U14).
Footnotes
References
- Atkinson M. M., Midland S. L., Sims J. J., Keen N. T. (1996). Syringolide 1 triggers Ca2 influx, K efflux, and extracellular alkalization in soybean cells carrying the disease-resistance gene Rpg4. Plant Physiol. 112 297–302. 10.1104/pp.112.1.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi M., Oelmüller R. (2014). WRKY transcription factors. Plant Signal. Behav. 9:e27700 10.4161/psb.27700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelson R. A., Qaisar U., Galbraith D. W. (2010). Functional analysis of the Gossypium arboreum genome. Plant Mol. Biol. Rep. 28 334–343. 10.1007/s11105-009-0157-5 [DOI] [Google Scholar]
- Birkenbihl R. P., Diezel C., Somssich I. E. (2012). Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159 266–285. 10.1104/pp.111.192641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges A., Melotto M., Tsai S. M., Caldas D. G. G. (2012). Changes in spatial and temporal gene expression during incompatible interaction between common bean and Anthracnose pathogen. J. Plant Physiol. 169 1216–1220. 10.1016/j.jplph.2012.04.003 [DOI] [PubMed] [Google Scholar]
- Broglie K., Chet I., Holliday M., Cressman R., Biddle P., Knowlton S., et al. (1991). Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254 1194–1197. 10.1126/science.254.5035.1194 [DOI] [PubMed] [Google Scholar]
- Chaman M. E., Copaja S. V., Argandoña V. H. (2003). Relationships between salicylic acid content, Phenylalanine Ammonia-Lyase (PAL) activity, and resistance of barley to aphid infestation. J. Agric. Food Chem. 51 2227–2231. 10.1021/jf020953b [DOI] [PubMed] [Google Scholar]
- Chen W., Provart N. J., Glazebrook J., Katagiri F., Chang H. S., Eulgem T., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14 559–574. 10.1105/tpc.010410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D. P., Lapthorn A., Edwards R. (2002). Plant glutathione transferases. Genome Biol. 3 3004.1–3004.10. 10.1186/gb-2002-3-3-reviews3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd C., Wilson I. W., McFadden H. (2004). Gene expression profile changes in cotton root and hypocotyl tissues in response to infection with Fusarium oxysporum f. sp. vasinfectum. Mol. Plant-Microbe Interact. 17 654–667. 10.1094/MPMI.2004.17.6.654 [DOI] [PubMed] [Google Scholar]
- Druzhinina I. S., Seidl-Seiboth V., Herrera-Estrella A., Horwitz B. A., Kenerley C. M., Monte E., et al. (2011). Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 9 749–759. 10.1038/nrmicro2637 [DOI] [PubMed] [Google Scholar]
- Durrant W. E., Dong X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42 185–209. 10.1146/annurev.phyto.42.040803.140421 [DOI] [PubMed] [Google Scholar]
- Gallou A., Cranenbrouck S., Declerck S. (2009). Trichoderma harzianum elicits defence response genes in roots of potato plantlets challenged by Rhizoctonia solani. Eur. J. Plant Pathol. 124 219–230. 10.1007/s10658-008-9407-x [DOI] [Google Scholar]
- Guerrero-González M. L., Rodríguez-Kessler M., Rodríguez-Guerra R., González-Chavira M., Simpson J., Sanchez F., et al. (2011). Differential expression of Phaseolus vulgaris genes induced during the interaction with Rhizoctonia solani. Plant Cell Rep. 30 1465–1473. 10.1007/s00299-011-1055-5 [DOI] [PubMed] [Google Scholar]
- Hermosa R., Viterbo A., Chet I., Monte E. (2012). Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158 17–25. 10.1099/mic.0.052274-0 [DOI] [PubMed] [Google Scholar]
- Huang F. C., Studart Witkowski C., Schwab W. (2010). Overexpression of hydroperoxide lyase gene in Nicotiana benthamiana using a viral vector system. Plant Biotechnol. J. 8 783–795. 10.1111/j.1467-7652.2010.00508.x [DOI] [PubMed] [Google Scholar]
- Innocenti G., Pucciariello C., Le Gleuher M., Hopkins J., de Stefano M., Delledonne M., et al. (2007). Glutathione synthesis is regulated by nitric oxide in Medicago truncatula roots. Planta 225 1597–1602. 10.1007/s00425-006-0461-3 [DOI] [PubMed] [Google Scholar]
- Kim D. S., Hwang B. K. (2014). An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 65 2295–2306. 10.1093/jxb/eru109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. C., Fan B., Chen Z. (2006). Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol. 142 1180–1192. 10.1104/pp.106.082487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen M. J., Somervuo P., Valkonen J. P. T. (2008). Infection with Rhizoctonia solani induces defense genes and systemic resistance in potato sprouts grown without light. Phytopathology 98 1190–1198. 10.1094/PHYTO-98-11-1190 [DOI] [PubMed] [Google Scholar]
- Lippok B., Birkenbihl R. P., Rivory G., Brümmer J., Schmelzer E., Logemann E., et al. (2007). Expression of AtWRKY33 encoding a pathogen-or PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol. Plant-Microbe Interact. 20 420–429. 10.1094/MPMI-20-4-0420 [DOI] [PubMed] [Google Scholar]
- Lodha T. D., Basak J. (2012). Plant-pathogen interactions: what microarray tells about it? Mol. Biotechnol. 50 87–97. 10.1007/s12033-011-9418-2 [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sánchez-Serrano J. J., Solano R. (2003). Ethylene response factor 1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 165–178. 10.1105/tpc.007468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscos J., Matamoros M. A., Becana M. (2008). Ascorbate and homoglutathione metabolism in common bean nodules under stress conditions and during natural senescence. Plant Physiol. 146 1282–1292. 10.1104/pp.107.114066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z.-X., Gaudet D. A., Frick M., Puchalski B., Genswein B., Laroche A. (2005). Identification and characterization of genes differentially expressed in the resistance reaction in wheat infected with Tilletia tritici, the common bunt pathogen. BMB Rep. 38 420–431. 10.5483/BMBRep.2005.38.4.420 [DOI] [PubMed] [Google Scholar]
- Mauch F., Dudler R. (1993). Differential induction of distinct glutathione-S-transferases of wheat by xenobiotics and by pathogen attack. Plant Physiol. 102 1193–1201. 10.1104/pp.102.4.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo S., Gutiérrez S., Malmierca M. G., Lorenzana A., Campelo M. P., Hermosa R., et al. (2015). Influence of Rhizoctonia solani and Trichoderma spp. in growth of bean (Phaseolus vulgaris L.) and in the induction of plant defense-related genes. Front. Plant Sci. 6:685 10.3389/fpls.2015.00685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat C. S., Ingle R. A., Wathugala D. L., Saunders N. J., Knight H., Knight M. R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7:e35995 10.1371/journal.pone.0035995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons A. (2005). Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs). Vitam. Horm. 72 155–202. 10.1016/S0083-6729(05)72005-7 [DOI] [PubMed] [Google Scholar]
- Narasimhan M. L., Bressan R. A., D’Urzo M. P., Jenks M. A., Mengiste T. (2009). Unexpected turns and twists in structure/function of PR-proteins that connect energy metabolism and immunity. Adv. Bot. Res. 51 439–489. 10.1016/S0065-2296(09)51011-7 [DOI] [Google Scholar]
- Noordermeer M. A., Veldink G. A., Vliegenthart J. F. G. (2001). Fatty acid hydroperoxide lyase: a plant cytochrome P450 enzyme involved in wound healing and pest resistance. Chembiochem 2 494–504. [DOI] [PubMed] [Google Scholar]
- Nugroho L. H., Verberne M. C., Verpoorte R. (2002). Activities of enzymes involved in the phenylpropanoid pathway in constitutively salicylic acid-producing tobacco plants. Plant Physiol. Biochem. 40 755–760. 10.1016/S0981-9428(02)01437-7 [DOI] [Google Scholar]
- Pandey S. P., Somssich I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150 1648–1655. 10.1104/pp.109.138990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J. L., Queiroz R. M. L., Charneau S. O., Felix C. R., Ricart C. A. O., Lopes Da Silva F., et al. (2014). Analysis of Phaseolus vulgaris response to its association with Trichoderma harzianum (ALL-42) in the presence or absence of the phytopathogenic fungi Rhizoctonia solani and Fusarium solani. PLoS ONE 9:e98234 10.1371/journal.pone.0098234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 e45–e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid K. E., Olsson N., Schlosser J., Peng F., Lund S. T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6:27 10.1186/1471-2229-6-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P. J., Somssich I. E. (1998). Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1 311–315. 10.1016/1369-5266(88)80052-9 [DOI] [PubMed] [Google Scholar]
- Rutledge R. G., Stewart D. (2008). Critical evaluation of methods used to determine amplification efficiency refutes the exponential character of real-time PCR. BMC Mol. Biol. 9:96 10.1186/1471-2199-9-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Lin W., Villamor J. G., Verslues P. E. (2013). Divergent low water potential response in Arabidopsis thaliana accessions Landsberg erecta and Shahdara. Plant. Cell Environ. 36 994–1008. 10.1111/pce.12032 [DOI] [PubMed] [Google Scholar]
- Shoresh M., Harman G. E., Mastouri F. (2010). Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 48 21–43. 10.1146/annurev-phyto-073009-114450 [DOI] [PubMed] [Google Scholar]
- Singh K. B., Foley R. C., Oñate-Sánchez L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5 430–436. 10.1016/S1369-5266(02)00289-3 [DOI] [PubMed] [Google Scholar]
- Son G. H., Wan J., Kim H. J., Nguyen X. C., Chung W. S., Hong J. C., et al. (2012). Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol. Plant-Microbe Interact. 25 48–60. 10.1094/MPMI-06-11-0165 [DOI] [PubMed] [Google Scholar]
- Upchurch R. G., Ramirez M. E. (2010). Defense-related gene expression in soybean leaves and seeds inoculated with Cercospora kikuchii and Diaporthe phaseolorum var. meridionalis. Physiol. Mol. Plant Pathol. 75 64–70. 10.1016/j.pmpp.2010.08.007 [DOI] [Google Scholar]
- Valenciano J. B., Casquero P. A., Boto J. A., Marcelo V. (2006). Evaluation of the occurrence of root rots on bean plants (Phaseolus vulgaris) using different sowing methods and with different techniques of pesticide application. New Zeal. J. Crop Hortic. Sci. 34 291–298. 10.1080/01140671.2006.9514419 [DOI] [Google Scholar]
- Vellicce G. R., Ricci J. C. D., Hernández L., Castagnaro A. P. (2006). Enhanced resistance to Botrytis cinerea mediated by the transgenic expression of the chitinase gene CH5b in strawberry. Transgenic Res. 15 57–68. 10.1007/s11248-005-2543-6 [DOI] [PubMed] [Google Scholar]
- Verhagen B. W. M., Van Loon L. C., Pieterse C. M. J. (2006). “Induced disease resistance signaling in plants,” in Floriculture, Ornamental and Plant Biotechnology ed. Teixeira da Silva J. A. (Utrecht: Global Science Books; ) 334–343. [Google Scholar]
- Wan X.-H., Chen S.-X., Wang C.-Y., Zhang R.-R., Cheng S.-Q., Meng H.-W., et al. (2013). Isolation, expression, and characterization of a hydroperoxide lyase gene from cucumber. Int. J. Mol. Sci. 14 22082–22101. 10.3390/ijms141122082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Huang S., Jia C., Liu J., Zhang J., Xu B., et al. (2013). Molecular cloning and expression of five glutathione S-transferase (GST) genes from Banana (Musa acuminata L. AAA group, cv. cavendish). Plant Cell Rep. 32 1373–1380. 10.1007/s00299-013-1449-7 [DOI] [PubMed] [Google Scholar]


